Abstract
Endoplasmic reticulum (ER) stress has been linked to the onset and progression of many diseases. SIL1 is an adenine nucleotide exchange factor of the essential ER lumen chaperone HSPA5/BiP that senses ER stress and is involved in protein folding. Mutations in the Sil1 gene have been associated with Marinesco–Sjögren syndrome, hallmarks of which include ataxia and cerebellar atrophy. We have previously shown that loss of SIL1 function in mouse results in ER stress, ubiquitylated protein inclusions, and degeneration of specific Purkinje cells in the cerebellum. Here, we report that overexpression of HYOU1/ORP150, an exchange factor that works in parallel to SIL1, prevents ER stress and rescues neurodegeneration in Sil1−/− mice, whereas decreasing expression of HYOU1 exacerbates these phenotypes. In addition, loss of DNAJC3/p58IPK, a co-chaperone that promotes ATP hydrolysis by BiP, ameliorates ER stress and neurodegeneration in Sil1−/− mice. These findings suggest that alterations in the nucleotide exchange cycle of BiP cause ER stress and neurodegeneration in Sil1-deficient mice. Our results present the first evidence of important genetic modifiers of Marinesco–Sjögren syndrome, and provide additional pathways for therapeutic intervention for this, and other ER stress-induced, diseases.
INTRODUCTION
Folding of transmembrane proteins and secreted proteins, which account for more than one third of newly synthesized proteins, occurs in the endoplasmic reticulum (ER) (1). HSPA5 (hereafter referred as BiP), the major ER-localized member of the HSP70 family of molecular chaperones, reversibly binds to contiguous segments of hydrophobic amino acids exposed in unfolded lumenal proteins to impede aggregation and promote folding (2). Properly folded proteins are exported out of ER, whereas terminally misfolded proteins are targeted to degradation by the ER-associated degradation (ERAD) pathway (3). Imbalances between the capacity to fold ER proteins and the cellular demands for ER-associated protein synthesis lead to ER stress, which diminishes fitness (4,5).
Binding of substrates to BiP and subsequent release from BiP is controlled by a continuous cycle of ATP hydrolysis and exchange of ATP for ADP that is regulated by cofactors. DNAJ domain proteins recruit substrates to ATP-bound BiP and stimulate its ATPase activity, which in turn converts ATP to ADP and results in high affinity, stable interactions of BiP with unfolded substrates. To complete the protein folding cycle, nucleotide exchange factors bind ADP-bound BiP to catalyze the release of ADP and rebinding of ATP. Thus, defects in the BiP ATP/ADP exchange cycle that result in altered substrate binding or release cause accumulation of unfolded proteins and ER stress (2,4).
A mutation of SIL1, a BiP co-chaperone, causes ataxia and neurodegeneration in the spontaneous mouse mutant, woozy (wz) (6). The majority of Purkinje cells in the wz mutant cerebellum develop ubiquitylated protein inclusions and degenerate between 3 and 4 months of age. Mutations in Sil1 are also associated with human Marinesco–Sjögren syndrome (MSS), an infantile-onset disease which exhibits cerebellar ataxia, cataracts, mental retardation, myopathy, delayed somatic maturation and short stature (7,8).
Sil1 encodes a BiP nucleotide exchange factor (9,10) suggesting that the neurodegeneration caused by Sil1 mutations in both mouse and human is due to alterations in the BiP nucleotide binding and hydrolysis cycle. Consistent with this hypothesis, death of Purkinje neurons in Sil1-deficient mice is preceded by signs of ER stress as revealed by upregulation of BiP and DDIT3/CHOP, which are well-validated markers of the mammalian ER stress response [also known as the unfolded protein response (UPR)] (11,12). If neurodegeneration induced by Sil1-deficiency is indeed due to defects in the BiP ATP/ADP cycle, changes in expression of other co-chaperones that regulate this cycle may also modify ER stress and neurodegeneration. Specifically, loss of function of other BiP nucleotide exchange factors may exacerbate the Sil1-deficiency phenotype. In contrast, decreased activities of DNAJ type co-chaperones of BiP, which have opposing functions to SIL1 in regulating the BiP ATP/ADP cycle, would be predicted to ameliorate the ER stress and cell death phenotype caused by Sil1 mutations.
In addition to SIL1, an atypical HSP70 protein, HYOU1/ORP150, also serves as a BiP nucleotide exchange factor (13,14). In support of nucleotide exchange factor function, partial redundancy of SIL1 and HYOU1 has been demonstrated in yeast. Deletion of both SIL1 and the yeast Hyou1 ortholog, LHS1, results in synthetic lethality. In addition, overexpression of SIL1 partially suppresses the severe growth phenotype of yeast lacking LHS1 and IRE1, the gene encoding an upstream regulator of the UPR (15). The interchangeability of the nucleotide exchange factor activity of Sil1p and Lhs1p is also supported by biochemical assays (13). In mammals, HYOU1 has been suggested to be a neuroprotective factor against ischemia and excitotoxicity, insults that potentially induce ER stress in neurons (16–18). Transgenic expression of Hyou1 in neurons driven by the platelet-derived growth factor (PDGF) promoter decreases Purkinje cell apoptosis during cerebellar development leading to a higher number of Purkinje cells at postnatal day 40 (19). No obvious phenotypes of the brain or other major organs have been reported in mice heterozygous for the Hyou1 targeted allele, whereas homozygosity is embryonic or perinatal lethal (19).
Multiple ER DNAJ domain proteins have been identified. These proteins may exist in distinct BiP complexes that function at various locations in the ER or have specific roles in the regulation of the different functions of BiP (20–25). For instance, yeast Sec63p binds to the translocon and together with Kar2p, the yeast ortholog of BiP, is involved in translocation of nascent peptides (26,27). DNAJB9 and DNAJC10 may be involved in ERAD, which also requires BiP (28,29). Recent findings suggest that another ER-associated J domain protein, DNAJC3/p58IPK, is localized to the lumen and serves as a co-chaperone of BiP to promote protein folding (30,31). Consistent with an important role in ER function, Dnajc3−/− mice have mild glucose intolerance due to defects in pancreas islet cells (32).
To test the hypothesis that altered BiP ATP/ADP cycle is the underlying mechanism of neurodegeneration caused by Sil1 mutations, we examined the possible functional redundancy of SIL1 and HYOU1, and genetic interactions between SIL1 and DNAJC3, in vivo. Our results demonstrate that reduction of the gene dosage of Hyou1 aggravates the temporal onset and the spatial specificity of the UPR and subsequent Purkinje cell death in the Sil1−/− cerebellum. In addition, overexpression of Hyou1 in the cerebellum completely suppresses Purkinje cell degeneration in Sil1−/− mice. These data suggest that Hyou1 and Sil1 have partially redundant functions in neurons. In contrast, homozygous deletion of Dnajc3 partially rescues the Sil1 null phenotype, consistent with the opposing functions of SIL1 and DNAJC3 in regulating the BiP ATP/ADP cycle. These results may lead to better understanding and diagnosis of MSS and shed light on the disease mechanisms of this, and other ER stress-related, diseases.
