Abstract
FokI is a type IIs restriction endonuclease comprised of a DNA recognition domain and a catalytic domain. The structural similarity of the FokI catalytic domain to the type II restriction endonuclease BamHI monomer suggested that the FokI catalytic domains may dimerize. In addition, the FokI structure, presented in an accompanying paper in this issue of Proceedings, reveals a dimerization interface between catalytic domains. We provide evidence here that FokI catalytic domain must dimerize for DNA cleavage to occur. First, we show that the rate of DNA cleavage catalyzed by various concentrations of FokI are not directly proportional to the protein concentration, suggesting a cooperative effect for DNA cleavage. Second, we constructed a FokI variant, FokN13Y, which is unable to bind the FokI recognition sequence but when mixed with wild-type FokI increases the rate of DNA cleavage. Additionally, the FokI catalytic domain that lacks the DNA binding domain was shown to increase the rate of wild-type FokI cleavage of DNA. We also constructed an FokI variant, FokD483A, R487A, which should be defective for dimerization because the altered residues reside at the putative dimerization interface. Consistent with the FokI dimerization model, the variant FokD483A, R487A revealed greatly impaired DNA cleavage. Based on our work and previous reports, we discuss a pathway of DNA binding, dimerization, and cleavage by FokI endonuclease.
The type IIs restriction endonuclease FokI, isolated from Flavobacterium okeanokoites, recognizes an asymmetric nucleotide sequence and cleaves both DNA strands outside of the recognition site: 5′-GGATG(N)9/13 (1). The fokIRM genes have been cloned and sequenced (2, 3). The endonuclease consists of 587 aa with a molecular mass of 65.4 kDa (2, 4). FokI has been shown to exist as a monomer in solution, based on gel filtration and sedimentation experiments (4). It has been concluded that, when bound to DNA and analyzed by gel-mobility shift experiments, only one monomer of FokI was bound to its DNA recognition sequence (5). The gel-mobility shift experiments, where a 1:1 complex was observed (5), were performed with a FokI precleaved DNA. Therefore this complex represents FokI bound to the DNA product not to the DNA substrate.
Unlike FokI, the typical type II restriction endonucleases, such as EcoRI or EcoRV, form a tight homodimer in solution and bind to DNA as a homodimer. Each monomeric subunit of homodimer contains one catalytic center. Mutational analysis by Waugh and Sauer (6) suggests that there is a single catalytic center per FokI monomer. This led them to conclude that either FokI must rearrange its catalytic center for sequential cleavage of each DNA strand or it must form a higher order complex to cleave both strands of DNA (6).
Based on proteolytic studies, it was shown that FokI endonuclease contains two separate structural domains, one for DNA recognition and one for DNA cleavage (7). A purified 41-kDa N-terminal proteolytic fragment bound the recognition sequence specifically but did not cleave DNA, whereas the 25-kDa C-terminal proteolytic fragment in the presence of Mg2+ cleaved DNA nonspecifically (7). Mutational analysis further supported a modular structure of FokI. A C-terminal deletion variant of FokI showed the same DNA binding properties as the wild-type enzyme (8). On the other hand, a single amino acid substitution in the C-terminal half of the FokI protein disabled endonuclease activity but did not affect the ability of the enzyme to bind specifically to DNA (6). Other studies have demonstrated that FokI catalytic domain can be fused to DNA binding proteins to yield catalytically active chimeric proteins with novel recognition specificities (9–12).
Yonezawa and Sugiura (13), by using four different footprinting techniques, showed that FokI interacts with the target recognition sequence from the major groove side of the DNA helix and that DNA protection at the site of cleavage in the absence of divalent metal ion is very weak. Waugh and Sauer (6) used DNase footprinting and methylation protection with cleavage-deficient variants of FokI in the presence of divalent metal ion to show also a weak protection pattern around the site of cleavage (6).
The three dimensional structure of FokI complexed to DNA confirmed that the protein is comprised of two functional domains, a N-terminal DNA recognition domain and a C-terminal catalytic domain (14). Together, the FokI structure in the presence of DNA (14) and in the absence of DNA (15) provides a basis for the biochemical and mutational studies reported here. The structure in the presence of DNA showed a single FokI molecule approaching DNA from the major groove side, with the recognition domain making all of the base-specific contacts at the recognition site. The catalytic domain of FokI revealed a structure remarkably similar to a monomer of the type II restriction endonuclease BamHI (14–16). Three catalytically important residues of FokI (Asp-450, Asp-467, and Lys-469) superimpose with the catalytic residues Asp-94, Glu-111, and Glu-113 of BamHI. Therefore, whether examined by mutational analysis (6) or by structural comparison (14), there appears to be only one catalytic site per FokI molecule. This raises the question, “How does monomeric FokI cleave both DNA strands by using a single catalytic center?” The FokI structure showed that the catalytic domain is sequestered alongside of the recognition domain and does not contact DNA at the cleavage site, in accordance with the earlier footprinting studies (6, 13). From the structure, the FokI catalytic domain can be relocated at the site of cleavage by a simple rotation around the linker segment so that the catalytic residues are positioned in the vicinity of the scissile phosphodiester bond located 13 nt away from the recognition site (14). However, for the same catalytic domain to cleave the other DNA strand, it would require the enzyme to adopt a second configuration, which is difficult to model without extensive refolding (14). In this paper, we provide evidence that FokI dimerization is required for DNA cleavage.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids.
