Abstract
The matrix metalloproteinases (MMPs) constitute a multigene family of over 25 secreted and cell surface enzymes that process or degrade numerous pericellular substrates. Their targets include other proteinases, proteinase inhibitors, clotting factors, chemotactic molecules, latent growth factors, growth factor–binding proteins, cell surface receptors, cell-cell adhesion molecules, and virtually all structural extracellular matrix proteins. Thus MMPs are able to regulate many biologic processes and are closely regulated themselves. We review recent advances that help to explain how MMPs work, how they are controlled, and how they influence biologic behavior. These advances shed light on how the structure and function of the MMPs are related and on how their transcription, secretion, activation, inhibition, localization, and clearance are controlled. MMPs participate in numerous normal and abnormal processes, and there are new insights into the key substrates and mechanisms responsible for regulating some of these processes in vivo. Our knowledge in the field of MMP biology is rapidly expanding, yet we still do not fully understand how these enzymes regulate most processes of development, homeostasis, and disease.
Keywords: proteinases, tissue inhibitors of metalloproteinases, extracellular matrix, proteolytic signaling
INTRODUCTION
Extracellular proteinases are required for numerous developmental and disease-related processes. The ability to degrade extracellular proteins is essential for any individual cell to interact properly with its immediate surroundings and for multicellular organisms to develop and function normally. This was obvious long before it was first shown that diffusible enzymes produced by fragments of involuting tadpole tail could degrade gels made of native fibrillar collagen (Gross & Lapiere 1962). Since then, a family of related enzymes has been identified in species from hydra to humans and collectively called matrix metalloproteinases (MMPs) because of their dependence on metal ions for catalytic activity, their potent ability to degrade structural proteins of the extracellular matrix (ECM), and specific evolutionary sequence considerations that distinguish them from other closely related metalloproteinases (Stöcker et al. 1995). In addition to their ECM substrates, MMPs also cleave cell surface molecules and other pericellular non-matrix proteins, thereby regulating cell behavior in several ways (Sternlicht et al. 2000). Thus like the many proteins they modify, the MMPs influence diverse physiologic and pathologic processes, including aspects of embryonic development, tissue morphogenesis, wound repair, inflammatory diseases, and cancer (Nelson et al. 2000, Sternlicht et al. 2001).
With the ability to alter cell fates and developmental outcomes comes the necessity for higher levels of control. Nevertheless, it took nearly a decade from the time collagenolytic activities were first demonstrated to realize that MMPs are synthesized as inactive zymogens that require activation (Harper et al. 1971), and even longer to demonstrate the existence of the first of at least four endogenous metalloproteinase inhibitors, now called tissue inhibitors of metalloproteinases, or TIMPs (Bauer et al. 1975). Since then, other levels of MMP regulation have been elucidated, although several layers of complexity are still left to unravel. In addition to being differentially regulated at the level of transcription, MMPs can be controlled at the protein level by their endogenous activators and inhibitors and by factors that influence their secretion, their cell surface localization, and their own degradation and clearance. Moreover, higher organisms express multiple MMPs, each with its own profile of expression, localization, activation, inhibition, and clearance, as well as its own, sometimes broad, range of preferred substrates. Thus multiple modifiers, each with its own regulatory inputs, control different MMP functions in vivo.
The multiplicity of MMPs with distinct but somewhat overlapping functions probably acts as a safeguard against any losses of regulatory control. Although such redundant and compensatory mechanisms are advantageous to the organism, they often confound efforts to fully understand how MMPs function in vivo. As the field has grown, the unforeseen complexity of MMPs has emerged, often revealing surprising mechanisms of action. Frequently, the belief that an MMP is involved in a given biologic process is based on the strength of its association with that process and the existence of plausible mechanisms and then tested using various experimental approaches. For example, MMPs are invariably upregulated in rheumatoid arthritis and malignant disease, with more severe increases often indicating a worse prognosis. Moreover, a major hallmark of these diseases is the capacity of cells to cross tissue boundaries and, in the case of cancer, spread to distant sites of the body. Thus ECM-degrading enzymes must be present to break down the structural barriers to invasion. Indeed, extensive experimental evidence supports this straightforward and conceptually appealing supposition, but the mechanisms may be more complex than originally thought. Furthermore, in vivo genetic approaches that test the consequences of selective gains or losses of MMP function have led to the surprising finding that MMPs promote the initial stages of cancer development itself but may decrease the severity of the ultimate malignancy (Coussens et al. 2000, Pozzi et al. 2000). In arthritis, the loss of certain MMPs surprisingly exacerbates rather than alleviates the disease (Mudgett et al. 1998). Considerable evidence implicates MMPs as important players in several biologic processes, yet the actual mechanisms underlying their influence are mostly unsolved. It is hoped that understanding these processes will result in a more rational approach toward reducing or entirely alleviating the ill effects of MMPs in disease while maintaining their necessary and beneficial functions.
OVERVIEW OF THE METALLOPROTEINASES
The Metzincin Superfamily
Proteolytic enzymes are classified as either exopeptidases or endopeptidases based on whether they cleave terminal or internal peptide bonds, respectively. Most endopeptidases are classified as serine, cysteine, aspartic or metalloproteinases based on their catalytic mechanism and inhibitor sensitivities, and the metalloproteinases are further separated into five superfamilies based on sequence considerations. Of these, the metzincin superfamily is distinguished by a highly conserved motif containing three histidines that bind zinc at the catalytic site and a conserved methionine turn that sits beneath the active site zinc (Stöcker et al. 1995). Their signature zinc-binding motif reads HEBXHXBGBXHZ, where histidine (H), glutamic acid (E) and glycine (G) residues are invariant, B is a bulky hydrophobic residue, X is a variable residue, and Z is a family-specific amino acid. The metzincins are further subdivided into four multigene families, the serralysins, astacins, ADAMs/adamalysins, and MMPs, based primarily on the identity of the Z residue, which is serine in all but a few MMPs (Stöcker et al. 1995).
The serralysins, which have a proline in the Z position, are large bacterial enzymes that often play an important role in bacterial virulence and pathogenicity (Sternlicht & Werb 1999). Astacins, on the other hand, contain glutamic acid in the Z position. They include bone morphogenetic protein-1 (BMP-1), which is the procollagen C-proteinase that removes the C-terminal propeptides of fibrillar procollagens, Drosophila and mammalian tolloid and tolloid-like proteins, which activate certain growth factors, and secreted and transmembrane meprins A and B that can process peptide hormones (reviewed in Sternlicht & Werb 1999).
The adamalysins, ADAMs and ADAMTSs have an aspartic acid in the Z position. The adamalysins are soluble snake venom enzymes with potent ECM-degrading activity. The ADAMs are transmembrane cell surface proteins that have a disintegrin and metalloproteinase domain (Primakoff & Myles 2000). Each of the ADAMs has an N-terminal signal sequence followed by a propeptide domain, a functional or nonfunctional metalloproteinase domain, a disintegrin-like domain that is similar to snake venom disintegrins but often lacks an Arg-Gly-Asp (RGD) sequence, a cysteine-rich domain, EGF-like repeats, a transmembrane domain, and a C-terminal cytoplasmic tail. Individual ADAMs may participate in proteolysis via their metalloproteinase domain, adhesion via their disintegrin domain, cell-cell fusion via a putative hydrophobic fusion peptide in their cysteine-rich domain, and cell signaling via SH3-recognition sequences that are sometimes present in their intracellular domain. Seventeen of the 30 known ADAMs have a functional zinc-binding motif, including ADAM17 (TNF-α converting enzyme, TACE), which cleaves membrane-bound TNF-α to generate active soluble TNF-α. TACE also probably contributes to the shedding of several other cell surface molecules and appears to be an essential activator of TGF-α in vivo (Peschon et al. 1998). Considering their localization, other ADAMs are also likely to regulate the shedding of several important cell surface molecules (Werb & Yan 1998).
The secreted ADAMTS proteins also have signal, propeptide, metalloproteinase, and disintegrin-like domains. However, unlike the ADAMs, their disintegrin domain is followed by a thrombospondin (TS) type I repeat, a cysteine-rich domain, one or more additional TS domains (except for ADAMTS-4, which lacks a second TS repeat), and, in some cases, a C-terminal domain of variable length (Tang & Hong 1999). They include ADAMTS-1 and ADAMTS-8, which potently inhibit angiogenesis via their TS repeats (Iruela-Arispe et al. 1999); ADAMTS-2, which is a procollagen amino-propeptidase that is required for the proper assembly of fibrillar collagens I and II (Colige et al. 1997); and ADAMTS-4 and ADAMTS-5/11 (aggrecanases 1 and 2, respectively), which can degrade the cartilage proteoglycan aggrecan (Abbaszade et al. 1999).
MMP Structure and Function
At present, 25 vertebrate MMPs and 22 human homologues have been identified (Nagase & Woessner 1999, Sternlicht & Bergers 2000, Lohi et al. 2001). In addition, several nonvertebrate MMPs have been identified, including the embryonic sea urchin hatching enzyme envelysin (Lepage & Gache 1990); Caenorhabditis elegans MMPs C31, H19, and Y19 (Wada et al. 1998); a Drosophila MMP (Llano et al. 2000); an MMP in hydra that regulates cell differentiation and foot process development (Leontovich et al. 2000); soybean leaf metalloendopeptidase-1 (McGeehan et al. 1992); an MMP in the flowering mustard plant Arabidopsis thaliana (Maidment et al. 1999); and gamete lytic enzyme from green alga (Kinoshita et al. 1992). Each of the vertebrate MMPs has distinct but often overlapping substrate specificities, and together they can cleave numerous extracellular substrates, including virtually all ECM proteins (reviewed in Sternlicht et al. 2001). In addition to their conserved zinc-binding motif (usually HEF/LGHS/ALGLXHS, where bold-noted amino acids are always present) and “Met turn” (usually ALMYP), the MMPs share added stretches of sequence homology, giving them a fairly conserved overall structure (Stöcker et al. 1995).
Individual MMPs are referred to by their common names or according to a sequential numeric nomenclature reserved for the vertebrate MMPs (Table 1). In addition, they are often grouped according to their modular domain structure (Figure 1). In this regard, all MMPs have an N-terminal signal sequence (or “pre” domain) that is removed after it directs their synthesis to the endoplasmic reticulum. Thus most MMPs are secreted; however, six display transmembrane domains and are expressed as cell surface enzymes. The pre domain is followed by a propeptide “pro” domain that maintains enzyme latency until it is removed or disrupted, and a catalytic domain that contains the conserved zinc-binding region (reviewed in Nagase & Woessner 1999). The catalytic domain dictates cleavage-site specificity through its active site cleft, through specificity sub-site pockets that bind amino acid residues immediately adjacent to the scissile peptide bond, and through secondary substrate-binding exosites located outside the active site itself (Overall 2001).
TABLE 1.
The vertebrate MMPs
| MMP | Common name(s) | Domain organizationa |
|---|---|---|
| MMP1 | Collagenase-1 | B |
| MMP2 | Gelatinase A | C |
| MMP3 | Stromelysin-1 | B |
| MMP7 | Matrilysin | A |
| MMP8 | Collagenase-2 | B |
| MMP9 | Gelatinase B | C |
| MMP10 | Stromelysin-2 | B |
| MMP11 | Stromelysin-3 | D |
| MMP12 | Macrophage metalloelastase | B |
| MMP13 | Collagenase-3 | B |
| MMP14 | MT1-MMP | E |
| MMP15 | MT2-MMP | E |
| MMP16 | MT3-MMP | E |
| MMP17 | MT4-MMP | F |
| MMP18 | Collagenase-4 (Xenopus) | B |
| MMP19 | RASI-1 | B |
| MMP20 | Enamelysin | B |
| MMP21 | XMMP (Xenopus) | G |
| MMP22 | CMMP (chicken) | B |
| MMP23 | H | |
| MMP24 | MT5-MMP | E |
| MMP25 | MT6-MMP | F |
| MMP26 | Endometase, Matrilysin-2 | A |
| MMP27 | B | |
| MMP28 | Epilysin | D |
See Figure 1 for domain arrangements.
Figure 1.
Domain structure of the MMPs. Pre, signal sequence; Pro, propeptide with a free zinc-ligating thiol (SH) group; F, furin-susceptible site; Zn, zinc-binding site; II, collagen-binding fibronectin type II inserts; H, hinge region; TM, transmembrane domain; C, cytoplasmic tail; GPI, glycophosphatidyl inositol-anchoring domain; C/P, cysteine/proline; IL-1R, interleukin-1 receptor. The hemopexin/vitronectin-like domain contains four repeats with the first and last linked by a disulfide bond.
With the exception of MMP7 (matrilysin), MMP26 (endometase/matrilysin-2), and MMP23, all MMPs have a hemopexin/vitronectin-like domain that is connected to the catalytic domain by a hinge or linker region. MMP7 and MMP26 merely lack these extra domains, whereas MMP23 has unique cysteine-rich, proline-rich, and IL-1 type II receptor-like domains instead of a hemopexin domain (Gururajan et al. 1998, Park et al. 2000). When present, the hemopexin domain influences TIMP binding, the binding of certain substrates, membrane activation, and some proteolytic activities. For example, chimeric enzyme studies indicate that both ends of MMP1 (collagenase-1) are required for it to cleave native fibrillar collagens (Murphy et al. 1992, Sanchez-Lopez et al. 1993). This collagenolytic activity requires the initial binding and orientation of the collagen fibril, local unwinding of its triple-helical structure, and sequential cleavage of each α-chain individually because the catalytic cleft is too narrow to accommodate the entire triple helix (Overall 2001). Apparently, the hemopexin domain participates in all but the last of these steps. The hinge region, in turn, varies in length and composition among the various MMPs and also influences substrate specificity (Knauper et al. 1997). Gelatinases A and B (MMP2 and MMP9, respectively) are further distinguished by the insertion of three head-to-tail cysteine-rich repeats within their catalytic domain. These inserts resemble the collagen-binding type II repeats of fibronectin and are required to bind and cleave collagen and elastin (Murphy et al. 1994, Shipley et al. 1996). In addition, MMP9 has a unique type V collagen-like insert of unknown importance at the end of its hinge region. Finally, the membrane-type (MT) MMPs have a single-pass transmembrane domain and a short cytoplasmic C-terminal tail (MMPs 14, 15, 16, and 24) or a C-terminal hydrophobic region that acts as a glycophosphatidyl inositol (GPI) membrane-anchoring signal (MMP17 and MMP25) (Itoh et al. 1999, Kojima et al. 2000). These domains play an essential role in the localization of several important proteolytic events to specific regions of the cell surface.
REGULATION OF MMP ACTIVITY
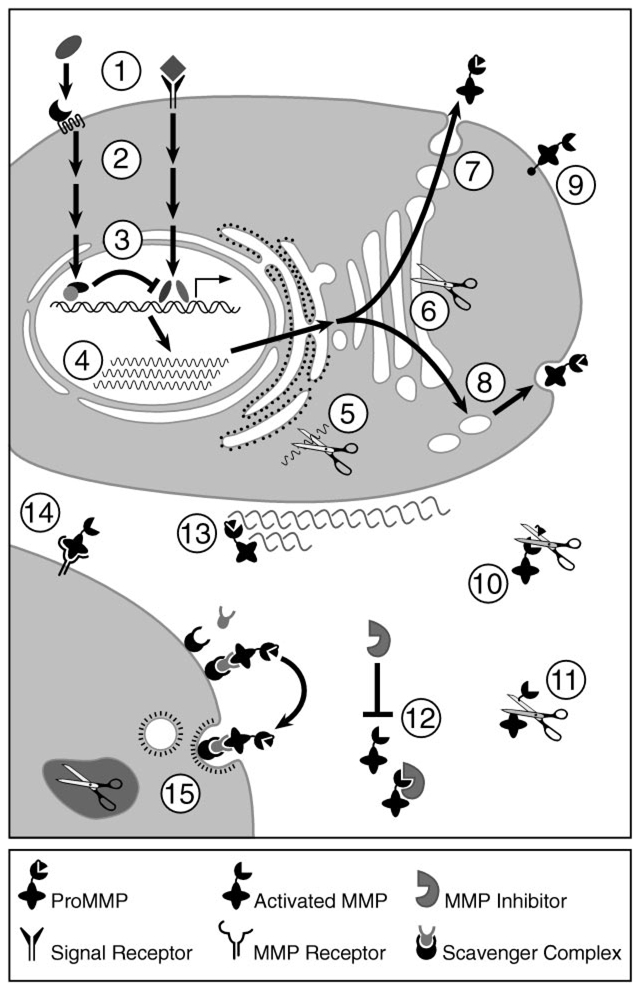
To accomplish their normal (or pathologic) functions, MMPs must be present in the right cell type and pericellular location, at the right time, and in the right amount, and they must be activated or inhibited appropriately. Thus MMPs are tightly regulated at the transcriptional and post-transcriptional levels and are also controlled at the protein level via their activators, their inhibitors, and their cell surface localization (Figure 2).
Figure 2.
Regulation of the MMPs. MMP regulatory mechanisms include inductive and suppressive signaling (1), intracellular signal transduction (2), transcriptional activation and repression (3), post-transcriptional mRNA processing (4), mRNA degradation (5), intracellular activation of furin-susceptible MMPs (6), constitutive secretion (7), regulated secretion (8), cell surface expression (9), proteolytic activation (10), proteolytic processing and inactivation (11), protein inhibition (12), ECM localization (13), cell surface localization (14), and endocytosis and intracellular degradation (15).
Transcriptional Regulation
Because MMP substrate specificities tend to overlap, the biologic function of individual MMPs is largely dictated by their differential patterns of expression. Indeed, differences in the temporal, spatial, and inducible expression of the MMPs are often indicative of their unique roles. Accordingly, most MMPs are closely regulated at the level of transcription, with the notable exception of MMP2, which is often constitutively expressed and controlled through a unique mechanism of enzyme activation (Strongin et al. 1995) and some degree of post-transcriptional mRNA stabilization (Overall et al. 1991). Nevertheless, data indicate that the basal expression of MMP2, MMP14 (MT1-MMP), and TIMP2 is co-regulated, which is consistent with their cooperation during MMP2 activation and with specific similarities in their gene promoters (Lohi et al. 2000). Otherwise, MMP gene expression is regulated by numerous stimulatory and suppressive factors that influence multiple signaling pathways (Fini et al. 1998). For example, the expression of various MMPs can be up- or downregulated by phorbol esters, integrin-derived signals, extracellular matrix proteins, cell stress, and changes in cell shape (Kheradmand et al. 1998; reviewed in Sternlicht & Werb 1999). Type I collagen acts as a ligand for discoidin domain-containing receptor-like tyrosine kinases that induce MMP1 expression when they are activated by intact collagen and become inactive when they bind MMP1-cleaved collagen (Vogel et al. 1997, Shrivastava et al. 1997). Thus MMP1 expression can be induced by its own substrate and specifically repressed once it cleaves that substrate and is no longer needed. In addition, MMP expression is regulated by several cytokines and growth factors, including interleukins, interferons, EGF, KGF, NGF, basic FGF, VEGF, PDGF, TNF-α, TGF-β, and the extracellular matrix metalloproteinase inducer EMMPRIN (Fini et al. 1998). Many of these stimuli induce the expression and/or activation of c-fos and c-jun proto-oncogene products, which heterodimerize and bind activator protein-1 (AP-1) sites within several MMP gene promoters.
Although AP-1 complexes play a critical role in the regulation of several MMP genes, other factors are also involved. In some cases, one signal may coordinately regulate some MMP genes and differentially regulate others. For example, TGF-β suppresses the transcription of the MMP1 and MMP3 (stromelysin-1) genes, but induces the expression of MMP13 (collagenase-3) (Uria et al. 1998). In addition, some MMPs are expressed in only a small repertoire of cell types, e.g., MMP20 expression appears to be confined to the enamel organ of developing teeth (Sternlicht et al. 2000), and normal MMP9 expression is largely limited to osteoclasts, macrophages, trophoblast cells, hippocampal neurons, and migrating keratinocytes at the margins of healing wounds (Mohan et al. 1998, Munaut et al. 1999). The use of mice carrying β-galactosidase reporter transgenes under the control of various portions of the MMP9 gene promoter have helped to identify the 5′ regions responsible for most of this cell-specific expression in vivo (Mohan et al. 1998). Cell-specific induction of MMP expression has also been observed in culture. For example, phorbol esters induce the expression of MMP3 rather than MMP10 in fibroblasts, yet the opposite occurs in keratinocytes (Windsor et al. 1993). Thus how an MMP gene responds to a given input depends on the organization of its transcriptional promoter and the presence or absence of other signals, i.e., on cellular context.
Several cis-regulatory elements influence MMP gene expression depending on their proximity to one another in the gene promoter. AP-1 sites give several MMP genes the ability to be induced by phorbol esters and act synergistically with adjacent Ets-binding sites in genes such as MMP1, but not in others such as MMP13 (Pendas et al. 1997). This difference is presumably because the Ets and AP-1 sites are 9 nucleotides apart in the MMP1 gene promoter and 20 nucleotides apart in the MMP13 promoter and because the distance between these two sites is critical (Gutman & Wasylyk 1990). As an indication of the importance of these sites, targeted disruption of the murine Ets2 transcription factor results in early embryonic lethality and deficient MMP9 expression, as well as the deficient induction of MMP3 and MMP13 in Ets2-deficient fibroblasts (Yamamoto et al. 1998). Moreover, their expression in the Ets2-deficient fibroblasts is restored by the artificial expression of Ets2. Several other putative regulatory elements have been identified within various MMP gene promoters, and many have been shown by functional analysis to regulate cell- and circumstance-specific gene expression. These include an osteoblast-specific element in the MMP13 gene promoter that responds to core-binding factor 1 (CBFA1) (Jimenez et al. 1999); a β-catenin-regulated LEF/TCF recognition site near the MMP7 transcription start site (Crawford et al. 1999); TGF-β inhibitory elements in several MMP genes; and AP-2, Sp1, Sp3, NF-κB, CCAAT/enhancer-binding protein-β, or retinoic acid response elements that are also found in several MMP genes (Fini et al. 1998, Lohi et al. 2000, Ludwig et al. 2000). In addition, a functional p53-binding site has been identified in the MMP2 gene promoter (Bian & Sun 1997), and wild-type p53 downregulates basal and inducible MMP1 gene expression in human fibroblasts and osteogenic sarcoma cells, whereas some mutant forms do not (Sun et al. 1999). On the other hand, the downregulation of p53 using SV40 T-antigen suppresses the expression of MMP2, MMP3, and MMP9 in human placental trophoblast-like cells (Logan et al. 1996). Despite numerous advances in our understanding of MMP gene regulation, the cross-talk between the many signaling pathways, nuclear factors, and gene regulatory elements that regulate MMP expression are barely understood.
