Abstract
Human alkyladenine DNA glycosylase initiates the repair of a wide variety of alkylated and deaminated purine lesions in DNA. In this study we take advantage of the natural fluorescence of the 1,N6-ethenoadenosine (εA) lesion and report a kinetic analysis of binding, nucleotide flipping, base excision, and product release. The transient changes in fluorescence of εA revealed the existence of two distinct complexes that are formed prior to the hydrolysis step. An initial recognition complex forms rapidly and is characterized by partial disruption of the stacking interactions of the lesioned base. Subsequently, a very stable extrahelical complex is formed in which the εA lesion is strongly quenched by interactions in the AAG active site pocket. Our results indicate that DNA binding and base flipping take place on the millisecond to second timescale. N-glycosidic bond cleavage is much slower, taking place on the minute time scale. A pulse-chase experiment was used to demonstrate that even for the tightly bound εA substrate, the extrahelical complex is not fully committed to excision. Nevertheless, flipping of εA is highly favorable and we calculate that the equilibrium constant for flipping is ~1300. This kinetic mechanism has important biological implications. First, two-step binding provides multiple opportunities to discriminate between damaged and undamaged nucleotides. Second, a rapid equilibrium flipping mechanism maximizes specificity for damaged versus undamaged bases, since undamaged bases generally form stronger base pairs than damaged bases. Finally, the highly favorable equilibrium for flipping of εA ensures that εA removal is independent of sequence context and highly efficient despite the relatively slow rate of N-glycosidic bond hydrolysis.
The nucleobases of DNA are susceptible to spontaneous chemical modification, including oxidation and alkylation. Failure to repair the resulting DNA lesions can result in mutations or in some cases cell death. The majority of simple base lesions are repaired via the base excision repair (BER) pathway (1). This pathway is initiated by a DNA repair glycosylase that is responsible for finding the damaged site and catalyzing the hydrolysis of the N-glycosidic bond. In the case of a monofunctional DNA glycosylase, this results in the release of the lesioned base and the formation of an abasic site in the DNA. The abasic site is further processed to nick the DNA backbone and remove the abasic sugar residue. Repair synthesis uses the intact strand as a template to ensure incorporation of the correct nucleotide and the nick is ultimately sealed by DNA ligase.
The human enzyme, alkyladenine DNA glycosylase (AAG), is responsible for repairing a diverse set of alkylated and oxidized purines, including purines alkylated at the N7 and N3 positions, and the deaminated purines hypoxanthine (Hx) and oxanine [(2, 3) and references therein]. Chemical structures of damaged and undamaged adenine bases are shown in Scheme 1. AAG also recognizes the bulkier alkylation adduct, 1,N6-ethenoadenosine (εA) that can be formed from lipid peroxidation products and exposure to exogenous agents such as urethane and vinyl chloride (4, 5). This lesion is highly mutagenic in mammalian cells and leads to misincorporation of all possible nucleotides if it is replicated prior to repair (6, 7). AAG is the only known human glycosylase that catalyzes the excision of εA, but an alternative direct reversal pathway has recently been discovered in which εA can be directly oxidized to restore the adenine base by AlkB family members (8). This backup pathway may account for the observation that mice lacking AAG remain viable (9, 10). Nevertheless, mice lacking AAG have decreased rates of repair of εA and are unable to tolerate the burden of increased levels of DNA alkylation (11, 12).
Scheme 1.
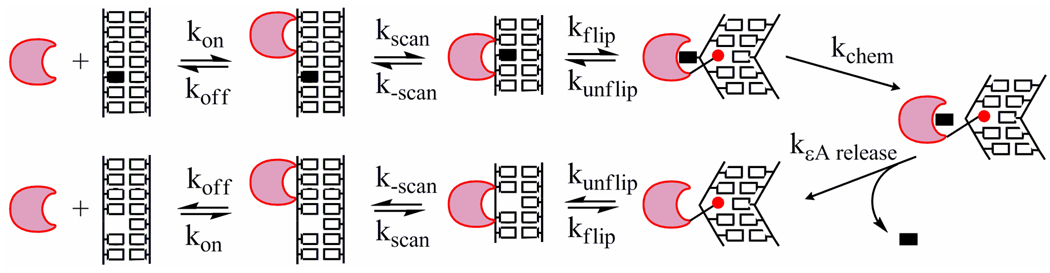
Structural and biochemical studies have provided considerable insight into the mechanism by which AAG recognizes and excises damaged bases and have suggested the minimal mechanism that is illustrated in Figure 1. Given the low frequency with which sites of DNA damage occur, initial binding is most likely to occur at an undamaged site. AAG uses nonspecific binding interactions to scan DNA in the search for sites of damage (13). Once a damaged nucleotide is encountered, it must be flipped out of the duplex by 180° into the enzyme active site where lesion recognition and N-glycosidic bond hydrolysis can occur (14–16). The excised base is released and the abasic nucleotide must disengage from the active site in the reverse of the flipping step. The nonspecifically bound AAG is able to diffuse away along the duplex in search of additional sites of damage and eventually can be released into solution. The rates of DNA binding and nucleotide flipping have not previously been measured for AAG, but are critical to understanding how the enzyme recognizes a wide variety of damaged nucleotides. Several previous studies have used intrinsic protein fluorescence or the incorporation of fluorescent reporter groups into DNA in order to investigate the kinetics of DNA binding and nucleotide flipping by other families of DNA glycosylases (17–22). Together with studies of other DNA modifying enzymes, these studies have begun to address the fundamental, common features of nucleotide flipping enzymes (14).
Figure 1.
Minimal mechanism for AAG-catalyzed nucleotide flipping and base excision. AAG (crescent) binds nonspecifically to DNA and rapidly slides in search of DNA damage. Once a lesion (filled rectangle) is encountered in an initial damage recognition complex it can be flipped out into the active site to form the specific recognition complex. In this specific complex the DNA is bent, the damaged nucleotide is flipped out by 180°, and Y162 (red circle) is intercalated into the DNA to take the place of the extrahelical lesion (15, 16). From this specific complex, AAG catalyzes hydrolysis of the N-glycosidic bond (kchem). The lesion base is subsequently released (kεA release). The abasic product is an inhibitor of AAG, so presumably the sugar can be stably bound in a flipped out conformation analogous to the abasic pyrrolidine conformation observed by x-ray crystallography (15). Dissociation of AAG from the abasic product requires rotation of the abasic sugar back into the duplex (kunflip). From the less tightly bound complex, AAG can translocate along the DNA in search of additional sites of damage and can ultimately dissociate (koff).
There is, however, circumstantial evidence that the requirement for nucleotide flipping provides a barrier to the excision of damaged bases by AAG (2, 23, 24). For example, the rate of excision of a given lesion paired with different opposing bases is inversely proportional to the base pairing stability [e.g., 7-methylguanine (7mG) is excised from a 7mG•T mismatch 50-fold faster than it is removed from a stable 7mG•C pair]. Since AAG does not make specific contacts with the opposing base, this observation is best explained by an AAG complex in which the lesion can still hydrogen bond with the opposing base (2). In addition, rates of repair vary widely for a given lesion base pair in different local sequence contexts (25). The effect of sequence context is most pronounced for excision of Hx (25, 26). For example, rates of excision of Hx from different regions of a polyA tract vary by almost two orders of magnitude (26). An early report mentioned that excision of εA is also sensitive to its position in a polyA tract (27), but subsequent studies found little or no sequence context effects for the excision of εA by AAG (25, 26). We were interested in learning whether nucleotide flipping is rapid, as has been observed for other DNA glycosylases (17, 19–22), or whether the flipping step might be significantly slower than the base excision step.
Since εA is known to be fluorescent and very sensitive to its local environment (28), we investigated the possibility of directly monitoring the dynamics for DNA binding and nucleotide flipping for this biologically relevant DNA adduct. We combined single turnover and multiple turnover glycosylase assays with stopped-flow fluorescence measurements to characterize the kinetic mechanism for AAG-catalyzed excision of εA. We show that AAG binding to the DNA is fast compared to the other steps and that binding is reversible. We present evidence that nucleotide flipping is rapid and highly favorable and that the excised lesion is rapidly released. These fast conformational changes, with relatively slow base excision are integral to the ability of AAG to discriminate between damaged and undamaged nucleotides.
MATERIALS AND METHODS
Purification of Human Recombinant Protein
The catalytic domain of human AAG that lacks the first 79 amino acids was expressed in E. coli and purified as previously described (29). The concentration of active AAG was determined by burst analysis, using an inosine-containing oligonucleotide substrate, as previously described (13, 30).
Synthesis and Purification of Oligodeoxynucleotides
The 25mer oligonucleotides or the constituent phosphoramidites were obtained from commercial sources and contained a central lesion that was placed opposite of a T. The sequences are given in Scheme 2. Oligonucleotides containing only normal deoxynucleotides or deoxyinosine were synthesized and deprotected with standard phosphoramidite chemistry, whereas those containing a single εA lesion were synthesized and deprotected using Ultra-Mild chemistry according to the supplier’s recommendations. The εA-containing oligonucleotides were either synthesized by the W.M. Keck Facility at Yale University or the phosphoramidite was obtained from Glen Research and incorporated using an ABI 394 DNA synthesizer. The oligonucleotides were desalted using Sephadex G-25 and purified using denaturing polyacrylamide gel electrophoresis as previously described (13). Oligonucleotides for gel-based assays were labeled on the lesion containing strands with a 5’-fluorescein (6-fam) label. The concentrations of the single stranded oligonucleotides were determined from the absorbance at 260 nm, using the calculated extinction coefficients for all oligonucleotides except those containing εA. For εA containing oligonucleotides, the extinction coefficient was calculated for the same sequence with an A in place of the εA and corrected by subtracting 9400 M−1cm−1 to account for the weaker absorbance of εA as compared to A (31). The lesion-containing oligonucleotides were annealed with a 1.1-fold excess of the complement by heating to 90°C and cooling slowly to 4°C.
