Abstract
Chronic generation of reactive nitrogen species (RNS) can cause DNA damage and may also directly modify DNA repair proteins. RNS-modified DNA is repaired predominantly by the base excision repair (BER) pathway, which includes the alkyladenine DNA glycosylase (AAG). The AAG active site contains several tyrosines and cysteines that are potential sites for modification by RNS. In vitro, we demonstrate that RNS differentially alter AAG activity depending on the site and type of modification. Nitration of tyrosine 162 impaired 1,N6-ethenoadenine (εA)-excision activity, whereas nitrosation of cysteine 167 increased εA excision. To understand the effects of RNS on BER in vivo, we examined intestinal adenomas for levels of inducible nitric oxide synthase (iNOS) and AAG. A striking correlation between AAG and iNOS expression was observed (r = 0.76, P = 0.00002). Interestingly, there was no correlation between changes in AAG levels and enzymatic activity. We found AAG to be nitrated in human adenomas, suggesting that this RNS modification is relevant in the human disease. Expression of key downstream components of BER, apurinic/apyrimidinic endonuclease 1 (APE1) and DNA polymerase β (POLβ), was also examined. POLβ protein was increased in nearly all adenomas compared with adjacent non-tumor tissues, whereas APE1 expression was only increased in approximately half of the adenomas and also was relocalized to the cytoplasm in adenomas. Collectively, the results suggest that BER is dysregulated in colon adenomas. RNS-induced posttranslational modification of AAG is one mechanism of BER dysregulation, and the type of modification may define the role of AAG during carcinogenesis.
Introduction
Sustained generation of elevated nitric oxide (•NO) is thought to be a key driving force in the association between chronic inflammation and increased cancer risk (1). Tissue damage manifests from the formation of reactive nitrogen species (RNS), such as peroxynitrite (ONOO−), nitrogen dioxide (•NO2) and nitrous anhydride (N2O3) (2,3). RNS can directly deaminate DNA bases (4,5). In addition, secondary lipid peroxidation products can react with DNA to form exocyclic DNA adducts such as 1,N6-ethenoadenine (εA) and N2,3-ethenoguanine (5,6). Repair of RNS-induced and oxidative DNA base adducts is initiated by several DNA glycosylases of the base excision repair (BER) pathway (7). Alkyladenine DNA glycosylase (AAG, also known as methylpurine glycosylase, alkylpurine DNA-N-glycosylase) appears to be the primary mammalian DNA glycosylase that removes hypoxanthine, εA and N2,3-ethenoguanine (8–13). We have focused on AAG-mediated BER because several reports have shown that levels of an AAG substrate, εA, increased under chronic inflammatory conditions in mouse models (5,13,14). Furthermore, elevated etheno adducts have been observed in colonic polyps from familial adenomatous patients (15). Examination of tissue biopsies from noncancerous colons of patients with ulcerative colitis has revealed that the activity of AAG and apurinic/apyrimidinic endonuclease 1 (APE1) increased and that patients with microsatellite instability have the highest increase in the levels of AAG and APE1 within inflamed colons (16).
In addition to reacting with DNA, RNS can modify proteins. Nitration and nitrosation are two types of posttranslational modifications that can regulate protein function and alter signaling pathways (17–20). Two other BER components, the OGG1 DNA glycosylase and APE1, have been shown to undergo posttranslational modification by RNS (21,22). Protein exposure to nitrating RNS, OONO− and •NO2, can result in formation of nitro-adducts on aromatic groups, such as tyrosyl residues. Nitrosating RNS, such as N2O3, attack nucleophilic sites, notably cysteinyl thiolates. In this regard, AAG provides an intriguing target because of the number of tyrosine and cysteine residues in close proximity to the active site (Figure 1). Here, we show that, depending on the specific modification, RNS may differentially modulate AAG activity. Moreover, we provide to our knowledge the first simultaneous examination of alterations in the first three enzymes in BER in intestinal adenoma samples and matched non-tumor controls. Lastly, demonstration of nitrated AAG in subjects with colon cancer suggests this RNS modification may be an important contributor to BER imbalance in adenocarcinomas.
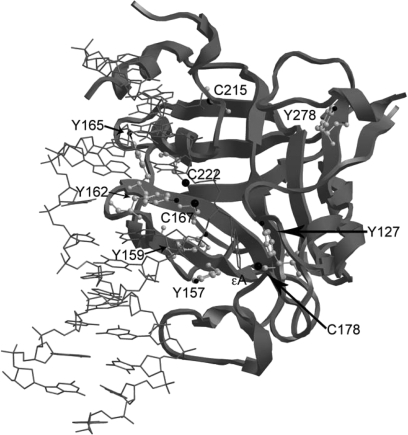
Fig. 1.
Crystal structure of AAG bound to εA-containing DNA. The structure was generated using the UCSF Chimera software package (23) and protein databank file 1F4R (24). Tyrosines and cysteines are represented by a gray ball and stick structure within the ribbon structure of AAG. The flipped εA is shown in gray as a stick structure.
Materials and methods
Generation of AAG variants and purification of proteins
Purification of wild-type and all variant AAG proteins was carried out as described previously (25,26). The AAG construct is Δ54-N-terminally truncated and contains all native tyrosines and six of seven cysteines (lacks C15). Greater stability in vitro was evident with the Δ54-AAG construct relative to longer variants. The Δ79-truncated protein crystallized lacks two tyrosines and three cysteines (24,27). Although many have reported that the N-terminus is not required for enzymatic activity, the N-terminus can influence the activity of AAG (11,28). Variants containing substitutions at tyrosine and cysteine positions were generated by site-specific mutagenesis with the appropriate primers. Generation of the Δ79 variant was carried out by polymerase chain reaction of the wild-type template using a primer to generate an N-terminal truncated fragment with a 5′-end cleavable by NdeI and the T7 terminator as the 3′-primer. The amplification product was cleaved by NdeI and XhoI and purified prior to ligation. The sequence of every variant was confirmed by sequencing in both directions. All AAG variants were expressed from the pET17 vector in Escherichia coli strain C41 (25,26). At a cell density of 0.5 (A600), the culture was induced with 0.4 mM isopropylthiogalacto-pyranoside for 18 h at 25°C. Cells were lysed on ice in sonication buffer containing, 4 mM β-mercaptoethanol, 0.5 M NaCl, 20 mM Tris–HCl pH 7.9 and protease inhibitor cocktail (Sigma, St Louis, MO). After sonication, the cells were spun down and the clear lysates were removed. The cell lysates were injected into a superloop and purified by fast protein liquid chromatography using a HiTrap Chelating HP affinity column (GE Amersham, Piscataway, NJ). Fractions containing purified AAG were determined by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and pooled. The pooled fractions were concentrated in a Millipore ultrafree-15 centrifugation filter device (10 kDa cutoff). Protein purification was confirmed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (supplementary Figure 1 is available at Carcinogenesis Online) and the protein concentration was determined by Bradford assay.