Basal and inducible levels of MMP gene expression can also be influenced by genetic variations that may, in turn, influence the development or progression of several diseases. Common bi-allelic single-nucleotide polymorphisms (SNPs) that influence the rate of transcription have been identified within several MMP gene promoters (Ye 2000). For example, an MMP1 SNP contains one or two guanidines 1607 basepairs (bp) upstream of the transcription start site (Rutter et al. 1998). Here, the insertion of an extra G creates a functional Ets-binding site immediately adjacent to an AP-1 site and, as a result, transcription is enhanced up to 37-fold. Interestingly, the high-expressing 2G allele has been associated with higher levels of MMP1 expression in vivo and is present more often in tumor cell lines and ovarian cancer patients than in the general population (Rutter et al. 1998, Kanamori et al. 1999). Moreover, an unusually large proportion of metastatic melanomas with loss of heterozygosity at this site retains the high-expressing 2G allele (Noll et al. 2001). Thus enhanced MMP1 transcription may contribute to human cancer susceptibility and progression as it does in mice (Di Colandrea et al. 2000). Another SNP located 1306 bp upstream of the MMP2 transcription start site contains either a cytidine or thymidine, such that the less common T allele disrupts an otherwise functional Sp1-binding site and diminishes promoter activity by about 50% (Price et al. 2001). An MMP3 SNP is located 1171 bp upstream of the transcription start site and contains a run of five or six adenosines (Ye et al. 2000). In this case, the 6A allele binds an 89-kDa nuclear factor more readily than the 5A allele and has 50% less transcriptional activity. Another SNP contains either a cytidine or thymidine 1562 bp upstream of the MMP9 transcription start site (Zhang et al. 1999). Here, nuclear protein complexes bind the C allele most readily, and the less common T allele is about 1.5-fold more potent than the C allele. Finally, an A-to-G transition exists 82 bp upstream of the MMP12 transcription start site, such that the A allele has higher AP-1 binding affinity and about 1.2-fold higher promoter activity than the less common G allele (Jormsjo et al. 2000). Most notably, however, the MMP3, MMP9, and MMP12 SNPs have each been associated with coronary artery disease progression despite their modest influence on gene transcription (Ye 2000).
Post-Transcriptional Regulation
Post-transcriptional mechanisms can also influence MMP expression. For example, mRNA transcripts that encode MMP1 and MMP3 are stabilized by phorbol esters and EGF, whereas MMP13 transcripts are stabilized by PDGF and glucocorticoids and destabilized by TGF-β (Delany et al. 1995, Vincenti 2001). The turnover of MMP1 mRNA is apparently regulated by AU-rich sequences in the 3′ untranslated region, and similar sequences may also regulate the stability of other MMP transcripts (Vincenti 2001). In addition, a soluble and proteolytically active form of MT3-MMP is generated by alternative mRNA splicing rather than membrane shedding (Matsumoto et al. 1997), whereas the multiple transcripts of MMP13, MMP17, and MMP20 probably result from alternative polyadenylation (reviewed in Sternlicht & Werb 1999).
Regulation of MMP Secretion
Although most MMPs are constitutively secreted once they become translated, conspicuous instances of secretory control do exist. MMP8 (collagenase-2, neutrophil collagenase) and MMP9 are synthesized by differentiating granulocytes in the bone marrow, stored in the specific and gelatinase (teriary) granules of circulating neutrophils, respectively, and released following neutrophil activation by inflammatory mediators (Hasty et al. 1990). In macrophages, plasmin and thrombin induce the secretion of MMP12, but do not alter its rate of transcription (Raza et al. 2000). Instead, post-translational release of pre-formed MMP12 from macrophages occurs in response to protein kinase C activation downstream of the G protein–coupled thrombin receptor PAR-1 (proteinase-activated receptor-1), which becomes activated after binding a ligand that is generated from its own N-terminal end by thrombin-dependent cleavage.
Activation of Latent Metalloproteinases
Like other proteolytic enzymes, MMPs are first synthesized as inactive proenzymes or zymogens. Their latency is maintained by an unpaired cysteine sulfhydryl group near the C-terminal end of the propeptide domain. This sulfhydryl acts as a fourth ligand for the active site zinc ion, and activation requires that this cysteine-to-zinc switch be opened by normal proteolytic removal of the propeptide domain or by ectopic perturbation of the cysteine-zinc interaction (Van Wart&Birkedal-Hansen 1990). Once displaced, the thiol group is replaced by a water molecule that can then attack the peptide bonds of MMP targets.
Although most MMPs are secreted as latent zymogens, MMP11 (stromelysin-3), MMP27 (epilysin), and the MT-MMPs contain an RXK/RR furin-like enzyme recognition motif between their propeptide and catalytic domains. This allows them to be activated by intracellular subtilisin-type serine proteinases before they reach the cell surface or are secreted (Pei & Weiss 1995). MMP23 also has a furin-susceptible cleavage site and is a likely target of intracellular proprotein convertases, but unlike all other MMPs, it lacks the conserved cysteine that is required for enzyme latency in the first place (Gururajan et al. 1998). All other MMPs lack a furin-susceptible insert and are thus activated outside the cell following their secretion.
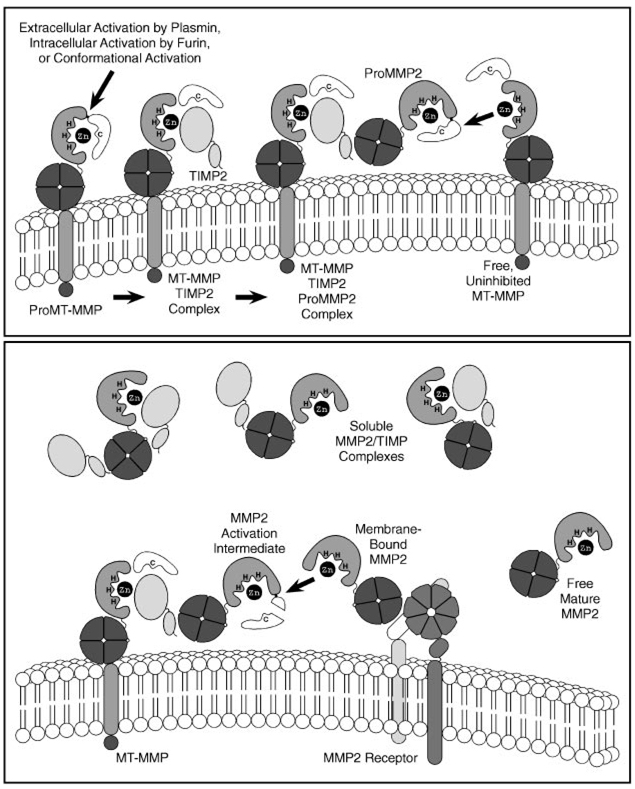
The extracellular activation of most MMPs can be initiated by other already activated MMPs or by several serine proteinases that can cleave peptide bonds within MMP prodomains (Woessner & Nagase 2000). However, MMP2 is refractory to activation by serine proteinases and is instead activated at the cell surface through a unique multistep pathway involving MT-MMPs and TIMP2 (Figure 3) (Strongin et al. 1995). Indeed, MT1-MMP is a particularly efficient MMP2 activator, whereas MT4-MMP and human (but not mouse) MT2-MMP are the only MT-MMPs that are unable to activate MMP2 (Zucker et al. 1998, Miyamori et al. 2000, English et al. 2000). First, a cell surface MT-MMP binds and is inhibited by the N-terminal domain of TIMP2, and the C-terminal domain of the bound TIMP2 acts as a receptor for the hemopexin domain of ProMMP2. Then, an adjacent, uninhibited MT-MMP cleaves and activates the tethered ProMMP2. Following the initial cleavage of ProMMP2 by MT1-MMP, a residual portion of the MMP2 propeptide is removed by another MMP2 molecule to yield a fully active, mature form of MMP2 (Deryugina et al. 2001).
Figure 3.
Cell surface activation of MMP2. A ProMT-MMP is activated during transport to the cell surface by an intracellular furin-like serine proteinase, at the cell surface by plasmin, or by non-proteolytic conformational changes. The activated MT-MMP is then inhibited by TIMP2 and the hemopexin domain of ProMMP2 binds to the C-terminal portion of TIMP2 to form a trimolecular complex. An uninhibited MT-MMP then partially activates the ProMMP2 by removing most of the MMP2 propeptide. The remaining portion of the propeptide is removed by a separate MMP2 molecule at the cell surface to yield fully active mature MMP2. Mature MMP2 can then be released from the cell surface or bound by another cell surface MMP2-docking protein. It can also be inhibited by another TIMP molecule or left in an uninhibited active state depending on local MMP:TIMP molar ratios.
It has been assumed that proteolytic removal of the MT-MMP prodomain by furin-like enzymes in the trans-Golgi network or by plasmin at the cell surface (Okumura et al. 1997) is required for MT-MMPs to activate MMP2. Such processing does appear to be necessary in some cell types (Strongin et al. 1995), but these cells display both processed and full-length MT-MMPs on their surface so that the function of either individual form can not be distinguished. However, removal of the prodomain is not required for MT1-MMP to bind TIMP2 or activate MMP2 in transfected COS-1 cells that lack endogenous MT1-MMP and are unable to process latent MT-MMPs (Cao et al. 1998). Even more surprisingly, COS-1 cells fail to bind TIMP2 or activate MMP2 if the MT1-MMP prodomain is absent or replaced by the MMP13 prodomain. However, COS-1 cells can activate MMP2 if they co-express a cDNA for prodomain-deleted MT1-MMP together with an independent cDNA for the MT1-MMP prodomain, but not if either cDNA is expressed alone (Cao et al. 2000). Therefore, the MT1-MMP prodomain is actually required for the cell surface activation of MMP2 to proceed, but it does not have to remain covalently attached. Furthermore, co-expression of the MMP1 prodomain instead of the MT1-MMP prodomain does not rescue the ability of prodomain-deleted MT1-MMP to activate MMP2, thus indicating that the MT1-MMP prodomain has specific attributes that enable it to do so. Direct exogenous addition of the MT1-MMP prodomain also fails to restore the function of processed MT1-MMP, and whereas full-length MT1-MMP is prominently expressed at the cell surface, much of the prodomain-deleted MT1-MMP is retained in the secretory pathway. Therefore, the MT1-MMP propeptide may act as an intramolecular chaperone that is necessary for the efficient trafficking of MT1-MMP to the cell surface. Although processed MT1-MMP is eventually expressed at the cell surface, it lacks the ability on its own to bind TIMP2 or activate MMP2. MT1-MMP may also be conformationally activated through interactions with the cell membrane, and its retained propeptide domain may facilitate the binding of TIMP2.
The role of TIMP2 in MMP2 activation is its dominant in vivo function, as shown by targeted mutagenesis in mice (Z. Wang et al. 2000). Nevertheless, while the C-terminal domain of TIMP2 participates in the cell surface docking and activation of MMP2, its N-terminal domain is an MMP inhibitor. Not surprisingly, low-to-moderate levels of TIMP2 promote the activation of MMP2, whereas higher levels inhibit its activation by saturating free MT-MMPs that are needed to remove the MMP2 prodomain (Strongin et al. 1995). TIMP2 protein levels are reduced and MMP2 activation is enhanced in the presence of the MMP2 substrate, type IV collagen (Maquoi et al. 2000). Furthermore, the ability of collagen to induce MMP2 activation on demand probably results from TIMP2 degradation because there are no accompanying changes in MMP2, MT1-MMP, or TIMP2 mRNA expression or in the synthesis or activation of MT1-MMP. Therefore, local accumulation of type IV collagen may trigger its own degradation by somehow lowering local TIMP2 concentrations to levels that favor MMP2 activation.
Endogenous Metalloproteinase Inhibitors
The TIMPs represent a family of at least four 20–29-kDa secreted proteins (TIMPs 1–4) that reversibly inhibit the MMPs in a 1:1 stoichiometric fashion (reviewed in Edwards 2001, Sternlicht & Werb 1999, Gomez et al. 1997). They share 37–51% overall sequence identity, a conserved gene structure, and 12 similarly separated cysteine residues. These invariant cysteines form six intrachain disulfide bridges to yield a conserved six-loop, two-domain structure. Truncated “tiny” TIMPs 1 and 2 retain their inhibitory activity despite containing only the first three loops, thus indicating that portions of the N-terminal domain interact with the MMP catalytic site (Murphy & Willenbrock 1995). Mutational analyses (O’Shea et al. 1992, Willenbrock & Murphy 1994, Huang et al. 1997) and peptide-and antibody-blocking experiments (Bodden et al. 1994) have helped to further specify which regions of the N-terminal domain influence inhibitory function. In addition, NMR (Williamson et al. 1997) and X-ray crystallographic studies (Gomis-Rüth et al. 1997) have revealed which TIMP residues interact directly with the MMP3 catalytic domain and how they inhibit MMP activity. Although these studies indicate that the inhibitory activity of the TIMPs resides almost entirely in the N-terminal domain alone, both domains influence enzyme-inhibitor binding (Willenbrock & Murphy 1994). For example, the C-terminal domain (loops 4–6) of TIMP1 binds the hemopexin domain of MMP9 more readily than it does the hemopexin domain of MMP2, whereas the C-terminal domain of TIMP2 preferentially binds the hemopexin domain of MMP2 (Murphy & Willenbrock 1995).
Individual TIMPs differ in their ability to inhibit various MMPs (reviewed in Woessner & Nagase 2000). For example, TIMP2 and TIMP3 inhibit MT1-MMP, whereas TIMP1 does not. Likewise, TIMP1 is a relatively poor inhibitor of MT3-MMP, and TIMP3 appears to be a more potent inhibitor of MMP9 than other TIMPs. TIMP3 is also unique in its ability to inhibit ADAMs-10 and -17, ADAMTS-4, and ADAMTS-5 (Kashiwagi et al. 2001), whereas TIMP1 can inhibit ADAMTS-1 (Tortorella et al. 1999). In addition, the TIMPs differ in terms of their gene regulation and tissue-specific patterns of gene expression (Edwards 2001). TIMP3 also has the unique ability to bind via its C-terminal domain to heparan sulfates proteoglycans within the ECM, thereby concentrating it to specific regions within tissues and basement membranes (Langton et al. 1998).
The particular importance of TIMP3 in the eye is indicated by its increased expression in various degenerative retinal diseases, its immunolocalization to Bruch’s membrane and drusen (deposits of extracellular matrix associated with macular degeneration), and the presence of TIMP3 mutations in patients with Sorsby’s fundus dystrophy (SFD), an autosomal-dominant form of early retinal degeneration (Langton et al. 1998). Several missense mutations that introduce an extra cysteine into the TIMP3 C-terminal domain, a nonsense mutation that truncates most of the C-terminal domain but leaves an unpaired cysteine, and a splice-site mutation that may also yield an unpaired cysteine have been found in the affected members of various SFD families (Langton et al. 2000). Although the presence of an unpaired cysteine could result in aberrant, function-perturbing disulfide bonds, SFD mutations apparently give rise to TIMP3 molecules that dimerize and show diminished turnover but still retain their MMP-inhibitory and ECM-binding properties (Langton et al. 2000).
There is a long history indicating that TIMPs exert growth-promoting activity independent of their metalloproteinase inhibitory activity. Indeed, TIMP1 was first cloned as EPA for its erythroid-potentiating activity, and TIMPs 1, 2, and 3 have since been shown to act as mitogens in several other cell types (Gomez et al. 1997). Moreover, their mitogenic activity persists despite the presence of added mutations that abolish their MMP inhibitory activity, thus indicating that these activities occur independently, perhaps as a distinct function of the C-terminal domain (Chesler et al. 1995, Wingfield et al. 1999). Although it is still unclear how TIMPs promote cell growth, this activity may explain several unexpected associations between TIMPs and cancer progression. However, TIMPs may also promote cell death or suppress mitogenic signals. For example, apoptosis is induced in various cell types by TIMP3, but not by TIMP1, TIMP2 or synthetic MMP inhibitors, thus suggesting that the mechanism may not rely on the inhibition of a metalloproteinase (Bond et al. 2000). However, the introduction of a mutation that abolishes the metalloproteinase-inhibitory activity of TIMP3 also abolishes its ability to promote apoptosis, indicating that its inhibitory activity is necessary and suggesting that its target may be an ADAM or ADAMTS rather than an MMP. TIMP2, on the other hand, can suppress growth factor-responsiveness by interfering with the activation of tyrosine kinase-type growth factor receptors, and its ability to block mitogenic signaling is independent of its MMP-inhibitory activity (Hoegy et al. 2001). Nevertheless, no TIMP receptors have yet been identified, suggesting that TIMPs may act as decoys for various signaling molecules. Moreover, these growth-promoting and growth-suppressive activities may not be entirely MMP-independent because the mutant TIMPs retain their ability to interact with MMPs via secondary non-inhibitory sites, such as those of their C-terminal domains (Howard & Banda 1991). Therefore, these growth-altering activities may still reflect the ability of TIMPs to indirectly modify MMP activity.
TIMPs are not the only endogenous MMP inhibitors. Indeed, α2-macroglobulin is a major endogenous inhibitor of the MMPs (Sottrup-Jensen & Birkedal-Hansen 1989), and its importance may have been overlooked due to the recent emphasis placed on the TIMPs. Because α2-macroglobulin is an abundant plasma protein, it represents the major inhibitor of MMPs in tissue fluids, whereas TIMPs may act locally. Moreover, because α2-macroglobulin/MMP complexes are removed by scavenger receptor-mediated endocytosis, α2-macroglobulin plays an important role in the irreversible clearance of MMPs, whereas TIMPs inhibit MMPs in a reversible manner.
Another, recently recognized class of MMP inhibitors, protein subdomains, have structural similarity to the TIMPs. For example, proteolytic processing of the procollagen C-terminal proteinase enhancer protein PCPE releases a C-terminal fragment with MMP inhibitory activity and structural similarity to the N-terminal domain of the TIMPs (Mott et al. 2000). A primary sequence alignment search also uncovered similarities between the TIMPs and the noncollagenous NC1 domain of type IV collagen (Netzer et al. 1998). Moreover, functional analyses indicate that the NC1 domain has MMP inhibitory activity (Netzer et al. 1998) and can inhibit angiogenesis and tumor growth (Petitclerc et al. 2000). Nevertheless, the physiologic targets of these inhibitory fragments remain uncertain. Although their activity against MMP2 is substantially lower than that of the TIMPs (Mott et al. 2000, Netzer et al. 1998), other MMPs or metzincins may be their true physiologic targets.
Pericellular Localization of Proteolytic Activity
Many of the extracellular signaling events that regulate cell behavior occur at or near the cell membrane, and many cellular signals are created or canceled via pericellular proteolysis (Werb 1997,Werb&Yan 1998). Thus because it is irreversible, the processing of pericellular proteins by proteolysis is an ideal means of regulating extracellular signal transduction. Although pericellular proteolysis may sometimes reflect the exclusive expression of a critical substrate at or near the cell surface, there are specific mechanisms that confine or concentrate proteinases in the immediate pericellular microenvironment. These mechanisms for localizing MMPs to the cell surface and to specific cell surface subdomains include the expression of membrane-bound MT-MMPs; the binding of MMPs to cell surface receptors; the presence of cell surface receptors for MMP-activating enzymes such as uPA, plasmin(ogen), thrombin, and elastase; and the concentration of MMPs on pericellular ECM molecules. These localization mechanisms often enhance MMP activation, limit the access of MMP inhibitors, concentrate MMPs within the vicinity of their targets, and limit the extent of proteolysis to discrete pericellular regions.
Transmembrane and GPI-linked MT-MMPs are the most obvious mediators of proteolytic activity at the cell surface. Removal of the transmembrane domains of MT1, MT2, and MT3-MMP abolishes their ability to promote cellular invasion (Hotary et al. 2000). Moreover, MT-MMPs can concentrate within specific cell surface domains such as cellular protrusions known as invadopodia (or invasive pseudopodia), where active ECM degradation takes place. Localization studies using chimeric constructs indicate that the cytoplasmic and transmembrane domains of MT1-MMP are required for it to cluster on invadopodia, and functional studies indicate that invadopodial recruitment of MT1-MMP is necessary for cellular invasion to take place (Nakahara et al. 1997). Because GPI-anchored proteins tend to partition into lipid rafts and caveolae where considerable signaling activity takes place (Brown & London 1998), the GPI-linked MT-MMPs should also be enriched in such regions.
Another means of localizing MMPs to the cell surface is via cell surface docking receptors. For example, activated MMP2 can bind to integrin αvβ3 on the surface of angiogenic endothelial cells and invasive cancer cells (Brooks et al. 1996). Because the C-terminal domain of MMP2 is required for the formation of αvβ3/MMP2 complexes in vitro, the catalytic domain probably remains exposed so that it can still carry out proteolysis. Interestingly, MT1-MMP generates only an MMP2 activation intermediate, and another already activated MMP2 is required to remove the residual portion of the MMP2 propeptide and achieve full MMP2 activation (Deryugina et al. 2001). Thus cell surface MMP2 receptors may cooperate with MT1-MMP to facilitate MMP2 maturation, and data suggest that integrin αvβ3 promotes such maturation by providing a platform for autocatalytic interactions between fully and partially activated MMP2 (Deryugina et al. 2001). Moreover, colocalization data suggest that integrin αvβ3 may also cooperate with MT1-MMP in the clustering of active MMP2 on invadopodia.
MMP1 can also interact with a cell surface integrin. In this case, active and inactive MMP1 binds to the I domain of α2 integrin, whereas MMPs 3 and 13 do not (Stricker et al. 2000). Optimal binding of chimeric constructs requires the MMP1 hinge and hemopexin domains, but not its propeptide or catalytic domains, so that the latter domain may remain available to interact with its targets. Binding of α2β1 integrin to type I collagen induces MMP1 expression in keratinocytes, and MMP1 is necessary for α2β1-dependent migration of keratinocytes over type I collagen (Pilcher et al. 1999). Thus α2β1 integrin can act both as an MMP1 inducer and as an MMP1 receptor, and the binding of MMP1 to α2β1 integrin may play an important role in the migration of keratinocytes that contact interstitial collagens during epidermal wound repair.