RESULTS
Transgenic expression of Hyou1 in Sil1−/− mice suppresses ER stress and Purkinje cell degeneration
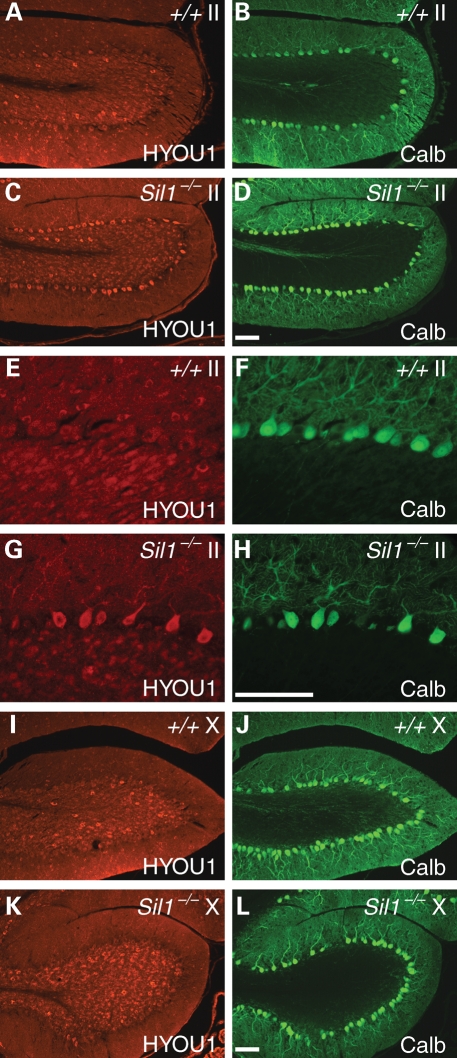
Deficiency of the BiP nucleotide exchange factor, Sil1, causes prolonged induction of the UPR indicative of ER stress, and eventual death of most Purkinje cells in the cerebellum (6). Interestingly, UPR is not induced in Purkinje cells in the vestibulocerebellum which includes lobule X and the caudal region of lobule IX, nor do these neurons degenerate, suggesting that this developmentally and functionally distinct region of the cerebellum may utilize other BiP co-chaperones (6). To test whether survival of Sil1-deficient Purkinje cells in these caudal lobules is compensated by HYOU1, the other known BiP nucleotide exchange factor, we examined expression of this protein in wild-type and Sil1−/− cerebella. HYOU1 was highly expressed in wild-type Golgi neurons in the granule cell layer with much lower levels of expression in Purkinje cells (Fig. 1A, B, E, F). In contrast to the low levels of expression in wild-type Purkinje cells, HYOU1 was moderately upregulated by 1-month of age in many Sil1−/− Purkinje cells located outside of the vestibulocerebellum (data not shown). In 2-month-old mice, HYOU1 upregulation in Sil1-deficient Purkinje cells in these lobules was quite obvious (Fig. 1C, D, G, H). Upregulation of HYOU1 expression was not observed in Sil1−/− Purkinje cells in either lobule X or those in caudal lobule IX (Fig. 1I–L), suggesting that HYOU1 is induced in response to ER stress. These data suggest that UPR-mediated HYOU1 induction in Sil1-deficient Purkinje cells is not sufficient to prevent Purkinje cell death.
Figure 1.
Loss of SIL1 function induces HYOU1 expression in Purkinje cells. Cerebellar sections from 2-month-old wild-type (+/+, A, B, E, F, I, J) or Sil1−/− (C, D, G, H, K, L) mice were immunostained with antibodies to HYOU1 (red) and calbindin-D28 (Calb; green). Images from lobules II (A–D) and lobules X (I–L) are shown. Higher magnification images of A–D are shown in E–H, respectively. Camera exposure times are equal for images of the same channel and magnification. Scale bar = 100 µm.
To test whether increased or earlier expression of HYOU1 can modulate the death of Sil1−/− Purkinje cells, we crossed Sil1−/− mice with transgenic mice overexpressing Hyou1 under the control of the CAG promoter, an artificial promoter composed of the chicken β-actin and minimal CMV promoters (33). CAG-Hyou1 transgenic mice do not exhibit any obvious brain pathology (data not shown). However, this transgene does cause vacuolar degeneration of cardiac and skeletal muscles, the latter of which is consistent with our observation that Hyou1 transgenic mice could not perform on treadmill tests for gait analysis (33,34) (data not shown). Unlike a previous study suggesting that mice overexpressing Hyou1 under the control of a PDGF promoter had reduced apoptosis of Purkinje cells resulting in 40% increase of Purkinje cell numbers before 40 days of age, we did not observe an increase in Purkinje cell numbers in Tg(CAG-Hyou1) mice [+/+, 190 ± 18 (×103); Tg-Hyou1, 180 ± 10 (×103)] and the transgenic mice do not have a visible ataxia phenotype or obvious brain pathology (data not shown).
The CAG promoter drives expression widely. In agreement, increased HYOU1 expression is detected in most, if not all, cells in the transgenic cerebellum, with levels 3-fold higher than those observed in 1-month-old wild-type and Sil1−/− cerebella (Fig. 2A). Although expression varies between Purkinje cells, immunofluorescence analysis revealed higher HYOU1 levels in these neurons in 1-month-old Sil1−/−; Tg-Hyou1 mice compared with levels in Purkinje cells of wild-type mice or Sil1−/− mice at 1 or 2 months of age (Fig. 2 B–G and Supplementary Material, Fig. S1). HYOU1 expression in the Purkinje cells of 2-month-old Sil1−/−; Tg-Hyou1 mice was similar to that observed at 1-month of age, indicating that the transgene was stably expressed (data not shown).
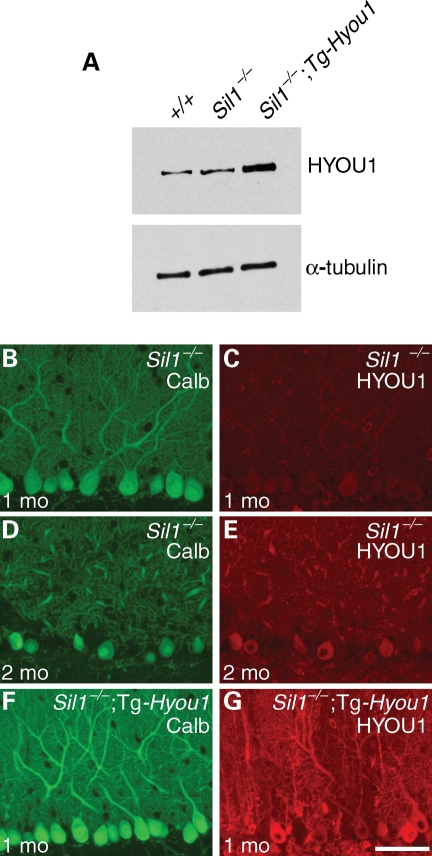
Figure 2.
A Hyou1transgene confers constitutively elevated expression. (A) Western blot of cerebellar protein extracts from 1-month-old wild-type (+/+), Sil1−/− and Sil1−/−; Tg-Hyou1 mice, probed with an antibody against HYOU1 and an α-tubulin antibody as loading control. (B–G) Comparison of HYOU1 expression in Purkinje cells from 1-month-old (B, C), 2-month-old (D, E) Sil1−/− and 1-month-old Sil1−/−; Tg-Hyou1 (F, G) mice. Camera exposure times are equal for images of the same channel. Scale bar = 50 µm.
