Abstract
Background and Purpose
Preliminary observations suggesting the presence of B and plasma cells and oligoclonality of immunoglobulin (Ig) G in cerebral cavernous malformations (CCMs) have motivated a systematic study correlating the infiltration of the immune cells with clinical activity and antigen-triggered immune response in surgically excised lesions.
Methods
Infiltration of plasma, B, T and HLA-DR expressing cells and macrophages within 23 excised CCMs was related to clinical activity. Relative amounts of Ig isotypes were determined. IgG clonality of mRNA from CCMs was assessed by spectratyping, cloning and sequencing.
Results
Infiltration of the immune cells ranged widely within CCM lesions and cells were generally co-expressed with each other. Immune cell infiltration did not associate with recent bleeding and lesion growth. Significantly more B lymphocytes in CCM lesions were associated with venous anomaly. More T cells were present in solitary lesions. More T cells and less macrophages were present in CCMs from younger subjects. IgG isotype was present in all CCM lesions. Most lesions also expressed IgM and IgA, with IgM predominance over IgA correlating with recent CCM growth. Oligoclonality was shown in IgG mRNA from CCMs, but not from peripheral blood lymphocytes, with only eight CDR3 sequences observed among 134 clones from two CCM lesions.
Conclusions
An antigen-directed oligoclonal IgG immune response is present within CCM lesions regardless of recent clinical activity. Apparent differences in immune response in younger patients and in lesions with recent growth will need confirmation in other series. The pathogenicity of oligoclonal immune response will require systematic hypothesis testing in recently available CCM murine models.
Keywords: Cerebral cavernous malformations, immunohistochemistry, lymphocytes, oligoclonality, immune response
Introduction
Cerebral cavernous malformation (CCM) is a prevalent vascular phenotype affecting up to 0.5% of the population, 1–5 predisposing patients to a lifetime risk of stroke and epilepsy. 4 The CCM lesions consist of clusters of dilated brittle capillaries that proliferate in the setting of repetitive hemorrhages. Lesions occur sporadically, in association with venous anomalies, 6 or may be inherited as an autosomal dominant trait, with three known gene loci, 7 but the genesis and proliferation of individual CCM lesions are largely unpredictable even among well-defined genotypes.8–11 There is no available therapy to prevent lesion genesis or clinical sequelae.
Immunoglobulin and other related genes are markedly unregulated within human CCM lesions. 12 Preliminary work suggests the infiltration of B and plasma cells within the lesions, and the presence of oligoclonal immunoglobulin (Ig) G in lesions but not in paired sera from the same patients. 13 A potential role of the immune response in CCMs has not been demonstrated previously, but would be compelling, given the unique antigenic milieu of CCM lesions with sequestered thrombi and leaky blood-brain barrier, and the numerous examples of immune modulation of angiogenesis in other disease states.
In this study we systematically assess the spectrum of immune cell infiltration and predominant Ig isotype in CCM lesions, in correlation with clinical behavior and other lesional features. And we aim to define the immune response in CCM lesions by assessing the distribution of lengths, cloning and sequences of complementary-determining regions 3 (CDR3) of the IgG variable heavy chain (IgGVH) genes in comparison to peripheral blood lymphocytes (PBLs) from the same patients.
Methods
Subjects
Patients were consecutive cases who had undergone CCM excision unrelated to this research. The diagnosis of CCM was established by typical histopathologic criteria in every case. The research was approved by the Institutional Review Board of Evanston Northwestern Healthcare, and the subjects gave informed consent.
Immunohistochemistry
Paraffin embedded sections of excised CCM lesions from 23 subjects were immunostained for the immune cells by a method previously described. 14 Anti-human primary antibodies included anti-CD138 (BC/B4), anti-CD20 (L26), anti-CD3 (PS1) and anti-HLA-DR (LN3) obtained from Biocare Medical, Concord, CA; anti-CD79α (JCB117) from Dako, Carpinteria, CA; anti-IgA, anti-IgG and anti-IgM from CellMarque Corporation, Rocklin, CA; and anti-CD45RO (UCHL-1) and anti-CD68 (KP-1) from Ventana Medical Systems, Inc., Tuscon, AZ. Tonsil was used as a positive control.
The number of clumps of cells/area and total cells/area for plasma cells (CD138), B lymphocytes (CD20, CD79α), T lymphocytes (CD3, CD45RO) and HLA-DR antigen presenting cells (HLA-DR) was determined in coded unidentified specimens.
For anti-CD68 (monocytes/macrophages), serial sections of stained and negative controls from up to ten representative fields for each specimen were captured in GrayScale using the Magnafire program at 10X magnification always using the white balance preset at 2800K and the exposure at 1.013 ms, avoiding dark stained areas in the negative control (i.e. blood filled caverns). The intensity for inverted images was acquired with an NIH Image J program. 15 The difference and ratio of intensity were calculated for each field on serial sections between anti-CD68 and negative control, and were averaged for each specimen.
The prevalence of IgG, IgA and IgM-stained lymphocytes was ranked blindly on a scale of 0–3, with 0 denoting a complete absence of Ig-stained cells and 1, 2 and 3 ranking the relative number of Ig-stained cells from lowest (1) to highest (3) in each specimen. Clinical and lesion features were categorized, blinded to immunohistochemical data (Table 1 online).
