Abstract
Many bacterial plasmids replicate by a rolling-circle mechanism that involves the generation of single-stranded DNA (ssDNA) intermediates. Replication of the lagging strand of such plasmids initiates from their single strand origin (sso). Many different types of ssos have been identified. One group of ssos, termed ssoA, which have conserved sequence and structural features, function efficiently only in their natural hosts in vivo. To study the host specificity of sso sequences, we have analyzed the functions of two closely related ssoAs belonging to the staphylococcal plasmid pE194 and the streptococcal plasmid pLS1 in Staphylococcus aureus. The pLS1 ssoA functioned poorly in vivo in S. aureus as evidenced by accumulation of high levels of ssDNA but supported efficient replication in vitro in staphylococcal extracts. These results suggest that one or more host factors that are present in sufficient quantities in S. aureus cell-free extracts may be limiting in vivo. Mapping of the initiation points of lagging strand synthesis in vivo and in vitro showed that DNA synthesis initiates from specific sites within the pLS1 ssoA. These results demonstrate that specific initiation of replication can occur from the pLS1 ssoA in S. aureus although it plays a minimal role in lagging strand synthesis in vivo. Therefore, the poor functionality of the pLS1 in vivo in a nonnative host is caused by the low efficiency rather than a lack of specificity of the initiation process. We also have identified ssDNA promoters and mapped the primer RNAs synthesized by the S. aureus and Bacillus subtilis RNA polymerases from the pE194 and pLS1 ssoAs. The S. aureus RNA polymerase bound more efficiently to the native pE194 ssoA as compared with the pLS1 ssoA, suggesting that the strength of RNA polymerase–ssoA interaction may play a major role in the functionality of the ssoA sequences in Gram-positive bacteria.
Keywords: single strand origin/single-stranded DNA promoter/RNA polymerase/in vitro replication
A large number of small, multicopy plasmids in Gram-positive bacteria and many in Gram-negative bacteria replicate by a rolling-circle mechanism (for reviews, see ref. 1–5). The initiator proteins encoded by these plasmids generate a site-specific nick within the double strand origin (dso) and leading strand replication proceeds by extension of the 3′-OH end generated by the initiator (4, 6). This results in the displacement of a single-stranded DNA (ssDNA) intermediate (7, 8), which is converted to the double-stranded (ds) form by lagging strand synthesis using the sso and the host proteins. The ssos are important in the stable maintenance of rolling-circle plasmids in bacterial hosts, and it has been suggested that they may play a role in plasmid promiscuity and horizontal transfer among related bacteria (4, 9–12). Four types of ssos have been described based on their sequence and structural similarities, namely ssoA, ssoU, ssoT, and ssoW (9, 13–16). The host RNA polymerase is involved in the initiation of lagging strand synthesis from all the four ssos, although limited initiation may occur in an RNA polymerase-independent manner from ssoW (12, 17–20). The ssoA-type origins contain extensive palindromic sequences that can form folded structures (9, 12, 13, 21, 22). An interesting aspect of the ssoA origins is that they function efficiently only in their natural hosts (3, 9, 23–25). Two regions, a 6-bp sequence, CS-6, located in the central loop of their palindromic structure and the recombination site B (RSB) are conserved in ssoAs (Fig. 1; ref. 9, 12, 13). Previously, we have shown that although RSB is important for binding by the RNA polymerase to generate a RNA primer for replication, the CS-6 region functions as the termination site of primer RNA synthesis (26, 27). To investigate the basis for host-specific function of the ssoA-type origins, we have studied the function of the ssoA sequences present in the Staphylococcus aureus plasmid pE194 and the streptococcal plasmid pLS1. Here, we demonstrate that the ssoA of pLS1 supports in vitro replication in cell-free extracts from S. aureus. Furthermore, although the pLS1 ssoA functions poorly in vivo in S. aureus, initiation from this ssoA is still specific. We also have mapped the initiation points of lagging strand synthesis in vivo from both the pE194 and pLS1 ssoA origins. Using the S. aureus and Bacillus subtilis RNA polymerases, we have identified the primer RNAs synthesized for lagging strand replication and mapped their initiation sites within the pE194 and pLS1 ssoAs. We also have found that the S. aureus RNA polymerase bound more efficiently to the ssoA of pE194 as compared with the pLS1 ssoA, suggesting that the strength of RNA polymerase–ssoA interaction may determine the functionality of the ssoA-type origins in vivo.
Figure 1.