Analysis of 4-month-old Sil1−/−; Tg-Hyou1 mice demonstrated that overexpression of Hyou1 greatly suppressed the ataxia and Purkinje cell death caused by Sil1-deficiency, as evidenced by immunostaining with an antibody against the Purkinje cell marker, calbindin-D28 (Fig. 3A–C, and data not shown). No obvious Purkinje cell loss was observed in adult Sil1−/−; Tg-Hyou1 mice, even those at 8 months of age, suggesting that rescue of cell death is not transient (data not shown). To determine if the transgene also blocks the development of ER stress in Sil1−/− Purkinje cells, we analyzed the expression of the ER stress-inducible proteins, BiP and CHOP (11,12). Although BiP and CHOP were upregulated in a few Purkinje cells in Sil1−/−; Tg-Hyou1 mice, overall expression of BiP and CHOP was lower than observed in Sil1−/− Purkinje cells, suggesting that transgenic overexpression of Hyou1 decreases ER stress caused by loss of SIL1 function (Fig. 3D–I and Supplementary Material, Fig. S2). Consistent with the lack of ER-stress marker upregulation, ubiquitylated protein inclusions were not observed in Purkinje cells of 3-month-old or older Sil1−/−; Tg-Hyou1 mice, compared with Purkinje cells from 3 month old Sil1−/− mice [Supplementary Material, Fig. S2 and data not shown; percentages of Purkinje cells harboring inclusions: 4.7 ± 2.2% (Sil1−/−); 0% (Sil1−/−; Tg-Hyou1)].
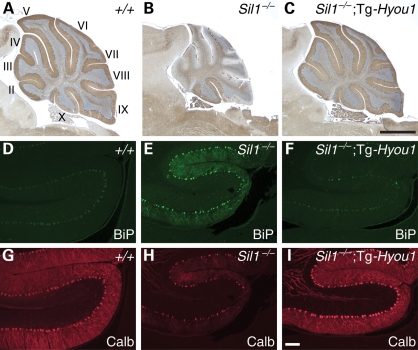
Figure 3.
Overexpression of HYOU1 in cerebella prevents Purkinje cell death and ER stress caused by loss of SIL1 function. (A–C) Immunohistochemistry using antibody to calbindin-D28 was performed on cerebella from 4-month-old wild-type (A), Sil1−/− (B) and Sil1−/−; Tg-Hyou1 (C) mice. Lobules are indicated by Roman numerals (A). (D–I) Expression of Hyou1 transgene suppresses BiP upregulation caused by Sil1 deficiency. Cerebellar sections from 3-month-old wild-type (+/+; D, G), Sil1−/− (E, H) and Sil1−/−; Tg-Hyou1 (F, I) mice were incubated with antibodies against BiP (D–F) and calbindin-D28 (Calb; G–I). Images of lobule II are shown. Camera exposure times are equal for images of the same channel. Scale bars = 1 mm (A–C) and 100 µm (D–I).
Decreased Hyou1 expression in Sil1-deficient background aggravates ER stress and Purkinje cell death
Purkinje cells in the caudal lobules of the cerebellum neither undergo neurodegeneration nor do they show signs of ER stress in Sil1-deficient mice (6). Rather than projecting their axons to form synapses on neurons of the deep cerebellar nuclei like other Purkinje cells, these caudal Purkinje cells synapse directly with the vestibular nuclei in the brainstem. These differences in connectivity suggest that these neurons may have metabolic differences, including differences in ER protein load, which would make them less sensitive to loss of Sil1 function. To test whether further reduction of BiP nucleotide exchange factor activity may induce Purkinje cell death in the caudal lobules, we crossed Sil1−/− mice to mice heterozygous for a targeted allele of Hyou1 (17).
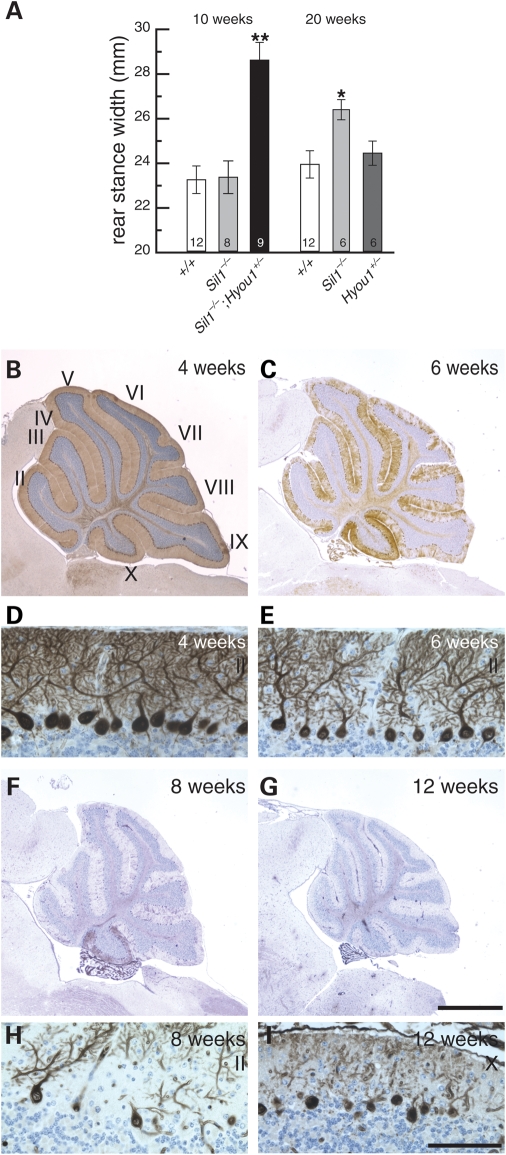
Sil1−/−; Hyou1+/− mice have visible ataxia by 2 months after birth. In agreement, treadmill tests demonstrated that 10-week-old Sil1−/−; Hyou1+/− mice have an abnormally wide stance that is characteristic of cerebellar dysfunction, which is not apparent in Sil1−/− mice until 20 weeks (Fig. 4A). By 20 weeks, Sil1−/−; Hyou1+/− mice are extremely ataxic and are unable to walk on the treadmill (data not shown). In agreement with the earlier onset of ataxia in Sil1−/−; Hyou1+/− mice, Purkinje cells begin to show characteristics of degenerating neurons between 4 and 6 weeks of age including shrunken soma and dendrite retraction (Fig. 4B–E). By 2 months, most Purkinje cells have degenerated (Fig. 4F and H). Most importantly, Purkinje cells in lobule X and caudal lobule IX, which are resistant to loss of Sil1, also die in Sil1−/−; Hyou1+/− mice by 3 months of age (Fig. 4G and I).
Figure 4.
Deletion of one copy of Hyou1 accelerates Purkinje cell death caused by loss of SIL1 function. (A) Rear stance (gait) on a treadmill test. Mean values ± SEM for the given genotypes and ages are shown. Numbers of mice tested are noted inside the columns for each genotype. *, P < 0.05; **, P < 0.001. (B, C, F, G) Calbindin-D28 immunohistochemistry of 4-week (B), 6-week (C), 8-week (F) or 12-week (G) old Sil1−/−; Hyou1+/− mice. Lobules are indicated by Roman numerals (B). Scale bar = 1 mm. (D, E, H, I). Details of Purkinje cell death in lobule II (D, E, H), and lobule X (I) at ages indicated are shown. Scale bar = 100 µm.