RNA isolation from CCM lesions and PBLs
CCM lesions were surgically excised from five subjects, rinsed with saline and snap-frozen in liquid nitrogen. The parts of the CCM lesions were homogenized separately with Zirconia/Silica beads. PBLs were isolated by a method published previously. 16 RNA was isolated from CCMs specimens and PBLs using TRIZOL® Reagent (Invitrogen, Carlsbad, CA) and purified using the RNeasy mini kit with DNase I digestion on columns (QIAGEN, Valenca, CA) following the manufacturer’s instructions.
cDNA synthesis and multiplex PCR
cDNA was generated from total RNA extracted from PBLs and CCM lesions using random hexamers, an IgG antisense C region primer CH5 conserved among all four IgG isotypes and Superscript III RT (Invitrogen) according to the manufacturer’s instructions. Table 2 (online) lists the primers used for cDNA synthesis, specific amplification of VH regions of IgG, and sequencing. 17, 18 Primary PCR was run with the conserved leader sequence primer for either VH family3 (VH3) or family 4 (VH4), the conserved C region primer for IgG (CHγ1) and Platinum® Taq DNA polymerase (Invitrogen). Cycling conditions included a single denaturation step at 94°C (5 min), followed by 34 cycles of 94°C (30 s), 55°C (30 s) and 72°C (1 min) ending with an incubation at 72°C (7 min). Nested PCR using VH3 framework region 3 or VH4 framework region 3 primer in conjunction with a pool of three J region primers was used to amplify the CDR3 regions of VH3 or VH4 family. Cycling conditions included a single denaturation step at 94°C (2 min), followed by 40 cycles of 94°C (1 min), 58°C (1min) and 72°C (2 min) ending with an incubation at 72°C (7 min).
Spectratyping and sequencing of CDR3 regions
Spectratyping and sequencing was conducted on an ABI 3730 Sequencer. The distribution of CDR3 sizes was assessed via a method previously reported 17 with modifications. GeneScan™500 LIZ™ Size Standard (Applied Biosystems) was run in the same well with the sample labeled with 6-FAM conjugated primers.
RNA from the parts of two CCM lesions shown to be oligoclonal (≤6 different sizes) for both VH3 and VH4 by spectratyping was sequenced. Nested PCR products for CDR3 were purified using the Wizard(R) SV Gel and PCR Clean-up System (Promega, Madison, WI) and ligated into pGEM®-T Easy Vector using T4 DNA ligase (Promega). Between 30 and 38 clones from the VH3 or VH4 family from CCM mRNA were selected. Recombinant plasmid DNA was purified with Wizard® Plus SV Minipreps DNA Purification System (Promega) and sequenced with T7 promoter primer. Alignment analysis of CDR3 sequences to every single clone obtained from VH3 and VH4 families and translation of amino acid was performed using Vector NTI 10.3.0 (Invitrogen).
Statistical analysis
Spearman correlations were computed and Wilcoxon two-sample test was applied for skew-distributed variables. Independent two-sample t test and linear regression were used in univariate analyses upon normal distributed variables and log-transformed values for skew-distributed variables. Multivariate analyses were conducted using mixed-effects models with McKeon F approximation via restricted maximum likelihood estimate. Two-sided P < 0.05 was considered significant, False Discovery Rate (FDR) adjusted P values from multivariate models were reported.
Results
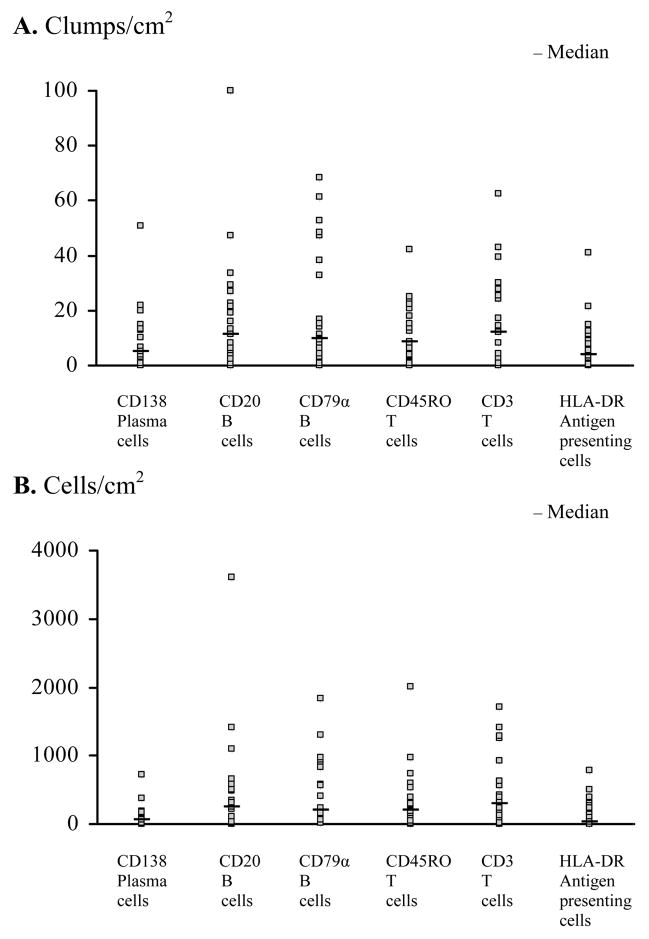
Immunostained B, T, plasma and HLA-DR antigen presenting cells were identified in nearly all specimens, predominantly in perivascular clumps around caverns and lesional vessels (Figure 1). There was a wide range of clumps/area and cells/area among lesions, with skew-distributions (Figure 2). Density of B cells/area and clumps/area were significantly correlated (Spearman Correlation Coefficients 0.441–0.788, P < 0.05) with T and plasma cells in the same lesions. Clumps/area of plasma cells correlated with HLA-DR antigen presenting cells (P = 0.0164). The mean difference and mean ratio for intensity of CD68 staining were inversely correlated with clumps/area of CD79α-stained B cells (Spearman Correlation Coefficients were -0.431, P = 0.0401 and -0.443, P = 0.0343, respectively).