Escherichia coli strain ER2566 [fhuA2, lacZ∷T7 gene1, Δ(mcrC-mrr) 114∷IS10, R(mcr-73∷ miniTn10– TetS)2, endA1] containing a chromosomal copy of the T7 RNA polymerase gene was constructed by E. Raleigh (New England Biolabs) and used for expression of FokI variants. Plasmid pAFP1 containing the cloned FokI endonuclease gene (2) and plasmid pAFD2 containing the FokI methyltransferase gene (2) were constructed by W. E. Jack (New England Biolabs). The pAFP1 derivative coding for the catalytically inactive D450A variant of FokI; the plasmid pSKFok1, a derivative of pBluescriptSK(−), containing a single FokI site; and the plasmid pSKFok0, the derivative of pSKFok1 in which deletion of the 445-bp PvuII fragment removed the single FokI recognition sequence were constructed by C. Noren (New England Biolabs; unpublished work). pTYB4 and pTYB2 are products of New England Biolabs.
Subcloning of the FokI Gene into the Impact One-Step Protein Purification System.
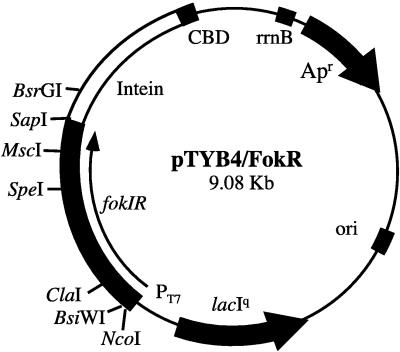
The full length FokI endonuclease gene and the truncated gene coding only for the catalytic domain of FokI endonuclease were amplified by PCR by using plasmid pAFP1 as a template. The 5′ primer, 5′- AGCCCATGGTTTCTAAAATACGTACGTTCGGTTGGG-3′, for amplication of the full length FokI endonuclease, was complementary to the fokIR gene except for silent mutations in the 6th and 7th codons to create a BsiWI site. The primer, 5′-GTGCATATGCAACTAGTCAAAAGTGAACTGG-3′, for amplification of the catalytic domain of FokI was flanked by the NdeI site. The rest of the primer sequence matched the fokIR gene sequence starting from the 384th codon. The 3′ reverse primer, 5′-CATGGCGCCAAAGTTTATCTCGCCGTTATTAA-3′, is complementary to the end of fokIR gene and is flanked by the EheI site. The resulting PCR products were agarose-gel purified and cleaved with restriction endonucleases: NcoI and EheI in the case of the full length FokI gene; or NdeI and EheI in the case of the gene coding for the FokI catalytic domain. The gel-purified restriction fragment containing the full length fokIR was ligated into the NcoI and SmaI digested vector pTYB4. This created the construct pTYB4/FokR (Fig. 1) where the C terminus of the fokIR gene was fused directly to the intein of the Impact Purification System (17, 18). The gel-purified restriction fragment containing the fokIR catalytic domain coding sequence was ligated into the NdeI and SmaI digested vector pTYB2. The resulting construct was designated pTYB2/FokCD. No sequence alterations other than desired mutations were detected by DNA sequence analysis.
Figure 1.
Plasmid map of the pTYB4/FokR. The fokIR gene is placed downstream of the T7 promoter, and fokIR is in-frame with the 5′ end of the intein coding sequence. CBD, chitin binding domain.
Construction of Catalytically Inactive FokI Variants.
The 1.13-kb ClaI–MscI fragment of pTYB4/FokR was replaced by the same fragment from the pAFP1/D450A construct (see above) to create the plasmid derivative pTYB4/FokDa450A coding for a catalytically inactive FokI variant. Similarly, the 0.5-kb SpeI–SapI fragment of pTYB2/FokCD was replaced by the SpeI–SapI fragment from pAFP1/D450A to create pTYB2/FokCD/D450A.
Construction of a Binding-Deficient FokI Variant.
To create a binding-deficient FokI variant, Asn13 was replaced by a bulky amino acid, tyrosine. The following mutagenic primers were used to amplify the 0.29-kb BsiWI–ClaI fragment of the FokI gene: 5′-ATACGTACGTTCGGTTGGGTTCAATATCCAGGTAATTTGAG-3′ (introduces N13Y) and 5′-GCCCAACGCAAAAAACCGTCAGATGA-3′.
The PCR product was cleaved with BsiWI and ClaI restriction endonucleases, and the gel-purified restriction fragment was ligated into the BsiWI and ClaI digested pTYB4/FokR vector. This created a new derivative, pTYB4/FokN13Y, coding for the FokI variant with altered base-specific contacts. The sequence of the replaced DNA fragment was verified by DNA sequence analysis.
Construction of Dimerization-Deficient FokI Variant.
