Abstract
Activation of the tumor suppressor p53 by stress and damage stimuli often correlates with induction of stress kinases, Jun-NH2 kinase (JNK). As JNK association with p53 plays an important role in p53 stability, in the present study we have elucidated the relationship between the JNK-signaling pathway and p53 stability and activity. Expression of a constitutively active form of JNKK upstream kinase, mitogen-activated protein kinase kinase kinase (ΔMEKK1), increased the level of the exogenously transfected form of p53 in p53 null (10.1) cells as well as of endogenous p53 in MCF7 breast cancer cells. Increased p53 level by forced expression of ΔMEKK1 coincided with a decrease in p53 ubiquitination in vivo and with prolonged p53 half-life. Computerized modeling of the JNK-binding site (amino acids 97–116; p7 region) enabled us to design mutations of exposed residues within this region. Respective mutations (p53101-5-8) and deletion (p53Δp7) forms of p53 did not exhibit the same increase in p53 levels upon ΔMEKK1 expression. In vitro phosphorylation of p53 by JNK abolished Mdm2 binding and targeting of p53 ubiquitination. Similarly, ΔMEKK1 expression increased p53 phosphorylation by immunopurified JNK and dissociated p53–Mdm2 complexes. Transcriptional activity of p53, as measured via mdm2 promoter-driven luciferase, exhibited a substantial increase in ΔMEKK1-expressing cells. Cotransfection of p53 and ΔMEKK1 into p53 null cells potentiated p53-dependent apoptosis, suggesting that MEKK1 effectors contribute to the ability of p53 to mediate programmed cell death. Our results point to the role of MEKK1-JNK signaling in p53 stability, transcriptional activities, and apoptotic capacity as part of the cellular response to stress.
Being a key target for inactivation in human cancer, the p53 tumor suppressor gene has been implicated in cell cycle control, DNA repair, replicative senescence, and programmed cell death (reviewed in refs. 1 and 2). The ability of p53 to elicit diverse regulatory functions is likely to depend on its phosphorylation pattern which is conformation dependent (3). p53 phosphorylation is mediated by several cellular kinases including casein kinase I, casein kinase II, protein kinase A, CDK7, DNA-activated protein kinase, and Jun-NH2 kinase (JNK) (3–8). Although kinases that efficiently phosphorylate p53 both in vitro and in vivo were successfully identified, the direct relationship between p53 phosphorylation by a specific signaling cascade and its ability to elicit its biologic activities in response to DNA damage is not fully understood. Among protein kinases that are expected to produce stress-activated phosphorylation of p53 are JNK and DNA-activated protein kinase, which reportedly phosphorylate residues within the amino terminal domain of p53 (4–6). DNA-activated protein kinase mediated phosphorylation of Ser-15, and Ser-37 has been shown to contribute to p53 accumulation (9). Ser-15 was identified as one of the major sites on p53 that is phosphorylated by cellular stress (10). Because the p53 response to stress and damage is preserved in cells from severe combined immunodeficient mice (10–12), we examined the role of JNK in this response.
JNKs are a family of stress kinases induced by change in redox potential, heat shock, osmotic shock, UV irradiation, and inflammatory cytokines (13–16). JNK activity requires mitogen-activated protein kinases kinase (MEKK) 1–4 which phosphorylates MKK4/7. MKK4/7, in turn, phosphorylates JNK on residues 183 and 185 (17–20). Activated JNK phosphorylates its substrates, c-Jun, ATF2, ELK1, and p53 (3, 13–14, 21). Although JNK activities in response to stress and damage have been implicated in cell growth control as well as in apoptosis (22–23), little is known about JNK effectors that elicit apoptosis.
JNK exerts an important regulatory function through its ability to target the ubiquitination of its unphosphorylated associated proteins (e.g., c-Jun, ATF2) in normal-growing cells. Upon its activation in response to stress, JNK mediates phosphorylation, which protects its substrates from ubiquitination and degradation (24–26). Similar to c-Jun, wild-type p53 has a short half-life, which is prolonged in response to stress or DNA damage (27). Key contributors to p53 half-life are its associated proteins Mdm2 and JNK (28, 29). Common to the ability of both Mdm2 and JNK to associate with p53 is their requirement for a specific conformation of the p53 protein (3, 9), which depends on its phosphorylation status and association with other cellular proteins. Changes in p53 conformation in response to stress-mediated phosphorylation of p53 have been documented (9, 30).
JNK association with p53 requires residues 97–155 of p53 as concluded from the observation that p53 deleted of the first 97 amino acids binds well to JNK, whereas p53 that lacks the first 155 amino acids fails to associate with JNK (3). Further analysis identified amino acids 97–116 as the primary p53 sequence domain required for JNK association (29). p53 deleted of amino acids 97–116 no longer associates with JNK (29). Identifying p53 as a JNK substrate led us to examine the biologic significance of p53 phosphorylation by JNK. Herein, we provide evidence that MEKK1/JNK signaling increases p53 stability and transcriptional activation and that MEKK1/JNK potentiates the ability of p53 to initiate programmed cell death. The mechanisms underlying the effect of MEKK1/JNK signaling on p53 stability and functions are discussed.
MATERIALS AND METHODS
Cells.