Another molecule that can both induce MMP1 expression and then localize it to the cell surface is CD147/EMMPRIN (Guo et al. 2000). EMMPRIN is enriched on the surface of cancer cells and induces adjacent fibroblasts to produce MMP1, which then apparently binds to the same cell surface EMMPRIN molecules that elicited its expression in the first place. Therefore, tumor-derived EMMPRIN may promote tumor cell invasion by both inducing fibroblasts to produce MMP1 and then concentrating it on the surface of the invasive cells. MMPs can also be bound to the cell surface by their own substrates. Latent ProMMP9, for example, binds with high affinity to type IV collagen α2 chains on the surface of several cell types, thus concentrating it at the cell-ECM interface in anticipation of any future need and in direct contact with its target (Olson et al. 1998, Toth et al. 1999). This interface is where MMP9 is found in nascent skin cancers (Coussens et al. 2000). Activated MMP9 binds to the cell surface hyaluronan receptor CD44 (Bourguignon et al. 1998). Moreover, the localization of MMP9 to the cell surface by CD44 appears to promote tumor cell invasion and angiogenesis and may mediate the activation of latent TGF-β by MMP9 (Yu & Stamenkovic 2000). Recent data also indicate that MMP7 binds to cell surface and ECM heparan sulfate moeities that may enhance the stability, activation, and activity of MMP7 (Yu & Woessner 2000). Moreover, cell surface heparan sulfates may act as docking molecules for other MMPs, as well as TIMP3.
Although many of the above docking mechanisms have not been definitively proven, they suggest the existence of localized pericellular feed-back networks that coordinate the need for a given MMP with its appropriate expression, activation, and physical placement. The specific cell surface molecules that mediate these interactions have not been definitively elucidated, in part, because such molecules are frequently enriched in specialized signaling domains such as caveolae and rafts. Furthermore, by combining many of the functions of a given feed-back loop within a few components (for instance by binding an MMP to its own inducer, its own substrate or a molecule that normally ligates its substrate), these signaling networks may be particularly compact and efficient. By the same token, the multifunctional nature of these interactions will undoubtedly make them all the more difficult to sort out in functional experiments.
MMP Catabolism and Clearance
An obvious means of regulating MMPs is via their own proteolytic inactivation and physical clearance. Although considerable progress has been made in understanding the progressive proteolytic processing of MMP propeptides, relatively little is known about the further autoproteolysis of active MMPs. Nevertheless, it is clear that some cleavages inactivate MMPs, whereas others, such as those that specifically remove the hemopexin domain, can generate truncated enzymes that lose their ability to cleave some substrates but retain their ability to cleave others (reviewed in Woessner & Nagase 2000). Such processing can also diminish their affinity for and ability to be inhibited by TIMPs, as occurs with C-terminally truncated MMP2 (Y. Itoh et al. 1998). Removal of the hemopexin-like domain also cancels the ability of certain MMPs to localize to the cell surface. In addition, MT-MMPs can be secreted if they are cleaved at a juxtamembrane site before or after they reach the cell surface (Imai et al. 1996). Thus factors that influence MMP degradation can alter the steady-state concentrations of MMPs, their substrate specificities, their localization, and their ability to be activated or inhibited.
Another means of regulating extracellular MMP levels is by the direct clearance of intact enzymes. Most MMPs cleave the bait region of α2-macroglobulin, thereby initiating a conformational change in the large tetrameric macroglobulin that irreversibly traps the enzyme (Sottrup-Jensen & Birkedal-Hansen 1989). Although the catalytic activity of the MMP is not inhibited per se, its physical entrapment keeps the enzyme from interacting with natural substrates, and the α2-macroglobulin/MMP complex is eventually endocytosed and permanently cleared.
Thrombospondin 2 (TS2) has also been implicated in the clearance of MMPs. Interestingly, TS2-deficient mice exhibit a number of connective tissue abnormalities, and their fibroblasts have an adhesion defect that is the result of increased MMP2 levels (Yang et al. 2000). The increased MMP2 levels occur because TS2 normally binds both latent and active MMP2 and because TS2 is normally endocytosed by the low-density lipoprotein receptor-related protein LRP and probably carries any bound MMP2 with it (Yang et al. 2001). The cellular internalization of TS2/MMP2 complexes by the LRP scavenger receptor may therefore play an important role in regulating MMP2 levels outside fibroblasts and other cells. Evidence also indicates that MMP13 is rapidly cleared after it binds to an MMP13-specific 170-kDa high-affinity receptor present on various cell types (Barmina et al. 1999). The binding requires calcium, and the subsequent internalization and degradation of MMP13 requires LRP because LRP-null cells bind MMP13 but fail to internalize it. Moreover, the internalization of both MMP13 and TS2/MMP2 complexes is inhibited by the 39-kDa receptor-associated protein RAP, which binds and inhibits LRP. Thus MMPs are tightly regulated by several variously characterized mechanisms during virtually every aspect of their life-span, from their induction to their ultimate destruction.
MMP Substrates
Although very few bona fide MMP substrates have been definitively identified in vivo, numerous candidates have been tested and identified in vitro (Table 2). For interstitial collagens, aggrecan and link protein, the cleavage sites identified in breakdown products isolated from tissue extracts match those that have been established in vitro; however, multiple MMPs can generate these same cleavages. In a limited number of cases, the MMP substrates and responsible MMPs have been identified genetically. The restricted expression pattern of an enzyme may also help to predict its proteolytic targets. For example, MMP20/enamelysin is principally expressed in the enamel organ of developing teeth, where ameloblast cells secrete amelogenin, the major protein that forms the organic matrix of tooth enamel. Amelogenin is continuously degraded and replenished as enamel formation proceeds, and it is hydrolyzed by recombinant MMP20 in vitro (Li et al. 2001). Moreover, an inherited missense mutation in the amelogenin gene is associated with X-linked amelogenesis imperfecta and alters an amelogenin cleavage site so that it is poorly hydrolyzed by MMP20 (Li et al. 2001). Therefore, amelogenin is almost certainly a natural substrate of MMP20, and inherited mutations that render it resistant to MMP20 cleavage may lead to defective tooth enamel formation. Nevertheless, such a concordance between the isolated temporal and spatial expression of an MMP and its substrate, together with the existence of disease-linked mutations that alter the proteolytic susceptibility of the substrate, is the exception. In virtually all other cases, the identification of key in vivo substrates and the responsible enzymes will require rigorous experimental approaches.
TABLE 2.
Common matrix metalloproteinase substratesa
| MMP | 1 | 2 | 3 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 16 | 18 | 19 | 26 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ECM Proteins: | |||||||||||||||
| Aggrecan | + | + | + | + | + | + | + | + | + | + | |||||
| Collagen I | + | + | − | + | + | − | + | + | + | + | + | − | |||
| Collagen II | + | + | − | + | + | − | |||||||||
| Collagen III | + | + | + | − | + | − | + | + | + | + | |||||
| Collagen IV | − | + | + | + | − | + | + | − | + | − | + | + | |||
| Collagen V | − | + | + | − | − | + | + | − | |||||||
| Collagen VI | − | − | − | − | + | ||||||||||
| Collagen VII | + | + | + | ||||||||||||
| Collagen VIII | + | ||||||||||||||
| Collagen IX | − | − | + | + | |||||||||||
| Collagen X | + | + | + | − | + | ||||||||||
| Collagen XI | + | + | + | + | − | ||||||||||
| Collagen XIV | − | − | − | + | + | ||||||||||
| Decorin | − | + | + | + | + | ||||||||||
| Elastin | − | + | + | + | + | + | + | − | |||||||
| Entactin/Nidogen | + | + | + | + | + | + | |||||||||
| Fibrillin | + | + | + | + | + | + | |||||||||
| Fibronectin | + | + | + | + | − | − | + | + | + | + | + | + | + | ||
| Fibulins | + | + | |||||||||||||
| Gelatin I | + | + | + | + | + | + | + | + | + | + | + | ||||
| IGFBPs | + | + | + | + | |||||||||||
| Laminin | + | + | + | + | + | − | + | + | − | ||||||
| Link Protein | + | + | + | + | + | + | |||||||||
| Myelin Basic | + | + | + | + | + | + | |||||||||
| Osteonectin | + | + | + | + | + | ||||||||||
| Tenascin | + | + | + | + | − | + | |||||||||
| Vitronectin | + | + | + | + | + | + | + | ||||||||
| Other Proteins: | |||||||||||||||
| α1-AC | + | + | + | − | |||||||||||
| α2-M | + | − | + | + | + | + | + | + | + | ||||||
| α1-PI | + | + | + | + | + | + | + | + | + | + | |||||
| Casein | + | − | + | + | + | + | − | + | + | − | |||||
| C1q | + | + | + | + | + | + | |||||||||
| E-cadherin | + | + | |||||||||||||
| Factor XII | − | + | + | + | |||||||||||
| Fibrin | + | + | + | + | + | ||||||||||
| Fibrinogen | + | + | + | + | + | + | + | + | + | + | + | ||||
| IL1α | − | − | − | − | |||||||||||
| IL1β | + | + | + | + | |||||||||||
| ProMMP2 | + | + | |||||||||||||
| ProTGFβ | + | + | |||||||||||||
| ProTNFα | + | + | + | + | + | + | + | ||||||||
| Plasminogen | + | + | + | + | + | − | |||||||||
| Substance P | + | + | + | + | |||||||||||
| T kininogen | − | + | − |
The symbols indicate whether the indicated protein is (+) or is not (−) digested by the indicated MMP.
Abbreviations: α1-AC, α1-antichymotrypsin; α2-M, α2-macroglobulin; α1-PI, α1-proteinase inhibitor. Adapted from Woessner & Nagase 2000 with additional data from Ashworth et al. 1999, Hiller et al. 2000, Hiraoka et al. 1998, Manes et al. 1997, Noe et al. 2001, Park et al. 2000, Stracke et al. 2000, Sternlicht et al. 2001, and Uria & Lopez-Otin 2000.
Screening methods are now emerging as another means of identifying potential physiologic MMP substrates. For example, monocyte chemoattractant protein-3 (MCP-3) was identified as an MMP2 substrate after using the MMP2 hemopexin domain as bait to search for potential binding partners in yeast two-hybrid screens (McQuibban et al. 2000). Thus the hemopexin domain of MMP2 contains at least one exosite that is required for the binding of MCP-3. A phage-displayed hexapeptide library has also been used to map the preferred substrate specificity of human MMP13 (Deng et al. 2000). Subsequent database searches for proteins with matching peptide sequences confirmed the ability of MMP13 to degrade aggrecan and collagens II and IV, but also identified a sequence within the prodomain of TGF-β3 as a potential target. This is particularly intriguing because cell surface-associated MMP2 and MMP9 can activate TGF-β and because TGF-β activation is blocked by metalloproteinase inhibitors (Yu & Stamenkovic 2000). Moreover, MMP13 can be activated at the cell surface by MT1-MMP, thus potentially placing it in the right location to activate TGF-β. As is so often the case, however, a determination of whether such processing actually occurs in vivo awaits further testing and will undoubtedly be difficult to prove. Therefore, although the list of potential substrates is long, the list of known in vivo substrates remains relatively short.
REGULATION OF CELLULAR SIGNALS BY MMPs
Although MMPs can cleave virtually all structural ECM molecules, they can also cleave several circulating, cell surface and pericellular proteins, which enables them to regulate cell behavior in numerous ways (Figure 4). These mechanisms include the alteration of cell-matrix and cell-cell interactions; the release, activation, or inactivation of autocrine or paracrine signaling molecules; and the potential activation or inactivation of cell surface receptors.
Figure 4.
Potential mechanisms of MMP-mediated cellular signaling.
Extracellular Matrix Remodeling and Cell Behavior
ECM-degradation can be viewed, on the one hand, as merely disrupting and remodeling structural barriers and, indeed, such degradative processes permit cellular invasion to take place. However, extracellular matrices are not just passive cellular scaffolds; they influence cell behavior by sequestering signaling molecules, such as growth factors and growth factor-binding proteins, and by acting as contextual ligands for cellular adhesion receptors, such as integrins, that transduce signals to the cell interior (Streuli 1999). Indeed, extracellular matrices regulate such basic processes as cell shape, movement, growth, differentiation, and survival by controlling cell adhesion and the cytoskeletal machinery (reviewed in Lukashev & Werb 1998). By extension, MMPs also influence these same processes by altering the composition and structural organization of the ECM, thereby altering matrix-derived signals. Moreover, proteolytic ECM remodeling results in the release of modular breakdown products with biologic activity. For example, the MMP-dependent cleavage of native fibrillar collagen exposes otherwise cryptic RGD sites that can then be ligated by αvβ3 integrin, and this interaction promotes the survival and growth of melanoma cells (Petitclerc et al. 1999). In addition, the cleavage of intact laminin-5 by MMP2 generates a γ2-chain fragment (γ2x) that induces epithelial cell motility, presumably by exposing an otherwise inaccessible site (Giannelli et al. 1997). ECM molecules also act as binding reservoirs for various growth factors and cytokines that are released once the ECM molecules are degraded. For example, the small collagen-associated proteoglycan decorin acts as a depot for TGF-β, and its degradation by various MMPs makes the otherwise sequestered TGF-β available to carry out its biologic functions (K Imai et al. 1997). One such function is to inhibit the expression of several MMP genes. Thus MMP-mediated activation and release of TGF-β may act as a negative feed-back mechanism to limit MMP expression and further TGF-β release. Likewise, MMP-cleaved collagen can feed back through discoidin domain receptor-2 to suppress collagenase expression and further collagen degradation, whereas intact collagen has the opposite effect (Vogel et al. 1997).
Most cells must adhere to a natural or provisional matrix to survive. Thus MMP-mediated disruption of subcellular matrices can induce apoptosis in anchorage-dependent cells and plays an important role in normal physiologic cell death in tissues such as the involuting mammary gland (Alexander et al. 1996). In addition, pericellular matrix destruction could defy cancer by raising the cell death rate, or promote it by selecting for anchorage-independent and apoptosis-resistant subclones.
Cell Surface Proteolysis and Cellular Signals
MMPs can potentially influence cell behavior by cleaving cell-cell adhesion proteins, by releasing bioactive cell surface molecules, or by cleaving cell surface molecules that transduce signals from the extracellular environment. For example, MMP3 and MMP7 both cleave the adherens junction protein E-cadherin, and the soluble extracellular fragment that is released disrupts cell aggregation and promotes cell invasion in a paracrine manner independent of the cleavage event itself (Lochter et al. 1997, Noe et al. 2001). It is tempting to speculate that the MMP-mediated cleavage of E-cadherin may abrogate its tumor suppressive activity and influence other aspects of cancer associated with E-cadherin and MMP alterations, such as epithelial-to-mesenchymal phenotypic conversions, invasion, and genomic instability (Sternlicht et al. 2000). Transmembrane MT-MMPs can also degrade cell surface tissue transglutaminase, an integrin-binding co-receptor that promotes the adhesion and spreading of cells on fibronectin and can thus regulate cancer cell migration either positively or negatively, depending on the type of ECM the cells encounter (Belkin et al. 2001).
MMPs can also release cell surface molecules. For example, MMP3 can release soluble L-selectin from leukocytes (Preece et al. 1996) and can release active heparin-binding EGF-like growth factor (HB-EGF) from cell surfaces by cleaving it at a site just outside the cell membrane (Suzuki et al. 1997). MMP7 can induce Fas receptor-mediated apoptosis by releasing active soluble Fas ligand from the surface of the same target cells, and this shedding appears to influence involution of the prostate following castration, since prostatic involution is significantly diminished in MMP7-deficient mice (Powell et al. 1999). The shedding of soluble Fas ligand could prevent cancer progression, or it could promote progression either by exerting pressure for the selection of subpopulations that are refractory to Fas-mediated cell death or by merely removing a critical component of the Fas-mediated apoptotic pathway. Indeed, the constitutive expression ofMMP7 in cells that express both Fas ligand and its receptor results in the emergence of clones that are resistant to Fas-mediated and chemotherapy-mediated apoptosis (Fingleton et al. 1999, Mitsiades et al. 2001). Cell surface localized MMP2 and MMP9 can activate latent TGF-β (Yu & Stamenkovic 2000). At least seven MMPs can activate recombinant soluble proTNF-α (Woessner & Nagase 2000) and, like ADAM17, GPI-linked MT4-MMP can cleave membrane-bound proTNF-α to generate active soluble TNF-α (English et al. 2000). Under the right circumstances, several MMPs may process TGF-β and TNF-α in vivo.
In addition to causing the activation and release of cytokines and growth factors, MMPs can also cleave their cell surface receptors. MMP2, for example, cleaves cell surface fibroblast growth factor (FGF) receptor 1 at a specific juxtamembrane site, thereby releasing a soluble receptor fragment that retains its ability to bind FGF (Levi et al. 1996). Soluble FGF receptor type 1 has been found in the circulation and in vascular basement membranes and may indirectly influence FGF availability. It is not known whether the removal of cell surface FGF receptors directly blocks FGF responsiveness in the receptor-depleted cells as occurs for other receptors. For example, MMP9 cleaves IL2 receptor α (IL2Rα) on T cells and significantly downregulates their proliferative response to IL2 in culture (Sheu et al. 2001). Furthermore, the in vivo responsiveness of tumor-infiltrating lymphocytes is suppressed by the metalloproteinase-dependent downregulation of IL2Rα protein (but not mRNA) levels (Sheu et al. 2001).
Regulation of Paracrine Signals
In addition to releasing several cell surface and matrix-bound growth factors and cleaving cell surface receptors to release soluble growth factor-binding proteins, MMPs may regulate the availability of paracrine signals either by inactivating them directly or by inactivating their soluble binding proteins. For example, MMPs can inactivate angiotensins I and II, bradykinin, and substance P, thereby limiting their respective physiologic effects directly (Woessner & Nagase 2000). On the other hand, the degradation of insulin-like growth factor (IGF) binding proteins (IGF-BPs) 3 and 5 by MMPs 1, 2 and 3, and of IGFBP-1 by MMP11, indirectly increases the availability of IGF survival and growth factors (Fowlkes et al. 1999, Manes et al. 1997). For example, IGFBP-1 inhibits the growth of human breast cancer cells, whereas the degradation of IGFBP-1 by MMP11 restores their IGF-1-dependent growth in culture and in vivo (Manes et al. 1997). Conversely, TIMP1 blocks hepatocyte growth and hepatocellular carcinogenesis in a bitransgenic carcinogenesis model by protecting IGFBP-3 from proteolysis, thereby limiting the availability of IGF-2 (Martin et al. 1999). Likewise, an MMP-resistant form of IGFBP-5 inhibits the responsiveness of smooth muscle cells to IGF-1 (Y Imai et al. 1997).
As inflammatory cell products, MMPs participate in and promote inflammatory processes, but they may also blunt such processes. For example, MMP2 can cleave monocyte chemoattractant protein-3, thereby inactivating it and generating a chemokine receptor-binding antagonist that further impedes inflammation (McQuibban et al. 2000) and, perhaps, immune surveillance against cancer. In a co-culture model of herniated disk resorption, MMP3 from chondrocytes is required to generate a macrophage chemotactic factor (Haro et al. 2000a), whereas macrophage MMP7 is required to release soluble TNF-α from the macrophages, thereby enabling them to infiltrate the cultured intervertebral discs (Haro et al. 2000b). In addition, MMPs can degrade the inflammatory cytokine IL1β, which is also a potent inducer of MMP expression, but not IL1α (Ito et al. 1996).
Generation and Inactivation of Bioactive Molecules
In addition to altering the activity and availability of various cytokines, growth factors, and hormones, MMPs can cleave a number of other nonmatrix proteins, sometimes abolishing their normal biologic function while generating breakdown products with entirely newbiologic activities. MMPs can cleave serine proteinases, such as plasminogen and urokinase-type plasminogen activator, as well as several serpin-type serine proteinase inhibitors, including α1-proteinase inhibitor (α1-PI), α1-antichymotrypsin, antithrombin III, and plasminogen activator inhibitor-2 (Sternlicht & Werb 1999). These cleavages often inactivate the proteinase or proteinase inhibitor and simultaneously create bioactive breakdown products. For example, MMPs 2, 7, 9, and 12 can cleave plasminogen to generate the angiogenesis inhibitor angiostatin (Patterson & Sang 1997, O’Reilly et al. 1999). Likewise, the inactivation of α1-PI by MMPs creates a cleavage product that acts as a potent chemoattractant for neutrophils (Banda et al. 1988) and assists tumor growth and invasion, possibly by modulating NK cell cytotoxicity (Kataoka et al. 1999). Virtually all MMPs can degrade fibrinogen, and MMPs 12, 13, and 14 can inactivate Factor XII (Hageman factor), thereby suppressing normal clotting mechanisms (Hiller et al. 2000). In addition, MMPs can degrade insoluble fibrin in a plasminin-dependent manner, and this fibrinolytic capacity plays an important role in new blood vessel formation, dermal wound repair, glomerular function, and other biologic processes (Hiraoka et al. 1998, Lund et al. 1999, Lelongt et al. 2001). Several MMPs can also cleave the collagen-like region of C1q, a subunit of the C1 component of the classic complement cascade; however, the cleaved collagen-like region still retains its ability to induce an oxidative burst in neutrophils (Ruiz et al. 1999). Undoubtedly, some of the MMP-mediated mechanisms outlined above contribute to a variety of biologic processes, but it is still largely unclear which mechanisms are crucial.
MMPs IN DEVELOPMENT AND DISEASE
Genetic Dissection of Biologic Roles and Critical In Vivo Substrates
One of the major challenges left in understanding the biology of the MMPs is the identification of their essential in vivo substrates. Even when a candidate protein can be cleaved by a given MMP in vitro and is, in fact, degraded during a particular biologic process; and even though its cleavage may make perfect mechanistic sense, an entirely different substrate may actually regulate the process. This is particularly well illustrated by the recent finding that α1-PI is the critical substrate for MMP9 in bullous pemphigoid, an autoimmune skin-blistering disease (Liu et al. 2000). Prior to this finding, evidence indicated that the key MMP9 substrate was the hemidesmosomal protein BP180 (collagen XVII), which helps anchor basal epidermal cells to the underlying basement membrane. BP180 is one of two major autoantigens recognized by the autoantibodies that initiate bullous pemphigoid, it is cleaved during blister formation, and it can be cleaved by MMP9 in solution. Moreover, MMP9 is upregulated in bullous pemphigoid, and knockout mice that lack MMP9 are resistant to the blistering that is elicited by BP180 antibodies in wild-type mice. The MMP9-deficient mice do, however, develop blisters if they are reconstituted with MMP9-expressing neutrophils. Thus it appears that MMP9 is responsible for BP180 cleavage and subsequent blister formation. However, neutrophil elastase (NE) can also cleave BP180; it too is upregulated in bullous pemphigoid, and mice lacking this enzyme are also refractory to antibody-induced blistering unless they are reconstituted with NE-competent neutrophils. Moreover, BP180 is cleaved and blisters do form if MMP9-null mice are reconstituted with neutrophils from NE-null mice, or if NE-null mice receive MMP9-null neutrophils, thus indicating that both enzymes participate and act along the same pathway. However, even though both enzymes can cleave BP180 in solution, only purified NE causes the blister-like separation of isolated skin sections. In addition, MMP9-null mice do form blisters if they are reconstituted with a large excess of MMP9-null (but NE-competent) neutrophils. Thus MMP9 acts upstream of NE and only indirectly participates in BP180 cleavage, whereas NE is absolutely required for BP180 cleavage and dermal-epidermal separation.