Scheme 2.
Steady-State Fluorescence Measurements
Fluorescence emission spectra were collected with a PTI QuantaMaster fluorometer controlled by FeliX software. For εA fluorescence, an excitation wavelength of 320 nm (6 nm band-pass) was used and the total fluorescence was measured at emission wavelengths from 350–500 nm (6 nm band-pass). Samples (200 µL) of 400 nM εA-containing DNA were prepared in the standard buffer (50 mM NaMES, pH 6.5, 100 mM NaCl, 1 mM EDTA, 1 mM DTT) and spectra were recorded at 25°C. To determine the steady-state fluorescence of εA-containing DNA bound to AAG, the spectra were recorded within 1 minute. No significant excision of εA occurs during this time.
Single turnover excision of εA monitored by fluorescence
Since εA is strongly quenched in duplex DNA and when bound to AAG, we were able to follow the release of εA into solution by the increase in fluorescence. Single turnover glycosylase assays were performed in the standard buffer at 25°C with 400 nM DNA duplex and 1.2–2.4 µM AAG. A full emission spectrum was recorded at various times between 2 and 120 minutes. The greatest change in fluorescence occurred at 410 nm, therefore this wavelength was chosen to follow the release of εA into solution. The fluorescence at 410 nm was used to determine the fraction of εA product (Fraction Product = (Ft–F0)/Fmax; Ft is the fluorescence at time = t, F0 is the initial fluorescence at time = 0 and Fmax is the maximal fluorescence change). This normalization gives fraction product values between 0 and 1. These data were fit by a single exponential equation using nonlinear least squares regression with Kaleidagraph (Synergy Software), in which kobs is the rate constant, t is the time, and A is the amplitude (Equation 1). The single turnover rate constant was determined at saturating concentration of AAG and this was confirmed by the observation of the same rate constant at two different concentrations of AAG (See Supporting Information). Under these conditions, the observed rate constant is equal to the maximal single turnover rate constant (kobs = kmax).
| (1) |
Glycosylase Activity Assay
We also measured single turnover glycosylase activity with the standard glycosylase activity assay that utilizes abasic site cleavage by NaOH followed by DNA separation on a denaturing PAGE gel (13, 29). Fluorescein labeled DNA substrates (20–100 nM) were prepared in the standard buffer with 0.1 mg/mL BSA added. The reactions were initiated with the addition of 100–400 nM AAG and incubated at 25°C. At various time points, a sample from the reaction was removed and quenched in 2 equivalents of 0.3 M NaOH, giving a final hydroxide concentration of 0.2 M. The abasic sites were cleaved by heating at 70°C for 10 minutes. Samples were mixed with an equal volume of formamide/EDTA loading buffer and heated 3 minutes at 70°C before loading onto a 12–15% polyacrylamide gel. Gels were scanned with a Typhoon Imager (GE Healthcare) to detect the fluorescein label by exciting at 488 nm and measuring emission with a 520BP40 filter. The gel bands were quantified using ImageQuant TL (GE Healthcare). The data was converted to fraction product [Fraction Product = product / (product + substrate)] and then fit by a single exponential (Equation 1). The observed rate constant for the single turnover reaction was independent of the concentration of AAG, indicating that the maximal rate constant was measured (kobs = kmax; see supporting information).
Stopped-Flow Kinetics
Pre-steady state kinetic experiments were performed on a Hi-Tech SF-61DSX2, controlled by Kinetic Studio (TgK Scientific). The fluorescence of εA was measured using an excitation wavelength of 320 nm and a WG360 long-pass emission filter. The concentrations that are given for enzyme and substrate are those obtained after mixing equal volumes in the stopped-flow. The reactions were followed for either 2 or 10 seconds to monitor initial binding events occurring before the base excision step could occur. Binding of AAG to εA containing DNA was measured by mixing a constant concentration of DNA with increasing concentrations of AAG. Experiments were performed with either 0.15 µM or 0.5 µM concentration of DNA after mixing, and final concentrations of AAG up to 8 µM. Three traces were averaged together at each concentration, which revealed two phases. The traces were fit by a double exponential (Equation 2), in which F is εA fluorescence as a function of time, C is the fluorescence of the free DNA, Y is the change in fluorescence of the first intermediate, and Z is the change in fluorescence of the second intermediate. The observed rate constants for the first and second steps are designated k1, obs and k2, obs (Equation 2).
| (2) |
It is possible to relate these observed rate constants to the microscopic rate constants in Scheme 3, with a bimolecular binding step followed by a unimolecular conformational change that occurs prior to the chemical step. The initial binding step appears to be reversible (see below), and therefore the observed rate constant is given by the sum of the association and dissociation steps in Scheme 3 (k1,obs = [AAG]k1 + k−1). It should be noted that this equation applies when [E]≫[S]. Under our conditions the enzyme was in only modest excess (2–7-fold). Nevertheless, the resulting values of k1 are in reasonably good agreement with the values measured by mixing equimolar solutions of enzyme and substrate (see below). We suggest that the unimolecular step corresponds to nucleotide flipping. Since this is an approach to equilibrium, the observed rate constant is equal to the sum of the rate constants for the forward and reverse flipping steps (k2,obs = k2 + k−2). Substrate dissociation experiments, measured by pulse-chase suggest that the value of k2 is much greater than k−2, indicating that k2,obs is approximately equal to k2, the microscopic rate constant for flipping.
Scheme 3.
Rapid binding and weak fluorescence made it difficult to fully analyze the binding step under conditions of true excess of enzyme over substrate, therefore we also performed mixing experiments in which a 1:1 ratio of DNA]: [AAG] was maintained (0.15–1.5 µM). Three traces were averaged together at each concentration and they were fit by equation 3, where F is εA fluorescence as a function of time, C is the fluorescence of the free DNA, Y and Z are the change in fluorescence of the first and second intermediates relative to the free DNA, E0 is the starting concentration of AAG and DNA ([AAG] = [DNA]), k1 is the bimolecular rate constant for binding, k2, obs is the observed unimolecular rate constant for the subsequent flipping step, and t is the time. The expression that corresponds to the initial binding step in equation 3 has been previously described (32), and the origin of this equation is provided in the Supporting Information.
| (3) |
The fits to equation 3 were excellent in all cases and the observed rate constants that were obtained were in good agreement with the values of k1 and k2,obs that were obtained from the binding experiments in which AAG was in excess over DNA.
Pulse-Chase Assay
To measure the rate constant for AAG dissociating from the εA-containing DNA substrate, pulse-chase experiments were performed. These assays were done at 25°C in the standard buffer, except that the concentration of NaCl was varied and 0.1 mg/mL BSA was added. In 20 µL reactions, 100 nM fluorescein labeled – TEC– DNA was mixed with 200 nM AAG for 20 seconds. A chase of 20 µM unlabeled – TEC– DNA was then mixed in. If AAG dissociates from the labeled DNA before the chemical cleavage step and then binds to the unlabeled DNA, less of the reaction will occur during the single turnover part of the curve as compared to the same experiment without chase. Samples were taken at the specified times and quenched as described above. The samples were run on sequencing gels and the fraction product was calculated as described above. It was expected that the production of product would follow a simple exponential (Equation 1), but under the conditions employed a very slow steady state reaction was also observed. This was accounted for by Equation 4, in which the burst phase gives the amplitude (A) and an observed rate constant (kobs). The steady state phase is dependent upon the steady state rate constant (kcat), the concentration of enzyme (E), and the total concentration of labeled and unlabeled DNA substrate.
| (4) |
According to the two step binding mechanism described in Scheme 3, two different partitioning equations can be written (33). Since all labeled substrate is initially bound, the fraction of product formed in is given by fraction that goes on to react. This is indicated by Equation 5, in which A is the burst amplitude (the fraction of product cleaved in the burst phase of the experiment), kmax is the maximal single turnover rate constant for formation of product, and koff,obs is the macroscopic rate constant for dissociation from the stable flipped out complex. This expression can be rearranged to solve for the desired dissociation rate constant (Equation 6). Similarly, for branched pathways, the observed rate constant for the burst phase of the pulse-chase experiment is given by the sum of the rate constants for the competing pathways, formation of product is given by kmax and the macroscopic dissociation of substrate is designated koff,obs (Equation 7). Solving for koff,obs give Equation 8.
| (5) |
| (6) |
| (7) |
| (8) |
Control reactions in which no chase was added provided the single turnover rate constant, kmax, and confirmed that these concentrations of AAG were saturating. From these values, the dissociation rate constant, koff, for AAG dissociating from εA-DNA was calculated by two different methods (Equation 6 and 8). Both methods gave essentially identical values for koff,obs and we report the average of the results obtained with both methods.
Since the εA-DNA binds in two steps, the observed rate constant for dissociation of substrate (koff, obs) could be limited by the unflipping rate (k−2) or dissociation from nonspecific DNA (k−1). According to Scheme 3, and assuming that the flipped out complex is stable (i.e., k2 ≫ k−2), this observed dissociation rate can be expressed in terms of the microscopic rate constants [Equation 9; (33)]. Stopped flow fluorescence suggests that dissociation from the initial AAG•DNA complex is rapid (k−1 ~30 s−1) relative to the forward flipping rate constant k2 = 2.6 s−1, and therefore the observed rate constant for substrate dissociation from the εA-DNA•AAG complex is approximately equal to the reverse rate constant for flipping.
| (9) |
The pulse chase experiment was performed at high and low ionic strength by varying the concentration of sodium chloride from 50–300 mM. It has previously been shown that the rate constant for dissociation of the abasic DNA product increases as ionic strength is increased, presumably due to the disruption of electrostatic interactions at higher ionic strength (13, 34).