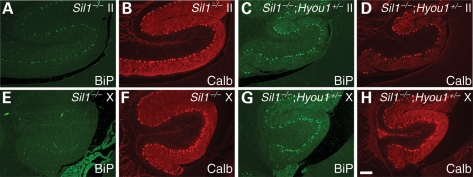
Consistent with signs of neuron damage, elevated levels of BiP and CHOP were observed in Purkinje cells of the rostral lobules in Sil1−/−; Hyou1+/− mice by 6 weeks after birth (Fig. 5 and Supplementary Material, Fig. S3). In addition, BiP and CHOP were clearly upregulated in caudal Purkinje cells in Sil1−/−; Hyou1+/− mice at this time. In contrast, BiP or CHOP expression was barely visible in Purkinje cells of Sil1−/− littermates at this time. These results suggest that the ER stress occurs earlier in Sil1−/−; Hyou1+/− Purkinje cells than in Sil1−/− neurons. In agreement with the earlier onset of ER stress, ubiquitin-positive protein inclusions are present in Purkinje cells of 6–8-week-old Sil1−/−; Hyou1+/− mice, but not until 10 weeks in Sil1−/− mice [Supplementary Material, Fig. S3 and data not shown; percentages of Purkinje cells harboring inclusions at 2 months of age: 0% (Sil1−/−); 10.7 ± 7.1% (Sil1−/−; Hyou1+/−)].
Figure 5.
Hyou1 heterozygosity aggravates ER stress in Sil1−/− Purkinje cells. BiP upregulation in lobule II (A–D) or lobule X (E–H) of cerebella from 6-week-old Sil1−/− (A, B, E, F) and Sil1−/−; Hyou1+/− mice (C, D, G, H). Sections were subjected to immunostaining with antibodies against BiP and calbindin-D28 (Calb). Camera exposure times are equal for images of the same channel. Scale bar = 100 µm.
Loss of Dnajc3 partially suppresses Sil1-deficiency in Purkinje cells
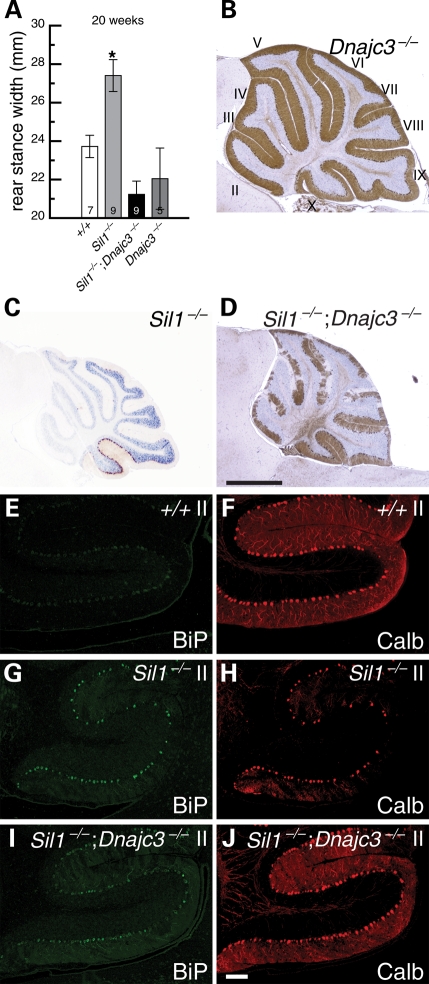
The synergism of the phenotype caused by mutations in the two exchange factors, SIL1 and HYOU1, indicates that diminished nucleotide exchange activity, which favors accumulation of ADP-bound BiP, might be implicated in neurodegeneration. Given its opposing role in promoting the ATPase activity of BiP, we tested whether loss of DNAJC3 function, which would be predicted to decrease the production of ATP-bound BiP, might restore ER balance and ameliorate neurodegeneration in Sil1−/− Purkinje cells. DNAJC3/p58IPK has been recently identified as an ER DnaJ protein that promotes BiP ATPase activity (30,31). Mice homozygous for a null allele of Dnajc3 gene do not display brain pathology or ataxia even when aged (Fig. 6A and B) (32). In contrast to Sil1−/− mice which develop ataxia between 3 and 4 months of age, Sil1−/−; Dnajc3−/− offspring did not exhibit obvious sign of ataxia even when aged to 8 months (data not shown). In agreement, gait analyses of 20-week-old mice demonstrated that the rear stance width of Sil1−/−; Dnajc3−/− mice were significantly different from the stance width of Sil1−/− mice, but similar to that of wild-type and Dnajc3−/− mice (Fig. 6A). Some signs of Purkinje cell degeneration start to appear at 4 months of age in Sil1−/−; Dnajc3−/− mice, however, the majority of Purkinje cells still remain intact at 8 months of age (Fig. 6B–D). Although BiP is still upregulated in Purkinje cells of 3-month-old Sil1−/−; Dnajc3−/− mice relative to levels in wild-type neurons, its levels were lower in most Purkinje cells than those observed in Sil1−/− neurons (Fig. 6E–J). This suggests that loss of Dnajc3 attenuates the level of ER stress observed in Sil1-deficient neurons. As expected, fewer Sil1−/−; Dnajc3−/− Purkinje cells harbor protein inclusions [Supplementary Material, Fig. S4 and data not shown; percentages of Purkinje cells harboring inclusions at 3 months of age: 3.5 ± 0.6% (Sil1−/−), 0.27 ± 0.07% (Sil1−/−; Dnajc3−/−)]. Taken together, these results indicate that Dnajc3 deficiency partially rescues ER stress in Sil1−/− Purkinje cells and their subsequent degeneration.
Figure 6.
Depletion of DNAJC3 activity partially suppresses Purkinje cell death and ER stress caused by SIL1 mutation. (A) Rear stance (gait) on a treadmill test. Mean values ± SEM for the given genotypes at 20 weeks of age are shown. Numbers of mice tested are noted inside the columns for each genotype. *, P < 0.05. (B–D) Immunohistochemistry of cerebellar sections from 4-month-old Dnajc3−/− (B), 4-month-old Sil1−/− (C) and 8-month-old Sil1−/−; Dnajc3−/− (D) mice with an antibody against calbindin-D28. Lobules are indicated by Roman numerals (B). (E–J) Expression of BiP (E, G, I) and calbindin-D28 (Calb, F, H, J) in cerebellar Purkinje cells of 3-month-old wild-type (+/+; E, F), Sil1−/− (G, H) and Sil1−/−; Dnajc3−/− (I, J) mice. Camera exposure times are equal for images of the same channel. Scale bars = 1 mm (B–D) and 100 µm (E–J).
