Abstract
Lateral inhibition is one of the key functions of Notch signaling during animal development. In the proneural clusters that give rise to Drosophila mechanosensory bristles, Delta (Dl) ligand in the sensory organ precursor (SOP) cell is targeted for ubiquitination by the E3 ligase Neuralized (Neur), resulting in activation of Dl’s capacity to signal to the Notch receptor on neighboring cells. The cells that receive this signal activate a genetic program that suppresses their SOP fate potential, insuring that only a single SOP develops within each cluster. Using multiple lines of investigation, we provide evidence that members of the Bearded family of proteins (BFMs) inhibit Dl activation in non-SOP cells by binding to Neur and preventing it from interacting with Dl. We show that this activity of BFMs is dependent on the conserved NXXN motif, and report the unexpected finding that several BFMs include multiple functional copies of this motif. We find that a conserved NXXN motif in the intracellular domain of Dl is responsible for its interaction with Neur, indicating direct competition between Dl and BFMs for binding to Neur, and we show that Neur-dependent endocytosis of Dl requires the integrity of its NXXN motif. Our results illuminate the mechanism of an important regulatory event in Notch signaling that appears to be conserved between insects and crustaceans.
Keywords: Bearded, Neuralized, Notch, Delta, endocytosis, lateral inhibition
Introduction
The evolutionarily conserved Notch cell-cell signaling pathway is utilized extensively for cell fate specification in developing metazoans. During peripheral nervous system (PNS) development in Drosophila, Notch-mediated lateral inhibition results in the specification of a single sensory organ precursor (SOP) cell from a field of cells, known as a proneural cluster (PNC), that all express proneural transcriptional activator proteins and hence have SOP cell fate potential. In the SOP, the Notch ligand Delta (Dl) is targeted by the E3 ubiquitin ligase Neuralized (Neur), which leads to Dl’s ubiquitination and endocytosis (Deblandre et al., 2001; Lai et al., 2001; Pavlopoulos et al., 2001), processes necessary to make the SOP an effective Notch pathway signaling cell (Pavlopoulos et al., 2001; Li and Baker, 2004; Wang and Struhl, 2004). In response to this signal from the SOP, the surrounding cells in the PNC activate a genetic program that suppresses their potential to become SOPs and commits them instead to an epidermal fate. Among the direct transcriptional targets of the Notch pathway in responding cells are the basic helix-loop-helix (bHLH) repressor genes of the Enhancer of split Complex [E(spl)-C] and members of the Bearded (Brd) family of genes, which reside in the E(spl)-C and in the Brd Complex (Brd-C) (Bailey and Posakony, 1995; Furukawa et al., 1995; Lecourtois and Schweisguth, 1995; Nellesen et al., 1999; Lai et al., 2000a; Lai et al., 2000b).
The Brd gene family was discovered through genetic and molecular analysis of a gain-of-function mutation of the Brd gene that confers mutant phenotypes in the adult PNS suggestive of a loss of Notch signaling capacity, including a bristle “tufting” effect resulting from the failure of lateral inhibition (Leviten and Posakony, 1996; Leviten et al., 1997). Indeed, it was subsequently shown that nearly all Brd family genes, including the E(spl)-C genes mα, m4, and m6, and the Brd-C genes Brd, Brother of Bearded (Bob), Twin of m4 (Tom), and Ocho, produce a similar Notch pathway loss-of-function phenotype when over- or misexpressed in PNCs (Apidianakis et al., 1999; Lai et al., 2000a; Lai et al., 2000b; Zaffran and Frasch, 2000). In contrast, the E(spl)-C Brd family gene m2 produces an oppositely directed phenotype (SOP loss) when misexpressed, reminiscent of Notch pathway hyperactivity (Lai et al., 2000b).
Brd family genes, which have thus far been found only in insects (Lai et al., 2000b; Lai et al., 2005; Schlatter and Maier, 2005), encode small proteins (70-218 a.a. in Drosophila) that are characterized by a predicted highly basic amphipathic alpha-helix located near the N terminus, termed the B domain (Leviten et al., 1997; Lai et al., 2000a; Lai et al., 2000b). The canonical members of the family in Drosophila, E(spl)mα, E(spl)m4, Tom, and Ocho, also share three additional conserved motifs (Lai et al., 2000a; Lai et al., 2000b): the N motif [NxANE(K/R)L], the G motif (VPVHFARTXXGTFFWT), and the D motif [DRW(A/V)QA]. The non-canonical family members [Brd, Bob, E(spl)m2, and E(spl)m6] contain one or two of these additional motifs, with E(spl)m2 being the only family member that does not bear an N motif (Leviten et al., 1997; Lai et al., 2000a; Lai et al., 2000b).