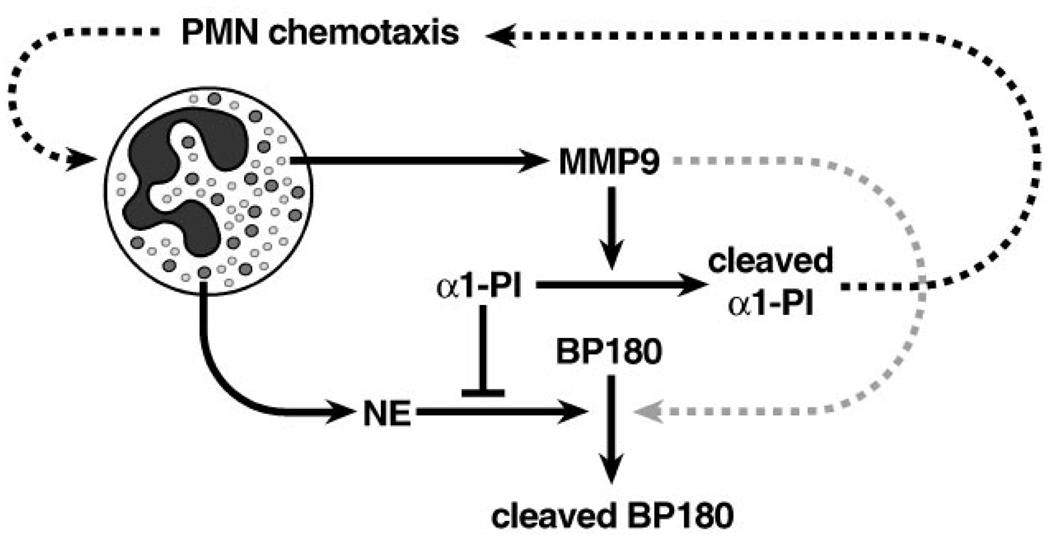
The principal endogenous inhibitor of NE, α1-PI, is also elevated in the skin of antibody-treated MMP9- and NE-deficient mice. This suggests that the role of MMP9 in blistering is to inactivate α1-PI, thereby removing the major constraint against NE and allowing it to cleave BP180. Indeed, α1-PI degradation was only absent in antibody-treated MMP9-deficient mice, even when they were given excess MMP9-null neutrophils. Nevertheless, BP180 degradation and blister formation did take place in MMP9-deficient mice that received excess MMP9-null neutrophils. Apparently, the additional MMP9-null neutrophils enable blister formation by providing enough NE to overcome the elevated levels of intact α1-PI. Moreover, the administration of excess α1-PI blocks antibody-induced blistering in wild-type mice, but not if the α1-PI is pre-treated with MMP9. Thus MMP9 indirectly contributes to antibody-induced skin blistering by inactivating α1-PI and allowing NE to cleave BP180 in an unrestricted manner (Figure 5), whereas direct cleavage of BP180 by MMP9, if it occurs at all, is probably minor.
Figure 5.
BP180 cleavage in bullous pemphigoid. Infiltrating neutrophils (PMNs) in and around bullous pemphigoid lesions release substantial amounts of neutrophil elastase (NE) and gelatinase B (MMP9) from large azurophilic primary granules and smaller tertiary granules, respectively. In addition, α1-proteinase inhibitor (α1-PI) is elevated, but because it is inactivated by MMP9, its ability to inhibit NE is abolished, thus enabling NE to cleave hemidesmosomal type XVII collagen (BP180) in an unrestricted manner. Furthermore, cleaved α1-PI fragments may exacerbate this process by enhancing PMN recruitment. Although MMP9 can also cleave BP180 in vitro, its direct contribution to BP180 cleavage in vivo is minimal or non-existent.
Similar interactions between MMPs, serine proteinases, and their inhibitors may also participate in other diseases. In pulmonary emphysema, for example, the irreversible enlargement of peripheral air spaces is thought to result from proteolytic degradation of interstitial elastic fibers, which otherwise have a cross-linked elastin core and a fibrillin-rich microfibrillar coat (Ashworth et al. 1999). The long-standing yet controversial elastase-antielastase theory states that the progressive erosion of alveolar walls results from inflammatory stimuli, such as cigarette smoke, that alter the balance between NE and its natural inhibitor, α1-PI. This theory derives primarily from the observations that (a) intrapulmonary instillation of NE causes experimental emphysema, (b) individuals with α1-PI deficiency are predisposed to develop emphysema, and (c) cigarette smoke can inactivate α1-PI (Dhami et al. 2000). Moreover, both exogenous α1-PI and an anti-neutrophil antibody protect against acute matrix degradation in mice exposed to cigarette smoke (Dhami et al. 2000). However, over 90% of the inflammatory cells in smokers’ lungs are macrophages rather than neutrophils, and macrophages can secrete MMPs 2, 7, 9, and 12, all of which can degrade elastin and inactivate α1-PI (Dhami et al. 2000). Moreover, MMPs 2, 9, and 12 (as well as NE and other enzymes) can degrade fibrillin within elastic fibers (Ashworth et al. 1999). Thus macrophage-derived proteinases that are increased in the lungs of smokers may be responsible for the matrix degradation of emphysema. Indeed, the chronic exposure of wild-type mice to cigarette smoke elicits significant accumulation of lung macrophages (but not neutrophils), induction and activation of MMP12 (but not NE), and emphysematous enlargement of pulmonary air spaces (Hautamaki et al. 1997). By comparison, smoke-exposed MMP12-deficient mice show no such pulmonary changes even after the forced recruitment of macrophages in response to monthly intratracheal administration of monocyte chemoattractant protein-1 (MCP-1), indicating that the lack of MMP12, rather than the lack of macrophages, protects them from smoke-induced pulmonary damage (Hautamaki et al. 1997). In addition, the relatively mild accumulation of neutrophils and the absence of NE in smoke-exposed wild-type and MMP12-deficient mice suggests the direct involvement of MMP12, rather than NE. Nevertheless, it remains unclear whether other enzymes participate or if elastin, fibrillin, α1-PI, or other substrates are critical to the natural development of emphysema.
The breakdown of elastic fibers has also been implicated in the formation and rupture of intracranial and aortic aneurysms. Elastolytic MMPs 2, 9, and 12 are prominent within human aortic aneurysms, and MMP12 immunolocalizes to residual elastic fiber fragments (Curci et al. 1998). In mice, the development of experimental aortic aneurysms is suppressed by the administration of a nonselective MMP inhibitor and by targeted disruption of the MMP9 gene, but not by selective disruption of the MMP12 gene (Pyo et al. 2000). Furthermore, resistance to aneurysms can be conferred by bone marrow transplantation from MMP9-deficient donors to wild-type hosts, and it can be abolished in MMP9-deficient mice by transplanting wild-type bone marrow cells (Pyo et al. 2000). Thus MMP9 appears to play an essential role in aneurysm formation, probably (though not conclusively) by virtue of its ability to cleave elastin and/or fibrillin.
One in vivo role for an MMP that has been definitively identified is the activation of intestinal crypt α-defensins by MMP7 (Wilson et al. 1999). The defensins, which play an important role in innate immunity, are antimicrobial (membrane-disrupting) peptides that are initially produced as inactive precursors by granulocytes and specific epithelial cells, including Paneth cells of the small intestine. In mice, crypt defensins (cryptdins) colocalize with MMP7 in apical Paneth cell granules, and the absence of MMP7 in gene-targeted mice has no effect on cryptdin production. MMP7 can, however, activate latent pro-cryptdins in vitro by removing their prodomain, and unlike wild-type mice, MMP7-deficient mice fail to process and instead accumulate inactive pro-cryptdins in their small intestines. As a result, intestinal extracts from MMP7-deficient mice have less antimicrobial activity than those of their wild-type counterparts, and orally administered bacteria show greater intestinal survival and overall virulence in MMP7-deficient mice than in wild-type controls. Thus MMP7 contributes to innate immunity by regulating cryptdin activation in the small intestine and may do so in other tissues as well.
These and other examples (Table 3) illustrate how genetically modified mice can reveal the proteinases and, if circumstances permit, the substrates that regulate physiologic and pathologic processes. Often, however, the conclusiveness of this approach is limited by the expression of redundant or compensatory enzymes with overlapping activity, the modest yet nonessential influence of an enzyme on the process in question, or the complete absence of any influence. For example, MMP3 is thought to be an important mediator of cartilage loss in rheumatoid and osteoarthritis, yet MMP3-deficient and wild-type mice are equally susceptible to collagen-induced arthritis, and both show cleavage of the aggrecan Asn341-Phe342 peptide bond (Mudgett et al. 1998). Thus even if MMP3 cleaves aggrecan, other redundant or compensatory enzymes are likely to participate and might mask the influence of MMP3, or MMP3 may not participate at all. Unmasking the responsible substrates and mechanisms may therefore require the generation of animals that are deficient in multiple enzymes. Functional overlap between distinct enzymes also probably explains the viability of most MMP-deficient mice and the subtle nature of their developmental phenotypes, with the notable exception of MT1-MMP null animals, which exhibit severe skeletal and connective tissue abnormalities and early postnatal lethality (Holmbeck et al. 1999, Zhou et al. 2000).
TABLE 3.
Influence of MMPs in genetically modified mice: non-malignant phenotypes
| Genotype | Phenotype | Reference |
|---|---|---|
| Haptoglobin-MMP1 | Emphysematous enlargement of pulmonary air spaces | (D’Armiento et al. 1992) |
| WAP-MMP3 | Increased mammary apoptosis in pregnancy | (Alexander et al. 1996) |
| Precocious mammary differentiation | (Sympson et al. 1994) | |
| MMTV-MMP7 | Precocious mammary differentiation, male infertility | (Rudolph-Owen et al. 1998a) |
| β-Actin-TIMP1 | Accelerated mammary adipogenesis | (Alexander et al. 2001) |
| MMP2−/− | Delayed mammary gland development | a |
| MMP3−/− | Hypomorphic mammary gland development | a |
| Accelerated mammary adipogenesis | (Alexander et al. 2001) | |
| Delayed incisional wound closure | (Bullard et al. 1999) | |
| Resistance to contact dermatitis | (Wang et al. 1999) | |
| Normal cardiac rupture following infarction | (Heymans et al. 1999) | |
| Normal collagen-induced arthritis development | (Mudgett et al. 1998) | |
| Impaired ex vivo herniated disk resorption | (Haro et al. 2000a) | |
| MMP7−/− | Deficient innate intestinal immunity | (Wilson et al. 1999) |
| Impaired tracheal wound reepithelialization | (Dunsmore et al. 1998) | |
| Diminished prostate involution after castration | (Powell et al. 1999) | |
| Impaired ex vivo herniated disk resorption | (Haro et al. 2000b) | |
| MMP9−/− | Delayed growth plate vascularization | (Vu et al. 1998) |
| Delayed endochondral ossification | (Vu et al. 1998) | |
| Defective osteoclast recruitment | (Engsig et al. 2000) | |
| Resistance to experimental bullous pemphigoid | (Liu et al. 2000) | |
| Resistance to experimental aortic aneurysms | (Pyo et al. 2000) | |
| Prolonged contact dermatitis | (Wang et al. 1999) | |
| Protection from ventricular enlargement after infarction | (Ducharme et al. 2000) | |
| Protection from cardiac rupture following infarction | (Heymans et al. 1999) | |
| Diminished motor defects following brain trauma | (Wang et al. 2000a) | |
| Diminished PMN infiltration in early glomerulonephritis | b | |
| Enhanced susceptibility to late-stage glomerulonephritis | (Lelongt et al. 2001) | |
| Abnormal decidual implantation sites | c | |
| Normal renal Alport syndrome progression | (Andrews et al. 2000) | |
| Normal LPS-induced lung injury | (Betsuyaku et al. 1999a) | |
| Normal neutrophil emigration | (Betsuyaku et al. 1999b) | |
| MMP11−/− | No overt developmental phenotype | (Masson et al. 1998) |
| Accelerated neointima formation after vessel injury | (Lijnen et al. 1999) | |
| MMP12−/− | Resistance to cigarette smoke-induced emphysema | (Hautamaki et al. 1997) |
| Normal susceptibility to experimental aortic aneurysms | (Pyo et al. 2000) | |
| Normal cardiac rupture following infarction | (Heymans et al. 1999) | |
| MMP14−/− | Severe bone and connective tissue abnormalities | (Holmbeck et al. 1999) |
| Inadequate collagen turnover, early postnatal death | (Holmbeck et al. 1999) | |
| Impaired endochondral ossification and angiogenesis | (Zhou et al. 2000) | |
| TIMP1−/− | Resistance to corneal Pseudomonas infections | (Osiewicz et al. 1999) |
| Normal renal interstitial fibrosis after proteinuria | (Eddy et al. 2000) | |
| TIMP2−/− | Deficient MMP2 activation; otherwise normal | (Wang et al. 2000b) |
| TIMP3−/− | Accelerated mammary gland involution | d |
| Emphysematous enlargement of pulmonary air spaces | d |
MD Sternlicht, BS Wiseman & Z Werb unpublished observations.
J-P Rougier & Z Werb unpublished observations.
N Rougier, JL Rinkenberger & Z Werb unpublished observations.
J Fata, K Leco & R Khokha personal communication.
Abbreviations: LPS, lipopolysaccharide; MMTV, mouse mammary tumor virus promoter; PMN, neutrophil; WAP, Whey acidic protein gene promoter.
Determination of Biologic Significance Using Substrate Cleavage Site Mutations
Mutations in amelogenin that decrease its susceptibility to MMP20 are associated with inherited defects in tooth enamal formation (Li et al. 2001). These data complement the enamel organ-specific expression of both MMP20 and amelogenin to strongly implicate amelogenin as a physiologic target of MMP20. A similar experimental approach is to generate animals that express mutant substrates that resist enzyme cleavage. For example, several collagenolytic MMPs cleave the α chains of native fibrillar collagens at a single peptide bond, such that type I collagen fibrils containing mutant α1(1) chains with an amino acid substitution at this site are resistant to cleavage. The introduction of this mutation into the endogenous COL1A1 gene of mice leads to marked dermal fibrosis resembling human scleroderma and impaired postpartum uterine involution (Liu et al. 1995). These studies suggest that collagen resorption can also be accomplished through collagenase cleavage at a novel site in the nonhelical N-telopeptide domain. Nevertheless, these studies do not identify which MMP cleaves type I collagen within its triple-helical domain during any specific processes. Moreover, considerable differences between collagenase expression patterns in humans and mice (Balbin et al. 2001) and differences between the catalytic activities of human and mouse MMP orthologues (Noel et al. 1995) may limit our ability to extrapolate murine data to humans and other species. Nevertheless, despite some inherent limitations, gene-targeting technologies provide the most powerful and potentially definitive means of determining how MMPs regulate biologic processes in vivo.
MMPs in Bone Development
Bone development can proceed in two distinct ways. Skeletal long bones and vertebrae form by the multistep process of endochondral ossification, whereas the flat bones of the skull develop by intramembranous ossification. In endochondral ossification, chondrocytes proliferate and differentiate to form a zone of hypertrophic cartilage that is then resorbed and replaced by mineralized bone matrix. It appears that resorption of the hypertrophic cartilage ECM is accomplished by chondroclasts that make way for invading capillary endothelial cells. Osteoblasts are then recruited to form trabecular bone that is subsequently remodeled by osteoclasts and maintained by the opposing but coordinated activities of bone-forming osteoblasts and bone-resorbing osteoclasts. With intramembranous ossification, osteoblasts deposit bone matrix onto a cartilage template that is later removed, and vascularization appears to be accomplished via osteoclast-mediated matrix remodeling. Bone development thus requires active remodeling of cartilage and bone matrices and the concerted differentiation and activity of several cell types. It is therefore not surprising that at least three MMPs, MMP2, MMP9, and MT1-MMP, influence bone development and homeostasis.
The only MMP gene mutations that are known to cause an inherited human disease were recently found in the MMP2 gene (Martignetti et al. 2001). These mutations abolish the expression of functional MMP2 and cause nodulosis, arthropathy, and osteolysis (NAO) syndrome, a rare autosomal recessive osteolysis (or “disappearing-bone”) disorder. Affected individuals present between one and ten years of age with subcutaneous fibrous nodules, arthritis, minor facial anomalies, sclerotic cranial sutures, generalized osteopenia, and multicentric osteolytic changes primarily involving the carpal and tarsal bones of the hands and feet. The seemingly counterintuitive finding that the absence of a matrix-degrading enzyme leads to progressive bone loss instead of an osteopetrotic increase in bone mass suggests that MMP2 is not a critical bone matrix-dissolving enzyme. Rather, it suggests that MMP2 promotes osteoblastic bone formation and/or inhibits osteoclastic bone resorption. Because TGF-β promotes osteoblastic activity (Filvaroff et al. 1999), and because mutations that favor unopposed TGF-β activity cause a human osteopetrotic disorder (Janssens et al. 2000), it is tempting to speculate that the absence of MMP2 might hinder bone deposition (and thus favor resorption) if MMP2 were a critical activator of TGF-β during bone remodeling. Alternatively, the lack of MMP2 might favor bone resorption if MMP2 is required to remove the collagen scaffold onto which osteoblasts lay new bone matrix and if such collagen removal were a prerequisite to the latter step of bone deposition. Although identifying the actual mechanisms clearly requires further experimentation, MMP2-deficient mice appear to develop normally and lack an overt osteolytic or arthritic phenotype. This may be due to compensatory mechanisms such as a lack of contributing factors that might reveal a phenotype that is otherwise too subtle to detect in mice, or the actual lack of a role for MMP2 in murine as opposed to human bone remodeling. Nevertheless, other MMP-deficient mice do exhibit skeletal phenotypes.
Mice lacking MMP9 accumulate hypertrophic cartilage within the growing region of their long bones and ultimately exhibit subtle bone shortening (Vu et al. 1998). This expansion of the hypertrophic cartilage zone results from deficient endochondral ossification rather than any changes in chondrocyte proliferation or differentiation. The apoptotic loss of hypertrophic chondrocytes, which only occurs near invading capillary endothelial cells, is delayed. This delay appears to be secondary to diminished vascular invasion into the hypertrophic cartilage. Indeed, MMP9-deficient hypertrophic cartilage fails to release normal levels of pro-angiogenic activity in culture. Thus the absence of MMP9 may diminish the release of an angiogenic factor from the hypertrophic cartilage ECM, thereby slowing vascular invasion and the subsequent death of hypertrophic chondrocytes. Several angiogenic factors are expressed in the growth plate of developing long bones, including vascular endothelial cell growth factor (VEGF), which is highly expressed in terminal hypertrophic chondrocytes (Gerber et al. 1999). Moreover, the inhibition of VEGF signaling with a soluble VEGF receptor chimeric protein (mFlt-IgG) inhibits capillary invasion into hypertrophic cartilage and leads to a bone phenotype that is similar to that seen in MMP9-null mice. In addition, mFlt-IgG inhibits the recruitment of chondroclasts, osteoblasts and osteoclasts (Gerber et al. 1999, Engsig et al. 2000). Thus MMP9 appears to play an essential role in releasing ECM-bound VEGF from hypertrophic cartilage stores and in the VEGF-mediated recruitment of several cell types that are necessary for proper bone development.
Whereas MMP9-null mice show a specific delay in endochondral ossification, targeted inactivation of the MT1-MMP gene in mice causes severe defects in both endochondral and intramembranous bone formation (Holmbeck et al. 1999, Zhou et al. 2000). Because MT1-MMP is a critical activator of MMP2, it is important to note that many of the features of the MT1-MMP-deficient mice (craniofacial dysmorphism, arthritis, osteopenia, and soft tissue fibrosis) resemble those described in MMP2-deficient humans with NAO syndrome (Martignetti et al. 2001). Similar defects have also been seen in mice harboring a mutation that renders their type I collagen fibrils resistant to collagenase cleavage (Liu et al. 1995), and cells isolated from MT1-MMP-null mice exhibit deficient collagenolytic activity in vitro (Holmbek et al. 1999). Thus impaired remodeling of collagen-containing skeletal and periskeletal matrices may lead to the defects seen in MT1-MMP-deficient mice. Bone marrow stromal cells from MT1-MMP-deficient mice show decreased bone formation, suggesting that MT1-MMP may influence the differentiation and activity of osteoblasts, and MT1-MMP-deficient mice have increased numbers of osteoclasts, suggesting that MT1-MMP may influence the differentiation and recruitment of the latter cell type as well. A better understanding of the mechanisms whereby MMPs influence skeletal development and homeostasis may have important implications in terms of avoiding any unwanted side-effects of clinical interventions aimed at inhibiting MMP activity.
MMPs in Vascular Remodeling
The formation of new blood vessels from existing vessels (angiogenesis) is required for tissue development and repair and for the growth of solid tumors. MMPs can contribute to angiogenesis in at least three ways: They can enable endothelial cell migration through surrounding tissues by disrupting ECM barriers, they can promote it by liberating sequestered angiogenic factors, or they can defy it by generating anti-angiogenic breakdown products. In addition, the balance between MMPs and their inhibitors must be restored during the maturation of newly formed blood vessels to favor basement membrane assembly and endothelial cell differentiation and quiescence (Kraling et al. 1999).
Insoluble fibrin is one of the barriers that angiogenic endothelial cells must penetrate during new blood vessel formation. Endothelial cell MMPs are required for neovascularization of fibrin to occur in culture and in vivo, whereas the fibrinolytic plasminogen/plasminogen activator system is surprisingly not required (Hiraoka et al. 1998). Moreover, the potent pericellular fibrinolysin MT1-MMP enables otherwise non-invasive cells to penetrate fibrin gels and form tubular structures. Invasion and tubulogenesis also proceed in the absence of MMP2 but are blocked by natural and synthetic MMP inhibitors and fail to take place if a transmembrane-deleted form of MT1-MMP is expressed instead of the full-length membrane-associated form. Thus membrane-associated MMPs play a critical role in angiogenesis by virtue of their pericellular fibrinoytic activity. Undoubtedly, they play a similar role in enabling endothelial cells to traverse other ECM barriers as well.