10.1 are p53 null mouse fibroblast cells (31), 293T are adenovirus-transformed human kidney cells that express the simian virus 40 large T antigen (32). Both 10.1 and 293T cells were maintained in DMEM (GIBCO) supplemented with 10% fetal bovine serum (GIBCO) and antibiotics in 5% CO2. MCF7 is a human breast cancer cell line maintained in RPMI 1640 supplemented with 10% fetal bovine serum and antibiotics.
Plasmids.
To generate a p53 expression vector that is His tagged, the cDNA of rat p53 was cloned into a pcDNA3 vector by PCR using a 5′ primer that encodes the N-terminal hexahistidine tag, thus generating hisp53 fusion protein. To generate p53 deleted of the p7 sequence, the rat cDNA of p53 (pCMV-hisp53) amino acids 95–114 (corresponding to amino acids 97–116 of human p53 which constitute the p7 domain) were deleted by using site-directed mutagenesis (Quick Change, Stratagene) resulting in the p53Dp7 construct. p53101–5-8 mutated within the p7 domain with substitutions of Y101A, Y105A, H108E (design based on computer modeling) as well as the p53Δp6 mutant lacking amino acids 68–91 were constructed using the same approach. In all cases, the integrity of the constructs was verified by dideoxy sequencing and immunoblotting. Additional expression vectors used include: a constitutively active ΔMEKK1, which lacks amino acids 1–351 and its catalytically inactive (K432M) counterpart TR-MEKK (18); JNKK wild type and its catalytically inactive mutant (19), Mdm2 expression vector and luciferase reporter construct under control of an Mdm2 promoter (33); hemagglutinin- (HA) tagged JNK2 expression vector (34); and wild-type human p53 and a p53 mutant lacking amino acids 13–52 (28).
Transfections.
To minimize stress caused by transfection, we have utilized Lipofection (FuGENE, Boehringer Mannheim). In all cases, cells were at 70% confluency at time of transfection. The total amount of plasmid DNA was kept constant by adding respective amounts of empty vector plasmids to transfection mixtures. Eighteen to 24 h after transfection cells were subjected to the respective treatment as indicated in Results. To ensure efficiency of transfection, we have used β-galactosidase assays. To monitor expression of the transfected constructs, respective Western blots were preformed as indicated in Figs. 1–5.
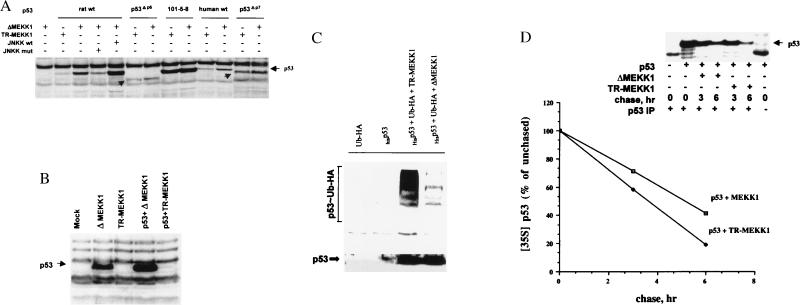
Figure 1.
(A) MEKK1 transfection leads to p53 accumulation. 10.1 cells were cotransfected with pCMV-β-galactosidase (100 ng) and various p53-expressing plasmids (1 μg), MEKK or TR-MEKK (30 ng), and JNKK [wild type (wt) or mutant, 100 ng) as indicated. The total amount of DNA was normalized to 1.23 μg/30-mm plate with pcDNA3 and pSRα3 plasmids. Immunoblot depicts the level of p53 proteins in the lysates prepared 24 h after transfection. Protein loading was normalized per β-galactosidase activity. (B) MEKK1 transfection leads to accumulation of endogenous p53 in MCF7 cells. p53 null cells were transfected with the indicated constructs and p53 expression level was determined by immunoblots performed on proteins prepared 24 h after transfections. (C) MEKK1 transfection inhibits ubiquitination of p53 in vivo. 10.1 cells were transfected as indicated. Hexahistidine-tagged p53 proteins were purified on nickel resins, separated by SDS-PAGE, and transferred onto nitrocellulose membrane. The membrane was cut above the 55-kDa marker. The lower and upper parts were probed with anti-p53 and anti-HA monoclonal antibodies, respectively. (D) MEKK1 transfection prolongs p53 half-life. 10.1 cells were transfected with indicated constructs, labeled with [35S]methionine and chased with cold methionine for 0–6 h as described in Materials and Methods. The results of pulse–chase were analyzed by autoradiography (inset) and quantified by using a Bio-Rad GS363 PhosphorImager (depicted in the graph, which represents values with error of <15%).
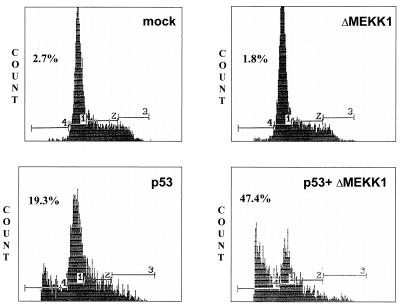
Figure 5.
ΔMEKK1 stimulates p53-mediated apoptosis in 10.1 cells. 10.1 cells were transfected as indicated. After 40 h, cells were stained with propidium iodide and analyzed by FACS. The distribution of stained cells and the percentage of apoptotic cells (<2n) is shown.
Antibodies.