Treatment of AAG with RNS agents
Prior to enzymatic assays or western analysis, AAG (5 μM) was treated with a nitrating agent, ONOO− or a nitrosating agent, spermine NONOate (SPER/NO), in 0.1 M sodium phosphate buffer (pH 7.0). Conditions were kept constant for pH and temperature among all experiments. Nitration reactions were carried out at 25°C for 5 min. Metal-catalyzed nitration of AAG was carried out by incubating Δ54 AAG (5 μM) with 100 μM NaNO2, 100 μM H2O2 and 2 μM FeSO4 at 37°C for 2 h (29). Unless otherwise noted, nitrosation reactions with SPER/NO were carried out at 30°C for 10 min. Nitrosothiols were converted to a stable modification prior to western analysis by substituting the NO-adduct with a biotin tag according to protocol using S-Nitrosylated Protein Detection Assay Kit (Cayman, Ann Arbor, MI).
Preparation of human and mouse tissue extracts
Human adenocarcinoma colon tissue was collected as described previously (30). Colon tissues from ApcMin/+ mice were kindly provided by Dr Marj Pena and Celestia Davis (Center for Colon Cancer Research, University of South Carolina). ApcMin/+ mice used in this study were males ∼9 months of age. Care and use of animals was overseen by the Animal Resource Facility of the University of South Carolina, under the direction of our veterinarian. The Animal Resource Facility is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care-International and is registered with the United States Department of Agriculture (56-R-003). The Animal Resource Facility also has an active letter of Assurance of Compliance on file at the National Institutes of Health. The use of human material was approved by the Institutional Review Board at the University of South Carolina. After the animals were killed, the tissues were split so that half of the samples were fixed in formalin and embedded in paraffin for immunohistochemistry, and the other half of the samples were processed for activity assays (8) and immunoprecipitations (31).
Western analysis
Ascites from the mouse monoclonal anti-human AAG antibody (clone 506-3D) were kindly provided by Dr S.J.Kennel (Oak Ridge National Laboratory) (32). Antibody was purified as described and diluted 1:1000 (33). Electrophoresis was carried out in denaturing 10% gels and proteins electrotransferred onto polyvinylidene difluoride membrane. 3-Nitrotyrosine was detected using anti-nitrotyrosine polyclonal antibody (Upstate, Billerica, MA, 06-284) diluted 1:1000 and incubated with anti-rabbit horseradish peroxidase-conjugated secondary antibody. Nitrosocysteine was detected using S-nitrosylation detection reagent I (avidin-conjugated horseradish peroxidase) as supplied in S-Nitrosylated Protein Detection Assay Kit. The bands were visualized by Supersignal West Pico chemiluminescent substrate (Pierce, Rockford, IL).
Immunoprecipitation
Whole human colonic epithelial tissue lysates and HT-29 colorectal adenocarcinoma cell lysates were obtained, and immunoprecipitation was carried out as described previously (31). Briefly, protein extracts (400 μg) were precleared by incubation with agarose beads (Calbiochem, Gibbstown, NJ) at 4°C while rocking for 1 h, followed by centrifugation. The precleared supernatant was incubated with the anti-AAG or anti-nitrotyrosine antibodies overnight at 4°C. Following overnight incubation, agarose beads were added to the supernatant and incubated for 1 h at 4°C. The pellet (agarose–antibody–antigen complex) was separated by centrifugation and washed (31). Immunoprecipitated protein was extracted from agarose–antibody complex by heat denaturation in Laemmli buffer, separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and electrotransferred onto polyvinylidene difluoride membrane for western blot using the anti-AAG or anti-nitrotyrosine antibodies. Nonspecific IgG was used as the negative control.
Immunohistochemistry analysis
Slides containing sections of tumor and non-tumor colon tissue from ApcMin/+ mice were analyzed by immunohistochemistry (IHC) as described (34). Tissues were incubated with antibodies against inducible nitric oxide synthase (iNOS; mouse monoclonal, clone 5D5-H7, cat# MC-5245; diluted 1 in 20 000, Research & Diagnostic Antibodies, North Las Vegas, NV), APE1 (mouse monoclonal, clone 13B8E5C2, cat# NB100-116; diluted 1 in 2 × 106, Novus, Littleton, CO), DNA polymerase β (POLβ; rabbit polycolonal, cat# AB53059; diluted 1 in 10 000, Abcam, Cambridge, MA) and AAG (as described above; diluted 1 in 1 × 106). Immunohistochemical staining was quantified blinded and independently by two experienced researchers (L.J.H. and A.B.H.). Expression was quantified by the percentage of epithelial cells staining positive and averaged.
Excision and binding assays
AAG activity was measured similarly to that described previously (26), except that the 25mer oligonucleotide containing εA (Midlands Certified Reagents, Midlands, TX) also contained a 5′-HEX. Single-turnover reactions were carried out by incubating 20 nM AAG and substrate at the time points shown. Multi-turnover reactions were carried out by incubating 1 nM AAG with increasing concentrations of DNA (5, 10, 15, 20 and 25 nM). Binding affinity of AAG for the εA-containing oligonucleotide was evaluated by electrophoretic mobility shift assays and analyzed using a one-site binding equation (Graphpad Prism, Graphpad, La Jolla, CA) as described (26).
Statistical analysis
SPSS (SPSS, Chicago, IL) and Graphpad Prism software were used for data analysis. A Pearson correlation coefficient (r) was applied for all comparisons of protein levels. The P value chosen for significance in this study was 0.05. The kinetic values, Vmax and Km, the activity of wild-type AAG and cysteine variants were obtained by linear regression analysis or using the Michaelis–Menten model and the equilibrium binding constants (Kd) were analyzed using the one-site binding model. The rate of catalysis (kcat) for wild-type AAG and cysteine variants were analyzed using the first order rate equation, [P]t = Ao[1−exp(kobst)], where Ao = amplitude of the exponential phase and kobs = observed rate constant.