During the early development of solid cancers, an “angiogenic switch” initiates the formation of new blood vessels prior to the emergence of invasive tumors (Hanahan & Folkman 1996). One possibility is that various stimuli induce the expression of pro-angiogenic genes thereby activating this switch. This appears not to be the case, however, in a transgenic model of pancreatic islet cell carcinogenesis (Bergers et al. 2000). In this model, VEGF, its receptors, and acidic FGF are all expressed at stable constitutive levels before and after angiogenesis begins, yet a synthetic inhibitor of VEGF signaling inhibits angiogenesis and tumor growth. On the other hand, MMPs 2 and 9 are upregulated in angiogenic islets, and synthetic inhibitors of MMP activity delay the onset of angiogenesis and inhibit tumor growth. The genetic ablation of MMP9 also suppresses angiogenesis and tumor growth, whereas the lack of MMP2 inhibits tumor formation, but does not alter the development of angiogenic islets. Moreover, exogenous MMP9 (but not MMP2) releases VEGF from cultured islets, thereby eliciting an angiogenic response in co-cultured endothelial cells. Therefore, MMP9 contributes to the induction of angiogenesis by releasing sequestered VEGF, although other mechanisms may also exist.
Although MMP2 does not appear to influence angiogenesis in the above model, its downregulation by targeted gene disruption and antisense methods does suppress tumor-induced angiogenesis in other models (T Itoh et al. 1998, Fang et al. 2000). Integrin αvβ3, which recognizes RGD sequences on vitronectin and other ECM molecules, is highly expressed on angiogenic endothelial cells, and antibodies and peptides that block its function inhibit angiogenesis in several model systems (Eliceiri & Cheresh 1999). Moreover, the few αv-deficient mice that survive to term die shortly thereafter with extensive vascular abnormalities in some tissues. The RGD-independent binding of MMP2 to integrin αvβ3 may influence angiogenesis by facilitating vascular invasion (Brooks et al. 1996). However, MMP2-deficient mice have no overt angiogenic phenotype but do display less tumor-induced angiogenesis than wild-type controls (T Itoh et al. 1998). Because inaccessible RGD sites on native collagen are unmasked by the MMP-dependent cleavage of collagen, focal MMP-mediated collagen degradation may not only remove a physical barrier to endothelial cell invasion, but may also foster αvβ3-mediated endothelial cell migration through the proteolyzed ECM. In favor of this hypothesis, the recombinant hemopexin domain of MMP2 prevents active MMP2 from binding to αvβ3 and inhibits angiogenesis in culture (Brooks et al. 1998) and in vivo (Pfeifer et al. 2000). An organic compound that blocks the ability of αvβ3 integrin to bind MMP2 (but not vitronectin) is also a potent inhibitor of angiogenesis and tumor growth in vivo (Silletti et al. 2001). Because this compound does not appear to alter the activation or activity of MMP2 directly, αvβ3 integrin may influence angiogenesis in part by concentrating MMP2 at the cell surface. However, other data indicate that αvβ3 integrin promotes the maturation of MMP2 to a fully active form following the MT1-MMP-mediated formation of an MMP2 activation intermediate (Deryugina et al. 2001).
Part of the anti-angiogenic activity ofTS1, TS2, and other TS domain-containing proteins may derive from their ability to downregulate MMP2 and MMP9 under certain conditions. However, even though TS1 generally acts as an angiogenesis inhibitor, it may also promote angiogenesis at low concentrations (Qian et al. 1997). Under these conditions, TS1 upregulates endothelial cell MMP9 activity in a dose-dependent manner, and endothelial cell tube formation is suppressed in culture by antibodies against either TS1 or MMP9 (Qian et al. 1997). By yeast two-hybrid and immunoprecipitaion analysis, MMP2 and MMP9 bind the pro-perdin-like type 1 repeats of TS1 and TS2 via their fibronectin type II gelatin-binding domains, but neither TS is degraded (Bein & Simons 2000). Purified TS type 1 repeats and intact TS1 also inhibit the degradation of type IV collagen and appear to suppress the activation of proMMP2 and proMMP9 in culture. However, the diminished MMP activity could also reflect reduced steady-state MMP levels. Indeed, TS2 binds both latent and active MMP2, but instead of directly inhibiting the activation or activity of MMP2, it apparently promotes MMP2 clearance through the LRP-dependent internalization of TS2/MMP2 complexes (Yang et al. 2001).
MMPs can also negatively regulate angiogenesis by generating anti-angiogenic peptides. Angiostatin is an N-terminal breakdown product of plasminogen that can be generated by MMPs 2, 3, 7, 9, and 12 (Patterson & Sang 1997, O’Reilly et al. 1999); endostatin and tumstatin are anti-angiogenic protein fragments from the C-terminal portions of collagens XVIII and IV, respectively (Maeshima et al. 2000); and the free hemopexin domain of MMP9 itself may act as an angiogenesis inhibitor (Brooks et al. 1998). In addition, by cleaving cell surface FGF receptor 1, MMP2 generates a circulating and matrix-associated binding protein that can modulate the potent angiogenic effects of FGF by altering its availability (Levi et al. 1996). FGF receptor shedding may also downregulate the FGF responsiveness of those cells that have lost their receptor. To date, however, the importance of MMPs in down-modulating angiogenesis is unclear, whereas they are clearly important positive regulators of angiogenesis.
MMPs in Cancer
MMPs are generally present in greater amounts and activated more often in and around malignant cancers than in normal, benign, or premalignant tissues, with the highest expression taking place in areas of active invasion at the tumor-stroma interface (reviewed in Sternlicht & Bergers 2000). Indeed, several MMPs were first cloned and have been repeatedly re-cloned as cancer-associated genes (reviewed in Sternlicht & Werb 1999). Significant positive correlations have been found between MMP expression and various indicators of a poor prognosis in virtually all types of cancer, and in some instances, increased MMP levels represent an independent predictor of shortened disease-free and overall survival (reviewed in Sternlicht &Bergers 2000). In epithelial cancers, however, most of the upregulated MMPs are expressed by the supporting stromal cells rather than by the carcinoma cells themselves (McKerrow et al. 2000). Thus MMPs from adjacent stromal cells are often induced and commandeered by the malignant epithelial cells.
In addition to the extensive correlative data linking MMP overexpression with more aggressive malignant behavior and poor clinical outcome, compelling experimental data show that MMPs actively contribute to cancer progression. Relatively benign cells acquire malignant properties when MMP activity is increased or TIMP activity diminished, and highly malignant cells are made less aggressive by reducing their MMP levels or suppressing their MMP activity. Several MMPs have been implicated as key agonists in tumor invasion, metastasis, and angiogenesis, including MMPs 1, 2, 3, 9, and 14 (reviewed in Sternlicht & Bergers 2000). Without the aid of ECM-degrading MMPs, endothelial cells would probably be unable to penetrate the ECM, and cancer cells would be unable to cross the matrix barriers that otherwise contain their spread. However, recent data indicate that MMPs do far more to influence cancer than merely remove the physical barriers to invasion and metastasis. A number of MMPs can promote early cancer development, and MMP3 can also promote late epithelial-to-mesenchymal phenotypic changes that are associated with more aggressive malignant behavior (Lochter et al. 1997, Sternlicht et al. 1999). These data support the notion that MMPs can contribute to virtually all stages of cancer evolution, both early and late, thus expanding the potential clinical utility of MMP inhibitors. Conversely, some MMPs may defy cancer progression, and others may not participate in cancer, but undoubtedly play normal physiologic roles. These possibilities must be considered and the mechanisms underlying the influence of MMPs in cancer must be understood in the design of therapeutic agents in order to optimize their efficacy and minimize their toxicity.
EARLY NEOPLASIA
A number of recent studies indicate that stromal influences promote cancer development (Barcellos-Hoff & Ravani 2000, Moinfar et al. 2000). Fibrotic and inflammatory conditions increase cancer risk, and some familial cancer syndromes are the result of gene defects that produce stromal changes before epithelial changes ever occur (Kinzler & Vogelstein 1998, Willenbucher et al. 1999). These are also conditions in which stromal MMPs are upregulated, and the attendant rise in MMP activity may contribute to cancer susceptibility (Sternlicht et al. 1999). Indeed, fibroblasts foster the growth of human cancer cells in nude mice, yet fibroblasts lacking MMP11 do not (Masson et al. 1998).
Several studies in genetically modified mice also support the notion that MMPs participate in early cancer development (Table 4). In general, MMP-overproducing transgenic mice develop spontaneous hyperproliferative lesions. Moreover, when MMP transgenic and knockout mice receive various oncogenic stimuli, the MMP-overexpressing mice develop more cancers than usual, whereas those that lack various MMPs or overproduce TIMP1 tend to get fewer cancers than normal. For instance, MMP1 transgenic mice show increased sensitivity to chemical carcinogens (D’Armiento et al. 1995, Di Colandrea et al. 2000), whereas MMP11-deficient mice and transgenic mice that have elevated levels of circulating TIMP1 show a diminished sensitivity to carcinogens (Masson et al. 1998, Buck et al. 1999). The overexpression of MMP7 accelerates mammary cancer development in mice carrying a mammary-targeted Her2/neu transgene (Rudolph-Owen et al. 1998b), whereas its absence in mice carrying the Apcmin mutation slows the formation of intestinal adenomas (Wilson et al. 1997). The absence of MMP9 defies the development but accelerates the progression of human papilloma virus-16-induced squamous cell carcinomas in transgenic mice, whereas normal carcinogenesis is restored by transplanting MMP9-expressing bone marrow cells to the MMP9-deficient transgenic mice, thus indicating the importance of inflammatory cell-derived MMP9 in skin carcinogenesis (Coussens et al. 2000). Most notably, however, independent lines of mammary-targeted MMP3 transgenic mice develop spontaneous premalignant lesions and malignant mammary cancers despite an absence of carcinogens or pre-existing gene mutations (Sternlicht et al. 1999, Matrisian 1999). In addition, these changes are blocked by the co-expression of a TIMP1 transgene (Sternlicht et al. 1999). Moreover, mammary-targeted MT1-MMP transgenic mice also develop spontaneous mammary cancers (Ha et al. 2001). Although each of these examples suggest that MMPs participate in early cancer development, the spontaneous development of premalignant and malignant lesions in the latter studies lends considerable support to the likelihood that MMPs act as natural tumor promoters and influence cancer susceptibility.
TABLE 4.
MMPs promote carcinogenesis in genetically modified mice
| MMP/TIMP genotype |
Oncogenic stimulus |
Phenotype | Reference |
|---|---|---|---|
| Haptoglobin-MMP1 | none | hyperkeratosis, acanthosis | (D’Armiento et al. 1995) |
| DMBA+ PMA | ↑skin carcinogenesis | (D’Armiento et al. 1995) | |
| DMBA+ chrysarobin | ↑skin papilloma formation | (DiColandrea et al. 2000) | |
| WAP-MMP3 | none | mammary hyperplasias and cancers | (Sternlicht et al. 1999) |
| MMTV-MMP3 | none | mammary hyperplasias and cancers | (Matrisian 1999) |
| MMTV-MMP7 | none | mammary hyperplasias | (Rudolph-Owen et al. 1998b) |
| MMTV-neu | ↑ mammary carcinogenesis | (Rudolph-Owen et al. 1998b) | |
| MMTV-MMP14 | none | mammary hyperplasias and cancers | (Ha et al. 2001) |
| MMP2−/− | RIP-TAg | ↓ pancreatic tumor growth | (Bergers et al. 2000) |
| Injected cells | ↓ angiogenesis and tumor growth | (Itoh et al. 1998a) | |
| MMP7−/− | Apcmin | ↓ intestinal adenoma formation | (Wilson et al. 1997) |
| MMP9−/− | K14-HPV16 | ↓ skin carcinogenesis | (Coussens et al. 2000) |
| RIP-TAg | ↓ pancreatic carcinogenesis | (Bergers et al. 2000) | |
| MMP11−/− | DMBA | ↓ mammary carcinogenesis | (Masson et al. 1998) |
| H2-TIMP1 | Crp-TAg | ↓ liver carcinogenesis | (Martin et al. 1996) |
| H2-TIMP1 (as) | Crp-TAg | ↑ liver carcinogenesis | (Martin et al. 1996) |
| WAP-TIMP1 | WAP-MMP3 | ↓ mammary neoplasia | (Sternlicht et al. 1999) |
| Albumin-TIMP1 | DMBA + PMA | ↓ mammary carcinogenesis | (Buck et al. 1999) |
| LFABP-TIMP1 | Apcmin | ↑ intestinal adenoma formationa | (Heppner-Goss et al. 1998) |
Intestinal adenoma formation was, however, inhibited by a broad-range synthetic MMP inhibitor.
Abbreviations: Apcmin, adenomatous polyposis coli gene with multiple intestinal neoplasia mutation; as, antisense; Crp, C-reactive protein gene promoter; DMBA, dimethylbenz-anthracine; H2, major histocompatability complex class I promoter; HPV16, human papilloma virus 16 early region; K14, keratin-14 promoter; LFABP, liver fatty acid binding protein promoter; MMTV, mouse mammary tumor virus promoter; PMA, phorbol myristate acetate; RIP, rat insulin promoter; TAg, SV40 T antigen; WAP, whey acidic protein promoter.
If, in fact, MMPs promote cancer development, then their inhibitors should defy it. However, this is not always the case (Sternlicht & Bergers 2000). Data favoring the likelihood that MMPs promote cancer include those indicating that murine 3T3 cells become tumorigenic after antisense depletion of TIMP1 (Khokha et al. 1989) and that TIMPs inhibit the transforming activity of phorbol esters (Shoji et al. 1997) and the growth of some cancer cells in vivo (Khokha 1994, Krüger et al. 1997). Moreover, TIMP1 overproduction slows chemical skin carcinogenesis (Buck et al. 1999) and SV40 large T antigen-induced liver carcinogenesis in transgenic mice (Martin et al. 1996). In the latter model, TIMP1 appears to protect IGFBP-3 thereby inhibiting IGF signaling and hepatocyte growth (Martin et al. 1999). Surprisingly, in Apcmin mice, TIMP1 overexpression had no effect on tumor development in one line and augmented intestinal adenoma formation in another, whereas a synthetic MMP inhibitor slowed adenoma formation (Heppner-Goss et al. 1998). This discrepancy may reflect the MMP-independent growth-promoting activity of TIMP1, which persists in mutant TIMPs that lack MMP-inhibitory activity (Chesler et al. 1995). TIMP1 may also confer a growth advantage by upregulating VEGF expression in vivo. Indeed, TIMP1-transfected human and rat mammary cancer cells show a direct relationship between TIMP1 levels and VEGF production, tumor growth, and tumor vascularization and proliferation in vivo, but are unaffected by TIMP1 in terms of their growth in culture (Yoshiji et al. 1998). TIMP1 may also act as an anti-apoptotic survival factor. For example, cultured B cells show increased survival in the face of various apoptotic stimuli if they are grown in the presence of TIMP1, but not if TIMP2 or a synthetic MMP inhibitor is added instead (Guedez et al. 1998). In another set of experiments, co-isogenic pairs of transformed fibroblasts were established from wild-type and TIMP1-deficient littermates and injected into the tail veins of wild-type and TIMP1-null recipients (Soloway et al. 1996). Interestingly, lung colonization depended on the TIMP1 status of the tumor cells rather than that of the host. As predicted, two TIMP1-deficient lines formed substantially more lung colonies than their respective wild-type partners, yet the opposite was true in two other independently isolated pairs, with the TIMP1-expressing cells instead showing enhanced colony formation. Thus TIMP1 may promote or suppress tumor growth depending on the context of the individual tumor cells and the potential for cross-talk with other signaling pathways. These studies may explain why TIMPs are often upregulated rather than downregulated in human cancers and why their upregulation is often associated with a poor prognosis (Sternlicht & Bergers 2000). In addition, TIMPs may inhibit the MMP-mediated infiltration of lymphocytes and immune surveillance. Alternatively, increased TIMP1 levels may merely reflect the increased matrix remodeling that occurs during invasion, and the critical parameter may be the balance between MMPs and their inhibitors rather than the expression of one component alone. For TIMP2, conflicting data from clinical samples may also reflect the fact that it participates both in the activation and inhibition of MMP2 in a dose-dependent manner. Thus increasing levels of TIMP2 should increase MMP2 activation to a point, after which the activation and proteolytic activity of MMP2 would be blocked by the inhibitory activity of TIMP2. Ultimately, the TIMPs may block some aspects of cancer by inhibiting certain metalloproteinases, yet promote other aspects of cancer by inhibiting different metalloproteinases and by influencing cells in a metalloproteinase-independent manner. Although it is clearly difficult to distinguish whether MMP-independent and -dependent TIMP activities are involved in a given process, it will be necessary to do so in order to gain a better understanding of their role in cancer.
MALIGNANT PROGRESSION
Malignant cancers are distinguished from in situ cancers and benign neoplasms by their ability to cross tissue boundaries and spread to other organs of the body. To do so, cancer cells must cross multiple ECM barriers as they traverse the epithelial basement membrane and interstitial stroma, enter blood vessels or lymphatics, and extravasate to form metastatic deposits at a distant site. Because of this requirement and because invasive behavior is associated with the upregulation of MMPs in virtually all human cancers, MMPs are thought to facilitate invasion and metastasis by degrading structural ECM components. However, although in vitro and in vivo cell invasion assays have generally confirmed that the levels of MMP activity influence the ability of cells to penetrate native and reconstituted basement membranes, the data supporting a positive role for MMPs in metastasis are limited. The MMP9 promoter is induced during progression from in situ to invasive disease in transgenic mice that carry an MMP9 promoter-driven β-galactosidase transgene (Kupferman et al. 2000), and mice that lack MMP2 show diminished lung colonization after intravenous administration of cancer cells (T Itoh et al. 1998). Moreover, PCR-based detection methods suggest that MMP9 and perhaps other MMPs are required for intravasation and hematogenous spread in vivo, since MMP inhibitors block intravasation, and only MMP9-expressing cells are able to enter the blood stream in this assay (Kim et al. 1998). Surprisingly however, intravital videomicroscopy indicates that TIMP1-overexpressing cancer cells exit the vasculature just as well as their parental counterparts, but yield fewer and smaller metastases owing to diminished tumor growth after they leave the bloodstream (Koop et al. 1994). Therefore, extravasation may not require MMP activity, and although it is generally accepted that MMPs contribute to invasion and metastasis, the underlying mechanisms appear to be more complex than originally thought. In other words, the MMP-dependent invasive capacity of malignant cells may rely on more than just the disruption of ECM barriers. For example, downregulation of E-cadherin is causally involved in the malignant progression of some cancers, and MMP-generated E-cadherin breakdown products promote tumor cell invasion (Noe et al. 2001). In addition, MMPs can trigger epithelial-to-mesenchymal phenotypic conversions, a process that is associated with more aggressive malignant behavior and one that occurs naturally during development and wound repair when stationary and otherwise adherent epithelial cells become migratory and invasive (Lochter et al. 1997, Sternlicht et al. 1999). Thus MMPs can potentially contribute, both negatively and positively, to all stages of cancer evolution by numerous distinct pathways.
PERSPECTIVES
Not long ago, MMPs and other ECM-degrading enzymes were thought to act by simply excavating the structural groundwork that supported cells and separated distinct tissue compartments. Their capacity to remodel the ECM and promote cellular invasion was thought to come only from their ability to destroy underlying cellular foundations and disrupt the barriers that otherwise keep cells within their own specialized compartments. This viewpoint changed with the realization that extracellular matrices convey their own inherent information and that these structural data act as critical modifiers of cell behavior and misbehavior. Moreover, the ECM harbors several other signaling molecules such as bound growth factors and growth factor-binding proteins. Thus MMP-mediated remodeling of the ECM alters the informational content seen by nearby cells because remodeling can destroy current structural inputs, unearth hidden structural information, and release otherwise sequestered signaling molecules. In addition, it is now clear that although MMPs were initially named for their potent matrix-degrading capacity, they do more than just hydrolyze ECM molecules. They can irreversibly cleave many cell surface and pericellular proteins. Thus they can activate or inactivate various signaling pathways and can increase or decrease the availability of signaling molecules by degrading their binding partners or generating new ones. Several mechanisms that dock MMPs at the cell surface have evolved that not only focus MMP activity to specific regions of the pericellular microenvironment, but also balance MMP supply and demand. The existence of multi-functional MMP docking proteins on the cell surface provides a versatile mechanism for establishing powerful local extracellular signaling networks.
Proteolytic mechanisms regulate many normal developmental and homeostatic processes, whereas inappropriate proteolysis causes or exacerbates a number of diseases. The challenge now is to determine exactly which of the many potential mechanisms and substrates worked out in in vitro systems actually regulate these physiologic and pathologic processes. Considerable headway has been made in elucidating the critical proteolytic pathways that regulate some of these processes, often with surprising results. However, far more mechanisms remain unidentified or unproven, thus leaving the field ripe for further progress and important new insights.
ACKNOWLEDGMENTS
This work was supported by grants from the National Cancer Institute (CA72006, CA58207 and CA37395).
Contributor Information
Mark D. Sternlicht, Email: sternli@itsa.ucsf.edu.
Zena Werb, Email: zena@itsa.ucsf.edu.