Antibodies against p53 and HA epitope were purchased (Ab-1, Oncogene Science, and HA11, BabCo, respectively). Antibodies to JNK (clone 666) were obtained from PharMingen and used according to the manufacturer’s recommendation. Anti-Mdm2 monoclonal antibody 2A10 was a gift from A. Levine (Princeton University, Princeton, NJ). Immunoblots were performed on 100 μg of whole-cell extract, whereas immunoprecipitations were performed on 1 mg of whole-cell extract. Antibody recognition was detected by using enhanced chemiluminescence (Amersham). Proteasome inhibitor lactacystin was purchased from Calbiochem.
Expression and Purification of Proteins.
Human p53 cDNA has been cloned into pET15b vector (Novagen) between NdeI and BamHI sites. Recombinant hisp53 proteins were expressed in BL21 bacterial strain and then purified and refolded as described (29). JNK was purified from 600 mg of protein extract prepared from UV-irradiated (60J/m2) BALB/3T3 cells 45 min after the treatment as described (3). Mdm2 was prepared from Sf9 cells infected with baculovirus-expressing human mdm2 cDNA. Sf9 lysates were purified on a 2A10 mdm2 antibody affinity column. The purity of Mdm2 and JNK was confirmed as a single band seen on silver-stained gels.
Immunokinase Reaction.
HA-tagged JNK2 was immunopurified from the lysates of 293T cells (pretreated with 40 J/m2 of UV irradiation or cotransfected with MEKK constructs) using antibodies to HA and protein A/G beads. Bound material was incubated with bacterially expressed hisp53 (29) in the presence of [γ-32P]ATP (50 cpm/fmol; Amersham) and 50 μM ATP in kinase buffer for 15 min at 30°C (15). The samples were boiled with Laemmli buffer, separated by 10% SDS-PAGE, and electrotransferred onto poly(vinylidene difluoride) membrane. The amount of JNK and p53 was analyzed by immunoblotting with respective antibodies, while p53 phosphorylation was assessed by autoradiography.
Solid-Phase Kinase Reaction.
Solid-phase kinase reaction with bacterially expressed hisp53 (2 μg) and JNK purified from UV-treated mouse fibroblasts was performed as described elsewhere (3).
In Vivo Ubiquitination.
Amino-terminal His-tagged p53 (4 μg) was cotransfected into mouse fibroblasts with ubiquitin HA expression vector (3 μg; ref. 26) and MEKK1 (0.25 μg) as indicated. Cells were lysed 24 h later with 6 M guanidinium HCl and the His-tagged proteins were purified by nickel resins as described (26), separated by 8% SDS-PAGE, and transferred to Hybond C nitrocellulose filter (Amersham). The filter was cut just above the 55-kDa protein molecular mass marker (Promega) and its lower part was analyzed by immunoblots using anti-p53 PAb421 antibody to identify nickel-purified his-p53. The upper part of the filter was probed with anti-HA antibody (BabCo) allowing detection of a smear, which represents slower-migrating ubiquitin HA conjugates.
In Vitro Ubiquitination.
The ability of Mdm2 to target p53 ubiquitination was assessed in an in vitro ubiquitination assay as previously described (25).
In Vivo Degradation.
10.1 cells were cotransfected with p53 and wild-type or mutant MEKK1 constructs. After 24 h, cells were metabolically labeled with [35S]methionine for 10 min and chased with 2 mM of unlabeled methionine for the indicated time periods. p53 was immunopurified from the extracts, immunoprecipitates were separated by SDS-PAGE, and analyzed via autoradiography.
Transcriptional Analysis.
Luciferase assays were performed using a kit (Promega) on whole-cell extract prepared from cells transfected with the mdm2 minimal promoter-driven luciferase gene (33).
Apoptosis Analysis.
Analysis was performed using a fluorescence cell sorter (FACS) on 10.1 cells stained with propidium iodide 40 h after transfection with p53 (1 μg), MEKK1 (20 ng), or empty vectors (pcDNA3 or pSRα3) as indicated.
Conformational Energy Calculations.
The coordinates for wild-type p53 and a mutant (R249T) (35) were subjected to energy minimizations as described previously (3). Electrostatically driven Monte Carlo and molecular dynamics calculations (36, 37) were performed on both structures as described previously (3). The convergence of electrostatically driven Monte Carlo runs was judged by convergence of the energy to a minimal value; runs were terminated where the backbone rms deviations of new structures with low energies were >2 Å from the energy-minimized x-ray structure and/or where the energy increased by >10 kcal/mol from the minimum energy value (3).
RESULTS
Computer Modeling Identifies Residues Important for JNK-p53 Association.
The results of the electrostatically driven Monte Carlo calculations are identical to those obtained in our previous computational study (3). Computations were performed for wild-type and the two mutant forms of p53 containing R249W and R249T. In both, the p53 domain containing residues 97–121 undergoes a major conformational change when the average structures of the two mutant forms are compared with that of the wild-type protein (3). As the 97–121 region was found to contain the JNK-binding site (3, 29), we aimed at identifying possible residues within this region that are important for association with JNK. Superimposing the average structures for the JNK-binding domain of the mutant and wild-type proteins identified Tyr-103, Tyr-107, and Arg-110, which were completely superimposable, while the conformations of all other residues deviated significantly. Therefore, these three superimposable residues were considered to be important in the interaction between p53 and JNK. Accordingly, a rat mutant form of p53 that contained Ala residues in place of Tyr-103 and Tyr-105 and Glu in place of His-108 (as per alignment of rat and human p53 protein sequences) was prepared. This mutant form of p53 was utilized in our subsequent studies as described below.