Results
Tyrosine nitration decreases the activity of AAG
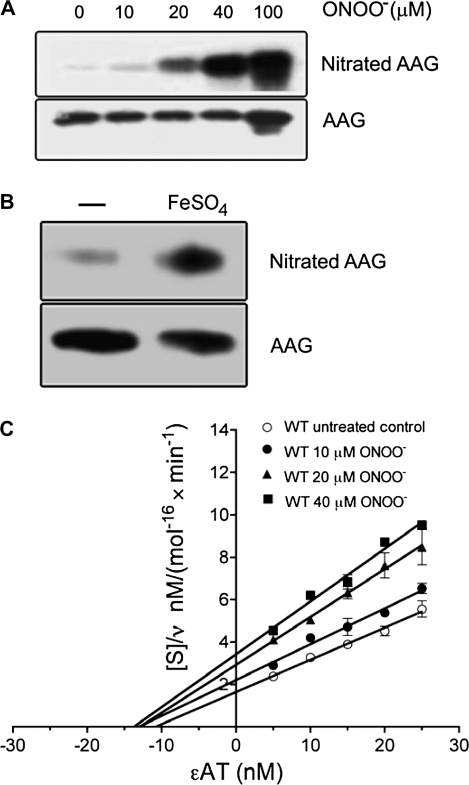
The potential for RNS to form posttranslational adducts on AAG protein and their influences on catalysis were investigated. Figure 1 was generated from the coordinates of the crystal structure of a Δ79-N-terminally truncated AAG bound to εA-containing DNA (24). As shown, five AAG protein tyrosyl residues are part of the active site or interact with the DNA structure immediately surrounding the damaged base to be excised, suggesting that nitration of these residues could affect AAG function. Our studies utilized a Δ54-N-terminally truncated protein that contains all native tyrosines and was stable under all assay conditions used (hereafter referred to as ‘wild-type’). A gel showing the wild-type and all mutants utilized in this study is shown in supplementary Figure 1 (available at Carcinogenesis Online). Purified AAG protein exposed to a single bolus of the nitrating agent ONOO− resulted in a dose-dependent increase in tyrosine nitration (Figure 2A, upper panel). Concurrent probing of these samples with AAG antibodies showed the protein was not degraded by nitrative conditions (Figure 2A, lower panel). Separately, AAG was exposed to nitrating species from a different source, namely •NO2 generated from iron-catalyzed oxidation of nitrite over a 2 h period (29,35). These conditions resulted in an 8.2-fold increase in nitrotyrosyl AAG formation (Figure 2B).
Fig. 2.
Tyrosine nitration of AAG decreases excision activity. (A) Tyrosine nitration of AAG following ONOO− treatment (Materials and Methods). Purified wild-type AAG was treated with ONOO− (10, 20, 40 and 100 μM). Nitrotyrosines and AAG were detected on separate membranes with an anti-nitrotyrosine (upper) and anti-AAG (lower) antibody, respectively. (B) Western blot showing metal-catalyzed tyrosine nitration of AAG. Purified wild-type AAG was incubated with NaNO2 and H2O2; the sample loaded in the left lane (−) lacked FeSO4, whereas the sample in the right lane was incubated with 2 μM FeSO4. (C) Hanes–Woolf plot of εA excision by ONOO−-treated AAG. After ONOO− treatment, AAG (1 nM) was incubated with εA-containing DNA (5–25 nM) for 60 min. Data are a mean of six independent experiments.
The influence of nitration on the activity of AAG was examined. Figure 2C shows that nitration decreased the maximum velocity (Vmax) of εA excision (1.4, 1.8 and 2.1-fold with 10, 20 and 40 μM ONOO−), while having a minimal effect on the Km up to 40 μM ONOO− (also shown in supplementary Table I, available at Carcinogenesis Online). In contrast, 100 μM ONOO− decreased activity to the extent that accurate Vmax and Km values could not be calculated under these conditions. Examination of substrate binding by AAG revealed that exposure to 40 μM ONOO− did not alter binding affinity to εA-containing DNA, whereas 100 μM ONOO− decreased binding by 2.1-fold (supplementary Figure 2 is available at Carcinogenesis Online). Collectively, these results suggest that moderate exposure to nitrative conditions altered substrate turnover, whereas extensive tyrosyl nitration decreased AAG substrate turnover and binding.
Tyrosine 162 is the primary site of tyrosine nitration
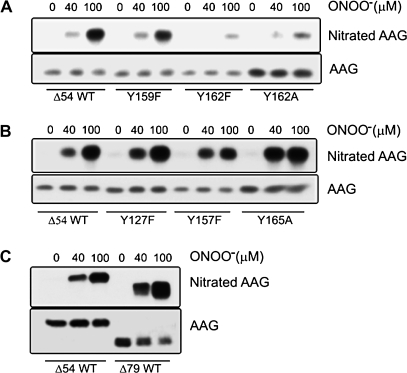
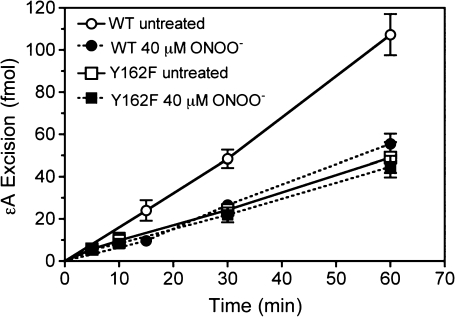
To determine potential nitration sites on AAG, site-specific mutagenesis was performed. Of particular interest were tyrosyl residues in close proximity to the active site (Y127, Y157, Y159, Y162 and Y165; shown in Figure 1). These residues were individually switched to a phenylalanine or an alanine initially. Figure 3 summarizes data examining seven of the eight native tyrosines, in which wild-type AAG and AAG site-specific variants were exposed to 40 or 100 μM ONOO−. Substitution of Y162 with alanine substantially decreased nitration by ONOO− (Figure 3A). Densitometry of western blots for 3-nitrotyrosine adducts indicated that ONOO−-induced nitration of the Y162A variant decreased by 1.6-fold (n = 3) and 3.7-fold (n = 3) at doses of 40 and 100 μM, respectively, compared with wild-type. Previous studies by us and others have shown that a Y162A mutant is severely compromised for activity, whereas the activity of a Y162F mutant is only mildly reduced (24,36). To examine a more conservative switch, Y162 was substituted with a phenylalanine. ONOO−-induced nitration of the Y162F variant decreased by 2.4-fold (n = 3) at 40 μM ONOO− and 4.3-fold (n = 3) at 100 μM ONOO−, similar to that seen for the Y162A mutant. Tyrosine nitration of the Y162 mutants was detectable under highly nitrative conditions (100 μM ONOO−), which is consistent with nitration of one or more of the remaining tyrosines. In contrast, the AAG variants Y127F, Y157F, Y159F and Y165A were nitrated to a similar or greater extent compared with wild-type (Figure 3B). A similar level of nitration was observed with the truncated Δ79-AAG variant (Figure 3C), which lacks Y71 and Y75, suggesting that these two tyrosines were not the target of nitration observed in the Δ54 wild-type AAG. Analysis of the enzymatic activity of the Y162F variant was also examined (Figure 4). Note that under non-nitrative conditions, the Y162F enzyme suffered an ∼2-fold drop in activity compared with wild-type (Figure 4, compare open circles to open squares), in agreement with a previous report (36). Following exposure to ONOO− (40 μM), the Y162F variant showed no decrease in εA excision activity (Figure 4, filled squares), which agreed with the lack of nitration seen at this dose (Figure 3A, middle lanes). Taken together, these data support Y162 as a major site for nitro-adduct formation on AAG.