LITERATURE CITED
- Abbaszade I, Liu RQ, Yang F, Rosenfeld SA, Ross OH, et al. Cloning and characterization of ADAMTS11, an aggrecanase from the ADAMTS family. J. Biol. Chem. 1999;274:23443–23450. doi: 10.1074/jbc.274.33.23443. [DOI] [PubMed] [Google Scholar]
- Alexander CM, Howard EW, Bissell MJ, Werb Z. Rescue of mammary epithelial cell apoptosis and entactin degradation by a tissue inhibitor of metalloproteinase-1 transgene. J. Cell Biol. 1996;135:1669–1677. doi: 10.1083/jcb.135.6.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander CM, Selvarajan S, Mudgett J, Werb Z. Stromelysin-1 regulates adipogenesis during mammary gland involution. J. Cell Biol. 2001;152:693–703. doi: 10.1083/jcb.152.4.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews KL, Betsuyaku T, Rogers S, Shipley JM, Senior RM, Miner JH. Gelatinase B (MMP-9) is not essential in the normal kidney and does not influence progression of renal disease in a mouse model of Alport syndrome. Am. J. Pathol. 2000;157:303–311. doi: 10.1016/S0002-9440(10)64541-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashworth JL, Murphy G, Rock MJ, Sherratt MJ, Shapiro SD, et al. Fibrillin degradation by matrix metalloproteinases: implications for connective tissue remodelling. Biochem. J. 1999;340:171–181. [PMC free article] [PubMed] [Google Scholar]
- Balbin M, Fueyo A, Knauper V, Lopez JM, Alvarez J, et al. Identification and enzymatic characterization of two diverging murine counterparts of human interstitial collagenase (MMP-1) expressed at sites of embryo implantation. J. Biol. Chem. 2001;276:10253–10262. doi: 10.1074/jbc.M009586200. [DOI] [PubMed] [Google Scholar]
- Banda MJ, Rice AG, Griffin GL, Senior RM. α1-Proteinase inhibitor is a neutrophil chemoattractant after proteolytic inactivation by macrophage elastase. J. Biol. Chem. 1988;263:4481–4484. [PubMed] [Google Scholar]
- Barcellos-Hoff MH, Ravani SA. Irradiated mammary gland stroma promotes the expression of tumorigenic potential by unir-radiated epithelial cells. Cancer Res. 2000;60:1254–1260. [PubMed] [Google Scholar]
- Barmina OY, Walling HW, Fiacco GJ, Freije JM, Lopez-Otin C, et al. Collagenase-3 binds to a specific receptor and requires the low density lipoprotein receptor-related protein for internalization. J. Biol. Chem. 1999;274:30087–30093. doi: 10.1074/jbc.274.42.30087. [DOI] [PubMed] [Google Scholar]
- Bauer EA, Stricklin GP, Jeffrey JJ, Eisen AZ. Collagenase production by human skin fibroblasts. Biochem. Biophys. Res. Commun. 1975;64:232–240. doi: 10.1016/0006-291x(75)90243-0. [DOI] [PubMed] [Google Scholar]
- Bein K, Simons M. Thrombospondin type 1 repeats interact with matrix metalloproteinase 2. Regulation of metalloproteinase activity. J. Biol. Chem. 2000;275:32167–32173. doi: 10.1074/jbc.M003834200. [DOI] [PubMed] [Google Scholar]
- Belkin AM, Akimov SS, Zaritskaya LS, Ratnikov BI, Deryugina EI, Strongin AY. Matrix-dependent proteolysis of surface transglutaminase by membrane-type metalloproteinase regulates cancer cell adhesion and locomotion. J. Biol. Chem. 2001;276:18415–18422. doi: 10.1074/jbc.M010135200. [DOI] [PubMed] [Google Scholar]
- Bergers G, Brekken R, McMahon G, Vu TH, Itoh T, et al. Matrix metalloproteinase-9 triggers the angiogenic switch during carcinogenesis. Nat. Cell Biol. 2000;2:737–744. doi: 10.1038/35036374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betsuyaku T, Shipley JM, Liu Z, Senior RM. Gelatinase B deficiency does not protect against lipopolysaccharide-induced acute lung injury. Chest. 1999a;116:17S–18S. [PubMed] [Google Scholar]
- Betsuyaku T, Shipley JM, Liu Z, Senior RM. Neutrophil emigration in the lungs, peritoneum, and skin does not require gelatinase B. Am. J. Respir. Cell. Mol. Biol. 1999b;20:1303–1309. doi: 10.1165/ajrcmb.20.6.3558. [DOI] [PubMed] [Google Scholar]
- Bian J, Sun Y. Transcriptional activation by p53 of the human type IV collagenase (gelatinase A or matrix metalloproteinase 2) promoter. Mol. Cell. Biol. 1997;17:6330–6338. doi: 10.1128/mcb.17.11.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodden MK, Harber GJ, Birkedal-Hansen B, Windsor LJ, Caterina NC, et al. Functional domains of human TIMP-1 (tissue inhibitor of metalloproteinases) J. Biol. Chem. 1994;269:18943–18952. [PubMed] [Google Scholar]
- Bond M, Murphy G, Bennett MR, Amour A, Knauper V, et al. Localization of the death domain of tissue inhibitor of metalloproteinase-3 to the N terminus. Metalloproteinase inhibition is associated with proapoptotic activity. J. Biol. Chem. 2000;275:41358–41363. doi: 10.1074/jbc.M007929200. [DOI] [PubMed] [Google Scholar]
- Bourguignon LY, Gunja-Smith Z, Iida N, Zhu HB, Young LJ, et al. CD44v(3, 8–10) is involved in cytoskeleton-mediated tumor cell migration and matrix metalloproteinase (MMP-9) association in metastatic breast cancer cells. J. Cell. Physiol. 1998;176:206–215. doi: 10.1002/(SICI)1097-4652(199807)176:1<206::AID-JCP22>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Brooks PC, Silletti S, von Schalscha TL, Fried-lander M, Cheresh DA. Disruption of angiogenesis by PEX, a noncatalytic metalloproteinase fragment with integrin binding activity. Cell. 1998;92:391–400. doi: 10.1016/s0092-8674(00)80931-9. [DOI] [PubMed] [Google Scholar]
- Brooks PC, Strömblad S, Sanders LC, von Schalscha TL, Aimes RT, et al. Localization of matrix metalloproteinase MMP-2 to the surface of invasive cells by interaction with integrin αvβ3. Cell. 1996;85:683–693. doi: 10.1016/s0092-8674(00)81235-0. [DOI] [PubMed] [Google Scholar]
- Brown DA, London E. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- Buck TB, Yoshiji H, Harris SR, Bunce OR, Thorgeirsson UP. The effects of sustained elevated levels of circulating tissue inhibitor of metalloproteinases-1 on the development of breast cancer in mice. Ann. NY Acad. Sci. 1999;878:732–735. doi: 10.1111/j.1749-6632.1999.tb07775.x. [DOI] [PubMed] [Google Scholar]
- Bullard KM, Lund L, Mudgett JS, Mellin TN, Hunt TK, et al. Impaired wound contraction in stromelysin-1-deficient mice. Ann. Surg. 1999;230:260–265. doi: 10.1097/00000658-199908000-00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Drews M, Lee HM, Conner C, Bahou WF, Zucker S. The propeptide domain of membrane type 1 matrix metalloproteinase is required for binding of tissue inhibitor of metalloproteinases and for activation of pro-gelatinase A. J. Biol. Chem. 1998;273:34745–34752. doi: 10.1074/jbc.273.52.34745. [DOI] [PubMed] [Google Scholar]
- Cao J, Hymowitz M, Conner C, Bahou WF, Zucker S. The propeptide domain of membrane type 1-matrix metalloproteinase acts as an intramolecular chaperone when expressed in trans with the mature sequence in COS-1 cells. J. Biol. Chem. 2000;275:29648–29653. doi: 10.1074/jbc.M001920200. [DOI] [PubMed] [Google Scholar]
- Chesler L, Golde DW, Bersch N, Johnson MD. Metalloproteinase inhibition and erythroid potentiation are independent activities of tissue inhibitor of metalloproteinase-1. Blood. 1995;86:4506–4515. [PubMed] [Google Scholar]
- Colige A, Li SW, Sieron AL, Nusgens BV, Prockop DJ, Lapiere CM. cDNA cloning and expression of bovine procollagen I N-proteinase: a new member of the superfamily of zinc-metalloproteinases with binding sites for cells and other matrix components. Proc. Natl. Acad. Sci. USA. 1997;94:2374–2379. doi: 10.1073/pnas.94.6.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Tinkle CL, Hanahan D, Werb Z. MMP-9 supplied by bone marrow-derived cells contributes to skin carcinogenesis. Cell. 2000;103:481–490. doi: 10.1016/s0092-8674(00)00139-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford HC, Fingleton BM, Rudolph-Owen LA, Goss KJ, Rubinfeld B, et al. The metalloproteinase matrilysin is a target of β-catenin transactivation in intestinal tumors. Oncogene. 1999;18:2883–2891. doi: 10.1038/sj.onc.1202627. [DOI] [PubMed] [Google Scholar]
- Curci JA, Liao S, Huffman MD, Shapiro SD, Thompson RW. Expression and localization of macrophage elastase (matrix metalloproteinase-12) in abdominal aortic aneurysms. J. Clin. Invest. 1998;102:1900–1910. doi: 10.1172/JCI2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Armiento J, Dalal SS, Okada Y, Berg RA, Chada K. Collagenase expression in the lungs of transgenic mice causes pulmonary emphysema. Cell. 1992;71:955–961. doi: 10.1016/0092-8674(92)90391-o. [DOI] [PubMed] [Google Scholar]
- D’Armiento J, Di Colandrea T, Dalal SS, Okada Y, Huang MT, et al. Collagenase expression in transgenic mouse skin causes hyperkeratosis and acanthosis and increases susceptibility to tumorigenesis. Mol. Cell. Biol. 1995;15:5732–5739. doi: 10.1128/mcb.15.10.5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delany AM, Jeffrey JJ, Rydziel S, Canalis E. Cortisol increases interstitial collagenase expression in osteoblasts by post-transcriptional mechanisms. J. Biol. Chem. 1995;270:26607–26612. doi: 10.1074/jbc.270.44.26607. [DOI] [PubMed] [Google Scholar]
- Deng SJ, Bickett DM, Mitchell JL, Lambert MH, Blackburn RK, et al. Substrate specificity of human collagenase 3 assessed using a phage-displayed peptide library. J. Biol. Chem. 2000;275:31422–31427. doi: 10.1074/jbc.M004538200. [DOI] [PubMed] [Google Scholar]
- Deryugina EI, Ratnikov B, Monosov E, Postnova TI, DiScipio R, et al. MT1-MMP initiates activation of pro-MMP-2 and integrin αvβ3 promotes maturation of MMP-2 in breast carcinoma cells. Exp. Cell Res. 2001;263:209–223. doi: 10.1006/excr.2000.5118. [DOI] [PubMed] [Google Scholar]
- Dhami R, Gilks B, Xie C, Zay K, Wright JL, Churg A. Acute cigarette smoke-induced connective tissue breakdown is mediated by neutrophils and prevented by α1-antitrypsin. Am. J. Respir. Cell Mol. Biol. 2000;22:244–252. doi: 10.1165/ajrcmb.22.2.3809. [DOI] [PubMed] [Google Scholar]
- Di Colandrea TD, D’Armiento J, Kesari KV, Chada KK. Collagenase induction promotes mouse tumorigenesis by two independent pathways. Mol. Carcinog. 2000;29:8–16. doi: 10.1002/1098-2744(200009)29:1<8::aid-mc2>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Ducharme A, Frantz S, Aikawa M, Rabkin E, Lindsey M, et al. Targeted deletion of matrix metalloproteinase-9 attenuates left ventricular enlargement and collagen accumulation after experimental myocardial infarction. J. Clin. Invest. 2000;106:55–62. doi: 10.1172/JCI8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunsmore SE, Saarialho-Kere UK, Roby JD, Wilson CL, Matrisian LM, et al. Matrilysin expression and function in airway epithelium. J. Clin. Invest. 1998;102:1321–1331. doi: 10.1172/JCI1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy AA, Kim H, Lopez-Guisa J, Oda T, Soloway PD. Interstitial fibrosis in mice with overload proteinuria: deficiency of TIMP-1 is not protective. Kidney Int. 2000;58:618–628. doi: 10.1046/j.1523-1755.2000.00208.x. [DOI] [PubMed] [Google Scholar]
- Edwards DR. The tissue inhibitors of metalloproteinases (TIMPs) In: Clendeninn NJ, Appelt K, editors. Matrix Metalloproteinase Inhibitors in Cancer Therapy. Totowa, NJ: Humana; 2001. pp. 67–84. [Google Scholar]
- Eliceiri BP, Cheresh DA. The role of αv integrins during angiogenesis: insights into potential mechanisms of action and clinical development. J. Clin. Invest. 1999;103:1227–1230. doi: 10.1172/JCI6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English WR, Puente XS, Freije JM, Knauper V, Amour A, et al. Membrane type 4 matrix metalloproteinase (MMP17) has tumor necrosis factor-α convertase activity but does not activate pro-MMP2. J. Biol. Chem. 2000;275:14046–14055. doi: 10.1074/jbc.275.19.14046. [DOI] [PubMed] [Google Scholar]
- Engsig MT, Chen QJ, Vu TH, Pedersen AC, Therkidsen B, et al. Matrix metalloproteinase 9 and vascular endothelial growth factor are essential for osteoclast recruitment into developing long bones. J. Cell Biol. 2000;151:879–890. doi: 10.1083/jcb.151.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Shing Y, Wiederschain D, Yan L, Butterfield C, et al. Matrix metalloproteinase-2 is required for the switch to the angiogenic phenotype in a tumor model. Proc. Natl. Acad. Sci. USA. 2000;97:3884–3889. doi: 10.1073/pnas.97.8.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filvaroff E, Erlebacher A, Ye JQ, Gitelman SE, Lotz J, et al. Inhibition of TGF-β receptor signaling in osteoblasts leads to decreased bone remodeling and increased trabecular bone mass. Development. 1999;126:4267–4279. doi: 10.1242/dev.126.19.4267. [DOI] [PubMed] [Google Scholar]
- Fingleton B, Vargo T, Crawford HC, Matrisian LM. Matrilysin (MMP-7) expression selects for apoptosis-resistant tumor cells. Clin. Exp. Metastasis. 1999;17:769. (Abstr.) [Google Scholar]
- Fini ME, Cook JR, Mohan R, Brinckerhoff CE. Regulation of matrix metalloproteinase gene expression. In: Parks WC, Mecham RP, editors. Matrix Metalloproteinases. New York: Academic; 1998. pp. 299–356. [Google Scholar]
- Fowlkes JL, Serra DM, Nagase H, Thrailkill KM. MMPs are IGFBP-degrading proteinases: implications for cell proliferation and tissue growth. Ann. NY Acad. Sci. 1999;878:696–699. doi: 10.1111/j.1749-6632.1999.tb07765.x. [DOI] [PubMed] [Google Scholar]
- Gerber HP, Vu TH, Ryan AM, Kowalski J, Werb Z, Ferrara N. VEGF couples hypertrophic cartilage remodeling, ossification and angiogenesis during endochondral bone formation. Nat. Med. 1999;5:623–628. doi: 10.1038/9467. [DOI] [PubMed] [Google Scholar]
- Giannelli G, Falk-Marzillier J, Schiraldi O, Stetler-Stevenson WG, Quaranta V. Induction of cell migration by matrix metalloprotease-2 cleavage of laminin-5. Science. 1997;277:225–228. doi: 10.1126/science.277.5323.225. [DOI] [PubMed] [Google Scholar]
- Gomez DE, Alonso DF, Yoshiji H, Thorgeirsson UP. Tissue inhibitors of metalloproteinases: structure, regulation and biological functions. Eur. J. Cell Biol. 1997;74:111–122. [PubMed] [Google Scholar]
- Gomis-Rüth FX, Maskos K, Betz M, Bergner A, Huber R, et al. Mechanism of inhibition of the human matrix metalloproteinase stromelysin-1 by TIMP-1. Nature. 1997;389:77–81. doi: 10.1038/37995. [DOI] [PubMed] [Google Scholar]
- Gross J, Lapiere CM. Collagenolytic activity in amphibian tissues: a tissue culture assay. Proc. Natl. Acad. Sci. USA. 1962;48:1014–1022. doi: 10.1073/pnas.48.6.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedez L, Stetler-Stevenson WG, Wolff L, Wang J, Fukushima P, et al. In vitro suppression of programmed cell death of B cells by tissue inhibitor of metalloproteinase-1. J. Clin. Invest. 1998;102:2002–2010. doi: 10.1172/JCI2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Li R, Zucker S, Toole BP. EMMPRIN (CD147), an inducer of matrix metalloproteinase synthesis, also binds interstitial collagenase to the tumor cell surface. Cancer Res. 2000;60:888–891. [PubMed] [Google Scholar]
- Gururajan R, Grenet J, Lahti JM, Kidd VJ. Isolation and characterization of two novel metalloproteinase genes linked to the CdC2Llocus on human chromosome 1p36.3. Genomics. 1998;52:101–106. doi: 10.1006/geno.1998.5401. [DOI] [PubMed] [Google Scholar]
- Gutman A, Wasylyk B. The collagenase gene promoter contains a TPA and oncogene-responsive unit encompassing the PEA3 and AP-1 binding sites. EMBO J. 1990;9:2241–2246. doi: 10.1002/j.1460-2075.1990.tb07394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha HY, Moon HB, Nam MS, Lee JW, Ryoo ZY, et al. Overexpression of membrane-type matrix metalloproteinase-1 gene induces mammary gland abnormalities and adenocarcinoma in transgenic mice. Cancer Res. 2001;61:984–990. [PubMed] [Google Scholar]
- Hanahan D, Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86:353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- Haro H, Crawford HC, Fingleton B, Mac-Dougall JR, Shinomiya K, et al. Matrix metalloproteinase-3-dependent generation of a macrophage chemoattractant in a model of herniated disc resorption. J. Clin. Invest. 2000a;105:133–141. doi: 10.1172/JCI7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haro H, Crawford HC, Fingleton B, Shinomiya K, Spengler DM, Matrisian LM. Matrix metalloproteinase-7-dependent release of tumor necrosis factor-α in a model of herniated disc resorption. J. Clin. Invest. 2000b;105:143–150. doi: 10.1172/JCI7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper E, Bloch KJ, Gross J. The zymogen of tadpole collagenase. Biochemistry. 1971;10:3035–3041. doi: 10.1021/bi00792a008. [DOI] [PubMed] [Google Scholar]
- Hasty KA, Pourmotabbed TF, Goldberg GI, Thompson JP, Spinella DG, et al. Human neutrophil collagenase. A distinct gene product with homology to other matrix metalloproteinases. J. Biol. Chem. 1990;265:11421–11424. [PubMed] [Google Scholar]
- Hautamaki RD, Kobayashi DK, Senior RM, Shapiro SD. Requirement for macrophage elastase for cigarette smoke-induced emphysema in mice. Science. 1997;277:2002–2004. doi: 10.1126/science.277.5334.2002. [DOI] [PubMed] [Google Scholar]
- Heppner-Goss KJ, Brown PD, Matrisian LM. Differing effects of endogenous and synthetic inhibitors of metalloproteinases on intestinal tumorigenesis. Int. J. Cancer. 1998;78:629–635. doi: 10.1002/(sici)1097-0215(19981123)78:5<629::aid-ijc17>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Heymans S, Luttun A, Nuyens D, Theilmeier G, Creemers E, et al. Inhibition of plasminogen activators or matrix metalloproteinases prevents cardiac rupture but impairs therapeutic angiogenesis and causes cardiac failure. Nat. Med. 1999;5:1135–1142. doi: 10.1038/13459. [DOI] [PubMed] [Google Scholar]
- Hiller O, Lichte A, Oberpichler A, Kocourek A, Tschesche H. Matrix metalloproteinases collagenase-2, macrophage elastase, collagenase-3, and membrane type 1-matrix metalloproteinase impair clotting by degradation of fibrinogen and factor XII. J. Biol. Chem. 2000;275:33008–33013. doi: 10.1074/jbc.M001836200. [DOI] [PubMed] [Google Scholar]
- Hiraoka N, Allen E, Apel IJ, Gyetko MR, Weiss SJ. Matrix metalloproteinases regulate neovascularization by acting as pericellular fibrinolysins. Cell. 1998;95:365–377. doi: 10.1016/s0092-8674(00)81768-7. [DOI] [PubMed] [Google Scholar]
- Hoegy SE, Oh H-R, Corcoran ML, Stetler-Stevenson WG. Tissue inhibitor of metalloproteinase-2 (TIMP-2) suppresses TKR-growth factor signaling independent of metalloproteinase inhibition. J. Biol. Chem. 2001;276:3203–3214. doi: 10.1074/jbc.M008157200. [DOI] [PubMed] [Google Scholar]
- Holmbeck K, Bianco P, Caterina J, Yamada S, Kromer M, et al. MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell. 1999;99:81–92. doi: 10.1016/s0092-8674(00)80064-1. [DOI] [PubMed] [Google Scholar]
- Hotary K, Allen E, Punturieri A, Yana I, Weiss SJ. Regulation of cell invasion and morphogenesis in a three-dimensional type I collagen matrix by membrane-type matrix metalloproteinases 1, 2, and 3. J. Cell Biol. 2000;149:1309–1323. doi: 10.1083/jcb.149.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard EW, Banda MJ. Binding of tissue inhibitor of metalloproteinases 2 to two distinct sites on human 72-kDa gelatinase. Identification of a stabilization site. J. Biol. Chem. 1991;266:17972–17977. [PubMed] [Google Scholar]
- Huang W, Meng Q, Suzuki K, Nagase H, Brew K. Mutational study of the amino-terminal domain of human tissue inhibitor of metalloproteinases 1 (TIMP-1) locates an inhibitory region for matrix metalloproteinases. J. Biol. Chem. 1997;272:22086–22091. doi: 10.1074/jbc.272.35.22086. [DOI] [PubMed] [Google Scholar]
- Imai K, Hiramatsu A, Fukushima D, Pierschbacher MD, Okada Y. Degradation of decorin by matrix metalloproteinases: identification of the cleavage sites, kinetic analyses and transforming growth factor-β1 release. Biochem. J. 1997;322:809–814. doi: 10.1042/bj3220809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Ohuchi E, Aoki T, Nomura H, Fujii Y, et al. Membrane-type matrix metalloproteinase 1 is a gelatinolytic enzyme and is secreted in a complex with tissue inhibitor of metalloproteinases 2. Cancer Res. 1996;56:2707–2710. [PubMed] [Google Scholar]