ΔMEKK1 Increases the Level of p53 Expression.
To determine the role of JNK in p53 stability and activity, plasmids expressing modulators of the JNK-signaling pathway were cotransfected with different p53 constructs into p53 null cells. Forced expression of JNKK upstream kinase-ΔMEKK1, led to a substantial increase in the level of both rat and human wild-type forms of p53 proteins (Fig. 1A). Conversely, a dominant negative JNKK construct attenuated the effect of ΔMEKK1, whereas a catalytically inactive MEKK1 (TR-MEKK1) failed to increase p53 expression levels (Fig. 1A). ΔMEKK1 overexpression also increased the amount of p53 from which its proline-rich region had been detected (p53Dp6). However, p53 deleted of (p53Dp7) or mutated within the JNK association site (p53101–5-8) exhibited only a marginal increase in response to ΔMEKK1 expression (Fig. 1A). Previous studies have demonstrated that p53Dp7 can no longer associate with JNK in vivo (29). That both p53Dp7 and p53101–5-8, which were designed to disrupt JNK association, could no longer respond to ΔMEKK1 suggests that JNK transduces a signal from MEKK1 to p53.
Using MCF7 breast cancer cells, we next determined whether ΔMEKK1 could also affect the level of endogenous p53. As shown in Fig. 1B, forced expression of ΔMEKK1 led to an increased level of endogenous p53 expression in MCF7 cells. Cotransfection of p53 with ΔMEKK1 caused further accumulation of p53, further illustrating that the results obtained with p53 null cells could be produced in a different cell system (Fig. 1B).
Northern blot analysis showed no difference in the steady-state level of p53 mRNA in the cells transfected either with ΔMEKK1 or TR-MEKK1 constructs (data not shown).
ΔMEKK1 Decreases p53 Ubiquitination and Prolongs Its Half-Life.
To explore how MEKK1/JNK phosphorylation of p53 contributes to its elevated expression level, we have determined possible changes in p53 ubiquitination and degradation. Cotransfection of ΔMEKK1 and p53 into p53 null cells caused a marked decrease in the amount of polyubiquitin chains attached to p53, as assessed by in vivo ubiquitination assay (Fig. 1C). To establish the relationship between the ΔMEKK1 effect on p53 ubiquitination and p53 degradation, we performed pulse–chase experiments. p53 half-life was prolonged to 5.5 h in cells that were cotransfected with ΔMEKK1 compared with a half-life of ≈3 h in TR-MEKK1 transfected cells (Fig. 1D). Altogether, these data suggest that JNK signaling protect p53 from ubiquitination, resulting in stabilization and accumulation of p53. These findings are in line with our previous observations that JNK-mediated phosphorylation of c-Jun and ATF2 protects them from ubiquitination (24, 25).
Since JNK is capable of phosphorylating p53 in vitro (3, 6), we next examined whether MEKK1 signaling would increase the efficiency of this phosphorylation. Using JNK immunokinase reactions, JNK immunoprecipitated from ΔMEKK1-expressing cells exhibited a noticeable increase in the ability to phosphorylate p53. ΔMEKK1 effect on JNK phosphorylation of p53 was comparable to that seen with the kinase purified from UV-irradiated cells (Fig. 2A). The background signal in the immunokinase reaction lacking p53 could be attributed to autophosphorylation of 54-kDa JNK2.
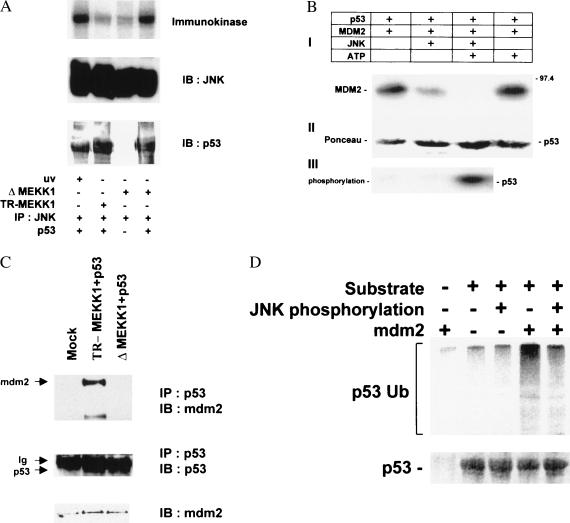
Figure 2.
(A) P53 is phosphorylated by JNK in the immunokinase assay. Soluble bacterially expressed p53 was subjected to phosphorylation by JNK immunopurified from 293T cells treated with UV irradiation or cotransfected with either TR-MEKK1 or ΔMEKK1 constructs as indicated. (Top) Autoradiograph. Immunoblots depicting the amounts of kinase (Middle) and substrate (Bottom) in the samples. (B) mdm2 binding to p53 is inhibited by JNK. Bead-bound human hisp53 was incubated with purified Mdm2 and JNK, as indicated, in the presence of [γ-32P]ATP (50 cpm/fmol; Amersham) and 50 μM ATP in kinase buffer for 15 min at 30°C. P53-associated proteins were extensively washed before analysis via immunoblots with antibodies to mdm2 (2A10) was performed (I). (II) Ponceau S stain of the same membrane revealing the amounts of hisp53 used throughout this experiment. (III) Autoradiograph depicting p53 phosphorylation. (C) In vivo association between p53 and Mdm2. p53 null cells (10.1) were transfected with wild-type p53 and either TR-MEKK1 or ΔMEKK1 constructs. Protein extracts were prepared 36 h after transfection and 8 h after treatment with lactacystin (5 μM). p53 proteins were immunoprecipitated with antibody to p53 and immunoprecipitated material was subjected to immunoblot with antibodies to Mdm2 (Top). (Middle) The same blot reprobed with p53 antibody. (Bottom) The level of Mdm2 expressed in whole-cell extracts. (D) Phosphorylation of p53 by JNK abrogates Mdm2-mediated targeting for p53 ubiquitination in vitro. Bead-bound hisp53 was phosphorylated by JNK, incubated with Mdm2 (as indicated), washed, and subjected to in vitro ubiquitination reaction in the presence of HA-tagged ubiquitin as described in Materials and Methods. (Top) Immunoblot with anti-HA antibody. (Bottom) The input of p53 detected by Ponceau S staining.