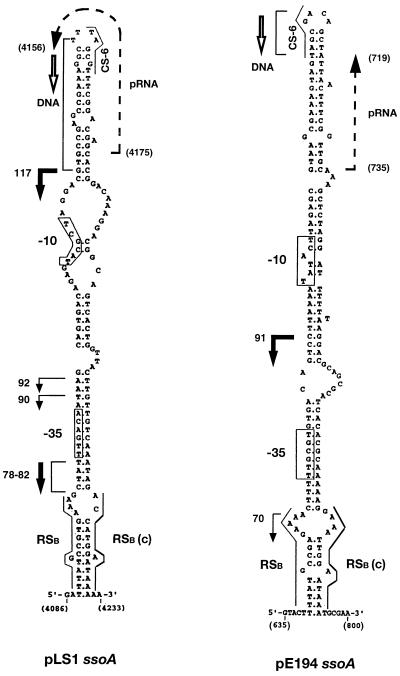
Proposed structures of the pLS1 and pE194 ssoAs. Possible −35 and −10 regions are boxed. The conserved RSB, its complement, RSB (c), and CS-6 sequences also are indicated. The initiation and termination sites (positions 735–719 in pE194 and 4175–4156 in pLS1) and direction of primer RNA (pRNA) synthesis are indicated by broken arrows. The initiation points of DNA synthesis (RNA–DNA transition points), mapped in vitro in Fig. 4A and ref. 12, are indicated by brackets and direction of DNA synthesis is indicated by open arrows. Initiation sites of lagging strand synthesis mapped in vivo are shown by bold arrows (major sites) or arrows (minor sites), and the numbers correspond to the size of bands in nt shown in Fig. 4B. Nucleotide coordinates of the plasmids are shown in brackets.
MATERIALS AND METHODS
Bacterial Strains and Plasmids.
S. aureus RN4220 has been described previously (12). The S. aureus plasmid pE194 specifies resistance to erythromycin and the streptococcal pMV158-derivative plasmid pLS1 (Δmob, ΔssoU), pLS1G3G7 (a pLS1 derivative containing mutations in the conserved CS-6 and RSB sequences) and pLS1DNA (a pLS1 derivative deleted for the ssoA) specify resistance to tetracycline (12, 23, 26, 28). A phagemid pALTER derivative containing the pLS1 ssoA (pA-pLS1ssoA) has been described (26). M13-pE194ssoA is a bacteriophage M13 derivative containing the pE194 ssoA (12).
Detection of Accumulated ssDNA and Mapping of Initiation Sites of Lagging Strand Synthesis in Vivo.
S. aureus harboring the desired plasmids were grown to middle exponential phase (≈2 × 108 colony forming units per ml of culture). ssDNAs from these cultures were analyzed by electrophoresis on 0.7% agarose gels of total DNA samples, followed by transfer to nitrocellulose filters with or without prior denaturation (7). Filters were hybridized by using a mixture of 32P-labeled pLS1 and pE194 DNAs as probes. The initiation points of lagging strand synthesis were mapped by using “repeat primer extension assays” (29, 30). Plasmid DNA containing both supercoiled DNA and trace amounts of single-stranded replicative intermediates was isolated from S. aureus and S. pneumoniae harboring pE194 or pLS1 (or its derivatives). These DNAs (50–200 ng) were used as templates in primer extension assays by using 5′-32P end- labeled primers (31) that are expected to hybridize downstream of the ssoA sequences. The primers used for DNA amplifications were: 5′-AGGCGATTAAGCCAATCGTG-3′, positions 4026–4045 for pLS1 and the negative control pLS1G3G7 and 5′-GGTCGATAGAAAGCGTGAGAA-3′, positions 585–605 for pE194. The repeat primer extension assays were carried out for 30 cycles, with each cycle including 1 min of incubation at 94°, 1 min at 48°, and 1 min at 72°.
Preparation of ssDNA Templates.
ssDNA for in vitro replication was isolated from Escherichia coli JM109 harboring the phagemid pALTER or bacteriophage M13 derivatives containing the pLS1 and pE194 ssoAs as described (12, 26). ssDNA containing the pE194 and pLS1 ssoAs (275 and 313 nt, respectively) for gel retardation and in vitro transcription assays was prepared by asymmetric PCR by using the pE194 and pLS1 DNAs and appropriate oligonucleotide primers as described (26). When required, ssDNA fragments were 5′ end-labeled with 32P (31).
In Vitro Replication.
Cell-free extracts were prepared from the restriction-deficient S. aureus strain RN4220 and in vitro-lagging strand synthesis was performed as described by Birch and Khan (17). The replication products were linearized by treatment with either HindIII or EcoRV, which cleave downstream of the pLS1 and pE194 ssoAs (12, 26). The reaction products were visualized by agarose gel electrophoresis and staining with ethidium bromide followed by autoradiography as described (12). Fine-structure mapping of the lagging strand initiation sites was performed by resolving products obtained from replication reactions on 8% polyacrylamide-8.3 M urea sequencing gels as described (12).
Binding of RNA Polymerase to the Single Strand Origins.
The interaction between host RNA polymerase and plasmid ssoA was studied by electrophoretic mobility-shift analysis (EMSA). S. aureus RNA polymerase was purified as described (32, 33). As templates, ssDNA containing the pLS1 or pE194 ssoAs or their complementary sequences were used. Each reaction (10 ml) containing 3 ng (20,000 cpm) of 5′-end-labeled ssDNA was incubated at room temperature for 15 min with increasing amounts of RNA polymerase in the binding buffer (50 mM potassium glutamate/25 mM Tris⋅HCl, pH 8/10 mM MgCl2/1 mM CaCl2/650 mM NaCl, 1 mM DTT/0.05% Nonidet P-40/40% sucrose/90 mM ammonium sulfate) in the presence of 1 μg poly(dA) and 100 μg/ml BSA. After addition of 2 μl of 30% glycerol, samples were loaded on a 3.8% native polyacrylamide gel and electrophoresed at 12.5 V/cm for 1 hr. The DNA bands were visualized by autoradiography of the gels and the bands quantified by using an AMBIS System 100 radioanalytic detector.