An interaction between Brd family proteins and Neur was first revealed in a comprehensive yeast two-hybrid screen, which detected Tom as a partner for Neur (Giot et al., 2003). Subsequent studies showed that, during the Notch-mediated specification of the mesoderm-ectoderm boundary in the Drosophila embryo, Tom acts as a Neur antagonist, capable of preventing the Neur-dependent endocytosis of Dl (Bardin and Schweisguth, 2006; De Renzis et al., 2006). It was also found that Tom can interfere with the co-immunoprecipitation of Dl and Neur in a cell culture assay, suggesting that Tom inhibits Dl-Neur binding (Bardin and Schweisguth, 2006). The N motif of Tom was shown to be important for its interaction with Neur, in that deletion or mutation of the motif weakened the interaction in both the yeast two-hybrid and co-immunoprecipitation assays. While these studies illuminated the interaction between Brd family proteins and Neur, the interaction between Neur and the Notch ligands Dl and Serrate (Ser) remains poorly understood. Furthermore, a requirement for the N motif in the inhibitory activities of Brd family proteins has not been demonstrated.
In this study, we have investigated the function of Brd proteins during lateral inhibition. We report the unexpected finding that the canonical Brd proteins E(spl)mα and E(spl)m4 contain multiple N motifs, and we show that these sequences are responsible for mediating the interaction with Neur. Integrity of these N motifs is also required for the capacity of E(spl)mα and E(spl)m4 to disrupt Neur-Dl binding in vitro and to interfere with lateral inhibition in vivo. Our definition of a more comprehensive consensus for the N motif permitted us to identify it as a conserved feature of the intracellular domains of arthropod Dl and Ser proteins. We show that, as for Brd proteins, Dl’s N motif is required for its binding to Neur in vitro, and we present in vivo evidence that the motif is also required for Neur-dependent endocytosis of Dl. We therefore propose that Brd family proteins antagonize Notch signaling by competing directly with Dl for N motif-mediated binding to Neur. Finally, we report the existence of a gene encoding a Brd family protein in the crustacean Daphnia pulex, pushing the known origin of this family back to more than 400 Mya. We show that this protein interacts specifically with Drosophila Neur in vitro, indicating the long-term evolutionary conservation of this key BFM activity.
Materials and methods
GAL4/UAS driver and responder lines
The following GAL4 driver lines were used for mis- or overexpression of UAS responder transgenes: yw; sca-GAL4 (Hinz et al., 1994; Nakao and Campos-Ortega, 1996); w1118 E(spl)mα-GAL4 (Castro et al., 2005); yw; neurP72-GAL4 UAS-PonGFP/TM6C (Bellaiche et al., 2001); and w1118; dpp-GAL4/CyO (kindly provided by Ethan Bier). UAS-neur and UAS-GFP (UAS-Stinger) have been described previously (Barolo et al., 2000; Lai and Rubin, 2001).
Generation of pUAST-V5-HIS
To create a UAS vector capable of C-terminally tagging expressed proteins with a V5 epitope and polyhistidine sequence, the multiple cloning site (MCS) and epitope region of the vector pAc5.1-V5-HIS-A (Invitrogen) was amplified using the forward primer GGCAATTGGGTACCTACTAGTCCAGT and the reverse primer GGGCTAGCCCTTAGAAGGCACAGTCGA, which introduce a 5′ MfeI site and 3′ NheI site. This amplicon was cloned into the pUAST vector (Brand and Perrimon, 1993) cut with EcoRI and XbaI, replacing the entire MCS of pUAST with this new sequence.
Misexpression constructs
FLAG-m4 constructs were generated by introducing the codons for a 1X FLAG tag (DYKDDDDK) after the starting M codon of E(spl)m4; 20 bp of the gene’s 3′ UTR sequence were also included in the construct. E(spl)mα constructs included 7 bp of 5′ UTR sequence along with the coding sequence. Dl and DlN constructs were generated using the full Dl coding sequence, isolated from w1118 embryo cDNA. DlN was mutated so as to encode the substitution NEQNAV→AAAAAA. These transgenes were cloned into the pUAST vector or the pUAST-V5-HIS vector and transformed into Drosophila using a standard P transposable element injection protocol (Rubin and Spradling, 1982).