MEKK1 Affects mdm2 Association with p53.
We previously showed that the conformation of p53 strongly affects JNK phosphorylation (3). Concurrently, we anticipate that p53 phosphorylated by JNK may undergo specific conformational changes, which would protect it from ubiquitination and increase its stability. An important determinant in p53 stability is Mdm2, which has been implicated in its degradation (28, 38, 39) through association with p53. Mdm2 targets p53 ubiquitination (ref. 39 and Fig. 2C), and its association with p53 is abrogated by stress stimuli (9). The changes seen in p53 ubiquitination and stability after MEKK1 expression (Fig. 1) led us to investigate MEKK1 effects on Mdm2-p53 association. First, we assessed whether JNK phosphorylation affects the association between p53 and mdm2 in vitro. Although JNK failed to phosphorylate Mdm2 in vitro (data not shown), phosphorylation of p53 by JNK abrogated the association of p53 with Mdm2 (Fig. 2B). Similarly, JNK phosphorylation of p53 inhibited Mdm2-mediated targeting of p53 for ubiquitination in vitro (Fig. 2C).
We demonstrated that activating JNK by overexpression of ΔMEKK1 inhibits p53 ubiquitination in vivo (Fig. 1C). To learn whether ΔMEKK1 overexpression alters p53–Mdm2 complex in vivo, 10.1 cells were cotransfected with p53 and ΔMEKK1 or its inactive counterpart TR-MEKK1. Cells were treated with the proteasome inhibitor lactacystin to enrich the otherwise unstable p53–Mdm2 complex. p53 was immunoprecipitated and the presence of Mdm2 in the immunoprecipitate was assessed by immunoblot. This analysis revealed that Mdm2 association with p53 in vivo is impaired by ΔMEKK1 expression (Fig. 2D). These data suggest that JNK signaling abrogates the association of p53 with mdm2 in vivo.
Further support for JNK’s effect on Mdm2’s ability to decrease p53 stability comes from the use of p53/mdm2 double null cells. Although cotransfection of p53 and mdm2 resulted in a marked decrease in p53 level (as compared with p53 level in cells transfected with p53), forced expression of ΔMEKK1 abolished Mdm2’s ability to degrade p53 (Fig. 3A).
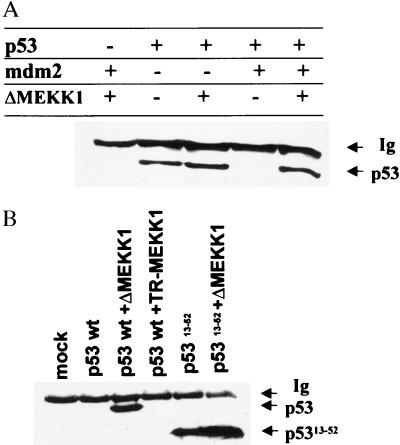
Figure 3.
(A) Cotransfection of ΔMEKK1 rescues p53 from mdm2-mediated degradation in p53/mdm2 null cells. Double null cells were cotransfected with wild-type rat p53 (200 ng), pCMV-mdm2 (800 ng), and ΔMEKK1. Immunoblot with antibody to p53 is shown. (B) Cotransfection of ΔMEKK1 increases the level of human mutant p53Δ13–52. Immunoblot analysis of lysates from 10.1 cells cotransfected with either human wild-type p53 or p53Δ13–52 and either TR-MEKK1 or ΔMEKK1 constructs probed with antibody to p53 are shown.
The effect of MEKK1 signaling on Mdm2-mediated p53 stability was also explored with the aid of a p53 mutant lacking the Mdm2-binding site (Δ13–52; ref. 28). Cotransfection of ΔMEKK1 with p5313–52 led to a minor (but still noticeable) increase in its level compared with the effect on the wild-type protein (Fig. 3B). These data suggest that, while Mdm2 dissociation is likely to serve as the primary mechanism of p53 stabilization by ΔMEKK1, other MEKK1-driven Mdm2-independent changes occur.
MEKK1 Signaling Increases p53 Transcriptional Activities.