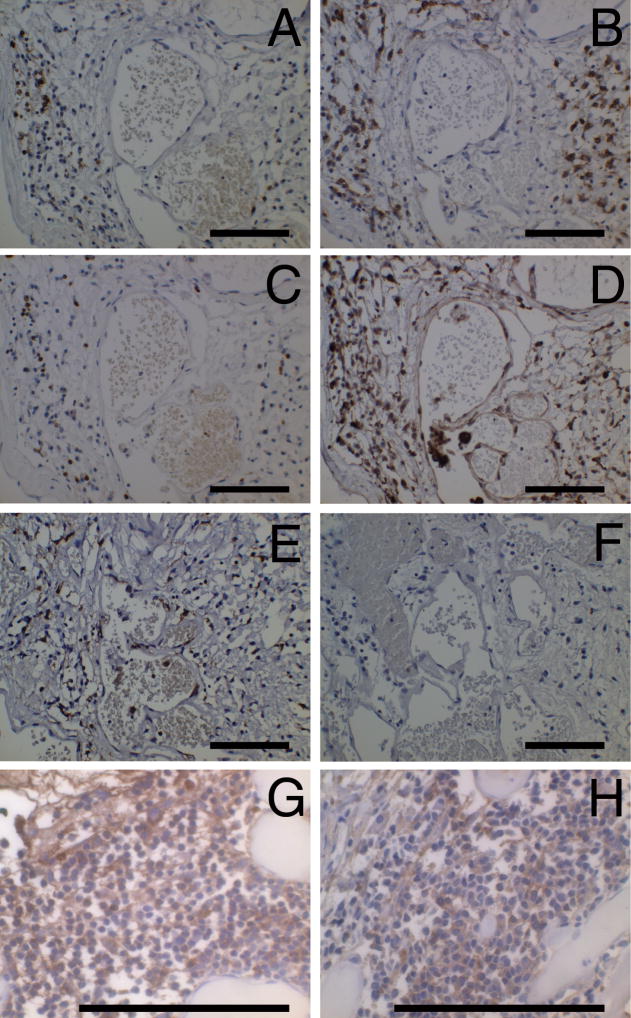
Figure 1.
Positive immune cells in CCM lesions. (A) B cells (CD20), (B) plasma cells (CD138), (C) T cells (CD3), (D) monocytes/macrophages (CD68), (E) antigen presenting cells (HLA-DR) and (F) negative control from the same area of the same specimen. (G) IgG positive cells and (H) IgM positive cells from two other specimens. Original magnification is 50 X (A–F) and 132 X (G, H). Scale bars are 100 μm.
Figure 2.
Spectrum of immunity in CCM lesions. Distribution of the number of (A) clumps/area and (B) cells/area for the indicated immune cells in CCM lesions.
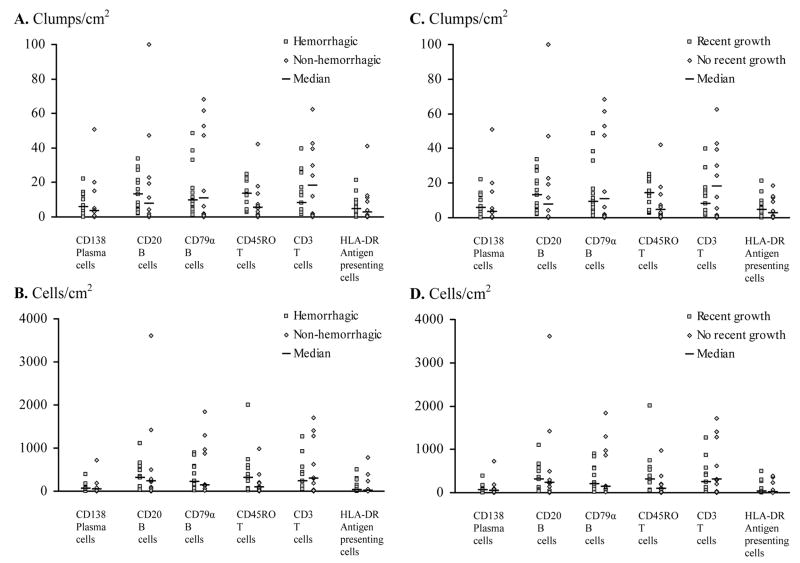
Recent bleeding and lesion growth did not correlate with density of any cell type (Figure 3). In univariate analysis, CCMs with associated venous anomaly had more clumps/sq cm (P = 0.0335) and cells/sq cm (P = 0.0408) of B lymphocytes stained with anti-CD20 antibody than in CCMs without this anomaly. Log transformation gave a similar result that CCMs with venous anomaly had more CD20- and CD79α-stained B cells/area (P = 0.018 and P = 0.024 respectively) and more clumps/area of CD20-stained cells (P = 0.026) in univariate analysis. A multivariate analysis with all six types of cells/area as dependent variables, further supported that CCMs with venous anomaly had more CD20- and CD79 α-stained B cells/area (FDR adjusted P = 0.0458). There were more clumps of CD3-stained T lymphocytes/area (in log value) in cases with single lesions than from subjects with multiple CCM lesions (P = 0.0071) in univariate analysis. Regression analysis revealed an inverse correlation between numbers of clumps of CD3 positive T cells/area (in log value) and the age at diagnosis (P = 0.0318) (regression coefficient = -0.0562, P = 0.0401). Regression analysis gave a positive correlation between the age at surgery and the mean difference and mean ratio for intensity of CD68-stained cells (regression coefficients were 0.6350, P = 0.0463 and 0.0069, P =0.0329, respectively), indicating greater CCM infiltration by those cells in older patients.