The FokI gene segment flanked by MscI and BsrGI restriction sites was amplified by PCR by using pTYB4/FokR as a template and a mutagenic primer that introduced substitutions D483A and R487A, 5′-CAATTGGCCAAGCAGCTGAAATGCAAGCATATGTCGAAGAAAATCAAACACG-3′. The second primer matched the DNA sequence downstream the fokIR gene and was flanked by BsrGI restriction site 5′- ACGCTGTACATAGTTTCTCTTCC-3′. The MscI–BsrGI fragment of pTYB4/FokR was replaced with MscI and BsrGI digested mutagenic PCR product. This created a new derivative, pTYB4/Fok483A,487A, which coded for a dimerization-deficient FokI variant.
Target Protein Expression and Purification.
Wild-type FokI restriction endonuclease was purified to homogeneity as described earlier (19). The concentration of wild-type FokI protein was determined spectrophotometrically at 280 nm by using the extinction coefficient of 72,520 M−1 cm−1 (20).
The One-Step Protein Purification System (Impact) (17) was used to facilitate purification of FokN13Y, FokCD, FokCD/D450A, and FokD483A, R487A variants. This technique is based on the self-catalyzed cleavage of the intein that can be controlled in vitro by DTT (18). The chitin binding domain is fused genetically to the C terminus of the intein and serves as an affinity tag to bind FokI–intein fusion onto a chitin column. The FokI is released from the fusion after the induction of the intein cleavage reaction by adding 50 mM DTT, while intein–chitin binding domain fusion remains bound to the column (17).
E. coli ER2566 cells containing mutant pTYB-Fok constructs were grown at 37°C in Luria–Bertani medium supplemented with 0.1 mg/ml ampicillin. Expression of the target protein was induced by adding isopropyl β-d-thiogalactoside to a final concentration of 0.1 mM at mid-log phase of growth (Klett = 90). After induction, the cultures were incubated overnight at 15°C. The cells were harvested, resuspended in column buffer (20 mM potassium phosphate, pH 7.4/0.1 mM EDTA/0.5 M NaCl) and disrupted by sonication. The crude extract containing FokI–intein fusion protein was loaded onto a chitin column (New England Biolabs) and washed with two column volumes, and the column was flushed quickly with 2 column volumes of 20 mM potassium phosphate buffer (pH 7.4), 0.1 mM EDTA, and 50 mM NaCl containing 50 mM DTT. The column was incubated at 4°C overnight to induce self-cleavage of the protein fusion (17). Next, the target protein was eluted from the column with column buffer and dialyzed to remove excess DTT. The purified proteins were concentrated by dialysis against 20 mM potassium phosphate (pH 7.4), 0.1 mM EDTA, 50 mM NaCl, and 50% glycerol and stored at −20°C. FokI variants purified via the Impact System have two additional glycine residues at their carboxyl termini.
DNA Cleavage Assays.
The cleavage activity of mutant FokI variants was assayed by incubating various amounts of protein at 37°C in either 30 or 50 μl of standard FokI reaction buffer (20 mM Tris-acetate, pH 7.9/10 mM magnesium acetate/50 mM potassium acetate/1 mM DTT/100 μg/ml BSA) containing 1 μg of λ or plasmid pSKFok1 DNA. The reaction was terminated by addition of 10 μl of stop solution (60 mM EDTA, pH 8.0/50% glycerol/1% SDS/0.02% bromphenol blue), and reaction products were analyzed by electrophoresis in 1% agarose gels.
For quantitative FokI activity assays, plasmid pSKFok1 was first linearized with the restriction endonuclease AhdI to make a linear substrate with the FokI recognition site positioned in the center of the DNA molecule. DNA then was purified by phenol-chloroform extraction followed by isopropanol/ethanol precipitation. DNA concentration was determined spectrophotometrically at 260 nm. FokI was added to either 70 or 200 μl of reaction buffer (see above) containing 6.3 nM substrate DNA. Samples were incubated at 37°C, 10-μl aliquots were removed at various time intervals, and the reaction products were analyzed by agarose gel electrophoresis. The ethidium bromide-stained gels were digitized with IS-500 Digital Imaging System (Alpha Innotech, San Leandro, CA). The intensity of the product band was quantitated by using the nih image program v. 1.61.
RESULTS
Rate of Cleavage vs. Enzyme Concentration.
The rate of DNA cleavage catalyzed by various FokI concentrations was determined to establish the relationship between the initial velocity of the reaction and the enzyme concentration. Linearized plasmid pSKFok1 DNA containing a single FokI recognition site positioned in the center of the DNA molecule was used as a substrate. Using such a DNA substrate simplified the evaluation of the concentration of reaction products that, because of their same size, run on the agarose gel as a single band.