In Vitro Transcription from ssoA.
The procedure of Kramer et al. (26) was followed essentially as described by using the B. subtilis (34) and S. aureus RNA polymerases. Reaction mixtures (50 μl) containing 0.1 pmol of ssDNA (313 and 275 nt long), 200 μM ATP, CTP, and GTP, 80 μM [γ-32P]-UTP (3,000 Ci/mmol), 5 μg of heparin, and 5 units of the RNase inhibitor from human placenta (Boehringer Mannheim) in the binding buffer were incubated at 37°C for 10 min with 0.2 μg of RNA polymerase (preincubated with saturating amounts of the purified σ factor). The DNA–RNA hybrid products were extracted with phenol-chloroform and precipitated with ethanol. Labeled transcripts were separated on a 15% sequencing gel (31) and visualized by autoradiography.
Identification of the 5′-Ends of Primer RNAs.
For this purpose, primer extension reactions were performed by using as template unlabeled de novo-synthesized primer RNAs obtained from in vitro transcription reactions (see above). After denaturation of the ssDNA-primer RNA hybrids at 65°C for 5 min, 1 pmol of the oligonucleotide (5′-TTAGCGTTTCGG-3′, positions 4158–4170, to map the 5′-end of the primer RNA generated from pLS1 ssoA, and 5′-ATTAACTTTCGGT-3′, positions 721–733, to map the 5′-end of the primer RNA synthesized from the pE194 ssoA) was used for reverse transcription as described earlier (26). Radiolabeled dNTPs used were: 10 μM [α-32P]-dATP (3,000 Ci/mmol) for pLS1 ssoA and [α-32P]-dTTP (3,000 Ci/mmol) for pE194 ssoA.
RESULTS
Establishment of Plasmid pLS1 in S. aureus.
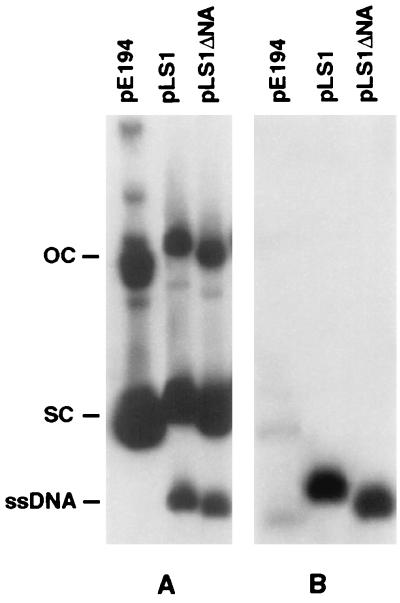
Because the ssoA present in the streptococcal plasmid pLS1 is very similar to those of many staphylococcal plasmids (see Fig. 8), we wished to determine the functionality of this ssoA in S. aureus. Plasmids were introduced into S. aureus by electroporation. Southern blot analysis indicated that plasmids containing the wild-type ssoA or lacking it (pLS1ΔNA) accumulated significant levels of ssDNA (Fig. 2A). This was seen more clearly when the DNA was transferred to the nitrocellulose paper without prior denaturation, which allows detection of only ssDNA (Fig. 2B). These results showed that the pLS1 ssoA does not function to a significant extent in vivo in S. aureus because a substantial proportion of the ssDNA replication intermediate was not converted to the dsDNA form. The plasmid copy number (measured as dsDNA) was reduced from 20 in S. pneumoniae to ≈5 per genome equivalent for both pLS1 and pLS1ΔNA plasmids (data not shown). The pE194 ssoA was recognized efficiently in S. aureus as evidenced by the accumulation of very little ssDNA (Fig. 2). These results indicate that the efficiency of ss → ds DNA conversion from the pLS1 ssoA is significantly impaired in S. aureus (and perhaps in other hosts in which ssDNA accumulates), and therefore no significant differences are observed between the pLS1 plasmid containing the wild-type ssoA or its derivatives lacking this origin.
Figure 8.
Comparison of the ssoA sequences of various S. aureus plasmids (21) with that of pLS1. The sequences conserved in all the ssoAs are shown in the upper case and are boxed. Sequences conserved in the plasmids of S. aureus, which differ from those in the pLS1 ssoA are shaded and shown in the upper case. Nucleotides that are not totally conserved are shown in the lower case. The conserved RSB sequence, its complement, RSB (c), and the CS-6 sequence are also shown.
Figure 2.
Intracellular ssDNA accumulated in S. aureus. Total DNA was isolated from plasmid-containing cultures, electrophoresed on 0.7% agarose gels, and transferred to nitrocellulose paper with (A) or without (B) prior denaturation. The Southern blots were probed and subjected to autoradiography. The positions of the supercoiled (SC), open circular (OC), and single-stranded (SS) forms of plasmid DNA are indicated.
In Vitro Replication Stimulated by the pLS1 ssoA.