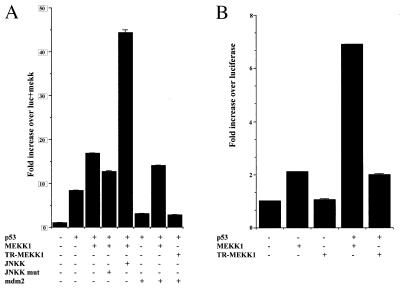
Upon the identification of the MEKK1 effect on p53 stability, we evaluated the possible changes by MEKK1 on p53 transcriptional activities. Currently available data suggest that different signaling pathways are capable of mediating p53 stabilization and transcriptional activity (9, 10). To this end, p53 ability to elicit the transactivation of mdm2 minimal promoter was monitored by using the luciferase reporter assay. Cotransfection of ΔMEKK1 with p53 increased p53-dependent transactivation of the mdm2 promoter (Fig. 4A). Mutant JNKK decreased the effect of ΔMEKK1, whereas, wild-type JNKK further stimulated mdm2-deriven luciferase activity. Furthermore, inhibition of p53 transactivation by Mdm2 was restored upon ΔMEKK1 expression (Fig. 4A). The ability of ΔMEKK1 to elicit p53 transcriptional activities was not confined to exogenously transfected p53 as they were also seen with endogenous p53 in MCF7 cells (Fig. 4B). These observations suggest that MEKK1/JNK signaling induces p53 transcriptional activity and further supports the previous suggestion that JNK elicited changes in p53 conformation make this tumor suppressor protein less susceptible to inactivation by mdm2.
Figure 4.
JNK signaling transactivates p53. (A) Luciferase reporter under control of p53-dependent mdm2 promoter was introduced into 10.1 cells with various constructs as indicated. Luciferase activity is normalized per β-galactosidase values. Each column represents an average value of three experiments (each in duplicate). (B) Transactivation of endogenous p53 in MCF7 cells stimulated by ΔMEKK1 is assessed as described in A.
MEKK1 Potentiates the Ability of p53 to Mediate Apoptosis.
Since induction of apoptosis in response to DNA damage and cellular stress is one of the main biologic functions of p53, we examined whether MEKK1/JNK signaling can alter this effect. Transfection of p53 null cells with ΔMEKK1 alone did not elicit any apoptosis, as assessed by FACS analysis of propidium iodide-stained cells (Fig. 5). Introduction of p53 increased the relative number of <2n fraction which is characteristic of apoptotic cells to 20%. Cotransfection of p53 with ΔMEKK1 led to a substantial increase in the number of apoptotic cells (to 40%). This increase suggests that JNK signaling contributes to stress-induced p53-dependent apoptosis. At high doses (>100 ng/60-mm plate), ΔMEKK1 abrogates p53-mediated apoptosis (data not shown), possibly due to the activation of NF-κB by very high expression of ΔMEKK1 (40), which could antagonize the p53 effect on cell death (41).
DISCUSSION
In the present studies, we have elucidated the role of JNK signaling in p53 stability and activities using JNKK upstream kinase MEKK1. With the aid of a constitutively active form of MEKK1, we demonstrated that the JNK-signaling pathway (i) stabilizes p53 by abrogating mdm2 association; (ii) increases p53 transactivation; and (iii) potentiates p53’s ability to elicit cellular apoptosis.
Important in our characterization of the MEKK1 effect on p53 has been the ability to abrogate JNK’s association with p53. Computer modeling identified residues representing exposed sites (amino acids 101, 105, and 108) which were respectively mutated to alter p53 interaction with JNK. This mutant form of p53 as well as p53, deleted of JNK association sites, provided direct support for the role of JNK in MEKK1 signaling to p53. Indeed, the levels of neither the p7 mutant p53Δp7 were affected by MEKK1 to the same degree as wild-type p53. That some residual MEKK1-mediated changes were still seen with p7 mutants suggests that MEKK1 may also mediate p53 phosphorylation by kinases other than JNK. Accordingly, other MEKK1-dependent kinases could affect proteins that regulate p53 stability.
The effects of MEKK1 on p53 are likely to be mediated by JNK since (i) JNKK is capable of enhancing these effects with regard to p53 stability and transcriptional activities; (ii) a catalytic inactive form of JNKK attenuates MEKK1’s effects; (iii) p53 that is mutated within or deleted of the p7 domain, which was mapped as a JNK-binding site on p53, are weakly affected by MEKK1; and (iv) JNK prepared from MEKK1-expressing cells efficiently phosphorylates p53 in immunokinase reactions.
Importantly, such changes (although to a much lesser extent) were also seen when we used a p53 that had been deleted of amino acids 13–52, which lacked amino-terminal phosphorylation sites and Mdm2 association, suggesting that (i) MEKK1-mediated p53 phosphorylation is likely to also occur outside of amino acids 13–52 and (ii) Mdm2 is not the only ubiquitination-targeting molecule whose binding might be affected by MEKK1 signaling. Among other possible sites for JNK phosphorylation are proline-driven serines and threonines, which flank the JNK association site. Here, we demonstrate that JNK phosphorylation of p53 is sufficient to abolish Mdm2 association with p53. These findings are in line with the observation that MEKK1 signaling causes Mdm2 dissociation from p53 in vivo. Parallel studies which monitored JNK association with p53 revealed that forced expression of MEKK1 was not sufficient to dissociate JNK from p53 (our unpublished data), suggesting that (i) changes elicited under these conditions effectively altered p53 conformation to affect JNK targeted ubiquitination, in addition to Mdm2, and (ii) other changes to p53 conformation, possibly via the phosphorylation of p53 by other stress-activated kinases are required for p53 dissociation from JNK.