Steady State Excision of Hypoxanthine
The maximal rate constant for excision of Hx (kcat) was determined for a 25 base pair oligonucleotide of the same sequence as the – TEC– DNA, but with deoxyinosine in place of the εA. Under multiple turnover conditions, the rate-limiting step is dissociation of the abasic DNA [kcat = koff for abasic DNA; (34, 35)]. The experiments were performed at 25°C and in the standard buffer that also contained 0.1 mg/mL BSA. In 20 µL reactions, 2 µM fluorescein labeled DNA was mixed with 20–50 nM AAG. Aliquots were removed at specific times and quenched as described above. These samples were diluted 10-fold into formamide/EDTA buffer and run on sequencing gels and quantified as described above. The fraction product was converted into the concentration of product ([P] = fraction product × [DNA]). The initial velocity (Vi) was determined from a linear fit to the time-dependent formation of abasic product over the first 15% of the reaction. The value of kcat was calculated by dividing the velocity by the concentration of enzyme (kcat = Vi/[E]).
RESULTS
Fluorescence of εA is a Sensitive Reporter of the Lesion Environment
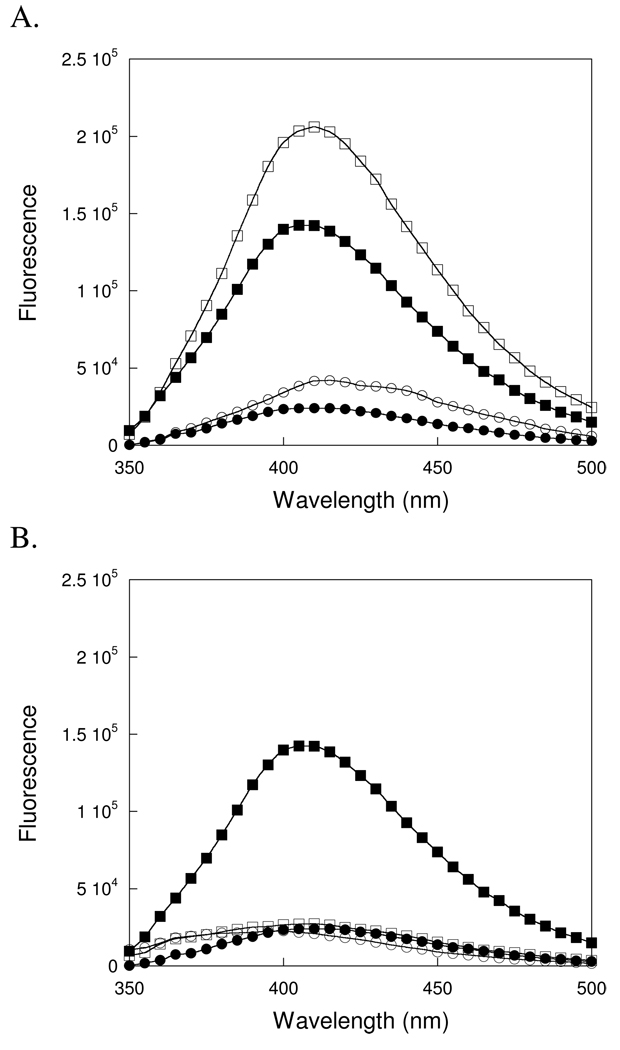
The feasibility of using the intrinsic fluorescence of εA to monitor the individual steps of the AAG-catalyzed reaction was evaluated for two different 25 base pair oligonucleotides that each contained a central εA lesion. These two sequences will be referred to by the central sequence of the lesion-containing strand (Scheme 2). Excitation of both single stranded oligonucleotides at 320 nm gave a broad fluorescence emission peak centered at 410 nm (Figure 2A). The oligonucleotide of sequence –TEC- gave ~4-fold lower fluorescence than free εA, consistent with quenching of εA fluorescence even in single-strand DNA. Surprisingly, the oligonucleotide of sequence –AEA- yielded much higher fluorescence, essentially identical to that of free εA. When a 1.1-fold excess of the appropriate complement was annealed to these sequences, then the fluorescence emission was decreased by a factor of ~1.7-fold for –TEC- and 1.4-fold for –AEA- (Table 1). This is consistent with increased quenching due to enhanced base stacking interactions in duplex DNA. Addition of a higher concentration of complement did not further quench the fluorescence (data not shown). It is not clear why these two sequence contexts show such different quenching, but it is possible that the lack of nearby G nucleotides can account for the high fluorescence of –AEA-. A previous study of charge transfer between εA and normal bases reported that G is the most efficient quencher of εA, with only modest quenching occurring between εA and C, T, or A bases (36). The trivial possibility that the concentration of oligonucleotides was incorrect could be ruled out by measuring the fluorescence of the εA base that is released upon complete hydrolysis by AAG (see below).2
Figure 2.
Fluorescence of εA-containing DNA in single-strand, duplex, and in complex with AAG. Fluorescence emission spectra of 400 nM DNA were collected with excitation at 320 nm, as described in the Materials and Methods. (A) Both single-strand (○) and duplex –TEC– (●) are compared to single-strand (□) and duplex (ν) –AEA– DNA. The εA in both sequences is more fluorescent in the single-strand and becomes quenched in the duplex. (B) Fluorescence emission spectra were collected in the presence of saturating concentration of AAG and compared to the spectra in the absence of AAG. The –TEC– duplex DNA exhibited very similar fluorescence in the absence (●) or presence (○) of AAG. In contrast, the –AEA– duplex DNA, which was strongly fluorescent in the absence of protein (ν), was strongly quenched in the presence of AAG (□). The spectra were taken within 1 minute of AAG addition (1 µM), during which time negligible excision can occur.
Table 1.
Sequence-dependent changes in εA fluorescence and rate constants for binding and excision by AAGa
| Sequence Context |
||
|---|---|---|
| –TEC– | –AEA– | |
| ΔF hybridizationb | 1.7 | 1.4 |
| ΔF bindingb | 1.1 | 5.2 |
| kon (M−1s−1)c | 2.1 ± 0.9 × 108 | 1.5 ± 0.4 × 108 |
| koff (s−1)d | 33 ± 15 | 48 ± 15 |
| kflip (s−1)e | 2.6 ± 0.1 | 4.3 ± 0.4 |
| kunflip (s−1)f | 2.0 ± 0.2 × 10−3 | NDk |
| Kflipg | 1300 | NDk |
| kchem (s−1)h | 5.7 ± 0.3 × 10−4 | 5.3 ± 0.2 × 10−4 |
| Kdns (nM)i | 160 | 320 |
| KdεA-DNA (nM)j | 0.1 | NDk |
Values are reported at 25 °C in a buffer containing 50 mM NaMES, pH 6.5, 100 mM NaCl, 1 mM EDTA, and 1mM DTT.
εA fluorescence was determined with 400 nM εA-containing DNA and a 3 mm pathlength cuvet with excitation at 320 nm and emission at 410 nm. The ΔF hybridization is the ratio of the fluorescence of single-stranded DNA divided by the fluorescence of the double-stranded DNA (Figure 2A). The ΔF binding to AAG is the ratio of the fluorescence of the double-stranded DNA divided by the fluorescence of the saturated AAG•DNA complex (Figure 2B).
The association rate constant (kon) was determined from the data in Figure 6.
The dissociation rate constant (koff) was determined from the data in Figure 5.
The rate constant for flipping was determined by stopped flow (Figure 7). As discussed in the text, the second phase of the change in εA fluorescence is simply kflip, since kunflip is much slower than kflip. Given the similar rate constants for the two sequences, we assumed that the observed rate constant for the –AEA– sequence is also the flipping rate constant.
The reverse rate constant for flipping (kunflip; Figure 1), is determined from the macroscopic rate constant for dissociation of the substrate (Figure 8), as described in the text.
The equilibrium for nucleotide flipping is defined as the ratio of the forward rate constant for flipping measured by stopped-flow and the reverse rate constant for flipping (Kflip = k2/k−2; Scheme 3).
The rate for the chemical step is simply the rate constant for the single-turnover excision of εA (Figure 3D), since the equilibrium constant for base flipping is highly favorable and the rate constant for flipping is much faster than the hydrolysis step. The values shown are determined from εA fluorescence. For the –TEC– DNA, a similar value of 6.8 ± 0.3 × 10−4 s−1 was determined from the gel-based assay.
The dissociation constant for the nonspecific AAG•DNA complex is given by the ratio of the off and on rates measured by stopped flow (Kdns = koff/kon).
The dissociation constant for the specific, flipped-out complex between AAG and εA-DNA is defined by KdεA-DNA = Kdns/Kflip.
ND, not determined.
Since the AAG-catalyzed excision of εA is relatively slow under these conditions, it was possible to examine the effect of AAG binding to each oligonucleotide in a conventional fluorometer. The addition of one equivalent of AAG resulted in a dramatic 5-fold decrease in the fluorescence of the –AEA– substrate, whereas that of the –TEC– substrate was only slightly quenched (Figure 2B). Addition of more than one equivalent of AAG had no further effect on either spectra (data not shown). The two oligonucleotides exhibit almost identical fluorescence emission when they are bound to AAG (Figure 2B). This observation strongly suggests that the εA is in a similar environment in both complexes, most likely flipped out of the duplex and bound in the AAG active site. Crystal structures of the AAG•εA-DNA complex indicate potential quenching interactions between the εA lesion and active site tyrosines Y127 and Y159 (16). The environment of the protein active site is more strongly quenching than the –AEA– sequence context, but very similar to that of the –TEC– sequence context. The strong quenching of both complexes relative to free εA in solution suggested that a continuous fluorescence-based assay could be used to follow the εA excision activity of AAG.