Figure 3.
CCM activity and immunity. Comparison of (A,C) clumps/area and (B,D) cells/area showing infiltration of the indicated immune cells in CCM lesions with and without (A,B) recent bleeding or (C,D) proliferation.
IgG isotype was expressed in lymphocytes in all lesions, and was predominant (IgG > IgA and/or IgM) in 15 of 23 lesions. IgM-stained cells were present in 18 of 23 lesions and IgA-stained cells were present in 19 of 23 lesions. IgM-stained cells predominated over IgA-stained cells in 15 of 23 lesions, and positively associated with recent growth of the CCM, where all ten CCMs with recent growth had more IgM- than IgA-stained cells and five of eight CCMs without recent growth had more IgM- than IgA-stained cells (P = 0.0065).
We subsequently analyzed B cell clonality by focusing on the CDR3 region in VH3 and VH4 families of IgGVH gene using the spectratyping, cloning and sequencing technology. In addition, to avoid IgG contamination from PBLs, we used paired PBLs from the same patient as an internal control. Our immunostaining results revealed B and plasma cells were predominantly identified in perivascular clumps. Therefore, to detect the clonality in the different lesional locations from the same patient, the CDR3 spectratyping from two different regions of the same patients was determined, except for one patient with a small tissue.
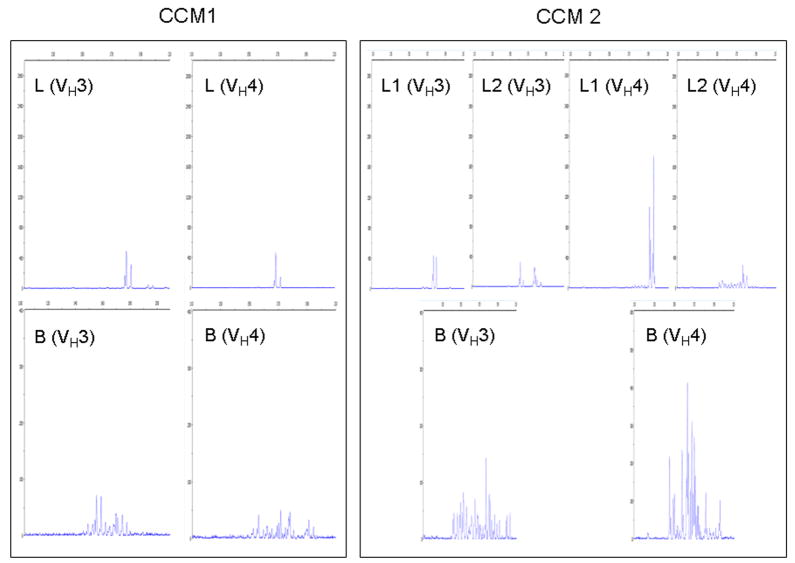
All extracts from paired PBL RNA showed a polyclonal CDR3 size distribution pattern with > 10 peaks, while spectratyping profiles of CDR3 of IgGVH from all five extracts of CCM lesions demonstrated oligoclonal patterns when the VH3 family was tested. CCM lesions showed only 2–5 sizes in the VH3 family. There was a significant difference in the number of peaks for CDR3 of IgGVH in the VH3 family between CCM lesions and PBLs (P < 0.0001). Three of four CCM lesions showed oligoclonal profiles in two different parts from the same lesions, while one lesion had an oligoclonality in one part and polyclonality in the other part. Likewise, similar polyclonal profiles of CDR3 size distribution from PBL RNA with almost similar number of peaks were observed in VH4 family. However, spectratyping of CDR3 mRNA from three of five CCM lesions gave an oligoclonal pattern, with 2–6 sizes in the VH4 family. One lesion had an oligoclonal pattern in two separate parts. Another lesion had oligoclonality in one part and polyclonality in the other part. The remaining two lesions had polyclonality in two separate parts.
There were no peaks with identical sizes for the VH3 and VH4 families in lesions from different CCM patients. There was also a significant difference in the number of peaks of CDR3 in the VH4 family between CCM lesions and PBLs (P < 0.0001). Representative spectratyping profiles for CCM lesions and PBLs from two different subjects are shown in Figure 4.
Figure 4.
Spectratyping of IgGVH CDR3 from CCM lesions. Dye-labeled runoff reaction of nested PCR products in VH3 and VH4 families of CCM lesions and PBLs from two CCM patients showing the oligoclonal CDR3 size usage in the lesions with the polyclonal CDR3 size distribution in the PBLs. (L: lesion; L1: lesion part 1; L2: lesion part 2 and B: PBLs; X axis: CDR3 size distribution, Y axis: Fluorescence intensity).