- Imai Y, Busby WH, Jr, Smith CE, Clarke JB, Garmong AJ, et al. Protease-resistant form of insulin-like growth factor-binding protein 5 is an inhibitor of insulin-like growth factor-I actions on porcine smooth muscle cells in culture. J. Clin. Invest. 1997;100:2596–2605. doi: 10.1172/JCI119803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iruela-Arispe ML, Vazquez F, Ortega MA. Antiangiogenic domains shared by thrombospondins and metallospondins, a new family of angiogenic inhibitors. Ann. NY Acad. Sci. 1999;886:58–66. doi: 10.1111/j.1749-6632.1999.tb09400.x. [DOI] [PubMed] [Google Scholar]
- Ito A, Mukaiyama A, Itoh Y, Nagase H, Thogersen IB, et al. Degradation of interleukin 1β by matrix metalloproteinases. J. Biol. Chem. 1996;271:14657–14660. doi: 10.1074/jbc.271.25.14657. [DOI] [PubMed] [Google Scholar]
- Itoh T, Tanioka M, Yoshida H, Yoshioka T, Nishimoto H, Itohara S. Reduced angiogenesis and tumor progression in gelatinase A-deficient mice. Cancer Res. 1998;58:1048–1051. [PubMed] [Google Scholar]
- Itoh Y, Ito A, Iwata K, Tanzawa K, Mori Y, Nagase H. Plasma membrane-bound tissue inhibitor of metalloproteinases (TIMP)-2 specifically inhibits matrix metalloproteinase 2 (gelatinase A) activated on the cell surface. J. Biol. Chem. 1998;273:24360–24367. doi: 10.1074/jbc.273.38.24360. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Kajita M, Kinoh H, Mori H, Okada A, Seiki M. Membrane type 4 matrix metalloproteinase (MT4-MMP, MMP-17) is a glycosylphosphatidylinositol-anchored proteinase. J. Biol. Chem. 1999;274:34260–34266. doi: 10.1074/jbc.274.48.34260. [DOI] [PubMed] [Google Scholar]
- Janssens K, Gershoni-Baruch R, Guanabens N, Migone N, Ralston S, et al. Mutations in the gene encoding the latency-associated peptide of TGF-β 1 cause Caurati-Engelmann disease. Nat. Genet. 2000;26:273–275. doi: 10.1038/81563. [DOI] [PubMed] [Google Scholar]
- Jimenez MJ, Balbin M, Lopez JM, Alvarez J, Komori T, Lopez-Otin C. Collagenase 3 is a target of Cbfa1, a transcription factor of the runt gene family involved in bone formation. Mol. Cell Biol. 1999;19:4431–4442. doi: 10.1128/mcb.19.6.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jormsjo S, Ye S, Moritz J, Walter DH, Dimmeler S, et al. Allele-specific regulation of matrix metalloproteinase-12 gene activity is associated with coronary artery luminal dimensions in diabetic patients with manifest coronary artery disease. Circ. Res. 2000;86:998–1003. doi: 10.1161/01.res.86.9.998. [DOI] [PubMed] [Google Scholar]
- Kanamori Y, Matsushima M, Minaguchi T, Kobayashi K, Sagae S, et al. Correlation between expression of the matrix metalloproteinase-1 gene in ovarian cancers and an insertion/deletion polymorphism in its promoter region. Cancer Res. 1999;59:4225–4227. [PubMed] [Google Scholar]
- Kashiwagi M, Tortorella M, Nagase H, Brew K. TIMP-3 is a potent inhibitor of ADAM-TS4 (aggrecanase 1) and ADAM-TS5 (aggrecanase 2) J. Biol. Chem. 2001;276:12501–12504. doi: 10.1074/jbc.C000848200. [DOI] [PubMed] [Google Scholar]
- Kataoka H, Uchino H, Iwamura T, Seiki M, Nabeshima K, Koono M. Enhanced tumor growth and invasiveness in vivo by a carboxyl-terminal fragment of α-proteinase inhibitor generated by matrix metalloproteinases: a possible modulatory role in natural killer cytotoxicity. Am. J. Pathol. 1999;154:457–468. doi: 10.1016/s0002-9440(10)65292-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradmand F, Werner E, Tremble P, Symons M, Werb Z. Role of Rac1 and oxygen radicals in collagenase-1 expression induced by cell shape change. Science. 1998;280:898–902. doi: 10.1126/science.280.5365.898. [DOI] [PubMed] [Google Scholar]
- Khokha R. Suppression of the tumorigenic and metastatic abilities of murine B16-F10 melanoma cells in vivo by the overexpression of the tissue inhibitor of metalloproteinases-1. J. Natl. Cancer Inst. 1994;86:299–304. doi: 10.1093/jnci/86.4.299. [DOI] [PubMed] [Google Scholar]
- Khokha R, Waterhouse P, Yagel S, Lala PK, Overall CM, et al. Antisense RNA-induced reduction in murine TIMP levels confers oncogenicity on Swiss 3T3 cells. Science. 1989;243:947–950. doi: 10.1126/science.2465572. [DOI] [PubMed] [Google Scholar]
- Kim J, Yu W, Kovalski K, Ossowski L. Requirement for specific proteases in cancer cell intravasation as revealed by a novel semi-quantitative PCR-based assay. Cell. 1998;94:353–362. doi: 10.1016/s0092-8674(00)81478-6. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Fukuzawa H, Shimada T, Saito T, Matsuda Y. Primary structure and expression of a gamete lytic enzyme in Chlamydomonas reinhardtii: similarity of functional domains to matrix metalloproteases. Proc. Natl. Acad. Sci. USA. 1992;89:4693–4697. doi: 10.1073/pnas.89.10.4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. Landscaping the cancer terrain. Science. 1998;280:1036–1037. doi: 10.1126/science.280.5366.1036. [DOI] [PubMed] [Google Scholar]
- Knauper V, Docherty AJ, Smith B, Tschesche H, Murphy G. Analysis of the contribution of the hinge region of human neutrophil collagenase (HNC, MMP-8) to stability and collagenolytic activity by alanine scanning mutagenesis. FEBS Lett. 1997b;405:60–64. doi: 10.1016/s0014-5793(97)00158-0. [DOI] [PubMed] [Google Scholar]
- Kojima S, Itoh Y, Matsumoto S, Masuho Y, Seiki M. Membrane-type 6 matrix metalloproteinase (MT6–MMP, MMP-25) is the second glycosyl-phosphatidyl inositol (GPI)-anchored MMP. FEBS Lett. 2000;480:142–146. doi: 10.1016/s0014-5793(00)01919-0. [DOI] [PubMed] [Google Scholar]
- Koop S, Khokha R, Schmidt EE, MacDonald IC, Morris VL, et al. Overexpression of metalloproteinase inhibitor in B16F10 cells does not affect extravasation but reduces tumor growth. Cancer Res. 1994;54:4791–4797. [PubMed] [Google Scholar]
- Kraling BM, Wiederschain DG, Boehm T, Rehn M, Mulliken JB, Moses MA. The role of matrix metalloproteinase activity in the maturation of human capillary endothelial cells in vitro. J. Cell Sci. 1999;112:1599–1609. doi: 10.1242/jcs.112.10.1599. [DOI] [PubMed] [Google Scholar]
- Krüger A, Fata JE, Khokha R. Altered tumor growth and metastasis of a T-cell lymphoma in Timp-1 transgenic mice. Blood. 1997;90:1993–2000. [PubMed] [Google Scholar]
- Kupferman ME, Fini ME, Muller WJ, Weber R, Cheng Y, Muschel RJ. Matrix metalloproteinase 9 promoter activity is induced coincident with invasion during tumor progression. Am. J. Pathol. 2000;157:1777–1783. doi: 10.1016/S0002-9440(10)64815-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langton KP, Barker MD, McKie N. Localization of the functional domains of human tissue inhibitor of metalloproteinases-3 and the effects of a Sorsby’s fundus dystrophy mutation. J. Biol. Chem. 1998;273:16778–16781. doi: 10.1074/jbc.273.27.16778. [DOI] [PubMed] [Google Scholar]
- Langton KP, McKie N, Curtis A, Goodship JA, Bond PM, et al. A novel tissue inhibitor of metalloproteinases-3 mutation reveals a common molecular phenotype in Sorsby’s fundus dystrophy. J. Biol. Chem. 2000;275:27027–27031. doi: 10.1074/jbc.M909677199. [DOI] [PubMed] [Google Scholar]
- Lelongt B, Bengatta S, Delauche M, Lund LR, Werb Z, Ronco PM. MMP9 protects mice from anti-glomerular basement membrane nephritis through its fibrinolytic activity. J. Exp. Med. 2001;193:793–802. doi: 10.1084/jem.193.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontovich AA, Zhang J, Shimokawa K, Nagase H, Sarras MP., Jr A novel hydra matrix metalloproteinase (HMMP) functions in extracellular matrix degradation, morphogenesis and the maintenance of differentiated cells in the foot process. Development. 2000;127:907–920. doi: 10.1242/dev.127.4.907. [DOI] [PubMed] [Google Scholar]
- Lepage T, Gache C. Early expression of a collagenase-like hatching enzyme gene in the sea urchin embryo. EMBO J. 1990;9:3003–3012. doi: 10.1002/j.1460-2075.1990.tb07493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi E, Fridman R, Miao HQ, Ma YS, Yayon A, Vlodavsky I. Matrix metalloproteinase 2 releases active soluble ectodomain of fibroblast growth factor receptor 1. Proc. Natl. Acad. Sci. USA. 1996;93:7069–7074. doi: 10.1073/pnas.93.14.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Gibson CW, Abrams WR, Andrews DW, DenBesten PK. Reduced hydrolysis of amelogenin may result in X-linked amelogenesis imperfecta. Matrix Biol. 2001;19:755–760. doi: 10.1016/s0945-053x(00)00121-9. [DOI] [PubMed] [Google Scholar]
- Lijnen HR, Van Hoef B, Vanlinthout I, Verstreken M, Rio MC, Collen D. Accelerated neointima formation after vascular injury in mice with stromelysin-3 (MMP-11) gene inactivation. Arteriosclerosis. 1999;19:2863–2870. doi: 10.1161/01.atv.19.12.2863. [DOI] [PubMed] [Google Scholar]
- Liu X, Wu H, Byrne M, Jeffrey J, Krane S, Jaenisch R. A targeted mutation at the known collagenase cleavage site in mouse type I collagen impairs tissue remodeling. J. Cell Biol. 1995;130:227–237. doi: 10.1083/jcb.130.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zhou X, Shapiro SD, Shipley JM, Twining SS, et al. The serpin α1-proteinase inhibitor is a critical substrate for gelatinase B/MMP-9 in vivo. Cell. 2000;102:647–655. doi: 10.1016/s0092-8674(00)00087-8. [DOI] [PubMed] [Google Scholar]
- Llano E, Pendas AM, Aza-Blanc P, Kornberg TB, Lopez-Otin C. Dm1-MMP, a matrix metalloproteinase from Drosophila with a potential role in extracellular matrix remodeling during neural development. J. Biol. Chem. 2000;275:35978–35985. doi: 10.1074/jbc.M006045200. [DOI] [PubMed] [Google Scholar]
- Lochter A, Galosy S, Muschler J, Freedman N, Werb Z, Bissell MJ. Matrix metalloproteinase stromelysin-1 triggers a cascade of molecular alterations that leads to stable epithelial-to-mesenchymal conversion and a premalignant phenotype in mammary epithelial cells. J. Cell Biol. 1997;139:1861–1872. doi: 10.1083/jcb.139.7.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan SK, Hansell EJ, Damsky CH, Werb Z. T-antigen inhibits metalloproteinase expression and invasion in human placental cells transformed with temperature-sensitive simian virus 40. Matrix Biol. 1996;15:81–89. doi: 10.1016/s0945-053x(96)90149-3. [DOI] [PubMed] [Google Scholar]
- Lohi J, Lehti K, Valtanen H, Parks WC, Keski-Oja J. Structural analysis and promoter characterization of the human membrane-type matrix metalloproteinase-1 (MT1-MMP) gene. Gene. 2000;242:75–86. doi: 10.1016/s0378-1119(99)00549-1. [DOI] [PubMed] [Google Scholar]
- Lohi J, Wilson CL, Roby JD, Parks WC. Epilysin: a novel human matrix met-alloproteinase (MMP-28) expressed in testis and keratinocytes and in response to injury. J. Biol. Chem. 2001;276:10134–10144. doi: 10.1074/jbc.M001599200. [DOI] [PubMed] [Google Scholar]
- Ludwig MG, Basset P, Anglard P. Multiple regulatory elements in the murine stromelysin-3 promoter. Evidence for direct control byCCAAT/enhancer-binding protein b and thyroid and retinoid receptors. J. Biol. Chem. 2000;275:39981–39990. doi: 10.1074/jbc.M007529200. [DOI] [PubMed] [Google Scholar]
- Lukashev ME, Werb Z. ECM signal-ling: orchestrating cell behaviour and misbehaviour. Trends Cell Biol. 1998;8:437–441. doi: 10.1016/s0962-8924(98)01362-2. [DOI] [PubMed] [Google Scholar]
- Lund LR, Romer J, Bugge TH, Nielsen BS, Frandsen TL, et al. Functional overlap between two classes of matrix-degrading proteases in wound healing. EMBO J. 1999;18:4645–4656. doi: 10.1093/emboj/18.17.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima Y, Colorado PC, Kalluri R. Two RGD-independent alpha v beta 3 integrin binding sites on tumstatin regulate distinct anti-tumor properties. J. Biol. Chem. 2000;275:23745–23750. doi: 10.1074/jbc.C000186200. [DOI] [PubMed] [Google Scholar]
- Maidment JM, Moore D, Murphy GP, Murphy G, Clark IM. Matrix metallo-proteinase homologues from Arabidopsis thaliana. Expression and activity. J. Biol. Chem. 1999;274:34706–34710. doi: 10.1074/jbc.274.49.34706. [DOI] [PubMed] [Google Scholar]
- Manes S, Mira E, Barbacid MM, Cipres A, Fernandez-Resa P et al. Identification of insulin-like growth factor-binding protein-1 as a potential physiological substrate for human stromelysin-3. J. Biol. Chem. 1997;272:25706–25712. doi: 10.1074/jbc.272.41.25706. [DOI] [PubMed] [Google Scholar]
- Maquoi E, Frankenne F, Noel A, Krell HW, Grams F, Foidart JM. Type IV collagen induces matrix metalloproteinase 2 activation in HT1080 fibrosarcoma cells. Exp. Cell Res. 2000;261:348–359. doi: 10.1006/excr.2000.5063. [DOI] [PubMed] [Google Scholar]
- Martignetti JA, Al Aqeel A, Al Sewaire W, Boumah CE, Kambouris M, et al. Matrix metalloproteinase 2 gene (MMP-2) mutations cause a multicentric osteolysis and arthritis syndrome. Nat. Genet. 2001;28:261–265. doi: 10.1038/90100. [DOI] [PubMed] [Google Scholar]
- Martin DC, Fowlkes JL, Babic B, Khokha R. Insulin-like growth factor II signaling in neoplastic proliferation is blocked by transgenic expression of the metalloproteinase inhibitor TIMP-1. J. Cell Biol. 1999;146:881–892. doi: 10.1083/jcb.146.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DC, Rüther U, Sanchez-Sweatman OH, Orr FW, Khokha R. Inhibition of SV40 T antigen-induced hepatocellular carcinoma in TIMP-1 transgenic mice. Oncogene. 1996;13:569–576. [PubMed] [Google Scholar]
- Masson R, Lefebvre O, Noël A, Fahime ME, Chenard MP, et al. In vivo evidence that the stromelysin-3 metalloproteinase contributes in a paracrine manner to epithelial cell malignancy. J. Cell Biol. 1998;140:1535–1541. doi: 10.1083/jcb.140.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrisian LM. Cancer biology: extracellular proteinases in malignancy. Curr. Biol. 1999;9:R776–R778. doi: 10.1016/S0960-9822(00)80011-1. [DOI] [PubMed] [Google Scholar]
- McGeehan G, Burkhart W, Anderegg R, Becherer JD, Gillikin JW, Graham JS. Sequencing and characterization of the soy-bean leaf metalloproteinase: structural and functional similarity to the matrix metalloproteinase family. Plant Physiol. 1992;99:1179–1183. doi: 10.1104/pp.99.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKerrow JH, Bhargava V, Hansell E, Huling S, Kuwahara T, et al. A functional proteomics screen of proteases in colorectal carcinoma. Mol. Med. 2000;6:450–460. [PMC free article] [PubMed] [Google Scholar]
- McQuibban GA, Gong JH, Tam EM, Mc-Culloch CA, Clark-Lewis I, Overall CM. Inflammation dampened by gelatinase A cleavage of monocyte chemoattractant protein-3. Science. 2000;289:1202–1206. doi: 10.1126/science.289.5482.1202. [DOI] [PubMed] [Google Scholar]
- Mitsiades N, Yu WH, Poulaki V, Tsokos M, Stamenkovic I. Matrix metalloproteinase-7-mediated cleavage of Fas ligand protects tumor cells from chemotherapeutic drug cytotoxicity. Cancer Res. 20011;61:577–581. [PubMed] [Google Scholar]
- Miyamori H, Takino T, Seiki M, Sato H. Human membrane type-2 matrix metalloproteinase is defective in cell-associated activation of progelatinase A. Biochem. Biophys. Res. Commun. 2000;267:796–800. doi: 10.1006/bbrc.1999.2050. [DOI] [PubMed] [Google Scholar]
- Mohan R, Rinehart WB, Bargagna-Mohan P, Fini ME. Gelatinase B/lacZ transgenic mice, a model for mapping gelatinase B expression during developmental and injury-related tissue remodeling. J. Biol. Chem. 1998;273:25903–25914. doi: 10.1074/jbc.273.40.25903. [DOI] [PubMed] [Google Scholar]
- Moinfar F, Man YG, Arnould L, Bratthauer GL, Ratschek M, Tavassoli FA. Concurrent and independent genetic alterations in the stromal and epithelial cells of mammary carcinoma: implications for tumorigenesis. Cancer Res. 2000;60:2562–2566. [PubMed] [Google Scholar]
- Mott JD, Thomas CL, Rosenbach MT, Takahara K, Greenspan DS, Banda MJ. Post-translational proteolytic processing of procollagen C-terminal proteinase enhancer releases a metalloproteinase inhibitor. J. Biol. Chem. 2000;275:1384–1390. doi: 10.1074/jbc.275.2.1384. [DOI] [PubMed] [Google Scholar]
- Mudgett JS, Hutchinson NI, Chartrain NA, Forsyth AJ, McDonnell J, et al. Susceptibility of stromelysin 1-deficient mice to collagen-induced arthritis and cartilage destruction. Arthritis Rheum. 1998;41:110–121. doi: 10.1002/1529-0131(199801)41:1<110::AID-ART14>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Munaut C, Salonurmi T, Kontusaari S, Reponen P, Morita T, et al. Murine matrix metalloproteinase 9 gene. 5′-upstream region contains cis-acting elements for expression in osteoclasts and migrating keratinocytes in transgenic mice. J. Biol. Chem. 1999;274:5588–5596. doi: 10.1074/jbc.274.9.5588. [DOI] [PubMed] [Google Scholar]
- Murphy G, Allan JA, Willenbrock F, Cockett MI, O’Connell JP, Docherty AJ. The role of the C-terminal domain in collagenase and stromelysin specificity. J. Biol. Chem. 1992;267:9612–9618. [PubMed] [Google Scholar]
- Murphy G, Nguyen Q, Cockett MI, Atkinson SJ, Allan JA, et al. Assessment of the role of the fibronectin-like domain of gelatinase A by analysis of a deletion mutant. J. Biol. Chem. 1994;269:6632–6636. [PubMed] [Google Scholar]
- Murphy G, Willenbrock F. Tissue inhibitors of matrix metalloendopeptidases. Methods Enzymol. 1995;248:496–510. doi: 10.1016/0076-6879(95)48032-3. [DOI] [PubMed] [Google Scholar]
- Nagase H, Woessner JF., Jr. Matrix metalloproteinases. J. Biol. Chem. 1999;274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- Nakahara H, Howard L, Thompson EW, Sato H, Seiki M, et al. Transmembrane/cytoplasmic domain-mediated membrane type 1-matrix metalloprotease docking to in-vadopodia is required for cell invasion. Proc. Natl Acad. Sci. USA. 1997;94:7959–7964. doi: 10.1073/pnas.94.15.7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson AR, Fingleton B, Rothenberg ML, Matrisian LM. Matrix metalloproteinases: biologic activity and clinical implications. J. Clin. Oncol. 2000;18:1135–1149. doi: 10.1200/JCO.2000.18.5.1135. [DOI] [PubMed] [Google Scholar]
- Netzer KO, Suzuki K, Itoh Y, Hudson BG, Khalifah RG. Comparative analysis of the noncollagenous NC1 domain of type IV collagen: identification of structural features important for assembly, function, and pathogenesis. Protein Sci. 1998;7:1340–1351. doi: 10.1002/pro.5560070610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noe V, Fingleton B, Jacobs K, Crawford HC, Vermeulen S, et al. Release of an invasion promoter E-cadherin fragment by matrilysin and stromelysin-1. J. Cell Sci. 2001;114:111–118. doi: 10.1242/jcs.114.1.111. [DOI] [PubMed] [Google Scholar]
- Noël A, Santavicca M, Stoll I, L’Hoir C, Staub A, et al. Identification of structural determinants controlling human and mouse stromelysin-3 proteolytic activities. J. Biol. Chem. 1995;270:22866–22872. doi: 10.1074/jbc.270.39.22866. [DOI] [PubMed] [Google Scholar]
- Noll WW, Belloni DR, Rutter JL, Storm CA, Schned AR, et al. Loss of heterozygosity on chromosome 11q22–23 in melanoma is associated with retention of the insertion polymorphism in the matrix metalloproteinase-1 promoter. Am. J. Pathol. 2001;158:691–697. doi: 10.1016/S0002-9440(10)64011-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly MS, Wiederschain D, Stetler-Stevenson WG, Folkman J, Moses MA. Regulation of angiostatin production by matrix metalloproteinase-2 in a model of concomitant resistance. J. Biol. Chem. 1999;274:29568–29571. doi: 10.1074/jbc.274.41.29568. [DOI] [PubMed] [Google Scholar]
- O’Shea M, Willenbrock F, Williamson RA, Cockett MI, Freedman RB, et al. Site-directed mutations that alter the inhibitory activity of the tissue inhibitor of metalloproteinases-1: importance of the N-terminal region between cysteine 3 and cysteine 13. Biochemistry. 1992;31:10146–10152. doi: 10.1021/bi00157a002. [DOI] [PubMed] [Google Scholar]
- Okumura Y, Sato H, Seiki M, Kido H. Proteolytic activation of the precursor of membrane type 1 matrix metalloproteinase by human plasmin. A possible cell surface activator. FEBS Lett. 1997;402:181–184. doi: 10.1016/s0014-5793(96)01523-2. [DOI] [PubMed] [Google Scholar]
- Olson MW, Toth M, Gervasi DC, Sado Y, Ninomiya Y, Fridman R. High affinity binding of latent matrix metalloproteinase-9 to the α2(IV) chain of collagen IV. J. Biol. Chem. 1998;273:10672–10681. doi: 10.1074/jbc.273.17.10672. [DOI] [PubMed] [Google Scholar]