By in situ hybridization to late third-instar wing discs, we verified that transcripts from the various E(spl)m4 and E(spl)mα UAS transgenes accumulate to comparable levels when driven by sca-GAL4 (see Supplementary Fig. S1).
In vitro constructs
Plasmid constructs encoding GST-tagged and His-tagged proteins were generated by cloning into pGEX-5X (Amersham Biosciences) and pRSET (Invitrogen) vectors, respectively. His-mα, His-m4, His-Dlintra, and His-DpBFM constructs all contained 7 bp of 5′ UTR from E(spl)mα along with their respective coding sequences. His-mα-N encodes a peptide that includes amino acids 63-80 of E(spl)mα, centered on the N motif (AEIDENAANEKLAQLAHS). His-mα-N mutant substitutes the two asparagine residues of the core NXXN motif with alanines (AEIDEAAAAEKLAQLAHS). His-Hairless148-311 (His-H148-311) encodes amino acids 148-311 of the Hairless protein, and was kindly provided by Feng Liu.
Bristle count assays
For Brd family gain-of-function phenotypes, 25 females per independent insertion line, from 2-4 representative lines per construct, were scored for the number of extra bristles present at 18 notum positions (notopleurals, presuturals, supra-alars, post-alars, dorsocentrals) and eight head positions (post-verticals, inner verticals, outer verticals, occellars), for a total of 26 bristle positions. The GAL4 drivers sca-GAL4, E(spl)mα-GAL4, and neur-GAL4 were used to direct expression in PNCs, non-SOPs of the cluster, and SOPs, respectively.
In vitro pulldown assays: Preparation of tagged proteins
Tagged proteins were expressed in E. coli strain BL21(DE3) using an IPTG-inducible T7 promoter. Bacterial cultures were grown at 37°C to OD600 = 0.6-0.7, induced with 0.8 mM IPTG, and incubated for 3 hours at 30°C. Bacteria were spun down at 6,000 X g for 15 minutes and pellets were frozen at -80°C.
Bacterial pellets for His-tagged proteins were resuspended in Cell Lysis Buffer (20 mM Tris-HCl pH 8.0; 200 mM NaCl; 0.5% Nonidet P-40; 2 μg/mL Aprotenin; 2 μg/mL Leupeptin; 0.2 mM PMSF; 1 μg/mL Pepstatin A) (2.5 mL per 40 mL culture) and lysed with 100 μg/mL lysozyme for 30 minutes on ice. 5 mM DTT was added, and the lysate was sonicated and centrifuged at 4°C for 25 minutes at 10,000 X g. Supernatant containing the His-tagged protein was saved and used directly for the pulldown assays. His-Neur lysate was not subjected to the last centrifugation step, and was instead run through a 25-gauge needle five times.
Bacterial pellets for GST-tagged proteins were resuspended in STE buffer [10 mM Tris-HCl pH 8.0; 150 mM NaCl; 1 mM EDTA; 2 μg/mL Aprotenin; 2 μg/mL Leupeptin; 0.2 mM PMSF; 1 μg/mL Pepstatin A; (Mercado-Pimentel et al., 2002)] (6 mL per 100 mL culture) and lysed with 100 μg/mL lysozyme for 30 minutes on ice. 1% Sarcosyl and 5 mM DTT were added, and the lysate was sonicated and centrifuged at 4°C for 25 minutes at 10,000 X g. The cleared lysate was incubated with Glutathione Sepharose 4B beads (GE Healthcare Life Sciences) for 3-4 hours at 4°C with rocking to bind the GST-tagged proteins. Beads were washed 4X with Cell Lysis Buffer and this purified sample was used for the pulldown assay.
In vitro pulldown assays: Assay conditions
For GST-Neur pulldowns, 25 μL of packed Glutathione Sepharose beads with bound GST-tagged protein was incubated with His-tagged protein lysate in Cell Lysis Buffer in a total volume of 400 μl for 2-4 hours at 4°C with rocking. Beads were spun down at 300 X g for 1.5 minutes and washed 3X, 1 mL each, with Cell Lysis Buffer minus protease inhibitors. Washed resin was resuspended in SDS-loading dye with 10 mM DTT, boiled for 6 minutes and Western blotted using standard procedures. Mouse anti-HisG antibody (Invitrogen) was used at a 1:5,000 dilution and goat-anti-mouse-HRP (Jackson Laboratories) was used at 1:10,000. Western Lightning Chemiluminescence Reagent Plus (NEL105, Perkin-Elmer) was used for detection.