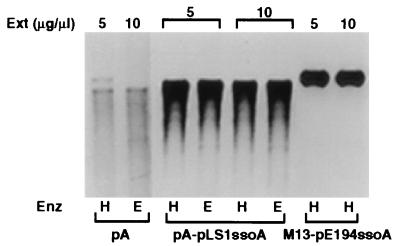
We studied the ability of the pLS1 ssoA to support lagging strand replication in cell-free extracts made from S. aureus. ssDNA containing the pLS1 ssoA (pA-pLS1ssoA) supported efficient replication, whereas ssDNA from the vector pALTER (pA) gave a weak signal (Fig. 3). These results demonstrate that DNA synthesis was mostly ssoA-specific. Under these conditions, the extent of synthesis with the pLS1 ssoA was comparable with that obtained with the pE194 ssoA (Fig. 3). The above results showed that in spite of the poor functionality of the pLS1 ssoA in vivo in S. aureus, it can be recognized by staphylococcal proteins and enzymes involved in lagging strand replication. In vitro replication of the pLS1 ssoA required RNA polymerase-dependent synthesis of (an) RNA primer(s) because the process was inhibited by rifampicin or the absence of rNTPs (not shown).
Figure 3.
In vitro replication of ssDNA in S. aureus extracts. ssDNA containing the pLS1 ssoA (pA-pLS1ssoA) or the pE194 ssoA (M13-pE194ssoA) was replicated for 60 min, and the reaction products were linearized with HindIII (H) or EcoRV (E). The reaction products were analyzed by agarose gel electrophoresis. The vector pALTER (pA) ssDNA was used as the negative control. The amounts of cell-free extract (Ext) used are indicated.
Initiation of in Vitro and in Vivo Replication from the pLS1 ssoA Is Site-Specific.
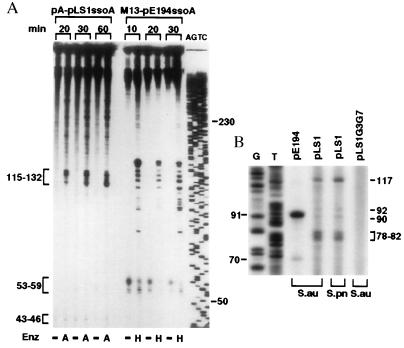
The above results provide information on the efficiency of ss → ds DNA synthesis but do not reveal the specificity of the initiation process from the pLS1 ssoA in a heterologous host. To address this question, we determined the start site(s) of lagging strand synthesis from the pLS1 ssoA in S. aureus cell-free extracts. ssDNA containing the pLS1 or pE194 ssoA was replicated in vitro and the reaction products were either treated or not treated with AflII or HindIII, enzymes that cleave immediately downstream of the pLS1 and pE194 ssoAs, respectively. This treatment should release single-stranded replication products whose size will correspond to the distance between the initiation site(s) and the enzyme cleavage site. Analysis of the replication products showed that the undigested samples consisted mainly of a series of large molecules (Fig. 4A). Faster migrating bands (43–46 nt and 53–59 nt) that were observed in the absence of restriction enzyme treatment (Fig. 4A) indicate random initiation and/or end points of ss → ds synthesis and do not reveal the specific initiation points (12). With the pLS1 ssoA DNA, samples digested with AflII generated specific bands in the range of 115–132 nt (Fig. 4A). We have shown previously that in the S. aureus in vitro system the RNA primers are removed from the replication products (12). Some of the 115 to 132 nt bands obtained with the pLS1 ssoA correspond to those seen in similar experiments done with streptococcal cell extracts (26, 27), and place the initiation site of DNA synthesis (major RNA-DNA transition points) immediately downstream of the CS-6 sequence (Fig. 1). The bands seen with the pE194 ssoA were the same as those reported (12). The above results demonstrate that replication from the pLS1 ssoA in staphylococcal extracts is specific and initiates from a few discrete sites. We also found that mutations in the conserved RSB and CS-6 sequences of pLS1 ssoA impair specific initiation in S. aureus extracts (data not shown).
Figure 4.
Determination of the initiation points of lagging-strand synthesis from the ssoA sequences in S. aureus. (A) In vitro mapping. The template ssDNAs containing the ssoA of either pLS1 or pE194 were replicated for the indicated times. Reaction products were digested with AflII (A), HindIII (H) or left undigested (−) before their separation on a denaturing gel. (B) In vivo mapping. The products of “repeat primer extension assays” using the indicated plasmids as templates purified from S. aureus (S. au) or S. pneumoniae (S. pn) were electrophoresed on a 8% polyacrylamide-urea gel. AGTC and GT, sequencing ladder generated by the Sanger’s method (31). Numbers indicate sizes in nucleotides.
Experiments also were done to identify the in vivo initiation points of lagging synthesis from the ssoA sequences. The rationale for these experiments was that although supercoiled DNA is the most abundant form of plasmids found in vivo, a small fraction of DNA my be present in partially single-stranded form in which the lagging strand synthesis has initiated but not been completed. Use of a primer corresponding to a sequence downstream of the ssoA in “repeat primer extension assays” is expected to identify the 5′ end(s) of the lagging strand. One major (91 nt) and one minor band (70 nt) was obtained with the pE194 plasmid in its native host S. aureus (Fig. 4B). In the case of pLS1, two major bands (117 nt, and closely spaced bands of 78–82 nt) and two minor bands (90 and 92 nt) were observed both in its native host S. pneumoniae and in nonnative S. aureus (Fig. 4B). The in vivo initiation sites from the pE194 and pLS1 ssoA sequences identified above are indicated in Fig. 1. To use the same primer with a negative control, we used the pLS1G3G7 mutant, which has been shown previously to lack specific initiation points in vitro in both streptococcal and staphylococcal cell-free extracts (26, 27, and data not shown). This mutant, which accumulates similar ssDNA levels and has the same copy number as pLS1 in S. aureus, did not generate any specific bands (Fig. 4B). The above results demonstrate that the pLS1 ssoA supports specific initiation of lagging strand synthesis in S. aureus.