Single Turnover Glycosylase Activity of AAG Monitored by εA Fluorescence and by Formation of the Abasic DNA Product
Hydrolysis of the N-glycosidic bond by AAG results in the formation of an abasic site and a free base (Figure 1). Early assays of AAG employed alkylated genomic DNA and relied upon directly detecting the released base by HPLC or scintillation counting (37, 38). More commonly, gel-based discontinuous assays are used to detect the formation of a single strand break (27, 29, 39). These assays can employ either a fluorescent or radioactive label, and the abasic product is converted into a single-strand nick by hydrolyzing the abasic site at alkaline pH. In contrast, the fluorescent properties of εA provide the opportunity for a continuous glycosylase assay. We directly compared the fluorescence-based assay to the traditional gel-based assay to determine whether εA release is fast or slow and to evaluate the potential of using fluorescence to monitor the glycosylase activity of AAG.
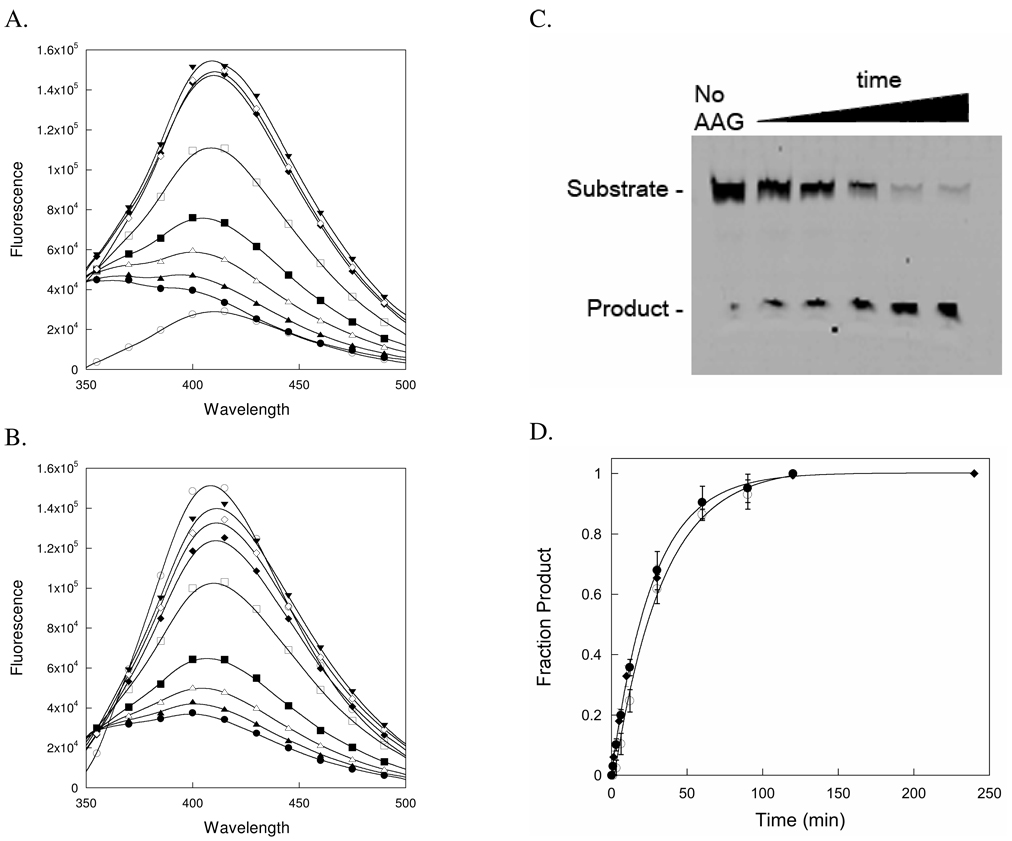
Under single turnover conditions, with enzyme in excess over the DNA substrate, AAG was incubated with oligonucleotide –TEC– and fluorescence emission spectra were collected at the indicated intervals (Figure 3A). The εA fluorescence (λmax = 410 nm) increased over time, reaching a maximum value at ~2 hours. Under identical conditions, a fluorescein-labeled oligonucleotide of the identical sequence (–TEC–) was incubated with AAG and aliquots were quenched in sodium hydroxide and analyzed by denaturing polyacrylamide gel electrophoresis (Figure 3C). The change in fluorescence was converted into the fraction of εA released into solution and the fraction of abasic product was quantified from the gel and these data were plotted together (Figure 3D; See Materials and Methods for details). The reaction progress curves were readily fit by a single exponential, and the observed rate constants for εA release (kmaxeA = 0.036 ± 0.002 min−1) and for abasic site formation (kmax = 0.038 ± 0.002 min−1) were identical within error. A simultaneous fit to both reaction progress curves gave the same value (kmax = 0.038 min−1; Figure 3D). Since the detection of the abasic DNA product by sodium hydroxide quench/hydrolysis does not require dissociation of the εA, the observation of the same rate constant for release of εA into solution is most simply consistent with rate-limiting N-glycosidic bond hydrolysis, followed by rapid release of εA.
Figure 3.
Comparison of εA release and N-glycosidic bond cleavage from single turnover assays. Fluorescence emission spectra monitoring the increase in fluorescence that is observed upon single turnover excision of εA from (A) –TEC– and (B) –AEA– DNA catalyzed by AAG. The spectra for samples containing 400 nM duplex DNA were collected prior to the addition of protein (○), and within 1 minute of the addition of 1.2 µM AAG (●). Additional spectra were collected after 3 (σ), 6 (∆), 12 (ν), 30 (□), 60 (υ), 90 (◊), and 120 (τ) minutes. Note that the high fluorescence of the –AEA– substrate is strongly quenched upon binding to AAG. Representative experiments are shown, but duplicate reactions gave essentially identical results, and the rate of reaction was independent of the concentration of AAG. (C) The gel-based glycosylase assay relies on selective hydroxide-catalyzed cleavage of abasic sites and the different mobility of intact substrate and nicked product to monitor formation of the abasic DNA product (See Materials and Methods for details). A representative denaturing polyacrylamide gel shows the separation of the fluorescein-labeled substrate (25-mer) and product (12-mer) and a time course for the formation of the abasic DNA product. The first lane shows a no enzyme control. The small amount of nicked DNA (~4%) results from nonenzymatic ring-opening and depurination during synthesis, purification, and the hydroxide heating step. The next 5 lanes indicate samples taken from a reaction containing 100 nM –TEC– DNA and 200 nM AAG that were quenched 2–120 minutes after the initiation of the reaction by the addition of AAG. (D) Quantification of the results from εA release and abasic site formation indicates that both assays are limited by a common step, presumably N-glycosidic bond cleavage. The fraction product was calculated from the εA fluorescence assay for –TEC– (●) and –AEA– (○) as well as the gel based excision assay for –TEC– (υ), as described in the Materials and Methods. The error bars indicate the standard deviation from the mean for 2–5 independent determinations. The reaction progress curves for –TEC– monitored by εA fluorescence and abasic site formation are fit by a single exponential (Equation 1; kmax = 0.038 min−1). The reaction of –AEA– is slightly slower (kmax = 0.031 min−1).
The single turnover assay for release of εA was also performed for the –AEA– DNA under the same conditions that were used for the –TEC– DNA (Figure 3B). The initial fluorescence of this DNA in the absence of AAG is much higher than for the – TEC– sequence, but upon the addition of AAG the fluorescence was strongly quenched. The observed increase in fluorescence was of similar magnitude to that which was observed for the –TEC– substrate, and a similar single turnover rate constant was observed (kmax = 0.031 ± 0.002 min−1; Figure 3D). 2 These data indicate that once AAG is bound, it shows very similar efficiency of excising εA from these two different sequence contexts.
Association and Nucleotide Flipping Monitored by Stopped-Flow Fluorescence
The quenching of εA fluorescence observed upon binding of AAG to εA-containing DNA suggested that εA would be a useful reporter to monitor association of AAG and DNA. Therefore, we performed stopped-flow fluorescence experiments to monitor the time-dependent changes in εA fluorescence. Two phases were observed when excess AAG was mixed with 500 nM –TEC– DNA, both of which occur well before the base excision step (Figure 4A). In the first 100 ms, there is a transient increase in fluorescence and this is followed by a decrease in fluorescence that is complete within a few seconds after mixing. The fluorescent traces were best fit by a double exponential equation, indicating a two-step model for DNA binding (Scheme 3). As increasing concentrations of AAG were mixed with a fixed concentration of –TEC– DNA, the rate of the first phase increased (Figure 4A and Figure 5). However, at concentrations above 1 uM AAG this initial phase occurred in the mixing time of the stopped flow. The dependence of the first phase of the fluorescence change upon the concentration of reactants indicates that a bimolecular event is being monitored. It should be noted that the fit of the two concentrations of AAG with the –TEC– substrate in Figure 5 is tenuous, however it was possible to measure binding over a wider concentration range for the more highly fluorescent –AEA– DNA.
Figure 4.
Stopped-flow fluorescence measurements of the binding of εA-DNA to AAG with excess protein. Representative data is shown from experiments in which a fixed concentration of DNA was mixed with increasing concentrations of AAG. Samples were excited at 320 nm and emission was collected with a 360 nm long-pass filter (See Materials and Methods for details). Several individual reactions were monitored at each concentration of AAG, and the traces shown are the averages of all reactions. Since the fluorescence changes were superimposed at the different concentrations of AAG, the traces are offset for clarity. (A) The binding of 0.5 µM –TEC– duplex DNA by 0.5 (bottom), 1, 2, 4, and 8 µM AAG (top). (B) The binding of 0.15 µM –AEA– duplex DNA by 0.5 (bottom), 1, and 2 µM (top) AAG. The lines indicate the best fits of equation 2 to each binding reaction.
Figure 5.