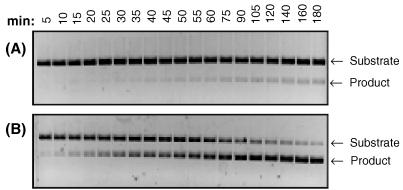
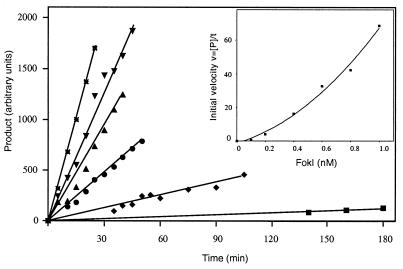
A limiting amount of FokI restriction endonuclease (from 0.1 to 1.0 nM final concentration) was added last to the reaction mixture, which was preheated at 37°C. The progress of each reaction was monitored by withdrawing 10-μl samples at timed intervals. Typical results of such cleavage assays, performed with 0.2 and 0.6 nM FokI, are shown in Fig. 2. The amount of cleaved DNA was expressed in arbitrary units after the quantitative evaluation of the intensity of the product band in an ethidium bromide-stained agarose gel. The plots showing the progress of the DNA product formation with time at different FokI concentrations are presented in Fig. 3. These plots were used to calculate the initial velocity (vo = d[P]/dt) of each reaction catalyzed by a given FokI concentration. When measured values of vo were plotted against the corresponding values of [FokI] a nonlinear plot was obtained (Fig. 3, Inset). This indicated that, at low FokI concentrations, the initial velocity of the reaction is not directly proportional to the enzyme concentration, suggesting that the FokI-catalyzed reaction is higher than first order with respect to [FokI]. Among the possible interpretations, the nonlinear relationship of vo vs. [FokI] can be explained by a cooperative binding of two FokI molecules to cleave both strands of DNA.
Figure 2.
Time course of the FokI-catalyzed DNA cleavage reaction. Linearized pSKFok1 DNA (6.3 nM) was incubated at 37°C with 0.2 (A) or 0.6 nM (B) FokI restriction endonuclease in 200 μl of reaction buffer (20 mM Tris-acetate, pH 7.9/10 mM magnesium acetate/50 mM potassium acetate/1 mM DTT/100 μg/ml BSA). At the time points indicated, 10-μl samples were withdrawn, immediately quenched by adding 5 μl of stop solution, and subjected to electrophoresis on 1% agarose-gel.
Figure 3.
Rate of DNA cleavage with various FokI concentrations. The conditions of DNA cleavage reaction are described in Fig. 2. Samples were withdrawn at the indicated times from the reactions that contained 0.1 nM FokI (■), 0.2 nM FokI (♦), 0.4 nM FokI (•), 0.6 nM FokI (▴), 0.8 nM FokI (▾), and 1.0 nM FokI (∗). The amount of cleaved DNA was determined as in Experimental Procedures. (Inset) Initial velocity vs. FokI concentration.
Characterization of FokN13Y Variant.
If the catalytically active FokI complex requires two FokI molecules, i.e., two catalytic domains, for double-stranded DNA cleavage, then the reaction at low concentrations of FokI protein may be stimulated by a binding-deficient variant of FokI that contains an active catalytic domain. The three-dimensional structure of the FokI endonuclease bound to the DNA substrate revealed that Asn13 makes bidentate hydrogen bonds with the central adenine base of the recognition sequence GGATG (14). Substitution of this residue with the bulky amino acid tyrosine should destroy base-specific contacts at the recognition interface. Such a mutant protein would be incapable of binding to the target DNA and therefore would be impaired for DNA cleavage. To test this, the pTYB4/FokN13Y derivative was constructed as described in Experimental Procedures. The mutant FokN13Y protein then was expressed and purified by a single chromatographic step from a chitin column. The purified protein was >95% pure as determined by SDS/PAGE. FokN13Y (200 nM) was incubated with supercoiled and linearized pSKFok1 DNA for 1 h at 37°C. At these conditions, no detectable double-strand or single-strand cleavage of DNA substrate was observed.
Stimulation of FokI Activity with FokN13Y.
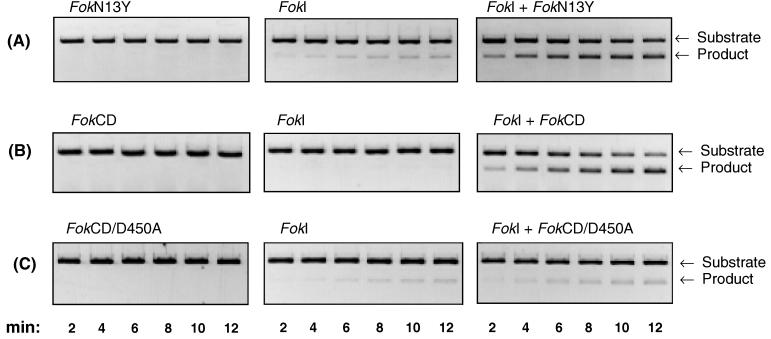
To test the FokN13Y variant for its ability to stimulate FokI restriction endonuclease, the substrate DNA was incubated with either 1 nM FokI alone, 500 nM FokN13Y protein alone, or the premixed FokI (1 nM) and FokN13Y (500 nM) protein combination. Fig. 4A reveals that the DNA cleavage reaction catalyzed by 1 nM FokI alone is slow. After 12 min only a small portion (≈5%) of the substrate DNA is cleaved. Under the same reaction conditions, 500 nM FokN13Y did not show any detectable DNA cleavage activity (Fig. 4A). However, when FokI endonuclease was supplemented with 500 nM FokN13Y protein, a 10–20-fold increase in the rate of DNA cleavage was observed relative to FokI alone (Fig. 4A). This suggests that wild-type FokI protein interacts with the FokN13Y molecule to facilitate site-specific cleavage.
Figure 4.