Synthesis of Specific RNA Primers by RNA Polymerase from the pE194 and pLS1 ssoA Templates.
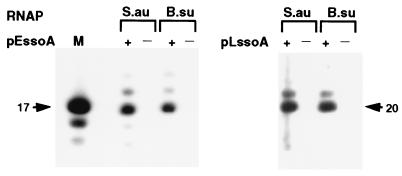
We tested whether the staphylococcal RNA polymerase synthesized a specific primer for lagging strand replication by using the pE194 and pLS1 ssoAs as the templates. The B. subtilis RNA polymerase also was used in these experiments because we have previously shown that it synthesizes a 20 nt primer RNA using the pLS1 ssoA as the template (26). In vitro transcription experiments with the pE194 ssoA showed that both the S. aureus and B. subtilis RNA polymerases synthesized a 17-nt long primer, along with a minor species of 18 nt (Fig. 5). With the pLS1 ssoA, both the RNA polymerases synthesized a predominantly 20-nt primer RNA (Fig. 5 and ref. 26). These results demonstrate that both homologous and heterologous RNA polymerases recognize sequence elements (promoters) in the ssoAs and are capable of synthesizing specific primers.
Figure 5.
RNA polymerase-directed synthesis of primer RNAs. ssDNA containing the pLS1 ssoA (pLssoA) and pE194 ssoA (pEssoA) in the functional (+) or nonfunctional (−) orientation were used as template. Reactions were carried out by using the S. aureus (S.au) or B. subtilis (B.su) RNA polymerase. A mixture of labeled oligonucleotides were used as size markers (M). The sizes of the major primer RNA products from the pE194 ssoA (17 nt) and the pLS1 ssoA (20 nt) are indicated by arrows.
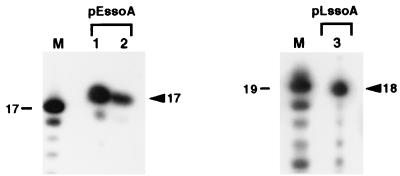
To map the initiation sites of primer RNA synthesis, primer extension experiments were carried out. Based on the results of previous in vitro replication experiments as well as results of in vitro transcription analysis with pLS1 ssoA by using the B. subtilis RNA polymerase (26), a few ss oligonucleotides were initially tested in the reverse transcriptase assay. These experiments identified the appropriate oligonucleotides for the mapping of the 5′ ends of the primer RNAs (see Materials and Methods). Unlabeled primer RNA was synthesized in vitro using a ssDNA template containing the pE194 ssoA and RNA polymerase from S. aureus and B. subtilis, and primer extension analysis was done by using a 13-mer oligonucleotide. A 17-nt band was observed with both the RNA polymerases (Fig. 6). This represents an incorporation of 4 nt to the 13-mer and places the initiation site of primer RNA synthesis at position 735 within the pE194 ssoA (Fig. 1). With the pLS1 ssoA using the S. aureus RNA polymerase, primer extension analysis with a 12-mer generated a 18-nt band (Fig. 6), placing the start-site of primer RNA synthesis at position 4175 of the pLS1 ssoA, which is identical to that seen with the B. subtilis RNA polymerase (Fig. 1 and ref. 26). The above results demonstrate that synthesis of the primer RNA initiates from a specific position within the ssoA sequences, and both homologous and heterologous RNA polymerases recognize the same promoter elements within the ssoAs.
Figure 6.
Determination of the 5′-end of the primer RNAs. In vitro-synthesized primer RNAs were used as a template for primer extension analysis. The positions of a major 17-nt band observed with the pE194 ssoA (pEssoA) and a 18-nt product obtained with the pLS1 ssoA (pLssoA) are indicated by arrowheads. Their sizes were corrected for the presence of a phosphate (31) in the mixture of labeled oligonucleotides markers (M). In samples 1 and 3, S. aureus RNA polymerase was used for the synthesis of RNA primers, whereas the B. subtilis RNA polymerase was used in sample 2.
Binding of ssoA Sequences to the RNA Polymerase from S. aureus and B. subtilis.