For GST-Dlintra pulldowns, 25 μl of packed Glutathione Sepharose beads with bound GST-tagged protein was incubated with His-Neur in Cell Lysis Buffer in a total volume of 400 μl for 2 hours at 4°C with rocking. Resin was spun down at 300 X g for 1.5 minutes, and the supernatant was removed. His-tagged competitors in Cell Lysis Buffer were added to a total volume of 400 μl and incubated at 4°C for 2 hours with rocking. Resin was spun, washed, resuspended, and blotted as above.
For all assays, we verified by Coomassie staining that the amount of control GST protein bound to the beads was at least equal to, and in most cases greatly exceeded, the amount of experimental GST-X protein.
Immunohistochemistry
Late third-instar larvae were dissected in phosphate-buffered saline (PBS) + 0.1% Triton X-100, fixed for 25 minutes with 4% paraformaldehyde in PBS + 0.3% Triton X- 100, and washed 5 × 10 minutes with PBS + 0.1% Triton X-100. Misexpressed mα-V5-HIS variants were visualized with mouse monoclonal anti-V5 (Invitrogen) diluted 1:400 and Alexa Fluor 488 donkey anti-mouse IgG (Molecular Probes) diluted 1:500. Misexpressed Dl and DlN were visualized with mouse monoclonal anti-Dl (C594-9B; Developmental Studies Hybridoma Bank) diluted 1:100 and Alexa Fluor 488 donkey anti-mouse IgG diluted 1:500. Images were acquired on a Leica TCS SP2 confocal microscope.
Identification and cloning of a Daphnia Brd family gene (Dp BFM)
Using a TBLASTN search with Drosophila bHLH repressor (bHLH-R) sequences on the Daphnia pulex genome (http://wfleabase.org/), we identified three bHLH-R genes on scaffold 170. Loading the scaffold sequence into GenePalette (Rebeiz and Posakony, 2004), we searched for conserved regulatory sequence motifs associated with Brd family genes in insects, including proneural protein (RCAGSTG) and Su(H) (YGTGDGAA) binding sites, as well as three 3′ UTR “seed” motifs that mediate miRNA recognition, the GY box (GTCTTCC), K box (TGTGAT), and Brd box (AGCTTTA). Scanning the scaffold for clusters of these motifs, we identified several regions with the potential to contain a Brd family gene; we then inspected the conceptual translations of these regions for the presence of conserved protein motifs typically found in Brd family proteins. The identified BFM was cloned from the Log50 strain of Daphnia pulex, obtained from Dr. Matthias Westphal at the Center for Genomics and Bioinformatics, Indiana University, Bloomington.
Results
Cell-type origin of the Brd family gain-of-function phenotype
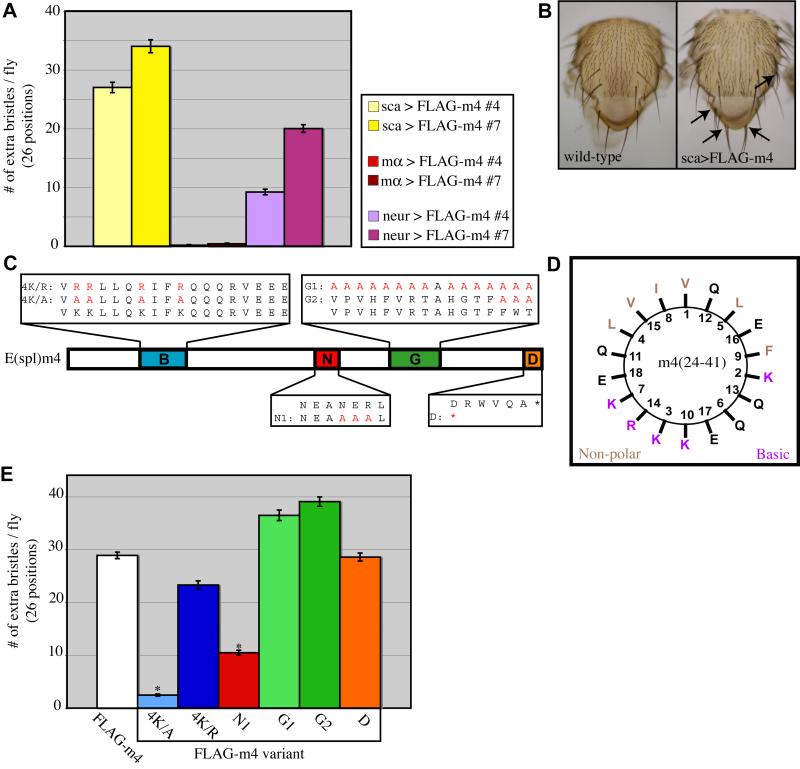
We have reported previously that over- or misexpression of seven of the eight Brd family genes in Drosophila [E(spl)m2 being the exception] causes developmental defects consistent with a loss of Notch signaling activity; i.e., failure of lateral inhibition in PNCs and cell fate transformations in the sensory organ lineage (Lai et al., 2000a; Lai et al., 2000b). These studies made use of the scabrous (sca)-GAL4 driver, which is active in both of the distinct cell populations within PNCs, the SOP and the surrounding non-SOP cells. The observed phenotypes could arguably be caused by overexpression — abnormally high levels of BFMs in their normal domain of expression (non-SOP cells), possibly producing a dominant-negative effect by sequestering some important factor(s) necessary for Notch signal transduction. Alternatively, these effects could be due to misexpression — BFMs mimicking