Stimulation of FokI endonuclease by (A) FokN13Y, (B) FokCD, or (C) FokCD/D450A. Reactions were performed at 37°C in 70 μl of 20 mM Tris-acetate buffer (pH 7.9), 10 mM magnesium acetate, 50 mM potassium acetate, 1 mM DTT, 100 μg/ml BSA, and 6.3 nM linearized pSKFok1 DNA; 10-μl samples were withdrawn at the indicated times from the reactions that contained: (A) either 500 nM FokN13Y, 1.0 nM FokI, or 1.0 nM FokI + 500 nM FokN13Y; (B) either 500 nM FokCD, 0.5 nM FokI, or 0.5 nM FokI + 500 nM FokCD; and (C) either 500 nM FokCD/D450A, 1.0 nM FokI, or 1.0 nM FokI + 500 nM FokCD/D450A. Samples were quenched by adding 5 μl of stop solution and were analyzed by electrophoresis on 1% agarose gel.
Characterization of FokCD and FokCD/D450A Variants.
The pTYB2/FokCD and pTYB2/FokCD/D450A derivatives code for the C-terminal 196 aa of FokI restriction endonuclease, constituting the 25-kDa catalytic domain. In addition, the derivative pTYB2/FokCD/D450A codes for a catalytically defective FokCD protein, FokCD/D450A. The purified FokCD and FokCD/D450A proteins were tested for the ability to cleave λ DNA. When <1 μM FokCD protein was incubated with 1 nM λ DNA for 1 h at 37°C, there was no detectable DNA cleavage. At >1 μM FokCD concentrations, nonspecific DNA cleavage was observed. In contrast, incubating 2.5 μM FokCD/D450A as long as 20 h with 1 nM λ DNA at 37°C did not show any detectable DNA cleavage.
Stimulation of FokI Activity with the Catalytic Domain.
Stimulation of FokI with the FokCD protein was performed as described for the FokN13Y variant. After 12 min, <1% of DNA substrate was cleaved by 0.5 nM FokI alone (Fig. 4B). No detectable DNA cleavage was observed with 500 nM FokCD alone (Fig. 4B). When 0.5 nM FokI was supplemented with 20 nM FokCD, no increase in the rate of DNA cleavage was observed (data not shown). However, with 0.5 nM FokI plus 500 nM FokCD, ≈50% of substrate DNA was cleaved after 8 min (Fig. 4B). This rate is ≈100-fold higher than with 0.5 nM FokI alone. In contrast, the catalytically inactive FokCD variant FokCD/D450A failed to activate FokI (Fig. 4C). These results indicate that two separate catalytic domains are necessary for site-specific, double-strand DNA cleavage by FokI.
Dimerization-Defective FokI Variant Displays Diminished Cleavage of Both DNA Strands.
In an accompanying paper in this issue of Proceedings (15), we report the structure of FokI in the absence of DNA. The structure reveals protein–protein interactions between two catalytic domains that mimic a BamHI homodimer. Close inspection of this interface shows that amino acids Asp-483 and Arg-487 are located within hydrogen bond distance with the corresponding residues in the other catalytic domain subunit. These residues were considered to be good candidates for disrupting the dimer interface while not affecting folding of the protein for the following reasons: (i) the residues are located on the surface of the domain; and (ii) these residues do not contact neighboring amino acid residues within the same subunit.
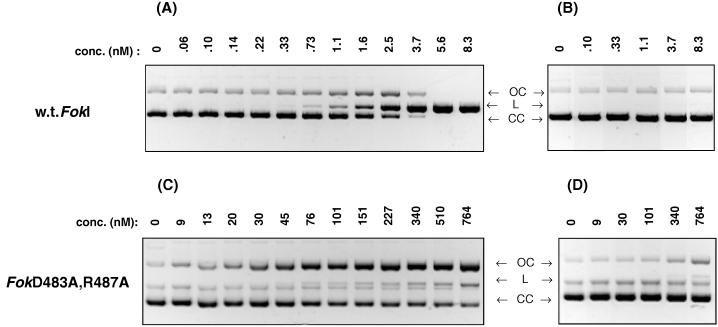
The pTYB4/FokD483A, R487A construct was made and the protein was expressed and purified as described in Experimental Procedures. Cleavage of supercoiled pSKFok1 plasmid by FokI endonuclease and FokD483A, R487A protein was compared. As shown in Fig. 5A, after 1 h at 37°C, 6 nM FokI completely converted 20 nM supercoiled pSKFok1 plasmid DNA to a full length linear product. Incubation of FokI with a plasmid DNA that does not contain a FokI recognition sequence, pSKFok0, showed that FokI cleavage reaction is sequence-specific (Fig. 5B). In contrast to the wild-type endonuclease, the FokD483A, R487A variant showed no detectable DNA cleavage at 9 nM (Fig. 5C). At a 10- to 80-fold higher concentration of FokD483A, R487A variant, the major product observed was the open circular form of plasmid DNA. The nicking activity was site-specific because the FokD483A, R487A variant did not convert plasmid pSKFok0 DNA to the open circular form (Fig. 5D). The results indicate that double-strand cleavage activity of the FokD483A, R487A variant is reduced three orders of magnitude as compared with the double-strand cleavage activity of wild-type enzyme (Fig. 5C).
Figure 5.