The presence of oligoclonal size distribution using spectratyping technique did not necessarily imply a predominant gene sequence in the individual fragment peak. Therefore, we cloned and sequenced the PCR products from two CCM patients with spectratyping oligoclonality. The cloning and sequencing results from Lesion 1 and Lesion 2 revealed eight different CDR3 sequences among 134 clones in VH3 and VH4 families. Identical sequences were found in 14 and 16 clones, respectively, from the VH3 family, and in all 30 clones from the VH4 family in Lesion 1 (Table 1). In Lesion 2, identical sequences were found in 3, 24 and 11 clones, respectively, from the VH3 family and in 12 and 24 clones, respectively, from the VH4 family (Table 1). For the VH3 family, the CDR3 sequences of the first two identical clones from Lesion 2 had only one base pair difference. Their putative amino acid sequence however is the same, indicating an identical protein function from the same clone (Table 1).
Table 1.
Identical nucleotide sequences of CDR3 in VH3 and VH4 families in two CCM lesions.
| Lesions | Families | No. of clones | Identical nucleotide sequence of clones | Genebank Acc # | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lesion 1 | VH3 | 14 | gat | gtc | cct | tat | tgg | agt | cga | cac | aac | tat | gcc | ccg | tac | tac | ttt | gac | ttc | FJ493233 | |||||
| Lesion 1 | VH3 | 16 | gac | cct | acc | cca | ttt | ggt | ggc | ctg | agg | tgg | gga | cgg | ccc | tat | ggt | tcg | ggg | agt | tct | aca | att | acg | FJ493234 |
| Lesion 1 | VH4 | 30 | gag | ggt | cgg | ggt | tat | agc | aac | aac | tgg | cac | ttt | gac | tac | FJ493235 | |||||||||
| Lesion 2 | VH3 | 3 | gac | agt | gtt | tgg | tac | gga | gac | tac | aga | tat | gac | gtc | tat | ttt | gac | tcc | FJ493236 | ||||||
| Lesion 2 | VH3 | 24 | gac | agt | gtt | tgg | tac | gga | gac | tac | aga | tat | gac | gtc | tac | ttt | gac | tcc | FJ493237 | ||||||
| Lesion 2 | VH3 | 11 | aga | tgg | ctg | aac | tac | cgt | gag | tcc | ctc | gtc | cgc | ggt | atg | gca | gcg | gcg | aca | tgt | atc | gac | gtc | FJ493238 | |
| Lesion 2 | VH4 | 12 | gcg | ccc | ccg | cag | ttt | atg | tat | tac | cgt | ggt | tcg | ggg | agt | tat | agg | FJ493239 | |||||||
| Lesion 2 | VH4 | 24 | gcg | ccc | tcg | cag | ttt | atg | tat | tac | cgt | ggt | tcg | ggg | agt | tat | agg | FJ493240 | |||||||
Discussion
A robust infiltration of the various immune cells in CCM lesions
Macrophages have long been recognized in CCM lesions, in presumed response to lesional hemorrhage. The presence of other immune cells in CCMs has only recently been described in preliminary observations 13, 19 but had not been previously examined systematically. Recent study of cerebral arteriovenous malformations revealed the presence of polymorphonuclear cells and macrophages, but no significant B-cell or plasma cell infiltration. 20 Our results confirm a robust infiltration of antibody producing B-lymphocytes and plasma cells in CCM lesions, with predominant IgG response, with occasional IgM and/or IgA expressing lymphocytes in several lesions. There was co-expression of T cells and antibody-producing cells in the same specimens.
Correlation with clinical activity and lesional features
Surgically removed CCMs may not reflect the biological spectrum of the majority of CCMs. However, this large cohort included cases with a variety of lesion phenotypes and clinical activity, allowing preliminary correlations with infiltration of immune cells and Ig isotypes (Table 1 online). Cell infiltration of the immune system studied, including antibody-producing cells, was not more prevalent in lesions with recent growth or hemorrhage. This finding is consistent with the hypothesis that cells of the immune system are a feature of CCM lesion phenotype regardless of recent clinical activity.
There was a greater T cell infiltration and less prevalent macrophages in younger patients. Although it was previously shown that the fraction of collagen IV expressing caverns in CCM lesions decreased with age, 14 an explanation for a possible association between immunity and collagen IV in CCMs is unknown. Other investigators found more macrophages/microglia were present inside and around a lesion caused by intracerebral hemorrhage in older than in younger mice.21 Microglial activation also has been shown to increase with age in human brains.22 In mice, increased prostaglandin E2 production by macrophages has been shown to be responsible for decreased T cell function with age. 23 These are consistent with the possibility that the immune response may shift from acquired to innate with age and growth of CCMs.