Determination of the forward and reverse rate constants for the initial binding of AAG to εA-DNA. The observed rate constant for DNA binding (k1, obs) is from the first phase of the binding reactions shown in Figure 4. The average and standard deviations are shown for 2–4 determinations. The dissociation rate constant for the initial AAG•DNA complex can be determined from the y-intercept of this plot (k1, obs = k−1 + k1[AAG]; Scheme 3). For –AEA– DNA (ν), the best linear fit (solid line) gives an intercept of 48 s−1 (k−1) and a slope of 1.5×108 M−1s−1 (k1). This second order rate constant is identical to the value determined from equimolar binding experiments (Table 1 and Figure 7). For –TEC– DNA (●), only two concentrations of protein gave reliable data due to the weak fluorescence and more rapid binding. A linear fit (dotted line) of the two data points that were obtained gives an intercept of 22 s−1 and a slope of 2.4 × 108 M−1s−1. By forcing the slope of the line to be 2.1× 108 M−1s−1 (dashed line), the value that was independently determined from equimolar binding reactions (Table 1 and Figure 7), an intercept of 43 s−1 was obtained. We take the average (33 ± 15 s−1) as a rough estimate for the k−1 value for this substrate.
Since initial binding is expected to be away from the site of damage, and the bimolecular step gives a detectable change in fluorescence, we interpret this behavior to reflect a rate-limiting binding step with no change in fluorescence, followed by a rapid sliding step that results in the formation of a complex with a detectable change in fluorescence. We infer that the scanning step is too fast to be directly observed in our experiments (Figure 1), with a lower limit of 250 s−1 indicated by the data in Figure 5. The increased fluorescence of this early intermediate is likely to reflect partial unstacking of the εA lesion during the initial damage recognition by AAG. The second phase of the binding reaction was insensitive to the concentration of AAG and we interpret this to be the flipping of the εA lesion into the AAG active site, where it is strongly quenched. For the –TEC– substrate, this AAG-bound complex happens to have very similar fluorescence to that of free DNA duplex in solution (Figure 2B).
We performed analogous stopped-flow binding experiments with the –AEA– substrate (Figure 4B). Binding of AAG to this sequence appears to be superficially different, without a transient increase in fluorescence, but is also best explained by a two-step binding mechanism. For this substrate, the fluorescence is initially high in the free DNA, is partially quenched in the first intermediate, and is more strongly quenched in the fully flipped complex. These traces could not be fit well by a single exponential (data not shown), but were well fit by a double exponential (Figure 4B). As the concentration of AAG was increased at a fixed concentration of DNA, the rate for the first exponential was proportional to the amount of AAG, and the second phase was not (Figure 4 and 5 and data not shown). The fits to the second phase of the reaction were excellent in all cases, but the fits to the first phase were more variable due to the very fast binding. As the fluorescence of εA is relatively weak, it was not possible to use significantly lower concentrations of DNA that would have allowed more accurate determination of the association and dissociation rate constants of this fast binding reaction.
Therefore, the stopped-flow experiments were repeated with equimolar amounts of enzyme and substrate, with concentrations ranging from 0.15–1.5 µM (Figure 6). This causes the binding reaction to slow as free ligand is consumed, allowing a time-dependent change in fluorescence to be observed even at high initial concentrations. Similar experiments have been used to study binding reactions of equimolar protein fragments (40, 41). Since the unimolecular flipping step also gives an observed fluorescence change, a second exponential term is also required (Equation 3). The fitting of this equation to the data in Figure 6 gave similar association rate constants of ~2 × 108 M−1s−1 for both sequences (Figure 7A; Table 1). Experiments carried out across a 10-fold concentration range gave the same rate constant. This better-defined value for the association rate constant (k1; Scheme 3) was used to fit the data from the concentration dependence of AAG when protein was in excess and provide an estimate of the dissociation rate constant (k−1 ≈ 30 s−1; Figure 5 and Table 1).
Figure 6.
Kinetics of DNA binding and nucleotide flipping, determined by stopped-flow fluorescence at equimolar concentrations of AAG and DNA. AAG was rapidly mixed with an equimolar solution of –TEC– (A) or –AEA– (B) DNA to yield final concentrations after mixing of 0.15 (bottom) to 1.5 µM (top). Each trace is the average of 3 independent binding reactions, and the lines indicate the best fits of equation 3. For the –TEC– substrate the raw data is plotted, indicating the increased signal observed at higher concentrations of DNA, but for the –AEA– substrate the larger fluorescence change required that the gain be adjusted between different concentrations of DNA and therefore the individual traces are arbitrarily offset.
Figure 7.
The rate constants for binding and nucleotide flipping by AAG in two different sequence contexts. The observed rate constants for –TEC– DNA (●, solid line) and – AEA– DNA (ν, dashed line) were taken from the fits of equation 3 to the data for equimolar binding reactions (Figure 6). (A) The observed bimolecular rate constant for the initial binding of AAG to DNA is very similar for both substrates (~2 × 108 M−1s−1; Table 1). According to the simple two-step binding mechanism depicted in Scheme 3, this observed rate constant is simply k1, the microscopic rate constant for binding. (B) The observed rate constant for the second step is independent of concentration, and yields an average value of 4.3 s−1 for the –AEA– sequence context compared to a value of 2.6 s−1 for the –TEC– sequence context. According to Scheme 3, this observed rate constant is the sum of the forward and reverse rate constants for flipping (k2, obs = k2 + k−2). Additional experiments described below indicate that the equilibrium constant for flipping is highly favorable (k2 ≫ k−2), and therefore the observed rate constant for flipping is simply the forward rate constant for flipping (see text for additional discussion).
The second phase of the binding reaction followed a single exponential and the same rate constant was observed across the concentration range tested (Figure 7B). The observed rate constant for flipping is an approach to equilibrium and therefore is the sum of the forward and reverse rate constants for flipping (k2, obs = k2 + k−2; Scheme 3). Below we describe a pulse-chase experiment that resolves this ambiguity by establishing that the reverse rate constant for flipping (k−2) is much slower than the rate constant measured by the stopped flow fluorescence, such that the observed rate constant is simply the forward rate constant for flipping (k2, obs = kflip). The nucleotide flipping step (kflip) is significantly faster for the –AEA– sequence than for the –TEC– sequence (4.3 ± 0.4 s−1 versus 2.6 ± 0.1 s−1). The difference in the rates of flipping in two different sequence contexts may reflect subtle differences in how AAG engages these different DNA molecules or sequence-dependent changes in the intrinsic motions of DNA. Regardless of the origin of the differences between the two sequence contexts that were examined, these data establish that the flipping step is much faster than the N-glycosidic bond hydrolysis step (Table 1).
A Pulse-Chase Assay to determine the macroscopic rate of substrate dissociation
In order to gain further insight into the two-step binding mechanism of AAG, we also examined the dissociation of substrate from AAG by a pulse-chase assay (30, 33). In this experiment, fluorescein-labeled –TEC– substrate was briefly incubated with AAG (t1 = 20 s) to give it time to bind, and then was chased with a 100-fold excess of –TEC– substrate lacking the fluorescein label (Figure 8A). Samples were taken over the normal single turnover time course to evaluate the partitioning of a bound E•S complex between N-glycosidic bond hydrolysis and dissociation. Figure 8B shows the εA-excision reaction in the presence and absence of chase. The addition of chase caused a decrease in the fraction of substrate hydrolyzed and an increase in the observed rate constant, as expected if the forward rate constant for reaction is similar to the rate constant for dissociation (see Methods for details). Under these conditions (I = 120 mM), the macroscopic rate constant for dissociation is 0.12 ± 0.02 min−1 (Table 1). Since the equilibrium for bound εA-DNA lies towards the fully flipped out complex, either k−1 or k−2 could in principle limit the observed rate constant for dissociation. However, the rapidly reversible initial binding step exhibits a much faster rate constant (k−1 ~ 30 s−1; Figure 5). Therefore, the observed rate constant for the dissociation of the substrate that was measured by the pulse-chase assay is simply the unflipping rate constant (k−2; Scheme 3; see the Materials and Methods for additional details).
Figure 8.
Pulse-chase experiment to measure the macroscopic dissociation rates for εA-containing DNA substrate. (A) The experimental design is indicated. Fluorescein (fam)-labeled DNA (100 nM –TEC–) was mixed with excess AAG (200 nM) for 20 seconds (incubation time, t1) then 20 µM unlabeled –TEC– DNA was added as a chase. The reactions were quenched at the indicated time points (t2), and the fraction of abasic DNA product determined by alkaline hydrolysis and gel electrophoresis (See Materials and Methods for additional details). (B) Single turnover excision of εA from –TEC– DNA by AAG in the absence (●) or presence (ν) of chase under the standard reaction conditions. In the absence of chase, the line indicates the best fit to a single exponential (Equation 1). In the presence of chase, the line indicates the best fit of an exponential burst followed by a slow steady state reaction (Equation 4). This gives a macroscopic dissociation rate constant of 0.12 ± 0.01 min−1. (C) Comparison of pulse chase experiments performed at concentrations of sodium chloride. The data from experiments at (●) 70 mM, (ν) 120 mM and (σ) 320 mM ionic strength were fit by equation 4 (lines). Analysis of the observed rate constant and the endpoint gave essentially identical macroscopic rate constants and the average and standard deviation of duplicate reactions determined by both methods yielded macroscopic dissociation rate constants of 0.08 ± 0.005, 0.12 ± 0.01, and 0.35 ± 0.05 min−1, respectively for 70, 120, and 320 mM ionic strength.