Fig. 3.
Modification of AAG variants by ONOO−. Purified wild-type and AAG variants (5 μM) were treated with ONOO− (40 and 100 μM) as described in Materials and Methods. (A) Blot shows tyrosine nitration of the Y159F, Y162F and Y162A AAG variants in comparison with wild-type. (B) Blot shows tyrosine nitration of the Y127F, Y157F and Y165A AAG variants in comparison with wild-type. (C) Blot shows tyrosine nitration of wild-type Δ54 and Δ79 AAG. Blots shown are representative of at least three independent experiments.
Fig. 4.
Activity of purified wild-type AAG (circles), Y162F variant (squares), untreated (open symbols) or treated with 40 μM ONOO− (filled symbols) as described in Materials and Methods. Data are a mean of six independent experiments.
Cysteine nitrosation increases AAG εA excision activity
The potential for nitrosation as a modulator of AAG activity was investigated. Thiolates are attractive targets for nitrosative modification resulting in the formation of S-nitrosothiol adducts (37). AAG contains seven cysteines, only four of which were present in the crystal structures of Δ79 AAG (Figure 1). The Δ54 wild-type AAG construct contains six of the seven native cysteines. Under aerobic conditions, formation of nitrosating species such as N2O3 increases proportionally with the concentration of NO (38). The effect of AAG cysteine nitrosation was examined using SPER/NO, a compound that hydrolyzes to yield NO at a defined rate under constant pH and temperature (39). Nitration of AAG tyrosines was not observed under these conditions (data not shown). Exposure of AAG to SPER/NO resulted in a concentration-dependent increase in nitrosation of wild-type AAG (supplementary Figure 3A is available at Carcinogenesis Online). Densitometry of western blots indicated that SPER/NO induced nitrosation by 1.8-fold (n = 3) and 2.3-fold (n = 3) at doses of 20 and 40 μM, respectively, compared with untreated AAG.
The effect of nitrosation on the DNA binding and enzymatic activity of AAG was determined. Nitrosation of AAG exposed to either 20 or 40 μM doses of SPER/NO increased the Vmax ∼1.5-fold (supplementary Table II is available at Carcinogenesis Online). Band shift assays demonstrated that exposure to either 1, 20 or 40 μM SPER/NO increased the binding affinity of AAG for εA by ∼1.3, 1.5 and 2.1-fold, respectively (supplementary Figure 3B is available at Carcinogenesis Online). In contrast to the profile observed with nitrative/oxidative RNS, these findings showed that nitrosative conditions promoted formation of nitrosothiol-AAG, which enhanced AAG efficiency.
Cysteine 167 is the primary site of nitrosation
The AAG crystal structure showed that C167 and C178 reside within 6 Å of the flipped out εA substrate-binding pocket (Figure 1), but to our knowledge, the roles of these cysteines have not been determined. Site-specific mutants (C167S and C178S) were generated and their εA-excision activity first characterized under non-nitrosative conditions. The Vmax/Km values were slightly lower for the C167S mutant (0.32 ± 0.015) and higher for the C178S mutant (0.76 ± 0.07) compared with wild-type (0.49 ± 0.09; supplementary Figure 3C is available at Carcinogenesis Online), demonstrating that these residues influence AAG activity. Treatment of C167S and C178S mutants with 20 μM SPER/NO did not significantly alter their εA excision activities (supplementary Table II is available at Carcinogenesis Online). However, following exposure to 40 μM SPER/NO for 10 min, a 2.0-fold increase in the Vmax for the C178S variant was observed (supplementary Table II is available at Carcinogenesis Online). The Vmax for the C167S variant was unchanged following SPER/NO treatment, suggesting C167 is a key residue in mediating enhanced AAG εA excision activity under nitrosative conditions. Both mutants were also analyzed by the biotin switch method to determine nitrosation modifications following treatment with SPER/NO. The C178S mutant showed an increase in nitrosation signal of 2.2-fold (n = 3) and 1.9-fold (n = 3) at doses of 20 and 40 μM, respectively, compared with untreated enzyme, which was similar to that seen for wild-type (supplementary Figure 3A is available at Carcinogenesis Online). Although there was a higher background with this assay for C167S in the absence of SPER/NO treatment (supplementary Figure 3A is available at Carcinogenesis Online), densitometry indicated that there was essentially no nitrosation for the C167S mutant induced by SPER/NO at doses of 20 and 40 μM.
AAG has altered expression and activity in mouse and human colon adenomas
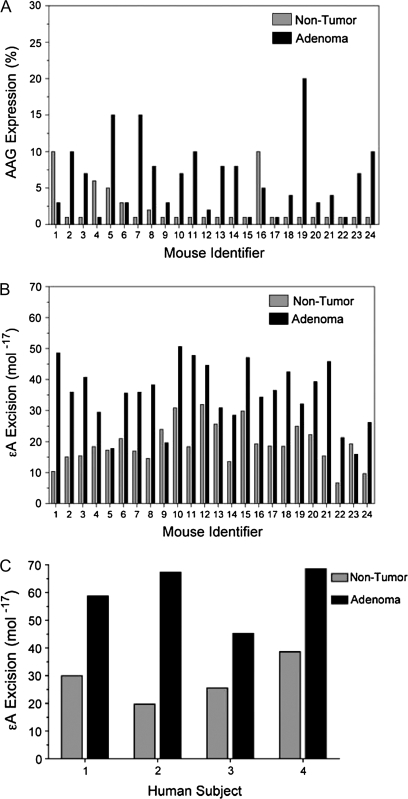
Changes in AAG levels and BER imbalance were examined in tissues from ApcMin/+ mice, a model for human familial adenomatous polyposis and colon cancer (40). Figure 5 shows representative photomicrographs, in which AAG immunoreactivity was elevated in adenomas compared with paired non-diseased tissue. AAG immunoreactivity in colon adenomas of ApcMin/+ mice was elevated in the adenomas compared with non-diseased tissue from the same animal in 17 of 24 cases (Figure 6A, supplementary Table III is available at Carcinogenesis Online). The mean percentage of tissue staining positive (±SEM) was 6.5 ± 1.0 in adenoma tissue compared with 2.25 ± 0.56 in paired non-tumor tissue (P = 0.0007). The enzymatic activity of AAG was higher in most (22 of 24) of the adenoma samples compared with adjacent non-diseased tissues (Figure 6B, supplementary Table III is available at Carcinogenesis Online). The mean activity was 19.1 × 10−17 mol/mg (±6.5) of protein for non-tumor tissue and 35.2 × 10−17 mol/mg (±10.1) of protein for tumor tissue (P = 3.5 × 10−8). However, a correlation was not observed when the change in AAG protein immunoreactivity between each paired adenoma and non-diseased tissue was compared with the change in AAG activity between the same pair (r = −0.1, P = 0.62). Figure 6C shows that activity is elevated in human adenomas compared with paired non-diseased tissue in four cases, although the sample number was too few to compare immunoreactivity and activity. The mean activity (±SEM) was 28.5 × 10−17 mol/mg (±4.0) of protein for non-tumor tissue and 60.0 × 10−17 mol/mg (±5.4) of protein for tumor tissue (P = 0.003).