Comparison of DNA cleavage by FokI and FokD483A, R487A. The cleavage reactions were performed for 1 h at 37°C in 30 μl of 20 mM Tris-acetate buffer (pH 7.9), 10 mM magnesium acetate, 50 mM potassium acetate, 1 mM DTT, and 100 μg/ml BSA, containing 20 nM of (A and C) supercoiled plasmid pSKFok1 or (B and D) supercoiled plasmid pSKFok0 and either (A and B) FokI or (C and D) FokD483A, R487A. The concentrations of FokI and FokD483A, R487A are indicated. (CC), covalently closed form of plasmid DNA; (OC), open circle form; (L), linear form.
DISCUSSION
Typical type II restriction endonucleases exist in solution as homodimers and are known to bind to and cleave DNA as homodimers. The molecular mass of the monomers are generally in the 20- to 40-kDa range. Each monomeric subunit is responsible for half of the DNA base-specific contacts, and each subunit contains one catalytic center that cleaves one strand of the duplex. The sites of cleavage are always found symmetrically positioned within the recognition sequence. The three-dimensional structures of the type II endonucleases show that the recognition and catalytic functions are well integrated into a single protein domain (21). On the other hand, the type IIs restriction endonuclease FokI is a larger, 65-kDa protein. It is monomeric in solution (4). The asymmetric recognition sequence and sites of cleavage are separated by one turn of the DNA helix. The DNA recognition function and single catalytic center of FokI reside on two separate domains of the protein. How does FokI cleave both strands of the DNA?
Unlike the type II endonucleases, the specific recognition of DNA duplex by FokI is mediated clearly by the single FokI recognition domain (14). However, the nature of the termini of the FokI-cleaved DNA is comparable to the termini of BamHI or EcoRI cleaved DNA because all three endonucleases cleave double-stranded DNA to leave a 4-base, 5′-extension. The three-dimensional structures of BamHI and EcoRI show a similar common core and positioning of the catalytic residues relative to the targeted scissile phosphate groups (22). The structural organization of the FokI catalytic domain displays a striking similarity to the monomer of BamHI (14). The two structures share a similar β-sheet core surrounded by α-helices. The active site residues of both endonucleases occur at one end of the β-sheet and are superimposed easily on each other (14). This data, together with the structural organization of FokI in the absence of DNA (15), led us to propose that FokI endonuclease may use a strategy similar to EcoRI and BamHI for DNA cleavage. From this perspective, we have postulated that (i) two FokI molecules are involved in double-stranded DNA cleavage; (ii) DNA cleavage reaction by FokI proceeds via an enzyme dimerization step that takes place in the presence of both the specific DNA and divalent metal ion; and (iii) dimerization occurs at the interface of two catalytic domains of FokI.
Measurement of the rate of DNA cleavage at different enzyme concentrations suggests that the FokI catalyzed reaction is higher than first order with respect to FokI. This is consistent with the cooperative binding of two FokI molecules. On the other hand, the nonlinear relationship between the initial velocity of the reaction and FokI at low enzyme concentrations also could be explained by other factors; for example, at low protein concentration, the enzyme may first be adsorbed to the side of the reaction vessel. Although the reaction buffer used in the DNA cleavage experiments contained 100 μg/ml BSA, which should alleviate such undesirable nonspecific binding of FokI, we cannot rule out that some analogous interaction did not take place.
To obtain additional evidence for FokI cooperativity, FokI variants with mutated (FokN13Y) or deleted (FokCD) recognition domains were constructed and were shown to stimulate FokI activity. When incubated alone with DNA, these variants were shown to be incapable of cleaving DNA. However, when FokN13Y was incubated together with a limiting amount of FokI, the rate of DNA cleavage was increased 10-fold. Likewise, in the presence of FokCD, FokI activity increased 100-fold. The inactive FokI catalytic domain FokCD/D450A failed to stimulate FokI endonuclease. Furthermore, the inactive FokI catalytic domain did not appear to inhibit the low level of FokI activity, indicating that the inactive catalytic domain is incapable of forming a complex with FokI. A precleavage complex probably relies on the binding energy derived from contributions of the appropriate ionic interactions between the magnesium coordinated with residues D450, D467, and the scissile phosphate. In the catalytically defective domain, D450 is replaced by alanine. It is expected that this will upset the coordination of the magnesium, hence the network of interactions that afford the transient binding energy to form a pre-cleavage complex. The stimulation of FokI by FokCD suggests that a FokI monomer must interact with a second catalytic center for double-stranded DNA cleavage to occur. Thus, during cleavage by wild-type FokI endonuclease, two monomeric FokI molecules interact with each other to align two catalytic centers for the cleavage of both DNA strands.
The cooperativity that FokI displays for DNA cleavage is apparent at a concentration range below 1 nM. Above 1 nM, the rate of DNA cleavage increases proportionately to the amount of FokI added to the reaction. This suggests that the equilibrium binding constant of FokI for a FokI/DNA complex is <1 nM. In the stimulation experiments, a >100 nM concentration of mutant FokI variant is required to observe the same rate of DNA cleavage as 3 nM FokI alone. These results indicate that the full length, wild-type FokI molecule is two orders of magnitude more efficient at facilitating DNA cleavage than the mutant proteins that had their DNA recognition domain mutated (FokN13Y) or deleted (FokCD). This suggests that the second recognition domain also plays a role in the formation and/or stability of the catalytically active FokI dimer/DNA complex.