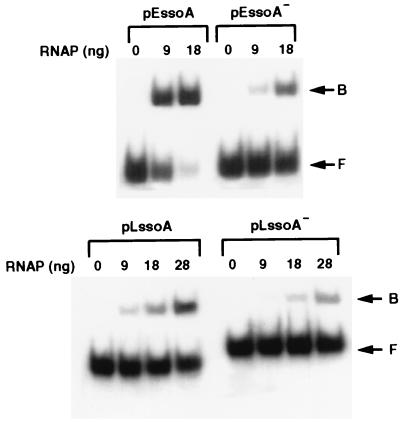
Because the pE194 ssoA but not the pLS1 ssoA functions efficiently in vivo in S. aureus, we wished to determine by EMSA whether this was caused by differences in their ability to bind to the RNA polymerase from S. aureus. The ssoA of pE194 in the functional orientation bound efficiently to the RNA polymerase, whereas the complementary ssDNA bound only weakly (Fig. 7). Measurement of the radioactivity showed that with the pE194 ssoA DNA in the functional orientation, 67% and 82% of the DNA was present in the bound form in the presence of 9 and 18 ng of RNA polymerase, respectively. Only 12% and 33% of the input DNA bound to the pE194 ssoA in the nonfunctional orientation at the above RNA polymerase concentrations. These results showed that the RNA polymerase from S. aureus is capable of specific interaction with the native pE194 ssoA. This RNA polymerase, however, bound poorly to the pLS1 ssoA (Fig. 7). In the presence of 9, 18, and 28 ng of the RNA polymerase, 15, 26, and 39%, respectively, of the input DNA containing the pLS1 ssoA in the functional orientation was bound. The percentages of binding observed with the pLS1 ssoA in the nonfunctional orientation in the presence of the above amounts of the RNA polymerase were 5, 8, and 17, respectively. These results demonstrate that the pE194 ssoA interacts more efficiently with the staphylococcal RNA polymerase than the pLS1 ssoA and suggest that the poor functionality of nonnative ssoA sequences in Gram-positive bacteria may be because of their weak interaction with the host RNA polymerase. Although the streptococcal RNA polymerase could not be used as a positive control in the above studies because it has not yet been purified, this conclusion is strengthened by the following observations. The specific activity of the pLS1 ssoA probe used in this experiment was similar to that of the pE194 probe, the unlabeled pLS1 ssoA DNA was a good substrate for in vitro transcription with saturating amounts of staphylococcal and B. subtilis RNA polymerases and the binding efficiency of the pLS1 ssoA to both of these polymerases is comparable (ref. 26 and data not shown).
Figure 7.
EMSA of S. aureus RNA polymerase bound to labeled ssDNA fragments. (Upper) EMSA with the pE194 ssoA in the functional (pEssoA) or nonfunctional (pEssoA−) orientation. (Lower) EMSA with the pLS1 ssoA in the functional (pLssoA) or nonfunctional (pLssoA−) orientation. The various ssDNAs were incubated with the indicated amounts of S. aureus RNA polymerase (in nanograms). Positions of the bound (B) and free (F) ssDNAs are indicated.
DISCUSSION
In this paper, we have investigated the role of ssoA origins in the lagging strand replication of rolling-circle plasmids. Previous studies have suggested that ssoAs function efficiently only in their natural hosts, and lagging strand replication of plasmids containing nonnative ssoAs may initiate from alternate or nonspecific sequences (3, 10, 23–25). Also, plasmids lacking ssoA are viable, suggesting that lagging strand replication in such cases must initiate from alternate sites. We have investigated the function of the ssoA of the streptococcal plasmid pLS1 in a nonnative host, S. aureus, as well as the pE194 ssoA in its natural host.
Plasmid pLS1 as well as its derivative lacking the ssoA (pLS1ΔNA) accumulated high but similar levels of ssDNA in vivo in S. aureus (Fig. 2). On the other hand, this ssoA supported efficient replication in staphylococcal extracts and the level of nucleotide incorporation was comparable with that obtained with the ssoA of pE194 that is native to S. aureus (Fig. 3). The differences between the ssoA activity in vivo and in vitro in a heterologous host may be caused by the presence of an excess of RNA polymerase and/or other host factors in cell-free extracts, which may be limiting in vivo (see below). We also have mapped the start-sites of DNA synthesis from the pE194 and pLS1 ssoA in vivo, which has previously not been reported for any rolling-circle plasmid. Although our results demonstrate poor functionality of the pLS1 ssoA in vivo in S. aureus, our data show that specific initiation does occur from this ssoA both in vivo and in vitro (Fig. 4). Based on the above data, we conclude that the poor functionality of a ssoA in vivo in a nonnative host is due to the low efficiency rather than a lack of specificity of the initiation process. However, replication initiating from this origin might not contribute significantly to the ss → dsDNA conversion in vivo in S. aureus.
We also have studied the interaction of the S. aureus RNA polymerase with the ssoA of pE194 that is native to this host, as well as with the pLS1 ssoA which is native to streptococci. DNA-binding experiments showed that the S. aureus RNA polymerase bound more efficiently to the pE194 ssoA as compared with the pLS1 ssoA (Fig. 7). Because the pLS1 ssoA functions poorly in vivo in S. aureus, our results suggest that this may be caused by the weak interaction between the nonnative ssoAs with the host RNA polymerase. The presence of sufficient quantities of RNA polymerase (and additional factors) may have allowed efficient replication from nonnative ssoA sequences in vitro.