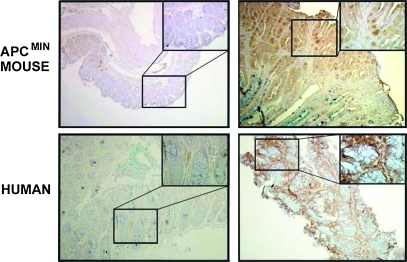
Fig. 5.
IHC images of AAG expression in non-tumor and adenoma tissues from a patient and from an ApcMin/+ mouse. The inset is ×400 magnification of the indicated area of the ×100 image.
Fig. 6.
AAG expression and activity are increased in colon adenomas from ApcMin/+ mice and humans. (A) The expression of AAG was analyzed in non-tumor (gray) and adenoma (black) pairs from 24 mice by IHC. Figure shows expression of AAG as a percent of stained tissue compared with that unstained. The mean percentage of tissue staining positive (±SEM) was 6.5 ± 1.0 in adenoma tissue compared with 2.25 ± 0.56 in paired non-tumor tissue (P = 0.0007). (B) Activity of AAG for εA was analyzed in non-tumor and adenoma tissues from the same mice as above. The mean activity was 19.1 × 10−17 mol/mg (±6.5) of protein for non-tumor tissue and 35.2 × 10−17 mol/mg (±10.1) of protein for tumor tissue (P = 3.5 × 10−8). (C) Human colon adenomas have elevated AAG activity (black) compared with normal colon tissues from the same patient (gray). The mean activity (±SEM) was 28.5 × 10−17 mol/mg (±4.0) of protein for non-tumor tissue and 60.0 × 10−17 mol/mg (±5.4) of protein for tumor tissue (P = 0.003). Protein extracts (2 μg) were incubated with εA-containing DNA (50 pM) for 2 h at 37°C (Materials and Methods).
The relationship between AAG and iNOS was examined by IHC to explore the link between RNS production and this DNA glycosylase. A striking correlation (r = 0.76, P = 0.00002) between AAG and iNOS was observed in ApcMin/+ mouse tissues (supplementary Figure 4A is available at Carcinogenesis Online). In human adenomas, a high level of iNOS activity (79.3 pmol citrulline/min/mg protein) was observed compared with non-diseased tissues (0.77 pmol citrulline/min/mg protein). These results confirm previous analysis of these tissues (30) and show a strong association between iNOS and AAG protein in colon adenomas. The relationship between AAG and two important BER pathway partners, APE1 and POLβ, was examined by IHC to determine whether a BER imbalance might be occurring. POLβ immunoreactivity was increased in all but one of the adenomas when compared with non-tumor tissues (supplementary Table III is available at Carcinogenesis Online). In contrast, only 11 of 24 tumor tissues displayed an increase in APE1 protein compared with non-tumor tissue (supplementary Table III is available at Carcinogenesis Online). A substantial change in staining from predominantly nuclear in non-tumor tissue to cytoplasmic staining for APE1 in tumor tissue was evident (supplementary Figure 4B is available at Carcinogenesis Online). Interestingly, there was no correlation between immunoreactivity of APE1 and POLβ (r = −0.074, P = 0.73), between AAG and APE1 (r = 0.099, P = 0.64) or between AAG and POLβ (r = 0.088, P = 0.68), when the change in expression between non-tumor and adenoma tissue for each animal was compared.
To test for the occurrence of nitrated AAG in vivo, immunoprecipitation experiments were performed in the paired human tissue samples (n = 4). Immunoprecipitation with AAG antibody was followed by blotting with the nitrotyrosine antibody (supplementary Figure 5A is available at Carcinogenesis Online) or in the reverse order (Supplementary Figure 5B is available at Carcinogenesis Online). Qualitatively, the immunoreactivity patterns were consistent with the presence of nitrated AAG in the human samples, although the level of nitrated AAG varied between individuals and was not exclusive to the adenoma relative to the surrounding tissue. Control experiments with rabbit IgG and purified AAG protein were run in parallel to confirm assay specificity. Treatment of human HT-29 colon adenocarcinoma cells in culture with SPER/NO (20 μM, 4 h) substantially increased the level of nitrated AAG observed following immunoprecipitation and blotting, relative to untreated control. Although immunoprecipitation is not a quantitative measurement of the extent of AAG nitration, the data in supplementary Figure 5 (available at Carcinogenesis Online) indicate that the nitration of AAG occurs in vivo.
Discussion
RNS can create DNA lesions at sites of intestinal inflammation, which may lead to mutagenic transformation and colon cancer if damaged nucleobases are not adequately repaired. We have focused on AAG-mediated BER because reports have shown that an AAG substrate, εA, was increased under chronic inflammatory conditions in mouse models (5,13,14). We examined the potential for RNS to posttranslationally modify certain AAG protein residues and the effect these modifications had on AAG catalytic function. The findings support the notion that BER imbalance plays a role in carcinogenesis; however, they also reveal a complexity for distinct RNS modifications in either decreasing or enhancing AAG catalytic function. These data point to a broader picture for RNS in not only causing DNA lesions directly but also potentially influencing repair activity depending on the nature of the chemistries involved.