Associated venous anomaly, as determined by contrast enhanced MRI, has been frequently documented with sporadic solitary lesions and is rare in familial CCM cases. 9, 24 B cell infiltration was greater in CCMs associated with venous anomaly. There was a greater T cell infiltration in solitary lesions, than in cases with multiple/familial CCMs. CCM lesions with recent growth were significantly associated with greater IgM isotype response than other lesions, suggesting a more acute antigenic response in these lesions than the more chronic response associated with IgG isotype. However, selected surgical cases and their clinical and imaging features may not reflect accurate lesion age or maturity. These findings will require confirmation in other clinical series and in murine models of early and late stage CCM lesions. 25
Antigen-driven immune response in CCM lesions but not PBLs
An antigen-directed IgG immune response in multiple sclerosis and antigen-directed IgA immune response in the vascular wall in acute Kawasaki disease have been described.17, 26 Antigen-driven immune response in CCMs could also be intriguing. Previous studies have suggested that oligoclonal bands from IgG proteins are present within CCM lesions, but not in serum from the same patients. 13, 19 However, it had not been previously determined if this reflects antigen-directed IgG immune response or an inflammation marker. To answer this question, we employed the techniques of spectratyping, cloning and sequencing to detect CDR3 region of the IgGVH gene. This region is unique to each B cell clone and specific to antigen recognition, 27 and CDR3 structure, independent from the VH framework, and is sufficient to define the antigen binding specificity of an antibody. 28 In addition, the sequence of CDR3 gene offers the useful clonal signature of an individual B cell. 27 The high frequency occurrence of a specific CDR3 sequence illustrates the expansion of its corresponding cell clone. The alternative method of CDR3 size distribution analysis is a quicker approach to characterize the presence of B or plasma clonal expansion.29
Clonal expansion may be caused by either specific antigen stimulation or inflammation (bystander effect). In this investigation, oligoclonality in CCM lesions, but not in PBLs from the same subjects, has been confirmed by size distribution and sequencing of the gene for the CDR3 region of IgGVH gene. Spectratyping profiles for CDR3 regions of ≤ six different sizes indicating oligoclonality for IgG in CCM lesions are comparable to those for IgA in the vascular wall from subjects with acute Kawasaki Disease. 17 Only eight different IgG CDR3 nucleotide sequences (coding for only seven amino acid sequences) were obtained from 134 clones from two CCM lesions in this study. These are fewer than the proportion of unique IgG CDR3 sequences found in multiple sclerosis plaques by other investigators using similar methods, 16 suggesting that fewer epitope selections are involved in CCM than in other diseases. Our findings demonstrated predominant and reproducible CDR3 amplification in CCM lesions, but significantly different from PBLs, suggesting CCM lesional specific B cell expansions, instead of a random B cell response or a random bystander response.
Further studies should determine if the oligoclonal antibody response is targeted against a foreign or self-antigen in the milieu of CCM lesions. Iron breakdown products, glial or neuronal cells are hypothetical candidates, as are potential infectious agents. The relationship between the antigen-triggered immune response and leaky blood-brain barrier also needs to be determined. These hypotheses are amenable to investigation by determining the putative antigen trigger in immunoprecipitation experiments with lesional antibodies.30, 31
One CCM patient showed the mixed oligoclonality and polyclonality of CDR3 from the different lesional locations for VH3 family. Likewise, another CCM patient also demonstrated the same mixed clonality of CDR3 from the different lesional locations for VH4 family. This may be related to the different stages of the disease, and consequently B and/or plasma cells in the different lesional areas reflecting distinct clonal expansion. Or it is possible that those heterogeneous clonal characteristics originated from PBL contamination. This discrepancy could be solved by capturing single or clumped plasma cells in CCM lesions via a technique of laser captured microdissection. The other patients showed identical clonality from the different lesion locations, indicating homogeneous distribution of B and/or plasma cell clonal expansion. From our immunostaining results of B and plasma cells, those cells were mostly distributed in clumps. Therefore, it is possible that each or several clumps may originate from clonal expansion of a few B and or plasma cells in homogenous clonal lesions. Heterogeneous clonality of IgG may be linked to B and or plasma cells in different clumps in CCM lesions. We have initiated studies examining clonality in microdissected plasma cell clumps from CCM lesions.
Among VH families (VH1–7), VH3 and VH4 are the largest, accounting for about 75% of repertoire in adult peripheral blood B cells and in the vascular wall in acute Kawasaki Disease. 17, 32 Our results seem to demonstrate that there is more frequent usage of oligoclonality for VH3 family than VH4 family in CCM lesions. The VH3 family had been reported as the most frequently used VH family in the synovial tissue of patients with rheumatoid arthritis,33 and VH1 and VH4 families in multiple sclerosis brain.34, 35
Pathobiologic implications and future directions
Taken together, these results suggest an antigen-directed oligoclonal IgG immune response in CCM lesions regardless of recent clinical activity. The role of this immune response in the genesis and progression of CCM lesions remains speculative.
Features of the immune response will be examined in recently characterized murine models, comparing primordial and more mature lesions 25, 36, 37 and the reaction to experimental intracerebral blood.38 If CCM genesis is affected by immunosuppression or immunomodulation in existing animal models, this would suggest that an immune response may be part of the cause of CCMs. Artificially created recombinant antibodies will be used to locate putative antigens in human specimens. This may offer novel strategies at therapeutic manipulation to modify CCM disease.
Supplementary Material
Acknowledgments
This study was partially supported by the National Institutes of Health (NS052285 to I.A.A.)
Footnotes
Conflicts of Interest Disclosures
None
Publisher's Disclaimer: This is an un-copyedited author manuscript that was accepted for publication in Stroke, copyright The American Heart Association. This may not be duplicated or reproduced, other than for personal use or within the “Fair Use of Copyrighted Materials” (section 107, title 17, U.S. Code) without prior permission of the copyright owner, The American Heart Association. The final copyedited article, which is the version of record, can be found at Stroke <http://stroke.ahajournals.org>. The American Heart Association disclaims any responsibility or liability for errors or omissions in this version of the manuscript or in any version derived from it by the National Institutes of Health or other parties.