Previously, CHOP has been reported to induce apoptosis during prolonged ER stress (35). Deletion of Ddit3, the gene encoding CHOP, has also been reported to reduce cell death in multiple tissues in several disease mouse models associated with ER stress including a neurotoxin-induced Parkinson disease model, the demyelinating Charcot–Marie–Tooth 1B mouse model, and a model of diabetes (36–38). We reasoned that perhaps CHOP is also responsible for Purkinje cell death in Sil1-deficient mice. Unexpectedly we found that Purkinje cell expression of CHOP was very similar in Sil1−/− and Dnajc3−/−; Sil1−/− mice (Supplementary Material, Fig. S4). Since most Purkinje cells are still alive after 8 months of age in Dnajc3−/−; Sil1−/− mice, this result suggested that prolonged expression of CHOP may not be sufficient for the induction of apoptosis in Purkinje cells. To further test this, we bred Sil1−/− mice with Ddit3−/− mice. Like Sil1−/− mice, Sil1−/−; Ddit3−/− mice still develop ataxia after 3 months after birth, with a nearly identical pattern of Purkinje cell loss after 4 months of age to that observed in age-matched Sil1−/− mice (Supplementary Material, Fig. S5). These results suggest that CHOP is not required for ER stress-induced Purkinje cell death in Sil1−/− mice.
Failure of ERAD may contribute to the accumulation of ER associated protein inclusions
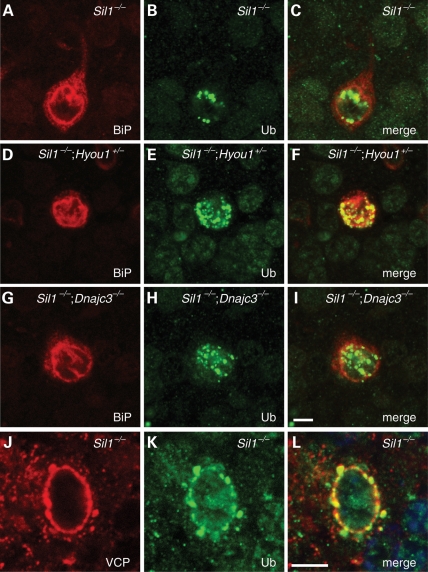
We next assessed the subcellular localization and protein composition of protein inclusions caused by loss of SIL1 function in the presence of different genetic modifiers. In Sil1−/− Purkinje cells, ubiquitylated protein inclusions are localized to ER and the nucleus (6). Similarly, the majority of inclusions are also localized to ER and the nucleus in Purkinje cells of Sil1−/−; Hyou1+/− or Sil1−/−; Dnajc3−/− mice (Fig. 7). Therefore, neither reduction of Hyou1 gene dosage, nor loss of Dnajc3, changes the localization of the inclusions, but rather changes the timing of protein aggregation.
Figure 7.
Ubiquitin-positive protein inclusions in Purkinje cells. (A–I) ER and nuclear localization of protein inclusions. Cerebellar sections from 3-month-old Sil1−/− (A–C), 2-month-old Sil1−/−; Hyou1+/− (D–F) and 3-month-old Sil1−/−; Dnajc3−/− (G–I) mice were immunostained with antibodies against BiP (A, D, G) and ubiquitin (Ub; B, E, H). Merged images are shown in C, F, I. (J–L) ERAD chaperone p97/VCP partially co-localizes with ubiquitylated protein inclusions in Sil1−/− Purkinje cells. Brain sections from 80-day-old Sil1−/− mice were subjected to immunohistochemistry with antibodies against p97/VCP (J) and ubiquitin (Ub, K). Merged image is shown in L. Scale bar = 5 µm.
Our previous studies demonstrated that protein inclusions in Sil1-deficient Purkinje cells contain the ER chaperones BiP, PDIA4/ERp72, HSP90B1/GRP94 and calreticulin (6). To access protein content of the ubiquitylated inclusions in Sil1−/−; Hyou1+/− or Sil1−/−; Dnajc3−/− Purkinje cells we performed immunofluorescence with antibodies to ubiquitin and ER chaperones. Like inclusions in Sil1−/− Purkinje cells, BiP, ERp72, GRP94 and calreticulin were found in protein inclusions in Purkinje cells of these mutant strains (Fig. 7 and data not shown). In addition to ER chaperones, we found that p97/VCP (valosin-containing protein), a cytoplasmic molecular chaperone, is also present in protein inclusions when the inclusions start to appear (Fig. 7). VCP is an AAA family protein that is essential for ERAD, binding ubiquitylated proteins as they are translocated in a retrograde fashion from the ER lumen (3). Therefore, our results indicate that failure of ERAD may contribute to the formation of protein inclusions when BiP nucleotide exchange factor function is impaired.
DISCUSSION
Protein quality control in the ER plays critical roles in the maturation of secreted and transmembrane proteins. With the assistance of molecular chaperones located at both the cytosolic and the lumenal faces of ER membrane, a delicate balance is achieved between protein maturation and degradation. As a result, only terminally misfolded proteins will be degraded, whereas partially unfolded proteins are subjected to retention in ER until being correctly folded. However, under certain disease conditions, ER folding capacity may be reduced, resulting in ER stress and accumulation of misfolded proteins. The UPR can be activated to counteract ER stress, but prolonged ER stress often causes cell death (4,5).
BiP, an HSP70 family molecular chaperone, regulates protein folding in the ER and the cellular response to ER stress. Mutation of the Sil1 gene, which encodes a BiP nucleotide exchange factor, causes ER stress in Purkinje cells. Our results indicate that Sil1 and Hyou1, another known nucleotide exchange factor for BiP, have partially redundant functions in neurons. Overexpression of Hyou1 prevents ER stress and degeneration of Sil1−/− Purkinje cells, whereas deletion of one copy of Hyou1 accelerates death of Purkinje cells in the Sil1−/− cerebellum. In addition, Purkinje cells that are normally resistant to loss of Sil1, exhibit signs of ER stress, develop ubiquitin-positive protein inclusions, and die in Sil1−/−; Hyou1+/− mice. However, we did not observe any other degenerating neurons in the brains of Sil1−/−; Hyou1+/− mice (other than the target-related death of granule cells which occurs secondarily after Purkinje cell death). Furthermore, although caudal Purkinje cells do die in Sil1−/−; Hyou1+/− mice, they are the last to mount an ER stress response and degenerate. These findings suggest that this subset of Purkinje cells is less sensitive to impaired BiP chaperoning activity, and these neurons may experience lower unfolded protein load. Although our results suggest that SIL1 and HYOU1 have partially redundant functions as BiP nucleotide exchange factors in Purkinje cells, it is formally possible that the synergism between the two reflects an indirect genetic interaction. Moreover, although SIL1 and HYOU1 are two alternative nucleotide exchange factors for BiP, only SIL1 has been associated with MSS (7,8,39). Since BiP has multiple roles in ER lumen, which include assisting protein translocation and folding, sensing ER stress, and promoting ERAD, perhaps SIL1 and HYOU1 perform BiP nucleotide exchange factor function under different scenarios (2). Further studies are necessary to elucidate BiP's special requirements for different nucleotide exchange factors and DNAJ proteins.