Analysis of the 2.1 Å crystal structure of AAG indicated that several tyrosine and cysteine (non-disulfide) residues comprise the catalytic pocket (Figure 1). These residues are chief sites for RNS adducts to occur through either nitrative or nitrosative reactions (37,41,42). Initial attempts to probe specific sites of RNS modifications on AAG by proteomic and mass spectroscopic analysis failed due to the close proximity of target residues (Y157, Y159, Y162, Y165, C167 and C178) in the enzyme core (32). Therefore, AAG site-directed mutagenesis coupled with western blotting for specific RNS adducts was utilized. This approach showed that AAG was susceptible to nitrating RNS (ONOO−, •NO2) in a dose-dependent manner and identified Y162 as a focal point for nitro-adduct formation. Y162 is critical for AAG activity by occupying the DNA site vacated by the flipped out nucleotide substrate and is located on an exposed flexible loop, which we suspect contributes to its susceptibility to nitration. Bi-directional intramolecular electron transfer reactions between juxtaposed residues are proposed to mediate tyrosine susceptibility to RNS modifications (43,44). In this regard, this stretch of AAG is quite remarkable in having a ‘hot spot’ of such residues: Y162, M164, Y165, F166, C167 and M168. In fact, increased nitration was seen for the Y165A mutant at 40 μM ONOO−, compared with wild-type (Figure 3B). Mutating Y162 to phenylalanine rescued εA excision activity under moderate nitrating conditions (40 μM ONOO−), whereas the Vmax of wild-type AAG was decreased 1.6-fold under these conditions relative to untreated controls. Therefore, these results show selective nitration of residues in the AAG active site diminishes its capacity to repair DNA damage. The presence of 3-nitrotyrosine immunoreactivity has been associated with colon adenocarcinoma and RNS-mediated DNA lesions (45–48).
In addition to nitration, nitrosation can also damage DNA and proteins. The nitrosating species N2O3 is formed during the autoxidation of •NO (38) and can cause nucleobase deamination (2). Interestingly, our study has revealed that AAG activity is enhanced by nitrosation. C167 and C178 comprise part of the AAG active site, flanking either side of the εA substrate. Characterization of C167S and C178S mutants showed a nearly 2-fold increase in both Km and Vmax for εA, which corresponded to the increase in wild-type AAG activity seen after nitrosation. The importance of C167 in the excision of methylated bases was demonstrated in a recent study that performed random mutagenesis of amino acids surrounding the AAG active site combined with a complementation assay in E.coli (49). The active site of AAG serves more to reject normal purines than to specifically recognize the diverse array of substrates it excises (26,50). The inherently modest catalytic activity of AAG also serves as a means of restricting the ability of AAG to inadvertently remove normal guanines and adenines. We speculate that these cysteines might contribute to restricting the activity of wild-type AAG and raise the intriguing possibility that the oxidation state of these residues might modulate AAG activity.
The biotin switch method detected an elevated background of S-nitrosation of the C167S mutant relative to either wild-type or C178S AAG in the absence of •NO exposure. This unexplained result may relate to altering the oxidation state of the neighboring C178 by this amino acid subsitution. Nonetheless, these results were consistent with the conclusion that the C167 and C178 residues play important previously unappreciated roles in AAG activity. RNS mediating an enhancement of BER is unprecedented; therefore, our finding that S-nitrosation of selective AAG residues augments activity adds a new dimension to the relationships between NOS and colon carcinogenesis.
The increased production of •NO and formation of RNS is thought to contribute to the progression from chronic inflammation to cancer because RNS can directly and indirectly damage DNA and proteins. A positive correlation between the expression of iNOS and AAG in tumor tissues analyzed from ApcMin/+ mice suggests an adaptive increase in AAG protein in response to RNS. A recent report demonstrated that Aag−/− mice are susceptible to tissue damage induced by RNS, consistent with AAG-mediated prevention of DNA damage and tumor initiation during RNS stress (14). However, the observed lack of correlation between AAG protein and activity in both ApcMin/+ mice and human colon tumors suggest that nitrative RNS predominate and may inhibit AAG in established adenomas. Analysis of colon tissues from subjects with adenoma demonstrated that nitration of AAG occurs in human cancer (supplementary Figure 5 is available at Carcinogenesis Online). These data when combined with our in vitro findings with purified AAG present a case for compromised AAG catalysis due an imbalance toward nitrative stress during in cancer.
The removal of mutagenic base adducts by AAG creates an abasic site in DNA that must be processed by the downstream components and it is well established that imbalances in AAG-mediated BER has negative consequences (51). Specifically, it has been shown that AAG can remove undamaged adenines and guanines (50,52) and overexpressing AAG caused mutations and microsatellite instability in the absence of exogenous DNA-damaging agents (16,53). Our analysis of the expression of key BER proteins by IHC revealed that APE1 was only increased in 46% of the tumors; we also observed a nuclear to cytoplasmic shift in tumor tissues. Indeed, S-nitrosation of cysteines on APE1 has been reported to increase the nuclear export of APE1 (22) and this subcellular relocalization has been observed in colon cancer cells (54) and in other tumor types (55). In contrast, POLβ is significantly higher in 96% of the adenomas compared with non-tumor tissues in ApcMin/+ mice. The observations suggest that although AAG upregulation is protective at the early stages of tumorigenesis, sustained upregulation during tumorigenesis may eventually drive a detrimental imbalance in the BER pathway.
Collectively, this study supports a role for imbalances in AAG-mediated BER as a contributor to colon tumorigenesis. Although we showed that nitration or nitrosation of AAG results in differing regulation of AAG activity, these chemical processes in a biological setting can be intertwined (35,43). We provide to our knowledge the first simultaneous examination of alterations in the first three enzymes in BER in intestinal adenoma samples and matched non-tumor controls. The results illustrate that it is not straightforward to equate the presence of NOS activity and RNS solely with DNA damage and carcinogenesis or cancer progression. Regardless, our study identifies key roles for specific AAG sites in controlling AAG activity that might prove to be a target for the prevention and/or treatment of diseases associated with BER imbalance.
Supplementary material
Supplementary Table I–III and Figures 1–5 can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health (R01 CA100450 to M.D.W. and P20 RR17698 to the Center for Colon Cancer Research); the Hollings Cancer Center/Medical University of South Carolina (DOD Grant GC-3319-05-4498CM); American Cancer Society (RSG-05-246-01-GMC) to R.W.S.; Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Supplementary Material
Acknowledgments
The authors gratefully thank Dr Marj Pena and Celestia Davis of the mouse core facility of the Center for Colon Cancer Research at the University of South Carolina (P20 RR17698).