We have identified a ssDNA promoter in the ssoA of pE194. In vitro transcription and primer extension analysis showed that the pE194 ssoA directed the synthesis of the same 17-nt primer RNA in the presence of both the S. aureus and B. subtilis RNA polymerases (Figs. 5 and 6) and clearly establish that both homologous and heterologous RNA polymerases recognize the same promoter elements. The first nucleotide of the primer RNA is expected to be a U residue, which is unusual and may reflect unique aspects of RNA transcription initiating from single-stranded origins. The coordinates of the primer RNA would position the start site of DNA synthesis (RNA–DNA transition) at position 718 of pE194, which is within a few nucleotides to that estimated from our previous in vitro replication experiments (12) and those shown in Fig. 4. An inspection of the sequence and predicted secondary structure of the pE194 ssoA revealed the presence of a putative promoter-like sequence in the vicinity of RSB (Fig. 1). This putative promoter contains sequences that resemble the consensus −35 (5′-TTGCGT-3′) and −10 (5′-TATACT-3′) regions. Such a putative promoter also has been identified in the ssoA of pLS1 (Fig. 1 and ref. 26), and ss-promoter regions are found in complementary strand origins of filamentous bacteriophages and coliphage N4 (36–39). A ss promoter (Frpo) that may support replication of the transferred ssDNA strand of the F plasmid during conjugation also has been described recently (40). In the predicted folded structure of the pE194 and pLS1 ssoAs, the putative −35 region is fully paired, whereas the −10 region has only partial pairing (Fig. 1). Interestingly, the single-strandedness of the −10 region appears to be critical for the function of the other ssDNA promoters (37, 40).
The S. aureus RNA polymerase synthesized a 20-nt long primer RNA from the pLS1 ssoA and transcription initiated at position 4175, which is identical to the results obtained using the B. subtilis RNA polymerase (Figs. 1 and 5 and ref. 26). Thus, the synthesis of the primer RNA for pLS1-lagging strand replication is initiated at the same position by two different heterologous RNA polymerases. This primer RNA is used as a substrate for DNA synthesis by the streptococcal DNA polymerase I, and this enzyme has been shown to be required for pLS1 replication in vivo in S. pneumoniae (26, 41).
The sequences and structures of various ssoAs are highly conserved, and an alignment of the ssoAs of several S. aureus plasmids and that of the streptococcal plasmid pLS1 is shown in Fig. 8. Three sequences, the RSB, its partial complement, and CS-6 are highly conserved among these ssoAs and are important for lagging strand synthesis (12, 26, 27). However, there are several regions (shaded nucleotides in Fig. 8) that are highly conserved in ssoAs native to S. aureus plasmids that are not conserved in the streptococcal pLS1 ssoA (Fig. 8). It is possible that these sequences are important for recognition of the ssos by the RNA polymerase of their respective hosts, and they may contribute to the efficiency of RNA polymerase binding and initiation of lagging strand synthesis.
Acknowledgments
We thank members of our laboratories for helpful discussions. Special thanks to R. Deora for providing the S. aureus RNA polymerase, M. Salas, M. Mencia, and J. M. Lazaro for their gift of B. subtilis RNA polymerase, and S. Lacks for providing streptococcal strains. We also thank Adam Zhao for advice in the preparation of cell-free extracts. This work was supported by grant GM31685 from the National Institutes of Health (to S.A.K.), Comision Interministerial de Ciencia y Tecnologia Grant BIO97-0347 (to M.E.), and by Campus Research Board and Benedict Research Foundation grants, University of Illinois at Chicago (to T.K.M.).
ABBREVIATIONS
- ss
single-stranded
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
- sso
single strand origin
- ds
double-stranded
- RSB
recombination site B
- EMSA
electrophoretic mobility-shift analysis
References
- 1.Baas P D, Jansz H S. Curr Top Microbiol Immunol. 1988;136:31–70. doi: 10.1007/978-3-642-73115-0_3. [DOI] [PubMed] [Google Scholar]
- 2.del Solar G, Moscoso M, Espinosa M. Mol Microbiol. 1993;8:789–796. doi: 10.1111/j.1365-2958.1993.tb01625.x. [DOI] [PubMed] [Google Scholar]
- 3.Gruss A D, Ehrlich S D. Microbiol Rev. 1989;53:231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan S A. Microbiol Mol Biol Rev. 1997;61:442–455. doi: 10.1128/mmbr.61.4.442-455.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zinder N, Horiuchi K. Microbiol Rev. 1985;49:101–106. doi: 10.1128/mr.49.2.101-106.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koepsel R R, Murray R W, Rosenblum W D, Khan S A. Proc Natl Acad Sci USA. 1995;82:6845–6849. doi: 10.1073/pnas.82.20.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.te Riele H, Michel B, Ehrlich S D. EMBO J. 1986;5:631–637. doi: 10.1002/j.1460-2075.1986.tb04257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray R W, Koepsel R R, Khan S A. J Biol Chem. 1989;264:1051–1057. [PubMed] [Google Scholar]