The phenomenon of “sequestration” of the FokI catalytic domain by the recognition domain that was observed in the three-dimensional structure (14) may exist to prevent the catalytic domain from interacting with the DNA while the FokI molecule scans the DNA in search of a new recognition site or searches for another monomer that already has bound to a FokI recognition sequence. The sequestering of the catalytic domain may explain how FokI manages to regulate its cleavage activity. The catalytic domain remains sequestered during binding to its DNA recognition sequence and is triggered for release by dimerization. When wild-type FokI is incubated with supercoiled DNA at low enzyme concentration and there is sufficient enzyme to bind only as monomer, it may be expected to only nick supercoiled DNA. This would result in the accumulation of the open circular form of DNA. However, the experimental results do not support such a model but rather support the view that the catalytic domain remains inactive. At a molar ratio of 1:20 of FokI:DNA, a slight excess of the nicked intermediate over the linear DNA was observed. At a 1:8 ratio of FokI:DNA, the linear product was predominant in the reaction (Fig. 5A). The behavior of the double mutant FokD483A, R487A also supports the view that the catalytic domain is inactive until dimerization occurs. The D483A+R487A mutations targeted the catalytic domain dimerization interface while leaving the recognition domain and catalytic center of the enzyme unchanged. However, by altering the catalytic domain dimerization interface, the FokI cleavage of either strand of the DNA duplex was impaired. Thus, neither the results for FokI or the FokD483A, R487A variant indicate that the FokI monomer can cleave one DNA strand. More likely, the FokI monomer complexed to the specific DNA is inactive because its catalytic domain is sequestered. Alternatively, the FokI catalytic domain may be able to dissociate from the recognition domain after the FokI makes specific contacts with its DNA substrate but may not be able to properly align its catalytic center for the phosphodiester bond cleavage without dimer formation.
Based on this work and previous reports, the following view of FokI endonuclease emerges:
FokI exists in solution as a monomer, and first binds to DNA as a monomer. The complex is catalytically inactive.
A second FokI monomer arrives either when: (i) DNA is scanned until the monomer collides with the first FokI molecule. Presumably, only monomers arriving in one of two orientations at the 3′ end of the recognition sequence can dimerize successfully; (ii) the monomeric FokI/DNA complex interacts with another monomeric FokI/DNA complex; or (iii) FokI monomer from solution collides with a FokI/DNA complex. This alternative seems the least likely because the binding-deficient FokI mutant proteins are relatively poor stimulators on a molar basis.
The catalytic domain of each FokI molecule swings away from the recognition domain to position its catalytic sites opposite the targeted phosphodiester bond. The residues at the catalytic domain dimerization interface interact with each other, forming a dimer that resembles a structure not unlike a prototypical type II restriction endonuclease bound to DNA. The formation of the dimer may require magnesium ion(s).
Cleavage of both DNA strands occurs after FokI dimerization in the presence of magnesium. The dimerization model offers two levels of control to prevent FokI from cleaving DNA nonspecifically. First, the release of the catalytic domain from its sequestered position may depend on sequence-specific DNA binding. Second, the dimerization of the catalytic domain may be required to achieve proper alignment of the two catalytic domains for phosphodiester bond cleavage.
Dimerization in the presence of specific DNA may be a strategy other type IIs restriction endonucleases use. To date, all type IIs endonucleases studied (MboII, MmeI, Tth111II, Eco57I) are monomers in solution (23–26). MboII binds to its recognition sequence as a monomer (27).
Several chimeric enzymes have been created by fusing the FokI catalytic domain with site-specific DNA binding domains. FokI catalytic domain was fused genetically to Drosophila Ubx homeodomain (9) and with Sp1-QNR and CP-QDR zinc finger proteins (10, 11). Recently, a Z-DNA-specific nuclease was constructed following the same strategy (12). However, the rate and efficiency of DNA cleavage by the hybrid endonucleases are much lower compared with the wild-type FokI (10). It is difficult to achieve complete site-specific cleavage with hybrid nucleases because high protein concentration and/or long incubation times usually lead to a nonspecific DNA cleavage. The dimerization of FokI suggests that cleavage rates and the specificity of chimeric endonucleases may be improved by adding excess exogenous FokI catalytic domain or by constructing a triple fusion where the heterologous DNA binding motif is linked to the covalently fused FokI catalytic domain dimer.
Acknowledgments
We thank W. Jack, C. Noren, and X. Xiong for providing plasmids and FokI constructs. We thank R. Whitaker for assistance with molecular display programs and technical advice in selecting substitutions in FokI that targeted DNA recognition, L. Dorner and R. Kucera for FokI purification, and M.-Q. Xu and S. Chong for assistance with Impact System. We are grateful to the personnel of NEB Organic Synthesis and Sequencing divisions for synthetic DNAs and sequencing of new DNA constructs. We thank S. Halford for helpful discussions and R. Roberts, C. Noren, and R. Whitaker for critical comments on the manuscript. The work in A. Aggarwal’s laboratory was supported by National Institutes of Health Grant GM 44006.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: FokN13Y, variant of FokI endonuclease containing Asn13Tyr substitution; FokCD, FokI catalytic domain representing C-terminal 196 aa of the endonuclease; FokCD/D450A, catalytically inactive catalytic domain containing Asp-450 → Ala substitution; FokD483A, R487A, variant of FokI endonuclease containing Asp-483 →Ala and Arg-487 → Ala substitutions.