It has previously been observed that the rate of dissociation of AAG from its abasic DNA product is extremely sensitive to the ionic strength, with a 25-fold increase in the rate of dissociation between 40 and 120 mM ionic strength (34). This effect of ionic strength is presumably due to electrostatic interactions between the positively charged protein and the negatively charged DNA that become weaker at higher ionic strength. In order to evaluate whether the unflipping rate constant or the DNA dissociation rate constant is rate-limiting, we varied the ionic strength with the expectation that the dissociation step should exhibit a large ionic strength effect. The results of pulse-chase experiments carried out at 70, 120, and 320 mM ionic strength are shown in Figure 8C. The pulse-chase experiment revealed a small decrease in amplitude and concomitant increase in the observed rate constant as the ionic strength is increased. Control reactions established that the single turnover rate constant in the absence of chase is independent of ionic strength under these conditions (data not shown), consistent with previous results (13, 34). Therefore, the ~4-fold change in the observed substrate dissociation rate constant between 70 and 320 mM is further evidence that k−2 and not k−1 is rate limiting for dissociation of εA-DNA. This moderate sensitivity to ionic strength indicates either a small effect of ionic strength on the flipping step, or a small contribution from the rate constant for dissociation from nonspecific DNA.
Rate constant for dissociation of the abasic DNA product
We measured the rate of dissociation of the abasic DNA product to compare it to the rate of εA product release. The steady state reaction of AAG with –TEC- substrate gave a kcat value that is essentially identical to the single turnover rate constant under these reaction conditions (data not shown), suggesting that the very slow rate of hydrolysis of the N-glycosidic bond is rate limiting for multiple turnover excision of εA at high ionic strength. Since AAG shows much faster rates of excision of Hx from inosine lesions, we measured the kcat value for multiple turnover on an inosine-containing substrate. It has previously been shown that AAG exhibits burst kinetics for the excision of Hx and that the steady state rate constant, kcat, reflects the rate of dissociation of the abasic DNA product (34). We determined that the rate constant for the dissociation of abasic DNA is 0.24 min−1 under these reaction conditions, ~6-fold faster than the rate constant for N-glycosidic bond cleavage (Scheme 4; see Supporting Information). This is only slightly faster than the observed rate constant for dissociation of the εA substrate (koff, obs = 0.12 min−1), suggesting that the abasic DNA product is also likely to be stably bound in an extrahelical conformation.
Scheme 4.
DISCUSSION
Minimal kinetic and thermodynamic mechanism for excision of εA
We have used the intrinsic fluorescence of the εA lesion to establish the kinetic mechanism for the AAG-catalyzed recognition and excision of this alkylated base. The fluorescence of εA is highly sensitive to the local environment of the lesion and it proved to be a sensitive reporter of three distinct steps in the AAG-catalyzed reaction; initial lesion recognition, nucleotide flipping, and εA product release. The rate constants that were determined from changes in εA fluorescence are in excellent agreement with the rate constants measured by the traditional glycosylase assay that detects the abasic DNA product, indicating that εA release is fast and that fluorescence provides a useful assay of AAG activity.
The kinetic mechanism for recognition and excision of εA is depicted for the – TEC– sequence in Scheme 4. Initial binding is near the diffusion-limited rate and is rapidly reversible. Given that a detectable change in εA fluorescence occurs as a bimolecular reaction, we cannot distinguish between the possibilities that initial binding by AAG occurs solely at the εA lesion or that AAG binds at another site but rapidly transfers to the εA lesion. We favor the latter model, since it is known that AAG can use nonspecific DNA binding interactions to translocate between sites (13). Since these data require the scanning step(s) to be very rapid (kscan; Figure 1), this step is not shown in Scheme 4. This initial recognition complex is rapidly reversible, with a dissociation rate constant of ~1800 min−1. This indicates a relatively weak complex for initial nonspecific DNA binding and we calculate a dissociation constant of ~150 nM from these data (Kdns; Table 1). Occasionally, AAG catalyzes full nucleotide flipping with a rate constant of ~160 min−1. The extrahelical complex is highly stable, with a calculated equilibrium constant of ~1300 for the flipping step, and this tight binding decreases the dissociation constant for binding of AAG to an εA lesion by several orders of magnitude to approximately 100 pM (Table 1). This exceptionally tight binding to the εA site is consistent with previous reports of sub-nanomolar binding by AAG under different experimental conditions (2, 42). The crystal structure of AAG bound to εA reveals a snug fit of the εA lesion into the AAG pocket where it accepts a hydrogen bond from the backbone amide of His136 (16). This interaction with His136 can also account for the discrimination against normal adenosine nucleotides (2, 16). Tight binding of the damaged nucleotide by AAG could protect it from DNA-templated activities such as transcription and replication.
Despite the rapid and highly favorable flipping of the εA lesion, AAG is limited by a very slow rate of N-glycosidic bond cleavage, such that the specific recognition complex is only partially committed to base excision. Release of εA, measured by the increased fluorescence of the free base, is much faster than the rate of N-glycosidic bond cleavage measured by formation of the abasic DNA product. At 25 °C, this rate constant for N-glycosidic bond cleavage is 0.04 min−1, 5-fold slower than is observed at 37 °C (2). Under these conditions, base excision is also slower than release of the abasic DNA product, such that no burst is observed (Table 1 and data not shown). Since N-glycosidic bond cleavage for inosine is much faster than for εA, the slow release of the abasic DNA product is rate-limiting for the multiple turnover reaction of AAG with inosine and other rapidly hydrolyzed lesions (34). Rate-limiting release of the abasic DNA product provides the opportunity for coordination of the steps of the BER pathway. It has previously been shown that the slow rate of dissociation of AAG from its abasic DNA product can be stimulated by APE1, the enzyme that acts next in the base excision repair pathway (34, 35, 43).
Recognition of εA in a polyA-tract
Earlier studies had investigated the AAG-catalyzed excision of εA from different sequence contexts. One study reported that the excision of εA by AAG is sensitive to the sequence context and that an εA lesion in the center of a polyA-tract is a particularly poor substrate for AAG (27). In contrast, a later report examined a number of similar sequence contexts and found that there did not appear to be but significant effects of sequence context on the excision of εA (26). However, Hx-containing substrates show large sequence context effects hinting at a fundamental difference between the recognition of εA and Hx (25, 26, 43, 44). In this study we performed a more thorough kinetic characterization to compare the recognition of εA in the polyA-tract (–AEA–) and in a heterogenous sequence (–TEC–).
Overall, the steps of εA recognition and excision by AAG are very similar for the two DNA substrates (Table 1). The rate constants for DNA binding and excision are identical within error, but the observed rate constant for flipping is ~1.7-fold higher for the polyA-tract sequence. Although this is a small change, the rate constant for flipping is independent of DNA and enzyme concentration and is sufficiently well defined by the stopped-flow data to indicate a real difference. This difference in rate could be due to changes in either the structure or the dynamics of different DNA sequences. Nevertheless, since nucleotide flipping is rapid and highly favorable, the difference in flipping rate does not translate into a significant difference in the rate of εA excision from these two sequences. The minimal kinetic and thermodynamic framework (Scheme 3) provides a molecular explanation for why AAG does not exhibit strong sequence-dependent differences in the rates of excision of εA, but does show strong effects for other lesions. By extension, it is very likely that other lesions that are recognized by AAG, such as the deaminated base lesion Hx, are not stably flipped and thus changes in stacking interactions have a larger effect on the flipping equilibrium and on the observed rates of excision (2, 25).
Comparison of AAG to other glycosylases
There are several distinct structural and evolutionary families of DNA repair glycosylases (45, 46). These enzymes differ in their chemical mechanisms and in the identity of the lesions that they recognize, but nonetheless all recognize damaged bases and catalyze the cleavage of the N-glycosidic bond (47, 48). The observation that all of these enzymes that recognize nucleotides in duplex DNA catalyze nucleotide flipping attests to the requirement of the enzyme to gain access to the N-glycosidic bond that is occluded in duplex DNA (26). Fluorescence methods have been used to study the DNA binding and nucleotide flipping steps for a few of these enzymes and the results from the literature are summarized in Table 2. Since each of these enzymes belongs to a different superfamily and therefore evolved independently, it is interesting to consider whether there are common functional constraints that have dictated the kinetic mechanism of the enzyme (46, 49, 50). One crude classification that can be used is whether the enzyme exhibits a narrow substrate range (recognizing one or a few structurally related lesions) or whether it exhibits a broad substrate range (recognizing a variety of structurally diverse lesions). The rate constant for flipping varies across two orders of magnitude from 3 s−1 to more than 1000 s−1. With these limited data, it seems that the enzymes with a narrow substrate range exhibit faster flipping rates than those with a broader substrate range. This could be related to the difficulty of optimizing the rate of flipping of distinct lesions by the enzymes with a broad substrate range, or it could simply reflect the slow rate of N-glycosidic bond cleavage since the flipping step is still much faster than the hydrolysis step. Indeed, the nucleotide flipping step is much faster than the N-glycosidic bond cleavage step for all of these enzymes (Table 2).
Table 2.
Comparison of nucleotide flipping by DNA repair glycosylases
| Enzyme | Superfamilya | Substrate | kflip (s−1) | kunflip (s−1) | Kflip | kchem (s−1)b | Reference |
|---|---|---|---|---|---|---|---|
| Narrow substrate range | |||||||
| UDG | UDG | 2’-fluoroU•G | 1260 | 38 | 33 | N/Ac | (17) |
| U•A | 1100 | 370 | 3 | 58 | (18) | ||
| U (–AUA–)d | 670 | 16 | 41 | 38 | (19) | ||
| U (–GUC–)d | 48 | 12 | 4 | NDe | (19) | ||
| T4 Pdg | HhH | D•2APf | ≥300 | NDe | ≥10 | N/Ac | (20) |
| Ogg1 | HhH | 8-oxo-G•C | 13 | 1 | 10 | 0.07 | (21) |
| Broad substrate range | |||||||
| Fpg | Fpg | 8-oxo-G•C | 12 | 12 | 1 | 0.16 | (22) |
| AAG | AAG | εA•T | 3 | 0.002 | 1300 | 0.0006 | This study |
HhH, helix-hairpin-helix superfamily. UDG (uracil DNA glycosylase), Fpg (formamidopyrimidine DNA glycosylase), and AAG are the founding members of distinct enzyme superfamilies. T4 Pdg, pyrimidine dimer DNA glycosylase from phage T4, and Ogg1, human 8-oxoguanine DNA glycosylase.
kchem is the rate constant for the first chemical step, hydrolysis of the N-glycosidic bond.