In contrast to the enhanced neurodegeneration observed in Sil1-deficient Purkinje cells when Hyou1 dosage is reduced, deletion of Dnajc3 delayed and attenuated Purkinje cell degeneration in the Sil1 null mouse. Early studies suggested DNAJC3 functions in the cytoplasm as a negative regulator for the PERK (EIF2AK3)/eIF2α branch of the UPR pathway or as a component of the preemptive ER quality control (pQC) system (40–42). However, recent data demonstrates that DNAJC3 is predominantly localized in the ER lumen where it acts as a BiP co-chaperone (30,31). In agreement with its role as a co-chaperone, Dnajc3−/− mice develop mild diabetes. In addition, homozygous deletion of Dnajc3 aggravated the diabetic phenotype caused by the ‘Akita’ allele of insulin 2 (Ins2C96Y), which causes misfolding of the insulin 2 protein in the ER (40). Like other DnaJ proteins, DNAJC3 binds hydrophobic regions of unfolded protein substrates, and transfers these substrates to ATP-bound BiP, activating the ATPase activity of BiP (30). The resulting ADP-bound BiP would be predicted to bind the unfolded substrate with high-affinity (2). Sil1-deficiency is predicted to cause accumulation of the ADP-bound form of BiP, which inefficiently releases substrate thus preventing completion of the folding process. It is likely that deletion of Dnajc3 would slow down (but given the redundancy of DnaJ proteins, not completely prevent) the conversion of ATP-bound BiP to the ADP-bound form. This attenuation of BiP ATP hydrolysis may allow time for HYOU1 to reduce the accumulation of ADP-bound BiP and partially restore the ATP/ADP exchange cycle and release of substrate, which in turn restores BiP's ability to buffer the unfolded and misfolded protein load in the ER.
Null mutation of CHOP suppresses death of various cells including pancreatic β-cells, dopaminergic neurons in substantia nigra, and Schwann cells, in several disease models (36–38,43). However, we observed persistent upregulation of CHOP in Sil1−/−; Dnajc3−/− Purkinje cells, although many of these neurons survived in 1-year-old mice. Furthermore, CHOP deletion had no apparent impact on the onset and progression of Purkinje cell death in the Sil1 mutant cerebellum. These results suggest that unlike other cells, CHOP expression may not be necessary or sufficient to induce cell death in ER-stressed Purkinje cells (36–38). Perhaps Purkinje cells are less sensitive to the effects of CHOP target genes. Alternatively, CHOP expression in Purkinje cells may not activate the full array of transcriptional target genes.
We observed that early stage protein inclusions in Sil1−/− Purkinje cells often contain VCP, an AAA+ family protein involved in extracting ubiquitylated ERAD substrates from ER, indicating that ERAD may be impaired in these neurons. Since some ER lumen chaperones are also co-localized with the protein inclusions, our results suggest that misfolded proteins cannot be efficiently released from ER in the absence of SIL1 function in Purkinje cells, resulting in accumulation and aggregation on the ER surface.
Although signs of UPR upregulation have been observed in postmortem tissue from patients with a variety of neurodegenerative diseases including Alzheimer's disease, Parkinson's disease, familial amyotrophic lateral sclerosis and polyglutamine expansion diseases, the nature of these studies makes it difficult to ascertain whether ER stress is a cause of neuron death or simply correlated with neuronal damage (44). Recent reports suggest that ER stress may also underlie motor neuron death in a mouse model of familial amyotrophic lateral sclerosis (45,46). Here, we demonstrate a direct relationship between the intensity of ER stress in Purkinje cells and timing of cell death. These data provide further evidence suggesting that ER stress is indeed likely the key inducer of Purkinje cell death, and suggest that therapies aimed at lowering ER stress may be beneficial for patients with MSS or other neurodegenerative diseases.
MATERIAL AND METHODS
Mouse strains
The Sil1−/− mouse strain B6.Sil1Gt(pGT2TMpfa)1Slac has been reported before and has been backcrossed onto the C57BL6/J strain background for over 10 generations (6). These mice were genotyped using LZO136 (5′-CACCGGATGCAGAAAAGCCACAAT-3′), LZO137 (5′-GCAACTCGCCGCACATCTGAACTT-3′), LZO487 (5′-TCACCTCCTGCTCCTTCTCATGC-3′) and LZO488 (5′-TGGATGTGAGAAGCCGTGAGTGA-3′). Hyou1tm1Oga, Dnajc3tm8663Wcl, 129S-Ddit3tm1Dron/J and Tg(CAG-Hyou1)Tam mice are all maintained on a C57BL6/J background (17,32,33,47). The Animal Care and Use Committee of The Jackson Laboratory approved all animal protocols.
Gait analysis
Measurement of rear stance-width was made using a commercially available treadmill system (Clever Sys Inc., Reston, VA, USA) as described previously, using a treadmill speed of 20 cm/s (48). Briefly, the paws of the mice were digitally recorded for a fixed number of frames whereas the animal walks on clear treadmill. Videos were analyzed using interactive analysis software, Treadscan™ 1.0 (Clever Sys, Inc., Reston, VA, USA) that precisely tracks the body and paw positions of the mice during locomotion. To measure rear stance width, the perpendicular distance between the right and left rear paws (measured at the midpoint pixel coordinate) was determined while the paw is in contact with the treadmill, i.e. during the stance phase. The speed of locomotion for analyzed strides was not different between groups (P > 0.05). An average of 19 strides were used to calculate the rear stance width for each mouse. Values were statistically analyzed by paired t-tests. A Bonferroni procedure (49) was used to adjust multiple comparisons.
Histology and immunohistochemistry
For determination of Purkinje cell numbers, neurons were counted from cresyl violet-stained serial sagittal sections from three mice of each genotypes as described previously (50). To count the numbers of Purkinje cells containing ubiquitylated protein inclusions, six matched parasagittal sections within 500 µm from the midline were used and three mice were counted for each genotype. The percentages of Purkinje cells containing inclusions were calculated as ratios of Purkinje cells with inclusions and total numbers of Purkinje cells positive for Calbindin-D28 immunofluoresence. For immunostaining of brain sections, mice were intracardially perfused with 4% paraformaldehyde and brains embedded in paraffin for sagittal sectioning. For Calbindin D-28 colorimetric staining and p97/VCP immunofluorescence, mice were perfused with acetic acid/methanol (1:3). After antigen retrieval, sections were incubated with antibodies against BiP/GRP78, DDIT3/CHOP, calbindin D-28, ubiquitin and GRP94 as previously described (6). Mouse antibody to p97/VCP (Novus Biologicals) and purified rabbit antiserum to HYOU1 was a used at 1:200 and 1:500 dilutions, respectively (51). Brains from at least three mice of each genotype and age tested were analyzed. Immunofluorescence images were obtained with either conventional epifluorescent microscopy, or with confocal microscopy for high magnification, and identical exposure times were used for imaging slides from different genotypes and ages in a given experiment. Images were processed using the same parameters of linear transformations (brightness and contrast only) in Photoshop.