References
- 1.Sugisaki H, Kanazawa S. Gene. 1981;16:73–78. doi: 10.1016/0378-1119(81)90062-7. [DOI] [PubMed] [Google Scholar]
- 2.Looney M, Moran L S, Jack W E, Feehery G R, Benner J S, Slatko B E, Wilson G G. Gene. 1989;80:193–208. doi: 10.1016/0378-1119(89)90284-9. [DOI] [PubMed] [Google Scholar]
- 3.Kita K, Kotani H, Sugisaki H, Takanami M. J Biol Chem. 1989;264:5751–5756. [PubMed] [Google Scholar]
- 4.Kaczorowski T, Skowron P, Podhajska A J. Gene. 1989;80:209–216. doi: 10.1016/0378-1119(89)90285-0. [DOI] [PubMed] [Google Scholar]
- 5.Skowron P, Kaczorowski T, Tucholski J, Podhajska A. Gene. 1993;125:1–10. doi: 10.1016/0378-1119(93)90738-o. [DOI] [PubMed] [Google Scholar]
- 6.Waugh D S, Sauer R T. Proc Natl Acad Sci USA. 1993;90:9596–9600. doi: 10.1073/pnas.90.20.9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, Wu L P, Chandrasegaran S. Proc Natl Acad Sci USA. 1992;90:2764–2768. doi: 10.1073/pnas.90.7.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L, Wu L P, Clarke R, Chandrasegaran S. Gene. 1993;133:79–84. doi: 10.1016/0378-1119(93)90227-t. [DOI] [PubMed] [Google Scholar]
- 9.Kim Y-G, Chandrasegaran S. Proc Natl Acad Sci USA. 1994;91:883–887. doi: 10.1073/pnas.91.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y-G, Cha J, Chandrasegaran S. Proc Natl Acad Sci USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang B, Schaeffer J, Li Q, Tsai M-D. J Protein Chem. 1996;15:481–489. doi: 10.1007/BF01886856. [DOI] [PubMed] [Google Scholar]
- 12.Kim Y-G, Kim P S, Herbert A, Rich A. Proc Natl Acad Sci USA. 1997;94:12875–12879. doi: 10.1073/pnas.94.24.12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yonezawa A, Sugiura Y. Biochim Biophys Acta. 1994;1219:369–379. doi: 10.1016/0167-4781(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 14.Wah D, Hirsch J A, Dorner L F, Schildkraut I, Aggarwal A K. Nature (London) 1997;388:97–100. doi: 10.1038/40446. [DOI] [PubMed] [Google Scholar]
- 15.Wah D A, Bitinaite J, Schildkraut I, Aggarwal A K. Proc Natl Acad Sci USA. 1998;95:10564–10569. doi: 10.1073/pnas.95.18.10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman M, Strzelecka T, Dorner L F, Schildkraut I, Aggarwal A K. Science. 1995;269:656–663. doi: 10.1126/science.7624794. [DOI] [PubMed] [Google Scholar]
- 17.Chong S, Mersha F B, Comb D G, Scott M E, Landry D, Vence L M, Perler F B, Benner J, Kucera R B, Hirvonen C A, et al. Gene. 1997;192:271–281. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- 18.Chong S, Yang S, Paulus H, Benner J, Perler F B, Xu M-Q. J Biol Chem. 1996;271:22159–22168. doi: 10.1074/jbc.271.36.22159. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch J, Wah D, Dorner L F, Schildkraut I, Aggarwal A K. FEBS Lett. 1997;403:136–138. doi: 10.1016/s0014-5793(97)00039-2. [DOI] [PubMed] [Google Scholar]
- 20.Wisconsin Package Version 9.1. Madison, WI: Genetics Computer Group; 1997. [Google Scholar]
- 21.Aggarwal A K. Curr Opin Struct Biol. 1995;5:11–19. doi: 10.1016/0959-440x(95)80004-k. [DOI] [PubMed] [Google Scholar]
- 22.Newman M, Strzelecka T, Dorner L F, Schildkraut I, Aggarwal A K. Nature (London) 1994;368:660–664. doi: 10.1038/368660a0. [DOI] [PubMed] [Google Scholar]
- 23.Sektas M, Kaczorowski T, Podhajska A J. Nucleic Acids Res. 1992;20:433–438. doi: 10.1093/nar/20.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tucholski J, Skowron P, Podhajska A J. Gene. 1995;157:87–92. doi: 10.1016/0378-1119(94)00787-s. [DOI] [PubMed] [Google Scholar]
- 25.Shinomiya T, Kobayashi M, Sato S. Nucleic Acids Res. 1980;8:3275–3285. doi: 10.1093/nar/8.15.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janulaitis A, Petrusyte M, Maneliene Z, Klimasauskas S, Butkus V. Nucleic Acids Res. 1992;20:6043–6049. doi: 10.1093/nar/20.22.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sektas M, Kaczorowski T, Podhajska A J. Gene. 1995;157:181–185. doi: 10.1016/0378-1119(94)00742-b. [DOI] [PubMed] [Google Scholar]