- 9.Gruss A D, Ross H F, Novick R P. Proc Natl Acad Sci USA. 1987;84:2165–2169. doi: 10.1073/pnas.84.8.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.del Solar G, Kramer M G, Ballester S, Espinosa M. Mol Gen Genet. 1993;241:97–105. doi: 10.1007/BF00280206. [DOI] [PubMed] [Google Scholar]
- 11.del Solar G, Alonso J C, Espinosa M, Díaz-Orejas R. Mol Microbiol. 1996;21:661–666. doi: 10.1046/j.1365-2958.1996.6611376.x. [DOI] [PubMed] [Google Scholar]
- 12.Dempsey L A, Zhao A C, Khan S A. Mol Microbiol. 1995;15:679–687. doi: 10.1111/j.1365-2958.1995.tb02377.x. [DOI] [PubMed] [Google Scholar]
- 13.del Solar G, Puyet A, Espinosa M. Nucleic Acids Res. 1987;15:5561–5580. doi: 10.1093/nar/15.14.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boe L, Gros M F, te Riele H, Ehrlich S D, Gruss A D. J Bacteriol. 1989;171:3366–3372. doi: 10.1128/jb.171.6.3366-3372.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bron S, Bosma P, van Belkum M J, Luxen E. Plasmid. 1987;18:8–15. doi: 10.1016/0147-619x(87)90073-4. [DOI] [PubMed] [Google Scholar]
- 16.van der Lelie D, Bron S, Venema G, Oskam L. Nucleic Acids Res. 1989;17:7283–7294. doi: 10.1093/nar/17.18.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birch P, Khan S A. Proc Natl Acad Sci USA. 1992;89:290–294. doi: 10.1073/pnas.89.1.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boe L, Nielsen T T, Madsen S M, Andrup L, Bolander G. Plasmid. 1991;25:190–197. doi: 10.1016/0147-619x(91)90012-l. [DOI] [PubMed] [Google Scholar]
- 19.Leenhouts K, Tolner J B, Bron S, Kok J, Venema G, Seegers J F M L. Plasmid. 1991;26:55–66. doi: 10.1016/0147-619x(91)90036-v. [DOI] [PubMed] [Google Scholar]
- 20.Seegers J F M L, Zhao A C, Meijer W J J, Khan S A, Venema G, Bron S. Mol Gen Genet. 1995;249:43–50. doi: 10.1007/BF00290234. [DOI] [PubMed] [Google Scholar]
- 21.Novick R P. Annu Rev Microbiol. 1989;43:537–565. doi: 10.1146/annurev.mi.43.100189.002541. [DOI] [PubMed] [Google Scholar]
- 22.Zaman S, Radnedge L, Richards H, Ward J M. J Gen Microbiol. 1993;139:669–676. doi: 10.1099/00221287-139-4-669. [DOI] [PubMed] [Google Scholar]
- 23.Kramer M G, del Solar G, Espinosa M. Microbiology. 1995;141:655–662. doi: 10.1099/13500872-141-3-655. [DOI] [PubMed] [Google Scholar]
- 24.Meijer W J J, van der Lelie D, Venema G, Bron S. Plasmid. 1995;33:79–89. doi: 10.1006/plas.1995.1010. [DOI] [PubMed] [Google Scholar]
- 25.Meijer W J J, van der Lelie D, Venema G, Bron S. Plasmid. 1995;33:91–99. doi: 10.1006/plas.1995.1011. [DOI] [PubMed] [Google Scholar]
- 26.Kramer M G, Khan S A, Espinosa M. EMBO J. 1997;16:5784–5795. doi: 10.1093/emboj/16.18.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer M G, Khan S A, Espinosa M. J Bacteriol. 1998;180:83–89. doi: 10.1128/jb.180.1.83-89.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lacks S A, López P, Greenberg B, Espinosa M. J Mol Biol. 1986;192:753–765. doi: 10.1016/0022-2836(86)90026-4. [DOI] [PubMed] [Google Scholar]
- 29.Bernander R, Krabbe M, Nordstrom K. EMBO J. 1992;12:4481–4487. doi: 10.1002/j.1460-2075.1992.tb05549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grohmann E, Zechner E L, Espinosa M. FEMS Microbiol Lett. 1997;152:363–369. doi: 10.1111/j.1574-6968.1997.tb10453.x. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 32.Deora R, Misra T K. Biochem Biophys Res Commun. 1995;208:610–616. doi: 10.1006/bbrc.1995.1382. [DOI] [PubMed] [Google Scholar]
- 33.Deora R, Misra T K. J Biol Chem. 1995;271:21828–21834. doi: 10.1074/jbc.271.36.21828. [DOI] [PubMed] [Google Scholar]
- 34.Mencía M, Monsalve M, Rojo F, Salas M. Proc Natl Acad Sci USA. 1996;93:6616–6620. doi: 10.1073/pnas.93.13.6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coverly D, Laskey R A. Annu Rev Biochem. 1994;63:745–776. doi: 10.1146/annurev.bi.63.070194.003525. [DOI] [PubMed] [Google Scholar]
- 36.Higashitani N, Higashitani A, Horiuchi K. J Virol. 1993;67:2175–2181. doi: 10.1128/jvi.67.4.2175-2181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Higashitani A, Higashitani N, Horiuchi K. Proc Natl Acad Sci USA. 1997;94:2909–2914. doi: 10.1073/pnas.94.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geider K, Kornberg A. J Biol Chem. 1974;249:3999–4005. [PubMed] [Google Scholar]
- 39.Glucksmann-Kuis M A, Dai X, Markiewicz P, Rothman-Denes L B. Cell. 1996;84:147–154. doi: 10.1016/s0092-8674(00)81001-6. [DOI] [PubMed] [Google Scholar]
- 40.Masai H, Arai K-I. Cell. 1997;89:897–907. doi: 10.1016/s0092-8674(00)80275-5. [DOI] [PubMed] [Google Scholar]
- 41.Díaz A, Lacks S A, López P. Mol Microbiol. 1994;14:773–783. doi: 10.1111/j.1365-2958.1994.tb01314.x. [DOI] [PubMed] [Google Scholar]