N/A, not applicable.
Rate constants for binding of UDG to single-stranded DNA with two different sequence contexts. It should be noted that a very different process of nucleotide selection is expected in single strand and duplex DNA, but the values are included here because they are remarkably similar to the values that were independently measured for the enzyme binding to duplex DNA.
ND, not determined.
The D•2AP (tetrahydrofuran•2-aminopurine) pair mimics an abasic site product.
The equilibrium constant for flipping is close to unity for formamidopyrimidine DNA glycosylase (Fpg), the only enzyme with a broad substrate range that has been previously characterized. It makes sense that an enzyme that can accommodate structurally diverse lesions might not be able to hold all lesions tightly. In contrast, the enzymes that recognize a narrow range of substrates all exhibit favorable equilibrium constants for flipping of ≥10. Interestingly, AAG has an exceptionally broad range of substrates and yet also has the most highly favorable equilibrium constant for flipping that has been measured. Biochemical evidence suggests that εA is unique among the well-characterized AAG substrates, because other lesions such as 7-methylguanosine and inosine appear to have unfavorable equilibrium constants for flipping by AAG (2, 25).
Implications of multi-stage recognition for the biological specificity of AAG
The findings presented above have several implications for the initiation of base excision repair by AAG. The most obvious is that the kinetic mechanism is optimized for specificity rather than speed. The vast excess of undamaged nucleotides in DNA provides a challenge to an enzyme that must recognize a broad range of structurally distinct substrates. Two-step binding is rapid and readily reversible at each step, which provides multiple opportunities for discrimination between damaged and undamaged bases. The multiplicative effect of multi-stage recognition allows even small differences between normal and damaged bases to be magnified to achieve the specificity that is required to recognize rare lesions that can occur throughout the genome.
Once the lesion is flipped, rate-limiting N-glycosidic bond cleavage allows for additional discrimination between damaged and undamaged nucleotides. Pyrimidines are selected against by the catalytic mechanism, which is optimized for purine nucleotides, and A and G are selected against by steric clashes with their exocyclic amino groups (2, 16, 29, 51). Since alkylated nucleotides that bear positive charge are activated for N-glycosidic bond cleavage, some alkylated purines are removed at biologically relevant levels despite the relatively poor rate enhancement (52, 53).
We propose that the highly favorable equilibrium constant for the flipping of εA by AAG is the exception that proves the rule. The opposing base effects that have previously been noted strongly suggest that the equilibrium constant for flipping methylated and deaminated purines is unfavorable (2, 25). In spite of this unfavorable equilibrium, many of these other lesions are excised by AAG more rapidly than the εA lesion is excised. The favorable equilibrium constant for εA helps to compensate for the very slow rate of base excision. If εA is not repaired, it is highly mutagenic, and therefore it must be removed from the genome. However, human cells have a relatively long cell cycle and this provides a wide window of time during which repair can be performed prior to DNA replication. Therefore DNA repair enzymes can afford to sacrifice speed (overall rate of reaction) for increased selectivity (ability to recognize subtle perturbations in the chemical structure of DNA). AAG is able to effectively locate rare sites of damage that are hidden in the sea of undamaged DNA by employing multiple DNA binding steps that are rapidly reversible and each provide specificity towards DNA damage.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dave Ballou and Bruce Palfey for advice and for the use of their stopped flow instruments, Carol Fierke for the suggestion of studying binding at equimolar concentrations, and Suzanne Admiraal, Alex Ninfa, and members of the O’Brien lab for helpful discussions and comments on the manuscript. We also thank an anonymous reviewer for careful critique of the kinetics.
Abbreviations
- AAG
Alkyladenine DNA glycosylase, also known as methylpurine DNA glycosylase (MPG) and 3-methyladenine DNA glycosylase
- BER
base excision repair
- BSA
bovine serum albumin
- εA
1,N6-ethenoadenine
- Fpg
formamidopyrimidine DNA glycosylase
- Hx
hypoxanthine (the base moiety of inosine)
- I
ionic strength
- NaHEPES
sodium N-(2-hydroxyethyl)piperazine-N′-(2-ethanesulfonate)
- NaMES
sodium 2-(N-morpholino)ethanesulfonate
- PAGE
polyacrylamide gel electrophoresis
Footnotes
This work was supported in part by a fellowship from the University of Michigan Chemical Biology Interface training grant to A.W., a Faculty Research Grant from the Horace H. Rackham School of Graduate Studies to P.O., and by a grant from the NIH to P.O. (CA122254).
Samples that contained 400 nM of either –TEC– or –AEA– duplex (determined using the UV-absorbance and the calculated extinction coefficients) gave identical fluorescence emission spectra upon the complete hydrolysis by AAG (Figure 3A and 3B). This eliminates the trivial possibility that different amounts of DNA were compared. Therefore, we conclude that the –AEA– sequence has significantly higher fluorescence than the –TEC– sequence in both single-strand and when annealed to its complement (Figure 2).
SUPPORTING INFORMATION AVAILABLE
Fluorescence excitation spectra of the εA-containing DNA, representative single turnover data showing that a saturating concentration of AAG was used to measure the single-turnover reaction, and representative multiple turnover data that was used to determine kcat for the excision of Hx. This material is available free of charge via the internet at http://pub.acs.org.
REFERENCES
- 1.Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Brien PJ, Ellenberger T. Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. J Biol Chem. 2004;279:9750–9757. doi: 10.1074/jbc.M312232200. [DOI] [PubMed] [Google Scholar]
- 3.Hitchcock TM, Dong L, Connor EE, Meira LB, Samson LD, Wyatt MD, Cao W. Oxanine DNA glycosylase activity from Mammalian alkyladenine glycosylase. J Biol Chem. 2004;279:38177–38183. doi: 10.1074/jbc.M405882200. [DOI] [PubMed] [Google Scholar]
- 4.Gros L, Ishchenko AA, Saparbaev M. Enzymology of repair of etheno-adducts. Mutat Res. 2003;531:219–229. doi: 10.1016/j.mrfmmm.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Speina E, Zielinska M, Barbin A, Gackowski D, Kowalewski J, Graziewicz MA, Siedlecki JA, Olinski R, Tudek B. Decreased repair activities of 1,N(6)-ethenoadenine and 3,N(4)-ethenocytosine in lung adenocarcinoma patients. Cancer Res. 2003;63:4351–4357. [PubMed] [Google Scholar]
- 6.Pandya GA, Moriya M. 1,N6-ethenodeoxyadenosine, a DNA adduct highly mutagenic in mammalian cells. Biochemistry. 1996;35:11487–11492. doi: 10.1021/bi960170h. [DOI] [PubMed] [Google Scholar]
- 7.Levine RL, Yang IY, Hossain M, Pandya GA, Grollman AP, Moriya M. Mutagenesis induced by a single 1,N6-ethenodeoxyadenosine adduct in human cells. Cancer Res. 2000;60:4098–4104. [PubMed] [Google Scholar]
- 8.Ringvoll J, Moen MN, Nordstrand LM, Meira LB, Pang B, Bekkelund A, Dedon PC, Bjelland S, Samson LD, Falnes PO, Klungland A. AlkB homologue 2-mediated repair of ethenoadenine lesions in mammalian DNA. Cancer Res. 2008;68:4142–4149. doi: 10.1158/0008-5472.CAN-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elder RH, Jansen JG, Weeks RJ, Willington MA, Deans B, Watson AJ, Mynett KJ, Bailey JA, Cooper DP, Rafferty JA, Heeran MC, Wijnhoven SW, van Zeeland AA, Margison GP. Alkylpurine-DNA-N-glycosylase knockout mice show increased susceptibility to induction of mutations by methyl methanesulfonate. Mol Cell Biol. 1998;18:5828–5837. doi: 10.1128/mcb.18.10.5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelward BP, Weeda G, Wyatt MD, Broekhof JL, de Wit J, Donker I, Allan JM, Gold B, Hoeijmakers JH, Samson LD. Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc Natl Acad Sci U S A. 1997;94:13087–13092. doi: 10.1073/pnas.94.24.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kisby GE, Olivas A, Park T, Churchwell M, Doerge D, Samson LD, Gerson SL, Turker MS. DNA repair modulates the vulnerability of the developing brain to alkylating agents. DNA Repair (Amst) 2009;8:400–412. doi: 10.1016/j.dnarep.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meira LB, Moroski-Erkul CA, Green SL, Calvo JA, Bronson RT, Shah D, Samson LD. Aag-initiated base excision repair drives alkylation-induced retinal degeneration in mice. Proc Natl Acad Sci U S A. 2009;106:888–893. doi: 10.1073/pnas.0807030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hedglin M, O'Brien PJ. Human alkyladenine DNA glycosylase employs a processive search for DNA damage. Biochemistry. 2008;47:11434–11445. doi: 10.1021/bi801046y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts RJ, Cheng X. Base flipping. Annu Rev Biochem. 1998;67:181–198. doi: 10.1146/annurev.biochem.67.1.181. [DOI] [PubMed] [Google Scholar]