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- AAG
alkyladenine DNA glycosylase
- APE1
apurinic/apyrimidinic endonuclease 1
- BER
base excision repair
- εA
1,N6-ethenoadenine
- IHC
immunohistochemistry
- iNOS
inducible nitric oxide synthase
- •NO2
nitrogen dioxide
- N2O3
nitrous anhydride
- ONOO−
peroxynitrite
- POLβ
DNA polymerase β
- RNS
reactive nitrogen species
- SPER/NO
spermine NONOate
References
- 1.Lu H, et al. Inflammation, a key event in cancer development. Mol. Cancer Res. 2006;4:221–233. doi: 10.1158/1541-7786.MCR-05-0261. [DOI] [PubMed] [Google Scholar]
- 2.Dedon PC, et al. Reactive nitrogen species in the chemical biology of inflammation. Arch. Biochem. Biophys. 2004;423:12–22. doi: 10.1016/j.abb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 3.Ohshima H, et al. Chemical basis of inflammation-induced carcinogenesis. Arch. Biochem. Biophys. 2003;417:3–11. doi: 10.1016/s0003-9861(03)00283-2. [DOI] [PubMed] [Google Scholar]
- 4.Dong M, et al. Relatively small increases in the steady-state levels of nucleobase deamination products in DNA from human TK6 cells exposed to toxic levels of nitric oxide. Chem. Res. Toxicol. 2006;19:50–57. doi: 10.1021/tx050252j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pang B, et al. Lipid peroxidation dominates the chemistry of DNA adduct formation in a mouse model of inflammation. Carcinogenesis. 2007;28:1807–1813. doi: 10.1093/carcin/bgm037. [DOI] [PubMed] [Google Scholar]
- 6.Bartsch H, et al. Accumulation of lipid peroxidation-derived DNA lesions: potential lead markers for chemoprevention of inflammation-driven malignancies. Mutat. Res. 2005;591:34–44. doi: 10.1016/j.mrfmmm.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 7.Almeida KH, et al. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst) 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelward BP, et al. Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc. Natl Acad. Sci. USA. 1997;94:13087–13092. doi: 10.1073/pnas.94.24.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hang B, et al. Targeted deletion of alkylpurine-DNA-N-glycosylase in mice eliminates repair of 1, N6 ethenoadenine and hypoxanthine but not of 3, N4 ethenocytosine or 8-oxoguanine. Proc. Natl Acad. Sci. USA. 1997;94:12869–12874. doi: 10.1073/pnas.94.24.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hitchcock TM, et al. Oxanine DNA glycosylase activity from mammalian alkyladenine glycosylase. J. Biol. Chem. 2004;279:38177–38183. doi: 10.1074/jbc.M405882200. [DOI] [PubMed] [Google Scholar]
- 11.Saparbaev MK, et al. 1, N2-Ethenoguanine, a mutagenic DNA adduct, is a substrate of Escherichia coli mismatch-specific uracil-DNA glycosylase and human alkylpurine-DNA-N glycosylase. J. Biol. Chem. 2002;277:26987–26993. doi: 10.1074/jbc.M111100200. [DOI] [PubMed] [Google Scholar]
- 12.Wuenschell GE, et al. Stability, miscoding potential, and repair of 2′-deoxyxanthosine in DNA: implications for nitric oxide-induced mutagenesis. Biochemistry. 2003;42:3608–3616. doi: 10.1021/bi0205597. [DOI] [PubMed] [Google Scholar]
- 13.Ringvoll J, et al. AlkB homologue 2-mediated repair of ethenoadenine lesions in mammalian DNA. Cancer Res. 2008;68:4142–4149. doi: 10.1158/0008-5472.CAN-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meira LB, et al. DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice. J. Clin. Invest. 2008;118:2516–2525. doi: 10.1172/JCI35073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmid K, et al. Increased levels of promutagenic etheno-DNA adducts in colonic polyps of FAP patients. Int. J. Cancer. 2000;87:1–4. doi: 10.1002/1097-0215(20000701)87:1<1::aid-ijc1>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 16.Hofseth LJ, et al. The adaptive imbalance in base excision-repair enzymes generates microsatellite instability in chronic inflammation. J. Clin. Invest. 2003;112:1887–1894. doi: 10.1172/JCI19757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benhar M, et al. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iyer AK, et al. Role of S-nitrosylation in apoptosis resistance and carcinogenesis. Nitric Oxide. 2008;19:146–151. doi: 10.1016/j.niox.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szabo C, et al. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat. Rev. Drug Discov. 2007;6:662–680. doi: 10.1038/nrd2222. [DOI] [PubMed] [Google Scholar]
- 20.Thomas DD, et al. The chemical biology of nitric oxide: implications in cellular signaling. Free Radic. Biol. Med. 2008;45:18–31. doi: 10.1016/j.freeradbiomed.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaiswal M, et al. Human Ogg1, a protein involved in the repair of 8-oxoguanine, is inhibited by nitric oxide. Cancer Res. 2001;61:6388–6393. [PubMed] [Google Scholar]
- 22.Qu J, et al. Nitric oxide controls nuclear export of APE1/Ref-1 through S-nitrosation of cysteines 93 and 310. Nucleic Acids Res. 2007;35:2522–2532. doi: 10.1093/nar/gkl1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 24.Lau AY, et al. Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG. Proc. Natl Acad. Sci. USA. 2000;97:13573–13578. doi: 10.1073/pnas.97.25.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connor EE, et al. Effects of substrate specificity on initiating the base excision repair of N-methylpurines by variant human 3-methyladenine DNA glycosylases. Chem. Res. Toxicol. 2005;18:87–94. doi: 10.1021/tx049822q. [DOI] [PubMed] [Google Scholar]
- 26.Connor EE, et al. Active-site clashes prevent the human 3-methyladenine DNA glycosylase from improperly removing bases. Chem. Biol. 2002;9:1033–1041. doi: 10.1016/s1074-5521(02)00215-6. [DOI] [PubMed] [Google Scholar]
- 27.Lau A, et al. Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanism for nucleotide flipping and base excision. Cell. 1998;95:249–258. doi: 10.1016/s0092-8674(00)81755-9. [DOI] [PubMed] [Google Scholar]
- 28.Adhikari S, et al. N-terminal extension of N-methylpurine DNA glycosylase is required for turnover in hypoxanthine excision reaction. J. Biol. Chem. 2007;282:30078–30084. doi: 10.1074/jbc.M704051200. [DOI] [PubMed] [Google Scholar]