their normal function in a cell type in which they are not normally expressed at any significant level (SOP cells). To distinguish between these possibilities, we made use of available GAL4 drivers with distinct expression specificities. When activated throughout the PNC using sca-GAL4, two independent insertions of a UAS construct expressing N-terminally FLAG-tagged E(spl)m4 (UAS-FLAG-m4) produce 27 and 34 extra bristles per fly, respectively, scored at 26 bristle positions on the notum and dorsal head of the fly (Fig. 1A,B). Driving high levels of FLAG-m4 expression specifically in non-SOP cells using E(spl)mα-GAL4 fails to produce any significant mutant phenotype, with 0.16 and 0.40 extra bristles per fly for the two insertions, respectively. By contrast, when FLAG-m4 is misexpressed solely in SOPs using a neur-GAL4 driver, a substantial disruption of lateral inhibition is observed (9.2 and 20 extra bristles per fly for the two UAS responder insertions, respectively), suggesting that misexpression of a BFM in the SOP disrupts the sending of the Dl signal from that cell, perhaps mimicking the normal function of BFMs in non-SOPs.
Fig. 1.
Integrity of the B domain and N motif of E(spl)m4 are important for the gain-of-function phenotype. (A) The Brd family gain-of-function phenotype results from misexpression in SOP cells. Expression of FLAG-m4 throughout PNCs (sca-GAL4; yellow bars) or specifically in SOPs (neur-GAL4; purple bars) results in the production of extra bristles on the notum and head. Overexpression in non-SOP cells of the PNC fails to produce a mutant phenotype (mα-GAL4; red bars). (B) Wild-type notum (left) shows the stereotypical pattern of macrochaete mechanosensory bristles, while flies expressing FLAG-tagged E(spl)m4 protein (FLAG-m4) under the control of the sca-GAL4 driver (right) show extra macrochaetes at multiple positions (arrows). (C) Domain/motif variants of E(spl)m4. The B domain (basic amphipathic alpha helix) and the N, G, and D motifs (Lai et al., 2000b) are indicated. Substituted residues are depicted in red. (D) Helical wheel plot of E(spl)m4’s B domain predicts the clustering of non-polar residues on one face of the helix and of basic residues on the opposite face. (E) Extra-bristle phenotypes resulting from misexpression of FLAG-m4 variants using the sca-GAL4 driver. Error bars indicate standard errors; asterisks denote statistical significance of differences from the wild-type (FLAG-m4) results (P<0.04; Mann-Whitney U test).
The basic amphipathic character of the B domain of E(spl)m4 is required for the gain-of-function phenotype
To gain a better understanding of the mechanism of Brd family protein activity during lateral inhibition, we sought to identify the properties of these proteins that are required to produce the characteristic gain-of-function phenotype (see Introduction and previous section). We had observed earlier that disrupting the helical nature of the B domain of Brd, via four proline substitutions on the hydrophobic helical face, has no significant effect on the protein’s ability to produce a neurogenic phenotype when misexpressed (Lai, 1999). By contrast, substituting neutral alanine residues for the basic lysine residues of the B domain in either Brd or Bob was found to eliminate the gain-of-function phenotype. In the present study, we extended these findings on non-canonical BFMs by testing several variants of the canonical BFM E(spl)m4 (Fig. 1C).