- Osiewicz K, McGarry M, Soloway PD. Hyper-resistance to infection in TIMP-1-deficient mice is neutrophil dependent but not immune cell autonomous. Ann. NY Acad. Sci. 1999;878:494–496. doi: 10.1111/j.1749-6632.1999.tb07706.x. [DOI] [PubMed] [Google Scholar]
- Overall CM. Matrix metalloproteinase substrate binding domains, modules and exosites. In: Clark IM, editor. Matrix Metalloproteinase Protocols. Totowa, NJ: Humana; 2001. pp. 79–120. [PubMed] [Google Scholar]
- Overall CM, Wrana JL, Sodek J. Transcriptional and post-transcriptional regulation of 72-kDa gelatinase/type IV collagenase by transforming growth factor-β 1 in human fibroblasts. Comparisons with collagenase and tissue inhibitor of matrix metalloproteinase gene expression. J. Biol. Chem. 1991;266:14064–14071. [PubMed] [Google Scholar]
- Park HI, Ni J, Gerkema FE, Liu D, Belozerov VE, Sang QX. Identification and characterization of human endometase (matrix metalloproteinase-26) from endometrial tumor. J. Biol. Chem. 2000;275:20540–20544. doi: 10.1074/jbc.M002349200. [DOI] [PubMed] [Google Scholar]
- Patterson BC, Sang QXA. Angiostatin-converting enzyme activities of human matrilysin (MMP-7) and gelatinase B/type IV collagenase (MMP-9) J. Biol. Chem. 1997;272:28823–28825. doi: 10.1074/jbc.272.46.28823. [DOI] [PubMed] [Google Scholar]
- Pei D, Weiss SJ. Furin-dependent intracellular activation of the human stromelysin-3 zymogen. Nature. 1995;375:244–247. doi: 10.1038/375244a0. [DOI] [PubMed] [Google Scholar]
- Pendas AM, Balbin M, Llano E, Jimenez MG, Lopez-Otin C. Structural analysis and promoter characterization of the human collagenase-3 gene (MMP13) Genomics. 1997;40:222–233. doi: 10.1006/geno.1996.4554. [DOI] [PubMed] [Google Scholar]
- Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, et al. An essential role for ectodomain shedding in mammalian development. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- Petitclerc E, Boutaud A, Prestayko A, Xu J, Sado Y, et al. New functions for non-collagenous domains of human collagen type IV. Novel integrin ligands inhibiting angiogenesis and tumor growth in vivo. J. Biol. Chem. 2000;275:8051–8061. doi: 10.1074/jbc.275.11.8051. [DOI] [PubMed] [Google Scholar]
- Petitclerc E, Stromblad S, von Schalscha TL, Mitjans F, Piulats J, et al. Integrin αvβ3 promotes M21 melanoma growth in human skin by regulating tumor cell survival. Cancer Res. 1999;59:2724–2730. [PubMed] [Google Scholar]
- Pfeifer A, Kessler T, Silletti S, Cheresh DA, Verma IM. Suppression of angiogenesis by lentiviral delivery of PEX, a noncatalytic fragment of matrix metalloproteinase 2. Proc. Natl. Acad. Sci. USA. 2000;97:12227–12232. doi: 10.1073/pnas.220399597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilcher BK, Wang M, Qin XJ, Parks WC, Senior RM, Welgus HG. Role of matrix metalloproteinases and their inhibition in cutaneous wound healing and allergic contact hypersensitivity. Ann. NY Acad. Sci. 1999;878:12–24. doi: 10.1111/j.1749-6632.1999.tb07671.x. [DOI] [PubMed] [Google Scholar]
- Powell WC, Fingleton B, Wilson CL, Boothby M, Matrisian LM. The metalloproteinase matrilysin proteolytically generates active soluble Fas ligand and potentiates epithelial cell apoptosis. Curr. Biol. 1999;9:1441–1447. doi: 10.1016/s0960-9822(00)80113-x. [DOI] [PubMed] [Google Scholar]
- Pozzi A, Moberg PE, Miles LA, Wagner S, Soloway P, Gardner HA. Elevated matrix metalloprotease and angiostatin levels in integrin α1 knockout mice cause reduced tumor vascularization. Proc. Natl. Acad. Sci. USA. 2000;97:2202–2207. doi: 10.1073/pnas.040378497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preece G, Murphy G, Ager A. Metalloproteinase-mediated regulation of L-selectin levels on leucocytes. J. Biol. Chem. 1996;271:11634–11640. doi: 10.1074/jbc.271.20.11634. [DOI] [PubMed] [Google Scholar]
- Price SJ, Greaves DR, Watkins H. Identification of novel, functional genetic variants in the human matrix metalloproteinase-2 gene: role of Sp1 in allele-specific transcriptional regulation. J. Biol. Chem. 2001;276:7549–7558. doi: 10.1074/jbc.M010242200. [DOI] [PubMed] [Google Scholar]
- Primakoff P, Myles DG. The ADAM gene family: surface proteins with adhesion and protease activity. Trends Genet. 2000;16:83–87. doi: 10.1016/s0168-9525(99)01926-5. [DOI] [PubMed] [Google Scholar]
- Pyo R, Lee JK, Shipley JM, Curci JA, Mao D, et al. Targeted gene disruption of matrix metalloproteinase-9 (gelatinase B) suppresses development of experimental abdominal aortic aneurysms. J Clin Invest. 2000;105:1641–1649. doi: 10.1172/JCI8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Wang TN, Rothman VL, Nicosia RF, Tuszynski GP. Thrombospondin-1 modulates angiogenesis in vitro by upregulation of matrix metalloproteinase-9 in endothelial cells. Exp. Cell Res. 1997;235:403–412. doi: 10.1006/excr.1997.3681. [DOI] [PubMed] [Google Scholar]
- Raza SL, Nehring LC, Shapiro SD, Cornelius LA. Proteinase-activated receptor-1 regulation of macrophage elastase (MMP-12) secretion by serine proteinases. J. Biol. Chem. 2000;275:41243–41250. doi: 10.1074/jbc.M005788200. [DOI] [PubMed] [Google Scholar]
- Rudolph-Owen LA, Cannon P, Matrisian LM. Overexpression of the matrix metalloproteinase matrilysin results in premature mammary gland differentiation and male infertility. Mol. Biol. Cell. 1998a;9:421–435. doi: 10.1091/mbc.9.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph-Owen LA, Chan R, Muller WJ, Matrisian LM. The matrix metalloproteinase matrilysin influences early-stage mammary tumorigenesis. Cancer Res. 1998b;58:5500–5506. [PubMed] [Google Scholar]
- Ruiz S, Henschen-Edman AH, Nagase H, Tenner AJ. Digestion of C1q collagen-like domain with MMPs-1,-2,-3, and -9 further defines the sequence involved in the stimulation of neutrophil superoxide production. J. Leukoc. Biol. 1999;66:416–422. doi: 10.1002/jlb.66.3.416. [DOI] [PubMed] [Google Scholar]
- Rutter JL, Mitchell TI, Butticè G, Meyers J, Gusella JF, et al. A single nucleotide polymorphism in the matrix metalloproteinase-1 promoter creates an Ets binding site and augments transcription. Cancer Res. 1998;58:5321–5325. [PubMed] [Google Scholar]
- Sanchez-Lopez R, Alexander CM, Behrendtsen O, Breathnach R, Werb Z. Role of zinc-binding- and hemopexin domain-encoded sequences in the substrate specificity of collagenase and stromelysin-2 as revealed by chimeric proteins. J. Biol. Chem. 1993;268:7238–7247. [PubMed] [Google Scholar]
- Sheu BC, Hsu SM, Ho HN, Lien HC, Huang SC, Lin RH. A novel role of metalloproteinase in cancer-mediated immunosuppression. Cancer Res. 2001;61:237–242. [PubMed] [Google Scholar]
- Shipley JM, Doyle GA, Fliszar CJ, Ye QZ, Johnson LL, et al. The structural basis for the elastolytic activity of the 92-kDa and 72-kDa gelatinases. Role of the fibronectin type II-like repeats. J. Biol. Chem. 1996;271:4335–4341. doi: 10.1074/jbc.271.8.4335. [DOI] [PubMed] [Google Scholar]
- Shoji A, Sakamoto Y, Tsuchiya T, Moriyama K, Kaneko T, et al. Inhibition of tumor promoter activity toward mouse fibroblasts and their in vitro transformation by tissue inhibitor of metalloproteinases-1 (TIMP-1) Carcinogenesis. 1997;18:2093–2100. doi: 10.1093/carcin/18.11.2093. [DOI] [PubMed] [Google Scholar]
- Shrivastava A, Radziejewski C, Campbell E, Kovac L, McGlynn M, et al. An orphan receptor tyrosine kinase family whose members serve as nonintegrin collagen receptors. Mol. Cell. 1997;1:25–34. doi: 10.1016/s1097-2765(00)80004-0. [DOI] [PubMed] [Google Scholar]
- Silletti S, Kessler T, Goldberg J, Boger DL, Cheresh DA. Disruption of matrix metalloproteinase 2 binding to integrin αvβ3 by an organic molecule inhibits angiogenesis and tumor growth in vivo. Proc. Natl. Acad. Sci. USA. 2001;98:119–124. doi: 10.1073/pnas.011343298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soloway PD, Alexander CM, Werb Z, Jaenisch R. Targeted mutagenesis of Timp-1 reveals that lung tumor invasion is influenced by Timp-1 genotype of the tumor but not by that of the host. Oncogene. 1996;13:2307–2314. [PubMed] [Google Scholar]
- Sottrup-Jensen L, Birkedal-Hansen H. Human fibroblast collagenase-α-macroglo-bulin interactions. Localization of cleavage sites in the bait regions of five mammalian α-macroglobulins. J. Biol. Chem. 1989;264:393–401. [PubMed] [Google Scholar]
- Sternlicht MD, Bergers G. Matrix metalloproteinases as emerging targets in anticancer therapy: status and prospects. Emerging Ther. Targets. 2000;4:609–633. [Google Scholar]
- Sternlicht MD, Bissell MJ, Werb Z. The matrix metalloproteinase stromelysin-1 acts as a natural mammary tumor promoter. Oncogene. 2000;19:1102–1113. doi: 10.1038/sj.onc.1203347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternlicht MD, Coussens LM, Vu TH, Werb Z. Biology and regulation of the matrix metalloproteinases. In: Clendeninn NJ, Appelt K, editors. Matrix Metalloproteinase Inhibitors in Cancer Therapy. Totowa, NJ: Humana; 2001. pp. 1–37. [Google Scholar]
- Sternlicht MD, Lochter A, Sympson CJ, Huey B, Rougier JP, et al. The stromal proteinase MMP-3/stromelysin-1 promotes mammary carcinogenesis. Cell. 1999;98:137–146. doi: 10.1016/s0092-8674(00)81009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternlicht MD, Werb Z. ECM proteinases. In: Kreis T, Vale R, editors. Guidebook to the Extracellular Matrix, Anchor and Adhesion Proteins. Oxford, UK: Oxford Univ. Press; 1999. pp. 503–562. [Google Scholar]
- Stöcker W, Grams F, Baumann U, Reinemer P, Gomis-Rüth FX. The metzincins—topological and sequential relations between the astacins, adamalysins, serralysins, and matrixins (collagenases) define a superfamily of zinc-peptidases. Protein Sci. 1995;4:823–840. doi: 10.1002/pro.5560040502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke JO, Hutton M, Stewart M, Pendas AM, Smith B, et al. Biochemical characterization of the catalytic domain of human matrix metalloproteinase 19. J. Biol. Chem. 2000;275:14809–14816. doi: 10.1074/jbc.275.20.14809. [DOI] [PubMed] [Google Scholar]
- Streuli C. Extracellular matrix remodelling and cellular differentiation. Curr. Opin. Cell Biol. 1999;11:634–640. doi: 10.1016/s0955-0674(99)00026-5. [DOI] [PubMed] [Google Scholar]
- Stricker TP, Dumin J, Chung L, Nagase H, Parks WC, Santoro SA. Structural analysis of the integrin α2:matrix metalloproteinase-1 interaction. Mol. Biol. Cell. 2000;11:258a. doi: 10.1074/jbc.M102217200. (Abstr.) [DOI] [PubMed] [Google Scholar]
- Strongin AY, Collier I, Bannikov G, Marmer BL, Grant GA, Goldberg GI. Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J. Biol. Chem. 1995;270:5331–5338. doi: 10.1074/jbc.270.10.5331. [DOI] [PubMed] [Google Scholar]
- Sun Y, Wenger L, Rutter JL, Brinckerhoff CE, Cheung HS. p53 down-regulates human matrix metalloproteinase-1 (collagenase-1) gene expression. J. Biol. Chem. 1999;274:11535–11540. doi: 10.1074/jbc.274.17.11535. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Raab G, Moses MA, Fernandez CA, Klagsbrun M. Matrix metalloproteinase-3 releases active heparin-binding EGF-like growth factor by cleavage at a specific juxtamembrane site. J. Biol. Chem. 1997;272:31730–31737. doi: 10.1074/jbc.272.50.31730. [DOI] [PubMed] [Google Scholar]
- Sympson CJ, Talhouk RS, Alexander CM, Chin JR, Clift SM, et al. Targeted expression of stromelysin-1 in mammary gland provides evidence for a role of proteinases in branching morphogenesis and the requirement for an intact basement membrane for tissue-specific gene expression. J. Cell Biol. 1994;125:681–693. doi: 10.1083/jcb.125.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang BL, Hong W. ADAMTS: a novel family of proteases with an ADAM protease domain and thrombospondin 1 repeats. FEBS Lett. 1999;445:223–225. doi: 10.1016/s0014-5793(99)00119-2. [DOI] [PubMed] [Google Scholar]
- Tortorella MD, Burn TC, Pratta MA, Abbaszade I, Hollis JM. Purification and cloning of aggrecanase-1:A member of the ADAMTS family of proteins. Science. 1999;284:1664–1666. doi: 10.1126/science.284.5420.1664. [DOI] [PubMed] [Google Scholar]
- Toth M, Sado Y, Ninomiya Y, Fridman R. Biosynthesis of α2(IV) and α1(IV) chains of collagen IV and interactions with matrix metalloproteinase-9. J. Cell Physiol. 1999;180:131–139. doi: 10.1002/(SICI)1097-4652(199907)180:1<131::AID-JCP15>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Uria JA, Jimenez MG, Balbin M, Freije JMP, Lopez-Otin C. Differential effects of transforming growth factor-β on the expression of collagenase-1 and collagenase-3 in human fibroblasts. J. Biol. Chem. 1998;273:9769–9777. doi: 10.1074/jbc.273.16.9769. [DOI] [PubMed] [Google Scholar]
- Uria JA, Lopez-Otin C. Matrilysin-2, a new matrix metalloproteinase expressed in human tumors and showing the minimal domain organization required for secretion, latency, and activity. Cancer Res. 2000;60:4745–4751. [PubMed] [Google Scholar]
- Van Wart HE, Birkedal-Hansen H. The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc. Natl. Acad. Sci. USA. 1990;87:5578–5582. doi: 10.1073/pnas.87.14.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincenti MP. The matrix metalloproteinase (MMP) and tissue inhibitor of metalloproteinase (TIMP) genes. In: Clark IM, editor. Matrix Metalloproteinase Protocols. Totowa, NJ: Humana; 2001. pp. 121–148. [DOI] [PubMed] [Google Scholar]
- Voge lW, Gish GD, Alves F, Pawson T. The discoidin domain receptor tyrosine kinases are activated by collagen. Mol. Cell. 1997;1:13–23. doi: 10.1016/s1097-2765(00)80003-9. [DOI] [PubMed] [Google Scholar]
- Vu TH, Shipley JM, Bergers G, Berger JE, Helms JA, et al. MMP-9/gelatinase B is a key regulator of growth plate angiogenesis and apoptosis of hypertrophic chondrocytes. Cell. 1998;93:411–422. doi: 10.1016/s0092-8674(00)81169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K, Sato H, Kinoh H, Kajita M, Yamamoto H, Seiki M. Cloning of three Caenorhabditis elegans genes potentially encoding novel matrix metalloproteinases. Gene. 1998;211:57–62. doi: 10.1016/s0378-1119(98)00076-6. [DOI] [PubMed] [Google Scholar]
- Wang M, Qin X, Mudgett JS, Ferguson TA, Senior RM, Welgus HG. Matrix metalloproteinase deficiencies affect contact hypersensitivity: stromelysin-1 deficiency prevents the response and gelatinase B deficiency prolongs the response. Proc. Natl. Acad. Sci. USA. 1999;96:6885–6889. doi: 10.1073/pnas.96.12.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Jung J, Asahi M, Chwang W, Russo L, et al. Effects of matrix metalloproteinase-9 gene knock-out on morphological and motor outcomes after traumatic brain injury. J. Neurosci. 2000;20:7037–7042. doi: 10.1523/JNEUROSCI.20-18-07037.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Juttermann R, Soloway PD. TIMP-2 is required for efficient activation of proMMP-2 in vivo. J. Biol. Chem. 2000;275:26411–26415. doi: 10.1074/jbc.M001270200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werb Z. ECM and cell surface proteolysis: regulating cellular ecology. Cell. 1997;91:439–442. doi: 10.1016/s0092-8674(00)80429-8. [DOI] [PubMed] [Google Scholar]
- Werb Z, Yan Y. A cellular striptease act. Cell. 1998;282:1279–1280. doi: 10.1126/science.282.5392.1279. [DOI] [PubMed] [Google Scholar]
- Willenbrock F, Murphy G. Structure-function relationships in the tissue inhibitors of metalloproteinases. Am. J. Respir. Crit. Care Med. 1994;150:S165–S170. doi: 10.1164/ajrccm/150.6_Pt_2.S165. [DOI] [PubMed] [Google Scholar]
- Willenbucher RF, Aust DE, Chang CG, Zelman SJ, Ferrell LD, et al. Genomic instability is an early event during the progression pathway of ulcerative-colitis-related neoplasia. Am. J. Pathol. 1999;154:1825–1830. doi: 10.1016/S0002-9440(10)65438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson RA, Carr MD, Frenkiel TA, Feeney J, Freedman RB. Mapping the binding site for matrix metalloproteinase on the N-terminal domain of the tissue inhibitor of metalloproteinases-2 by NMR chemical shift perturbation. Biochemistry. 1997;36:13882–13889. doi: 10.1021/bi9712091. [DOI] [PubMed] [Google Scholar]
- Wilson CL, Heppner KJ, Labosky PA, Hogan BL, Matrisian LM. Intestinal tumorigenesis is suppressed in mice lacking the metalloproteinase matrilysin. Proc. Natl. Acad. Sci. USA. 1997;94:1402–1407. doi: 10.1073/pnas.94.4.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, Lopez-Boado YS. Regulation of intestinal α-defensin activation by the metalloproteinase matrilysin in innate host defense. Science. 1999;286:113–117. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- Windsor LJ, Grenett H, Birkedal-Hansen B, Bodden MK, Engler JA, Birkedal-Hansen H. Cell type-specific regulation of SL-1 and SL-2 genes. Induction of the SL-2 gene but not the SL-1 gene by human keratinocytes in response to cytokines and phorbolesters. J. Biol. Chem. 1993;268:17341–17347. [PubMed] [Google Scholar]
- Wingfield PT, Sax JK, Stahl SJ, Kaufman J, Palmer I. Biophysical and functional characterization of full-length, recombinant human tissue inhibitor of metalloproteinases-2 (TIMP-2) produced in Escherichia coli. Comparison of wild type and amino-terminal alanine appended variant with implications for the mechanism of TIMP functions. J. Biol. Chem. 1999;274:21362–21368. doi: 10.1074/jbc.274.30.21362. [DOI] [PubMed] [Google Scholar]
- Woessner JF, Nagase H. Matrix Metalloproteinases and TIMPs. New York: Oxford Univ. Press; 2000. [Google Scholar]
- Yamamoto H, Flannery ML, Kupriyanov S, Pearce J, McKercher SR, et al. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 1998;12:1315–1326. doi: 10.1101/gad.12.9.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Kyriakides TR, Bornstein P. Matricellular proteins as modulators of cell-matrix interactions: adhesive defect in thrombospondin 2-null fibroblasts is a consequence of increased levels of matrix metalloproteinase-2. Mol. Biol. Cell. 2000;11:3353–3364. doi: 10.1091/mbc.11.10.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Strickland DK, Bornstein P. Extracellular MMP2 levels are regulated by the low-density lipoprotein-related scavenger receptor and thrombospondin 2. J. Biol. Chem. 2001;276:8403–8408. doi: 10.1074/jbc.M008925200. [DOI] [PubMed] [Google Scholar]
- Ye S. Polymorphism in matrix metalloproteinase gene promoters: implication in regulation of gene expression and susceptibility of various diseases. Matrix Biol. 2000;19:623–629. doi: 10.1016/s0945-053x(00)00102-5. [DOI] [PubMed] [Google Scholar]
- Yoshiji H, Harris SR, Raso E, Gomez DE, Lindsay CK, et al. Mammary carcinoma cells over-expressing tissue inhibitor of metalloproteinases-1 show enhanced vascular endothelial growth factor expression. Int. J. Cancer. 1998;75:81–87. doi: 10.1002/(sici)1097-0215(19980105)75:1<81::aid-ijc13>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Yu Q, Stamenkovic I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-β and promotes tumor invasion and angiogenesis. Genes Dev. 2000;14:163–176. [PMC free article] [PubMed] [Google Scholar]
- Yu WH, Woessner JF. Heparan sulfate proteoglycans as extracellular docking molecules for matrilysin (matrix metalloproteinase 7) J. Biol. Chem. 2000;275:4183–4191. doi: 10.1074/jbc.275.6.4183. [DOI] [PubMed] [Google Scholar]
- Zhang B, Ye S, Herrmann SM, Eriksson P, de Maat M, et al. Functional polymorphism in the regulatory region of gelatinase B gene in relation to severity of coronary atherosclerosis. Circulation. 1999;99:1788–1794. doi: 10.1161/01.cir.99.14.1788. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Apte SS, Soininen R, Cao R, Baaklini GY, et al. Impaired endochondral ossification and angiogenesis in mice deficient in membrane-type matrix metalloproteinase I. Proc. Natl. Acad. Sci. USA. 2000;97:4052–4057. doi: 10.1073/pnas.060037197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker S, Drews M, Conner C, Foda HD, DeClerck YA, et al. Tissue inhibitor of metalloproteinase-2 (TIMP-2) binds to the catalytic domain of the cell surface receptor, membrane type 1-matrix metalloproteinase 1 (MT1-MMP) J. Biol. Chem. 1998;273:1216–1222. doi: 10.1074/jbc.273.2.1216. [DOI] [PubMed] [Google Scholar]