References
- 1.Del Curling O, Jr, Kelly DL, Jr, Elster AD, Craven TE. An analysis of the natural history of cavernous angiomas. J Neurosurg. 1991;75:702–708. doi: 10.3171/jns.1991.75.5.0702. [DOI] [PubMed] [Google Scholar]
- 2.Gunel M, Laurans MS, Shin D, DiLuna ML, Voorhees J, Choate K, Nelson-Williams C, Lifton RP. Krit1, a gene mutated in cerebral cavernous malformation, encodes a microtubule-associated protein. Proc Natl Acad Sci U S A. 2002;99:10677–10682. doi: 10.1073/pnas.122354499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Otten P, Pizzolato GP, Rilliet B, Berney J. 131 cases of cavernous angioma (cavernomas) of the cns, discovered by retrospective analysis of 24,535 autopsies. Neurochirurgie. 1989;35:82–83. 128–131. French. [PubMed] [Google Scholar]
- 4.Robinson JR, Awad IA, Little JR. Natural history of the cavernous angioma. J Neurosurg. 1991;75:709–714. doi: 10.3171/jns.1991.75.5.0709. [DOI] [PubMed] [Google Scholar]
- 5.Whitehead KJ, Plummer NW, Adams JA, Marchuk DA, Li DY. Ccm1 is required for arterial morphogenesis: Implications for the etiology of human cavernous malformations. Development. 2004;131:1437–1448. doi: 10.1242/dev.01036. [DOI] [PubMed] [Google Scholar]
- 6.Guclu B, Ozturk AK, Pricola KL, Seker A, Ozek M, Gunel M. Cerebral venous malformations have distinct genetic origin from cerebral cavernous malformations. Stroke. 2005;36:2479–2480. doi: 10.1161/01.STR.0000183616.99139.d3. [DOI] [PubMed] [Google Scholar]
- 7.Revencu N, Vikkula M. Cerebral cavernous malformation: New molecular and clinical insights. J Med Genet. 2006;43:716–721. doi: 10.1136/jmg.2006.041079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Battistini S, Rocchi R, Cerase A, Citterio A, Tassi L, Lando G, Patrosso MC, Galli R, Brunori P, Sgro DL, Pitillo G, Lo Russo G, Marocchi A, Penco S. Clinical, magnetic resonance imaging, and genetic study of 5 italian families with cerebral cavernous malformation. Arch Neurol. 2007;64:843–848. doi: 10.1001/archneur.64.6.843. [DOI] [PubMed] [Google Scholar]
- 9.Gault J, Sain S, Hu LJ, Awad IA. Spectrum of genotype and clinical manifestations in cerebral cavernous malformations. Neurosurgery. 2006;59:1278–1285. doi: 10.1227/01.NEU.0000249188.38409.03. [DOI] [PubMed] [Google Scholar]
- 10.Gault J, Sarin H, Awadallah NA, Shenkar R, Awad IA. Pathobiology of human cerebrovascular malformations: Basic mechanisms and clinical relevance. Neurosurgery. 2004;55:1–17. [PubMed] [Google Scholar]
- 11.Denier C, Labauge P, Bergametti F, Marchelli F, Riant F, Arnoult M, Maciazek J, Vicaut E, Brunereau L, Tournier-Lasserve E. Genotype-phenotype correlations in cerebral cavernous malformations patients. Ann Neurol. 2006;60:550–556. doi: 10.1002/ana.20947. [DOI] [PubMed] [Google Scholar]
- 12.Shenkar R, Elliott JP, Diener K, Gault J, Hu LJ, Cohrs RJ, Phang T, Hunter L, Breeze RE, Awad IA. Differential gene expression in human cerebrovascular malformations. Neurosurgery. 2003;52:465–478. doi: 10.1227/01.NEU.0000044131.03495.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi C, Shenkar R, Batjer HH, Check IJ, Awad IA. Oligoclonal immune response in cerebral cavernous malformations. Laboratory investigation. J Neurosurg. 2007;107:1023–1026. doi: 10.3171/JNS-07/11/1023. [DOI] [PubMed] [Google Scholar]
- 14.Shenkar R, Sarin H, Awadallah NA, Gault J, Kleinschmidt-DeMasters BK, Awad IA. Variations in structural protein expression and endothelial cell proliferation in relation to clinical manifestations of cerebral cavernous malformations. Neurosurgery. 2005;56:343–354. doi: 10.1227/01.neu.0000148903.11469.e9. [DOI] [PubMed] [Google Scholar]
- 15.NIH. Image j program website. http://rsb.Info.Nih.Gov/ij/
- 16.Owens GP, Burgoon MP, Anthony J, Kleinschmidt-DeMasters BK, Gilden DH. The immunoglobulin g heavy chain repertoire in multiple sclerosis plaques is distinct from the heavy chain repertoire in peripheral blood lymphocytes. Clin Immunol. 2001;98:258–263. doi: 10.1006/clim.2000.4967. [DOI] [PubMed] [Google Scholar]
- 17.Rowley AH, Shulman ST, Spike BT, Mask CA, Baker SC. Oligoclonal iga response in the vascular wall in acute kawasaki disease. J Immunol. 2001;166:1334–1343. doi: 10.4049/jimmunol.166.2.1334. [DOI] [PubMed] [Google Scholar]
- 18.Owens GP, Ritchie AM, Burgoon MP, Williamson RA, Corboy JR, Gilden DH. Single-cell repertoire analysis demonstrates that clonal expansion is a prominent feature of the b cell response in multiple sclerosis cerebrospinal fluid. J Immunol. 2003;171:2725–2733. doi: 10.4049/jimmunol.171.5.2725. [DOI] [PubMed] [Google Scholar]