Having used a constitutively active form of MEKK1, changes in the degree of MEKK1 activation in response to diverse biologic stimuli was not addressed. MEKK1 is known to be a specific JNKK kinase, although, when highly overexpressed, this construct is also capable of activating mitogen-activated protein kinase and NF-κB pathways (18, 40). Overexpression of constitutively active MEK1 mutant did not affect the p53 level (data not shown). Among multiple upstream signals implicated in the activation of MEKK1 (42), which require phosphorylation on Thr residues 560 and 572 are Pak3 and protein kinase C (43). That MEKK1 is found in complex with p21ras places MEKK1 in proximity to JNK, which was previously identified as p21ras-associated protein (44). p21ras is also required for UV-mediated JNK activation (45). Along these lines, while the response to UV irradiation is expected to resemble the effect of a constitutively active MEKK1, γ irradiation, which in many cell systems is a poor activator of JNK, is expected to use a different pathway to activate p53. Indeed, unlike UV irradiation, γ irradiation was shown to stabilize p53 without inhibiting its ubiquitination in vivo (46). Moreover, p53 phosphorylation in response to γ irradiation takes place on amino-terminal residues including Ser-15 (10), which is dispensable for MEKK1-mediated p53 transactivation (our unpublished data).
These results also provide direct evidence for the role of MEKK1/JNK in acquiring p53 transcriptional activities in vivo. Although MEKK1 effects were shown using mdm2 minimal promoter as a target for p53 transcriptional activities, a similar degree of activation is expected on other p53 target genes that share the same promoter sequences.
The ability of MEKK1 to potentiate p53-mediated programmed cell death provides the underlying mechanism for a documented role of JNK in contributing to programmed cell death (22, 23). Because our studies were limited to p53 null cells that underwent apoptosis in response to p53’s expression, one cannot exclude the possibility that MEKK1 represents only part of the signaling cascades required for p53 apoptotic potential.
Identifying the contribution of MEKK1 to p53 transcriptional activation and p53-mediated apoptosis by JNK phosphorylation points to the functional role of JNK signaling in p53’s ability to elicit a response to DNA damage and identifies new targets for the regulation of this regulatory cascade.
Acknowledgments
We thank D. Bohmann, M. Karin, A. Lin, A. Minden, M. Oren, and B. Vogelstein for kindly providing us with expression vectors, A. Levine for Mdm2 antibody and baculovirus expression vector, X. Wu for the 10.1 cells and mdm2 reporter construct, E. Spanopoulou for 293T cells, S. Jones for mdm2/p53 double null cells, and C. Monnel for JNK antibodies. We are grateful to J. Chen for assistance in computer calculations. This study was supported by National Cancer Institute Grants CA59908 and CA78419 (to Z.R.) and CA42500 (to M.R.P.).
ABBREVIATIONS
- MEKK1
mitogen-activated protein kinases kinase 1 (also known as JNKKK)
- ΔMEKK1
a constitutively active form of MEKK1 which lacks the first 350 amino acids
- JNK
Jun-NH2-terminal kinase
- HA
hemagglutinin tag
References
- 1.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Haffner R, Oren M. Curr Opin Genet Dev. 1995;5:84–90. doi: 10.1016/s0959-437x(95)90058-6. [DOI] [PubMed] [Google Scholar]
- 3.Adler V, Pincus M R, Minamoto T, Fuchs S Y, Bluth M J, Brandt-Rauf P W, Friedman F K, Robinson R C, Chen J M, Wang X W, Harris C C, Ronai Z. Proc Natl Acad Sci USA. 1997;94:1686–1691. doi: 10.1073/pnas.94.5.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lees-Miller S P, Sakaguchi K, Ullrich S J, Appella E, Anderson C W. Mol Cell Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Eckhart W. Proc Natl Acad Sci USA. 1992;89:4231–4235. doi: 10.1073/pnas.89.10.4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milne D M, Campbell L E, Campbell D G, Meek D W. J Biol Chem. 1995;270:5511–5518. doi: 10.1074/jbc.270.10.5511. [DOI] [PubMed] [Google Scholar]
- 7.Knippschild U, Milne D M, Campbell L E, DeMaggio A J, Christenson E, Hoekstra M F, Meek D W. Oncogene. 1997;15:1727–1736. doi: 10.1038/sj.onc.1201541. [DOI] [PubMed] [Google Scholar]
- 8.Ko L J, Shieh S Y, Chen X, Jayaraman L, Tamai K, Taya Y, Prives C, Pan Z Q. Mol Cell Biol. 1997;17:7220–7229. doi: 10.1128/mcb.17.12.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shieh S Y, Ikeda M, Taya Y, Prives C. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 10.Siliciano J D, Canman C E, Taya Y, Sakaguchi K, Appella E, Kastan M B. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fried L M, Koumenis C, Peterson S R, Green S L, van Zijl P, Allaunis-Turner J, Chen D J, Fishel R, Giaccia A J, Brown J M, Kirchgessner C U. Proc Natl Acad Sci USA. 1996;93:13825–13830. doi: 10.1073/pnas.93.24.13825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guidos C J, Williams C J, Grandal I, Knowles G, Huang M T F, Danska J S. Genes Dev. 1996;10:2038–2054. doi: 10.1101/gad.10.16.2038. [DOI] [PubMed] [Google Scholar]