- 15.Lau AY, Scharer OD, Samson L, Verdine GL, Ellenberger T. Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision. Cell. 1998;95:249–258. doi: 10.1016/s0092-8674(00)81755-9. [DOI] [PubMed] [Google Scholar]
- 16.Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T. Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase. AAG, Proc Natl Acad Sci U S A. 2000;97:13573–13578. doi: 10.1073/pnas.97.25.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stivers JT, Pankiewicz KW, Watanabe KA. Kinetic mechanism of damage site recognition and uracil flipping by Escherichia coli uracil DNA glycosylase. Biochemistry. 1999;38:952–963. doi: 10.1021/bi9818669. [DOI] [PubMed] [Google Scholar]
- 18.Wong I, Lundquist AJ, Bernards AS, Mosbaugh DW. Presteady-state analysis of a single catalytic turnover by Escherichia coli uracil-DNA glycosylase reveals a "pinch-pull-push" mechanism. J Biol Chem. 2002;277:19424–19432. doi: 10.1074/jbc.M201198200. [DOI] [PubMed] [Google Scholar]
- 19.Bellamy SR, Krusong K, Baldwin GS. A rapid reaction analysis of uracil DNA glycosylase indicates an active mechanism of base flipping. Nucleic Acids Res. 2007;35:1478–1487. doi: 10.1093/nar/gkm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker RK, McCullough AK, Lloyd RS. Uncoupling of nucleotide flipping and DNA bending by the t4 pyrimidine dimer DNA glycosylase. Biochemistry. 2006;45:14192–14200. doi: 10.1021/bi060802s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuznetsov NA, Koval VV, Zharkov DO, Vorobjev YN, Nevinsky GA, Douglas KT, Fedorova OS. Pre-steady-state kinetic study of substrate specificity of Escherichia coli formamidopyrimidine--DNA glycosylase. Biochemistry. 2007;46:424–435. doi: 10.1021/bi060787r. [DOI] [PubMed] [Google Scholar]
- 22.Kuznetsov NA, Koval VV, Zharkov DO, Nevinsky GA, Douglas KT, Fedorova OS. Kinetics of substrate recognition and cleavage by human 8-oxoguanine-DNA glycosylase. Nucleic Acids Res. 2005;33:3919–3931. doi: 10.1093/nar/gki694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vallur AC, Feller JA, Abner CW, Tran RK, Bloom LB. Effects of hydrogen bonding within a damaged base pair on the activity of wild type and DNA-intercalating mutants of human alkyladenine DNA glycosylase. J Biol Chem. 2002;277:31673–31678. doi: 10.1074/jbc.M204475200. [DOI] [PubMed] [Google Scholar]
- 24.Biswas T, Clos LJ, 2nd, SantaLucia J, Jr, Mitra S, Roy R. Binding of specific DNA base-pair mismatches by N-methylpurine-DNA glycosylase and its implication in initial damage recognition. J Mol Biol. 2002;320:503–513. doi: 10.1016/S0022-2836(02)00519-3. [DOI] [PubMed] [Google Scholar]
- 25.Vallur AC, Maher RL, Bloom LB. The efficiency of hypoxanthine excision by alkyladenine DNA glycosylase is altered by changes in nearest neighbor bases. DNA Repair (Amst) 2005;4:1088–1098. doi: 10.1016/j.dnarep.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 26.Wyatt MD, Samson LD. Influence of DNA structure on hypoxanthine and 1,N(6)-ethenoadenine removal by murine 3-methyladenine DNA glycosylase. Carcinogenesis. 2000;21:901–908. doi: 10.1093/carcin/21.5.901. [DOI] [PubMed] [Google Scholar]
- 27.Hang B, Sagi J, Singer B. Correlation between sequence-dependent glycosylase repair and the thermal stability of oligonucleotide duplexes containing 1, N6-ethenoadenine. J Biol Chem. 1998;273:33406–33413. doi: 10.1074/jbc.273.50.33406. [DOI] [PubMed] [Google Scholar]
- 28.Leonard NJ. Etheno-substituted nucleotides and coenzymes: fluorescence and biological activity. CRC Crit Rev Biochem. 1984;15:125–199. doi: 10.3109/10409238409102299. [DOI] [PubMed] [Google Scholar]
- 29.O'Brien PJ, Ellenberger T. Human alkyladenine DNA glycosylase uses acid-base catalysis for selective excision of damaged purines. Biochemistry. 2003;42:12418–12429. doi: 10.1021/bi035177v. [DOI] [PubMed] [Google Scholar]
- 30.Rose IA, O'Connell EL, Litwin S. Determination of the rate of hexokinase-glucose dissociation by the isotope-trapping method. J Biol Chem. 1974;249:5163–5168. [PubMed] [Google Scholar]
- 31.Secrist JA, 3rd, Barrio JR, Leonard NJ, Weber G. Fluorescent modification of adenosine-containing coenzymes. Biological activities and spectroscopic properties. Biochemistry. 1972;11:3499–3506. doi: 10.1021/bi00769a001. [DOI] [PubMed] [Google Scholar]
- 32.Fersht A. Structure and Mechanism in Protein Science. New York: W.H. Freeman and Company; 1999. pp. 200–201. [Google Scholar]
- 33.Hsieh J, Walker SC, Fierke CA, Engelke DR. Pre-tRNA turnover catalyzed by the yeast nuclear RNase P holoenzyme is limited by product release. RNA. 2009;15:224–234. doi: 10.1261/rna.1309409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baldwin MR, O'Brien PJ. Human AP Endonuclease I Stimulates Multiple-Turnover Base Excision by Alkyladenine DNA Glycosylase. Biochemistry. 2009;48:6022–6033. doi: 10.1021/bi900517y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maher RL, Vallur AC, Feller JA, Bloom LB. Slow base excision by human alkyladenine DNA glycosylase limits the rate of formation of AP sites and AP endonuclease 1 does not stimulate base excision. DNA Repair (Amst) 2007;6:71–81. doi: 10.1016/j.dnarep.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 36.Kelley SO, Barton JK. Electron transfer between bases in double helical DNA. Science. 1999;283:375–381. doi: 10.1126/science.283.5400.375. [DOI] [PubMed] [Google Scholar]
- 37.Riazuddin S, Lindahl T. Properties of 3-methyladenine-DNA glycosylase from Escherichia coli. Biochemistry. 1978;17:2110–2118. doi: 10.1021/bi00604a014. [DOI] [PubMed] [Google Scholar]
- 38.O'Connor TR. Purification and characterization of human 3-methyladenine-DNA glycosylase. Nucleic Acids Res. 1993;21:5561–5569. doi: 10.1093/nar/21.24.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wyatt MD, Allan JM, Lau AY, Ellenberger TE, Samson LD. 3-methyladenine DNA glycosylases: structure, function, and biological importance. Bioessays. 1999;21:668–676. doi: 10.1002/(SICI)1521-1878(199908)21:8<668::AID-BIES6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 40.de Prat Gay G, Ruiz-Sanz J, Fersht AR. Generation of a family of protein fragments for structure-folding studies. 2. Kinetics of association of the two chymotrypsin inhibitor-2 fragments. Biochemistry. 1994;33:7964–7970. doi: 10.1021/bi00191a025. [DOI] [PubMed] [Google Scholar]
- 41.Schreiber G, Fersht AR. Interaction of barnase with its polypeptide inhibitor barstar studied by protein engineering. Biochemistry. 1993;32:5145–5150. doi: 10.1021/bi00070a025. [DOI] [PubMed] [Google Scholar]
- 42.Scharer OD, Verdine GL. A designed inhibitor of base excision repair. J. Am. Chem. Soc. 1995;117:10781–10782. [Google Scholar]
- 43.Xia L, Zheng L, Lee HW, Bates SE, Federico L, Shen B, O'Connor TR. Human 3-methyladenine-DNA glycosylase: effect of sequence context on excision, association with PCNA, and stimulation by AP endonuclease. J Mol Biol. 2005;346:1259–1274. doi: 10.1016/j.jmb.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 44.Abner CW, Lau AY, Ellenberger T, Bloom LB. Base excision and DNA binding activities of human alkyladenine DNA glycosylase are sensitive to the base paired with a lesion. J Biol Chem. 2001;276:13379–13387. doi: 10.1074/jbc.M010641200. [DOI] [PubMed] [Google Scholar]
- 45.Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 46.O'Brien PJ. Catalytic promiscuity and the divergent evolution of DNA repair enzymes. Chem Rev. 2006;106:720–752. doi: 10.1021/cr040481v. [DOI] [PubMed] [Google Scholar]
- 47.Berti PJ, McCann JA. Toward a detailed understanding of base excision repair enzymes: transition state and mechanistic analyses of N-glycoside hydrolysis and N-glycoside transfer. Chem Rev. 2006;106:506–555. doi: 10.1021/cr040461t. [DOI] [PubMed] [Google Scholar]
- 48.Stivers JT, Jiang YL. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 49.Gerlt JA, Babbitt PC. Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu Rev Biochem. 2001;70:209–246. doi: 10.1146/annurev.biochem.70.1.209. [DOI] [PubMed] [Google Scholar]
- 50.Glasner ME, Gerlt JA, Babbitt PC. Evolution of enzyme superfamilies. Curr Opin Chem Biol. 2006;10:492–497. doi: 10.1016/j.cbpa.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 51.Connor EE, Wyatt MD. Active-site clashes prevent the human 3-methyladenine DNA glycosylase from improperly removing bases. Chem Biol. 2002;9:1033–1041. doi: 10.1016/s1074-5521(02)00215-6. [DOI] [PubMed] [Google Scholar]
- 52.O'Brien PJ, Ruiz-Sanz J. The Escherichia coli 3-methyladenine DNA glycosylase AlkA has a remarkably versatile active site. J Biol Chem. 2004;279:26876–26884. doi: 10.1074/jbc.M403860200. [DOI] [PubMed] [Google Scholar]
- 53.Berdal KG, Johansen RF, Seeberg E. Release of normal bases from intact DNA by a native DNA repair enzyme. Embo J. 1998;17:363–367. doi: 10.1093/emboj/17.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.