- 19.Shenkar R, Shi C, Check IJ, Lipton HL, Awad IA. Concepts and hypotheses: Inflammatory hypothesis in the pathogenesis of cerebral cavernous malformations. Neurosurgery. 2007;61:693–703. doi: 10.1227/01.NEU.0000298897.38979.07. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Zhu W, Bollen AW, Lawton MT, Barbaro NM, Dowd CF, Hashimoto T, Yang G, Young WL. Evidence for inflammatory cell involvement in brain arteriovenous malformations. Neurosurgery. 2008;62:1340–50. doi: 10.1227/01.neu.0000333306.64683.b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JC, Cho GS, Choi BO, Kim HC, Kim YS, Kim WK. Intracerebral hemorrhage-induced brain injury is aggravated in senescence-accelerated prone mice. Stroke. 2006;37:216–222. doi: 10.1161/01.STR.0000195151.46926.7b. [DOI] [PubMed] [Google Scholar]
- 22.Streit WJ, Sparks DL. Activation of microglia in the brains of humans with heart disease and hypercholesterolemic rabbits. J Mol Med. 1997;75:130–138. doi: 10.1007/s001090050097. [DOI] [PubMed] [Google Scholar]
- 23.Beharka AA, Wu D, Han SN, Meydani SN. Macrophage prostaglandin production contributes to the age-associated decrease in t cell function which is reversed by the dietary antioxidant vitamin e. Mech Ageing Dev. 1997;93:59–77. doi: 10.1016/s0047-6374(96)01819-2. [DOI] [PubMed] [Google Scholar]
- 24.Abdulrauf SI, Kaynar MY, Awad IA. A comparison of the clinical profile of cavernous malformations with and without associated venous malformations. Neurosurgery. 1999;44:41–47. doi: 10.1097/00006123-199901000-00020. [DOI] [PubMed] [Google Scholar]
- 25.Shenkar R, Venkatasubramanian PN, Wyrwicz AM, Zhao J, Shi C, Akers A, Marchuk DA, Awad IA. Advanced magnetic resonance imaging of cerebral cavernous malformations: Part II. Imaging of lesions in murine models. Neurosurgery. 2008;63:790–798. doi: 10.1227/01.NEU.0000315862.24920.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bar-Or A. The immunology of multiple sclerosis. Semin Neurol. 2008;28:29–45. doi: 10.1055/s-2007-1019124. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz RS. Shattuck lecture: Diversity of the immune repertoire and immunoregulation. N Engl J Med. 2003;348:1017–1026. doi: 10.1056/NEJMsa022766. [DOI] [PubMed] [Google Scholar]
- 28.Seidl KJ, Wilshire JA, MacKenzie JD, Kantor AB, Herzenberg LA. Predominant vh genes expressed in innate antibodies are associated with distinctive antigen-binding sites. Proc Natl Acad Sci U S A. 1999;96:2262–2267. doi: 10.1073/pnas.96.5.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miqueu P, Guillet M, Degauque N, Dore JC, Soulillou JP, Brouard S. Statistical analysis of cdr3 length distributions for the assessment of t and b cell repertoire biases. Mol Immunol. 2007;44:1057–1064. doi: 10.1016/j.molimm.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 30.Williamson RA, Burgoon MP, Owens GP, Ghausi O, Leclerc E, Firme L, Carlson S, Corboy J, Parren PW, Sanna PP, Gilden DH, Burton DR. Anti-DNA antibodies are a major component of the intrathecal b cell response in multiple sclerosis. Proc Natl Acad Sci U S A. 2001;98:1793–1798. doi: 10.1073/pnas.031567598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rowley AH, Baker SC, Shulman ST, Garcia FL, Guzman-Cottrill JA, Chou P, Terai M, Kawasaki T, Kalelkar MB, Crawford SE. Detection of antigen in bronchial epithelium and macrophages in acute kawasaki disease by use of synthetic antibody. J Infect Dis. 2004;190:856–865. doi: 10.1086/422648. [DOI] [PubMed] [Google Scholar]
- 32.Brezinschek HP, Brezinschek RI, Lipsky PE. Analysis of the heavy chain repertoire of human peripheral b cells using single-cell polymerase chain reaction. J Immunol. 1995;155:190–202. [PubMed] [Google Scholar]
- 33.Kim HJ, Berek C. B cells in rheumatoid arthritis. Arthritis Res. 2000;2:126–131. doi: 10.1186/ar77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baranzini SE, Jeong MC, Butunoi C, Murray RS, Bernard CC, Oksenberg JR. B cell repertoire diversity and clonal expansion in multiple sclerosis brain lesions. J Immunol. 1999;163:5133–5144. [PubMed] [Google Scholar]
- 35.Owens GP, Kraus H, Burgoon MP, Smith-Jensen T, Devlin ME, Gilden DH. Restricted use of vh4 germline segments in an acute multiple sclerosis brain. Ann Neurol. 1998;43:236–243. doi: 10.1002/ana.410430214. [DOI] [PubMed] [Google Scholar]
- 36.Plummer NW, Gallione CJ, Srinivasan S, Zawistowski JS, Louis DN, Marchuk DA. Loss of p53 sensitizes mice with a mutation in ccm1 (krit1) to development of cerebral vascular malformations. Am J Pathol. 2004;165:1509–1518. doi: 10.1016/S0002-9440(10)63409-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plummer NW, Squire TL, Srinivasan S, Huang E, Zawistowski JS, Matsunami H, Hale LP, Marchuk DA. Neuronal expression of the ccm2 gene in a new mouse model of cerebral cavernous malformations. Mamm Genome. 2006;17:119–128. doi: 10.1007/s00335-005-0098-8. [DOI] [PubMed] [Google Scholar]
- 38.Xue M, Del Bigio MR. Intracerebral injection of autologous whole blood in rats: Time course of inflammation and cell death. Neurosci Lett. 2000;283:230–232. doi: 10.1016/s0304-3940(00)00971-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.