Western blot analysis
For western blot analyses, 20 µg of cerebellar extract was subjected to SDS-acrylamide gel electrophoresis and transferred to nitrocellulose according to standard protocols (52). Blots were probed with an antibody to HYOU1 (1:2000) and signal was detected by the ECL method (Amersham). The results were analyzed with ImageJ (53).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (NS042617 to S.L.A); The Howard Hughes Medical Institute; and The Jackson Laboratory CORE (CA34196). Funding to pay the Open Access publication charges for this article was provided by the Howard Hughes Medical Institute.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Satoshi Ogawa for the Hyou1 targeted and transgenic mouse strains and the HYOU1 antibody. We also thank Dr Michael Katze for the Dnajc3 targeted strain. We thank Jennifer Cook and Thomas Jucius for their superb technical assistance and Drs Robert Burgess and Wayne Frankel for their critical comments on the manuscript. S.L.A. is an investigator of the Howard Hughes Medical Institute.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Kaufman R.J. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes. Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 2.Gething M.J. Role and regulation of the ER chaperone BiP. Semin. Cell. Dev. Biol. 1999;10:465–472. doi: 10.1006/scdb.1999.0318. [DOI] [PubMed] [Google Scholar]
- 3.Meusser B., Hirsch C., Jarosch E., Sommer T. ERAD: the long road to destruction. Nat. Cell. Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 4.Zhao L., Ackerman S.L. Endoplasmic reticulum stress in health and disease. Curr. Opin. Cell. Biol. 2006;18:444–452. doi: 10.1016/j.ceb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Schroder M., Kaufman R.J. ER stress and the unfolded protein response. Mutat. Res. 2005;569:29–63. doi: 10.1016/j.mrfmmm.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 6.Zhao L., Longo-Guess C., Harris B.S., Lee J.W., Ackerman S.L. Protein accumulation and neurodegeneration in the woozy mutant mouse is caused by disruption of SIL1, a cochaperone of BiP. Nat. Genet. 2005;37:974–979. doi: 10.1038/ng1620. [DOI] [PubMed] [Google Scholar]
- 7.Anttonen A.K., Mahjneh I., Hamalainen R.H., Lagier-Tourenne C., Kopra O., Waris L., Anttonen M., Joensuu T., Kalimo H., Paetau A., et al. The gene disrupted in Marinesco–Sjogren syndrome encodes SIL1, an HSPA5 cochaperone. Nat. Genet. 2005;37:1309–1311. doi: 10.1038/ng1677. [DOI] [PubMed] [Google Scholar]
- 8.Senderek J., Krieger M., Stendel C., Bergmann C., Moser M., Breitbach-Faller N., Rudnik-Schoneborn S., Blaschek A., Wolf N.I., Harting I., et al. Mutations in SIL1 cause Marinesco–Sjogren syndrome, a cerebellar ataxia with cataract and myopathy. Nat. Genet. 2005;37:1312–1314. doi: 10.1038/ng1678. [DOI] [PubMed] [Google Scholar]
- 9.Kabani M., Beckerich J.M., Gaillardin C. Sls1p stimulates Sec63p-mediated activation of Kar2p in a conformation-dependent manner in the yeast endoplasmic reticulum. Mol. Cell. Biol. 2000;20:6923–6934. doi: 10.1128/mcb.20.18.6923-6934.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung K.T., Shen Y., Hendershot L.M. BAP, a mammalian BiP-associated protein, is a nucleotide exchange factor that regulates the ATPase activity of BiP. J. Biol. Chem. 2002;277:47557–47563. doi: 10.1074/jbc.M208377200. [DOI] [PubMed] [Google Scholar]
- 11.Kozutsumi Y., Segal M., Normington K., Gething M.J., Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- 12.Wang X.Z., Lawson B., Brewer J.W., Zinszner H., Sanjay A., Mi L.J., Boorstein R., Kreibich G., Hendershot L.M., Ron D. Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153) Mol. Cell. Biol. 1996;16:4273–4280. doi: 10.1128/mcb.16.8.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steel G.J., Fullerton D.M., Tyson J.R., Stirling C.J. Coordinated activation of Hsp70 chaperones. Science. 2004;303:98–101. doi: 10.1126/science.1092287. [DOI] [PubMed] [Google Scholar]
- 14.Weitzmann A., Volkmer J., Zimmermann R. The nucleotide exchange factor activity of Grp170 may explain the non-lethal phenotype of loss of Sil1 function in man and mouse. FEBS Lett. 2006;580:5237–5240. doi: 10.1016/j.febslet.2006.08.055. [DOI] [PubMed] [Google Scholar]
- 15.Tyson J.R., Stirling C.J. LHS1 and SIL1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. Embo. J. 2000;19:6440–6452. doi: 10.1093/emboj/19.23.6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamatani M., Matsuyama T., Yamaguchi A., Mitsuda N., Tsukamoto Y., Taniguchi M., Che Y.H., Ozawa K., Hori O., Nishimura H., et al. ORP150 protects against hypoxia/ischemia-induced neuronal death. Nat. Med. 2001;7:317–323. doi: 10.1038/85463. [DOI] [PubMed] [Google Scholar]
- 17.Kitao Y., Ozawa K., Miyazaki M., Tamatani M., Kobayashi T., Yanagi H., Okabe M., Ikawa M., Yamashima T., Stern D.M., et al. Expression of the endoplasmic reticulum molecular chaperone (ORP150) rescues hippocampal neurons from glutamate toxicity. J. Clin. Invest. 2001;108:1439–1450. doi: 10.1172/JCI12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyazaki M., Ozawa K., Hori O., Kitao Y., Matsushita K., Ogawa S., Matsuyama T. Expression of 150-kd oxygen-regulated protein in the hippocampus suppresses delayed neuronal cell death. J. Cereb. Blood Flow Metab. 2002;22:979–987. doi: 10.1097/00004647-200208000-00009. [DOI] [PubMed] [Google Scholar]
- 19.Kitao Y., Hashimoto K., Matsuyama T., Iso H., Tamatani T., Hori O., Stern D.M., Kano M., Ozawa K., Ogawa S. ORP150/HSP12A regulates Purkinje cell survival: a role for endoplasmic reticulum stress in cerebellar development. J. Neurosci. 2004;24:1486–1496. doi: 10.1523/JNEUROSCI.4029-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu M., Haslam R.H., Haslam D.B. HEDJ, an Hsp40 co-chaperone localized to the endoplasmic reticulum of human cells. J. Biol. Chem. 2000;275:24984–24992. doi: 10.1074/jbc.M000739200. [DOI] [PubMed] [Google Scholar]
- 21.Brightman S.E., Blatch G.L., Zetter B.R. Isolation of a mouse cDNA encoding MTJ1, a new murine member of the DnaJ family of proteins. Gene. 1995;153:249–254. doi: 10.1016/0378-1119(94)00741-a. [DOI] [PubMed] [Google Scholar]
- 22.Shen Y., Meunier L., Hendershot L.M. Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J. Biol. Chem. 2002;277:15947–15956. doi: 10.1074/jbc.M112214200. [DOI] [PubMed] [Google Scholar]
- 23.Kurisu J., Honma A., Miyajima H., Kondo S., Okumura M., Imaizumi K. MDG1/ERdj4, an ER-resident DnaJ family member, suppresses cell death induced by ER stress. Genes Cells. 2003;8:189–202. doi: 10.1046/j.1365-2443.2003.00625.x. [DOI] [PubMed] [Google Scholar]
- 24.Nakanishi K., Kamiguchi K., Torigoe T., Nabeta C., Hirohashi Y., Asanuma H., Tobioka H., Koge N., Harada O., Tamura Y., et al. Localization and function in endoplasmic reticulum stress tolerance of ERdj3, a new member of Hsp40 family protein. Cell Stress Chaperones. 2004;9:253–264. doi: 10.1379/CSC-52.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cunnea P.M., Miranda-Vizuete A., Bertoli G., Simmen T., Damdimopoulos A.E., Hermann S., Leinonen S., Huikko M.P., Gustafsson J.A., Sitia R., et al. ERdj5, an endoplasmic reticulum (ER)-resident protein containing DnaJ and thioredoxin domains, is expressed in secretory cells or following ER stress. J. Biol. Chem. 2003;278:1059–1066. doi: 10.1074/jbc.M206995200. [DOI] [PubMed] [Google Scholar]