- 13.Kyriakis J M, Avruch J. J Biol Chem. 1996;271:24313–24316. doi: 10.1074/jbc.271.40.24313. [DOI] [PubMed] [Google Scholar]
- 14.Deng T, Karin M. Genes Dev. 1993;7:479–490. doi: 10.1101/gad.7.3.479. [DOI] [PubMed] [Google Scholar]
- 15.Adler V, Schaffer A, Kim J, Dolan L, Ronai Z. J Biol Chem. 1995;270:26071–26077. doi: 10.1074/jbc.270.44.26071. [DOI] [PubMed] [Google Scholar]
- 16.Galcheva-Gargova Z, Derijard B, Wu I H, Davis R J. Science. 1994;265:806–808. doi: 10.1126/science.8047888. [DOI] [PubMed] [Google Scholar]
- 17.Westwick J K, Weitzel C, Minden A, Karin M, Brenner D A. J Biol Chem. 1994;269:26396–26401. [PubMed] [Google Scholar]
- 18.Minden A, Lin A, McMahon M, Lange-Carter C, Derijard B, Davis R J, Johnson G L, Karin M. Science. 1994;266:1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 19.Lin A, Minden, Martinetto H, Clarett F-X, Lange-Carter C, Mercurio F, Johnson G L, Karin M. Science. 1995;268:286–290. doi: 10.1126/science.7716521. [DOI] [PubMed] [Google Scholar]
- 20.Yang D, Tournier C, Wysk M, Lu H T, Xu J, Davis R J, Flavell R A. Proc Natl Acad Sci USA. 1997;94:3004–3009. doi: 10.1073/pnas.94.7.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta S, Campbell D, Derijard B, Davis R J. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y R, Meyer C F, Tan T H. J Biol Chem. 1996;271:631–634. doi: 10.1074/jbc.271.2.631. [DOI] [PubMed] [Google Scholar]
- 23.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 24.Fuchs S Y, Dolan L, Davis R J, Ronai Z. Oncogene. 1996;13:1531–1535. [PubMed] [Google Scholar]
- 25.Fuchs S Y, Xie B, Adler V, Fried V A, Davis R J, Ronai Z. J Biol Chem. 1997;272:32163–32166. doi: 10.1074/jbc.272.51.32163. [DOI] [PubMed] [Google Scholar]
- 26.Musti A M, Treier M, Bohmann D. Science. 1997;275:400–402. doi: 10.1126/science.275.5298.400. [DOI] [PubMed] [Google Scholar]
- 27.Maltzman W, Czyzyk L. Mol Cell Biol. 1984;4:1689–1694. doi: 10.1128/mcb.4.9.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 29.Fuchs, S. Y., Adler, A., Buschmann, T., Yin, Z., Wu, X., Jones, S. N. & Ronai, Z. (1998) Genes Dev., in press. [DOI] [PMC free article] [PubMed]
- 30.Hupp T R, Sparks A, Lane D P. Cell. 1995;83:237–245. doi: 10.1016/0092-8674(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 31.Livingstone L R, White A, Sprouse J, Livanos E, Jacks T, Tlsty T D. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 32.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X, Levine A J. Proc Natl Acad Sci USA. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kallunki T, Su B, Tsigelny I, Sluss H K, Derijard B, Moore G, Davis R, Karin M. Genes Dev. 1994;8:2996–3007. doi: 10.1101/gad.8.24.2996. [DOI] [PubMed] [Google Scholar]
- 35.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 36.Ripoll D, Scheraga H A. Biopolymers. 1988;27:1283–1303. doi: 10.1002/bip.360270808. [DOI] [PubMed] [Google Scholar]
- 37.Dauber-Osguthorpe P, Roberts V A, Osguthorpe D J, Wolff J, Genest M, Hagler A T. Proteins. 1988;4:31–47. doi: 10.1002/prot.340040106. [DOI] [PubMed] [Google Scholar]
- 38.Kubbutat M H, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 39.Fuchs, S. Y., Adler, V., Buschmann, T., Wu, X. & Ronai, Z. (1998) Oncogene, in press. [DOI] [PubMed]
- 40.Lee F S, Hagler J, Chen Z J, Maniatis T. Cell. 1997;88:213–222. doi: 10.1016/s0092-8674(00)81842-5. [DOI] [PubMed] [Google Scholar]
- 41.Mayo M W, Wang C Y, Cogswell P C, Rogers-Graham K S, Lowe S W, Der C J, Baldwin A S., Jr Science. 1997;278:1812–1815. doi: 10.1126/science.278.5344.1812. [DOI] [PubMed] [Google Scholar]
- 42.Fanger G R, Johnson N, L, Johnson G L. EMBO J. 1997;16:4961–4972. doi: 10.1093/emboj/16.16.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siow Y L, Kalmar G B, Sanghera J S, Tai G, Oh S S, Pelech S L. J Biol Chem. 1997;272:7586–7594. doi: 10.1074/jbc.272.12.7586. [DOI] [PubMed] [Google Scholar]
- 44.Adler V, Pincus M R, Brandt-Rauf P W, Ronai Z. Proc Natl Acad Sci USA. 1995;92:10585–10589. doi: 10.1073/pnas.92.23.10585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adler V, Pincus M R, Polotskaya A, Montano X, Friedman F K, Ronai Z. J Biol Chem. 1996;271:23304–23309. doi: 10.1074/jbc.271.38.23304. [DOI] [PubMed] [Google Scholar]
- 46.Maki C G, Howley P M. Mol Cell Biol. 1997;17:355–363. doi: 10.1128/mcb.17.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]