- 29.Thomas DD, et al. Protein nitration is mediated by heme and free metals through Fenton-type chemistry: an alternative to the NO/O2- reaction. Proc. Natl Acad. Sci. USA. 2002;99:12691–12696. doi: 10.1073/pnas.202312699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ambs S, et al. Relationship between p53 mutations and inducible nitric oxide synthase expression in human colorectal cancer. J. Natl Cancer Inst. 1999;91:86–88. doi: 10.1093/jnci/91.1.86. [DOI] [PubMed] [Google Scholar]
- 31.Ying L, et al. Chronic inflammation promotes retinoblastoma protein hyperphosphorylation and E2F1 activation. Cancer Res. 2005;65:9132–9136. doi: 10.1158/0008-5472.CAN-05-1358. [DOI] [PubMed] [Google Scholar]
- 32.Roy R, et al. The domains of mammalian base excision repair enzyme N-methylpurine-DNA glycosylase. Interaction, conformational change, and role in DNA binding and damage recognition. J. Biol. Chem. 1996;271:23690–23697. doi: 10.1074/jbc.271.39.23690. [DOI] [PubMed] [Google Scholar]
- 33.Trivedi RN, et al. Human methyl purine DNA glycosylase and DNA polymerase {beta} expression collectively predict sensitivity to temozolomide. Mol. Pharmacol. 2008;74:505–516. doi: 10.1124/mol.108.045112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotakadi VS, et al. Ginkgo biloba extract EGb 761 has anti-inflammatory properties and ameliorates colitis in mice by driving effector T cell apoptosis. Carcinogenesis. 2008;29:1799–1806. doi: 10.1093/carcin/bgn143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Espey MG, et al. Direct real-time evaluation of nitration with green fluorescent protein in solution and within human cells reveals the impact of nitrogen dioxide vs. peroxynitrite mechanisms. Proc. Natl Acad. Sci. USA. 2002;99:3481–3486. doi: 10.1073/pnas.062604199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vallur AC, et al. Effects of hydrogen bonding within a damaged base pair on the activity of wild-type and DNA-intercalating mutants of human alkyladenine DNA glycosylase. J. Biol. Chem. 2002;277:31673–31678. doi: 10.1074/jbc.M204475200. [DOI] [PubMed] [Google Scholar]
- 37.Hogg N. Biological chemistry and clinical potential of S-nitrosothiols. Free Radic. Biol. Med. 2000;28:1478–1486. doi: 10.1016/s0891-5849(00)00248-3. [DOI] [PubMed] [Google Scholar]
- 38.Espey MG, et al. Distinction between nitrosating mechanisms within human cells and aqueous solution. J. Biol. Chem. 2001;276:30085–30091. doi: 10.1074/jbc.M101723200. [DOI] [PubMed] [Google Scholar]
- 39.Maragos CM, et al. Complexes of .NO with nucleophiles as agents for the controlled biological release of nitric oxide. Vasorelaxant effects. J. Med. Chem. 1991;34:3242–3247. doi: 10.1021/jm00115a013. [DOI] [PubMed] [Google Scholar]
- 40.Luongo C, et al. Loss of Apc+ in intestinal adenomas from Min mice. Cancer Res. 1994;54:5947–5952. [PubMed] [Google Scholar]
- 41.Espey MG, et al. Mechanisms of cell death governed by the balance between nitrosative and oxidative stress. Ann. N. Y. Acad. Sci. 2000;899:209–221. doi: 10.1111/j.1749-6632.2000.tb06188.x. [DOI] [PubMed] [Google Scholar]
- 42.Schopfer FJ, et al. NO-dependent protein nitration: a cell signaling event or an oxidative inflammatory response? Trends Biochem. Sci. 2003;28:646–654. doi: 10.1016/j.tibs.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 43.Zhang H, et al. Intramolecular electron transfer between tyrosyl radical and cysteine residue inhibits tyrosine nitration and induces thiyl radical formation in model peptides treated with myeloperoxidase, H2O2, and NO2-: EPR SPIN trapping studies. J. Biol. Chem. 2005;280:40684–40698. doi: 10.1074/jbc.M504503200. [DOI] [PubMed] [Google Scholar]
- 44.Zhang H, et al. The effect of neighboring methionine residue on tyrosine nitration and oxidation in peptides treated with MPO, H2O2, and NO2(-) or peroxynitrite and bicarbonate: role of intramolecular electron transfer mechanism? Arch. Biochem. Biophys. 2009;484:134–145. doi: 10.1016/j.abb.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kojima M, et al. Nitric oxide synthase expression and nitric oxide production in human colon carcinoma tissue. J. Surg. Oncol. 1999;70:222–229. doi: 10.1002/(sici)1096-9098(199904)70:4<222::aid-jso5>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 46.Seril DN, et al. Inhibition of chronic ulcerative colitis-associated colorectal adenocarcinoma development in a murine model by N-acetylcysteine. Carcinogenesis. 2002;23:993–1001. doi: 10.1093/carcin/23.6.993. [DOI] [PubMed] [Google Scholar]
- 47.Yerushalmi HF, et al. The role of NO synthases in arginine-dependent small intestinal and colonic carcinogenesis. Mol. Carcinog. 2006;45:93–105. doi: 10.1002/mc.20168. [DOI] [PubMed] [Google Scholar]
- 48.Burney S, et al. The chemistry of DNA damage from nitric oxide and peroxynitrite. Mutat. Res. 1999;424:37–49. doi: 10.1016/s0027-5107(99)00006-8. [DOI] [PubMed] [Google Scholar]
- 49.Chen CY, et al. Substrate binding pocket residues of human alkyladenine-DNA glycosylase critical for methylating agent survival. DNA Repair (Amst) 2008;7:1731–1745. doi: 10.1016/j.dnarep.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Brien PJ, et al. Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. J. Biol. Chem. 2004;279:9750–9757. doi: 10.1074/jbc.M312232200. [DOI] [PubMed] [Google Scholar]
- 51.Wyatt MD, et al. Methylating agents and DNA repair responses: methylated bases and sources of strand breaks. Chem. Res. Toxicol. 2006;19:1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berdal KG, et al. Release of normal bases from intact DNA by a native DNA repair enzyme. EMBO J. 1998;17:363–367. doi: 10.1093/emboj/17.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Glassner BJ, et al. Generation of a strong mutator phenotype in yeast by imbalanced base excision repair. Proc. Natl Acad. Sci. USA. 1998;95:9997–10002. doi: 10.1073/pnas.95.17.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kakolyris S, et al. Human apurinic endonuclease 1 expression in a colorectal adenoma-carcinoma sequence. Cancer Res. 1997;57:1794–1797. [PubMed] [Google Scholar]
- 55.Tell G, et al. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid. Redox Signal. 2005;7:367–384. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.