- 26.Riezman H. The ins and outs of protein translocation. Science. 1997;278:1728–1729. doi: 10.1126/science.278.5344.1728. [DOI] [PubMed] [Google Scholar]
- 27.Young B.P., Craven R.A., Reid P.J., Willer M., Stirling C.J. Sec63p and Kar2p are required for the translocation of SRP-dependent precursors into the yeast endoplasmic reticulum in vivo. Embo. J. 2001;20:262–271. doi: 10.1093/emboj/20.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong M., Bridges J.P., Apsley K., Xu Y., Weaver T.E. ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. Mol. Biol. Cell. 2008;19:2620–2630. doi: 10.1091/mbc.E07-07-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ushioda R., Hoseki J., Araki K., Jansen G., Thomas D.Y., Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–572. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- 30.Rutkowski D.T., Kang S.W., Goodman A.G., Garrison J.L., Taunton J., Katze M.G., Kaufman R.J., Hegde R.S. The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol. Biol. Cell. 2007;18:3681–3691. doi: 10.1091/mbc.E07-03-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrova K., Oyadomari S., Hendershot L.M., Ron D. Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. Embo. J. 2008;27:2862–2872. doi: 10.1038/emboj.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ladiges W.C., Knoblaugh S.E., Morton J.F., Korth M.J., Sopher B.L., Baskin C.R., MacAuley A., Goodman A.G., LeBoeuf R.C., Katze M.G. Pancreatic beta-cell failure and diabetes in mice with a deletion mutation of the endoplasmic reticulum molecular chaperone gene P58IPK. Diabetes. 2005;54:1074–1081. doi: 10.2337/diabetes.54.4.1074. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi T., Ohta Y. Enforced expression of oxygen-regulated protein, ORP150, induces vacuolar degeneration in mouse myocardium. Transgenic Res. 2003;12:13–22. doi: 10.1023/a:1022176004928. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi T., Takita Y., Suzuki A., Katsu Y., Iguchi T., Ohta Y. Vacuolar degeneration of skeletal muscle in transgenic mice overexpressing ORP150. J. Vet. Med. Sci. 2008;70:115–118. doi: 10.1292/jvms.70.115. [DOI] [PubMed] [Google Scholar]
- 35.Marciniak S.J., Yun C.Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H.P., Ron D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silva R.M., Ries V., Oo T.F., Yarygina O., Jackson-Lewis V., Ryu E.J., Lu P.D., Marciniak S.J., Ron D., Przedborski S., et al. CHOP/GADD153 is a mediator of apoptotic death in substantia nigra dopamine neurons in an in vivo neurotoxin model of parkinsonism. J. Neurochem. 2005;95:974–986. doi: 10.1111/j.1471-4159.2005.03428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pennuto M., Tinelli E., Malaguti M., Del Carro U., D'Antonio M., Ron D., Quattrini A., Feltri M.L., Wrabetz L. Ablation of the UPR-mediator CHOP restores motor function and reduces demyelination in Charcot–Marie–Tooth 1B mice. Neuron. 2008;57:393–405. doi: 10.1016/j.neuron.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song B., Scheuner D., Ron D., Pennathur S., Kaufman R.J. Chop deletion reduces oxidative stress, improves beta cell function, and promotes cell survival in multiple mouse models of diabetes. J. Clin. Invest. 2008;118:3378–3389. doi: 10.1172/JCI34587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anttonen A.K., Siintola E., Tranebjaerg L., Iwata N.K., Bijlsma E.K., Meguro H., Ichikawa Y., Goto J., Kopra O., Lehesjoki A.E. Novel SIL1 mutations and exclusion of functional candidate genes in Marinesco–Sjogren syndrome. Eur. J. Hum. Genet. 2008;16:961–969. doi: 10.1038/ejhg.2008.22. [DOI] [PubMed] [Google Scholar]
- 40.Oyadomari S., Yun C., Fisher E.A., Kreglinger N., Kreibich G., Oyadomari M., Harding H.P., Goodman A.G., Harant H., Garrison J.L., et al. Cotranslocational degradation protects the stressed endoplasmic reticulum from protein overload. Cell. 2006;126:727–739. doi: 10.1016/j.cell.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 41.Yan W., Frank C.L., Korth M.J., Sopher B.L., Novoa I., Ron D., Katze M.G. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc. Natl. Acad. Sci. USA. 2002;99:15920–15925. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Huizen R., Martindale J.L., Gorospe M., Holbrook N.J. P58IPK, a novel endoplasmic reticulum stress-inducible protein and potential negative regulator of eIF2alpha signaling. J. Biol. Chem. 2003;278:15558–15564. doi: 10.1074/jbc.M212074200. [DOI] [PubMed] [Google Scholar]
- 43.Oyadomari S., Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11:381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- 44.Kim I., Xu W., Reed J.C. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat. Rev. Drug Discov. 2008;7:1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 45.Saxena S., Cabuy E., Caroni P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat. Neurosci. 2009;12:627–636. doi: 10.1038/nn.2297. [DOI] [PubMed] [Google Scholar]
- 46.Nishitoh H., Kadowaki H., Nagai A., Maruyama T., Yokota T., Fukutomi H., Noguchi T., Matsuzawa A., Takeda K., Ichijo H. ALS-linked mutant SOD1 induces ER stress- and ASK1-dependent motor neuron death by targeting Derlin-1. Genes Dev. 2008;22:1451–1464. doi: 10.1101/gad.1640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oyadomari S., Takeda K., Takiguchi M., Gotoh T., Matsumoto M., Wada I., Akira S., Araki E., Mori M. Nitric oxide-induced apoptosis in pancreatic beta cells is mediated by the endoplasmic reticulum stress pathway. Proc. Natl. Acad. Sci. USA. 2001;98:10845–10850. doi: 10.1073/pnas.191207498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wooley C.M., Xing S., Burgess R.W., Cox G.A., Seburn K.L. Age, experience and genetic background influence treadmill walking in mice. Physiol. Behav. 2009;96:350–361. doi: 10.1016/j.physbeh.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris R.J. A primer of multivariate statistics. New York: Academic Press; 1975. [Google Scholar]
- 50.Park C., Finger J.H., Cooper J.A., Ackerman S.L. The cerebellar deficient folia (cdf) gene acts intrinsically in Purkinje cell migrations. Genesis. 2002;32:32–41. doi: 10.1002/gene.10024. [DOI] [PubMed] [Google Scholar]
- 51.Kuwabara K., Matsumoto M., Ikeda J., Hori O., Ogawa S., Maeda Y., Kitagawa K., Imuta N., Kinoshita T., Stern D.M., et al. Purification and characterization of a novel stress protein, the 150-kDa oxygen-regulated protein (ORP150), from cultured rat astrocytes and its expression in ischemic mouse brain. J. Biol. Chem. 1996;271:5025–5032. doi: 10.1074/jbc.271.9.5025. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook J., Fritsch E., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Edition. USA: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 1989. New York 11724. [Google Scholar]
- 53.Abramoff M., Magelhaes P., Ram S. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.