When misexpressed using the sca-GAL4 driver, FLAG-m4 produces an average of 28.9 extra bristles per fly (Fig. 1B,E). The FLAG-m44K/A variant eliminates the four basic lysine residues of the B domain, substituting them with alanines, which should not disrupt the helical nature of this region (Fig. 1C,D). This mutation nearly abolishes the protein’s ability to produce extra bristles when misexpressed (2.46 extra bristles per fly; Fig. 1E), phenocopying the results obtained with the corresponding mutants of Brd and Bob (Lai, 1999). To test whether the lysine residues of the B domain per se are required to produce the misexpression phenotype, the FLAG-m44K/R variant was created, replacing the lysine residues with arginines while retaining both the helical nature and the strong basic amphipathicity of the domain (Fig. 1C,D). Misexpression of FLAG-m44K/R produces 23 extra bristles per fly, indicating a retained capacity to disrupt lateral inhibition (Fig. 1E). These data indicate that the basic amphipathic nature of E(spl)m4’s B domain is required to disrupt lateral inhibition when misexpressed, while the lysine residues themselves are dispensable.
The N motif of E(spl)m4 contributes to its misexpression phenotype
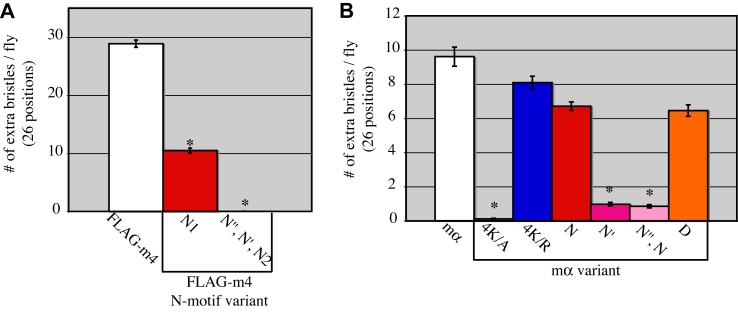
Outside of the four conserved domains/motifs found in canonical Brd family proteins, overall sequence similarity between the various family members is low (Lai et al., 2000a; Lai et al., 2000b). To assess the importance of the other conserved motifs in producing a misexpression phenotype, additional variants of FLAG-m4 were created that disrupt the N, G, and D motifs (Fig. 1C). The FLAG-m4N1 variant mutates the core of the N motif to alanines (NEANERL to NEAAAAL; Fig. 1B). When misexpressed, this variant produces an average of 10.5 additional macrochaete bristles per fly, compared with the wild-type FLAG-m4 phenotype of 28.9 extra bristles (Fig. 1E). This weakening of the misexpression phenotype was consistently observed with four independent insertions of the construct, and points to a role for the N motif in disrupting lateral inhibition, though it seems not to be strictly required.
Two different G-motif variants were constructed, FLAG-m4G1 mutating all 16 amino acids of the extended G motif (VPVHFVRTAHGTFFWT) to alanines, and FLAG-m4G2 mutating only the more highly conserved region (FWT) to alanines (Fig. 1C). Both FLAG-m4G1 and FLAG-m4G2 produce strong misexpression phenotypes, an average of 36 and 39 extra macrochaetes per fly, respectively (Fig. 1E). We conclude that the G motif of E(spl)m4 is not required for the misexpression phenotype.
The D motif consists of six amino acids found at the C terminus of the protein (DRWVQA); its disruption was accomplished with a stop-codon truncation of the protein just N-terminal to this motif (Fig. 1C). We find that the FLAG-m4D variant produces a misexpression phenotype of 29 extra bristles per fly (Fig. 1E), very similar to that of wild-type FLAG-m4, indicating that the D motif is likewise not required for the disruption of lateral inhibition in this assay.
The recognized conserved motifs of E(spl)mα are not required for interaction with Neur in vitro
It has been reported that the interaction of the Brd protein Tom with the E3 ubiquitin ligase Neur is mediated by the N motif, and it was suggested that this interaction is the basis of Notch signaling inhibition in vivo (Bardin and Schweisguth, 2006). However, the fact that mutating the N motif of E(spl)m4 only reduced, but did not eliminate, the protein’s ability to disrupt lateral inhibition when misexpressed in the SOP led us to believe that another, unidentified, motif may participate in this function.
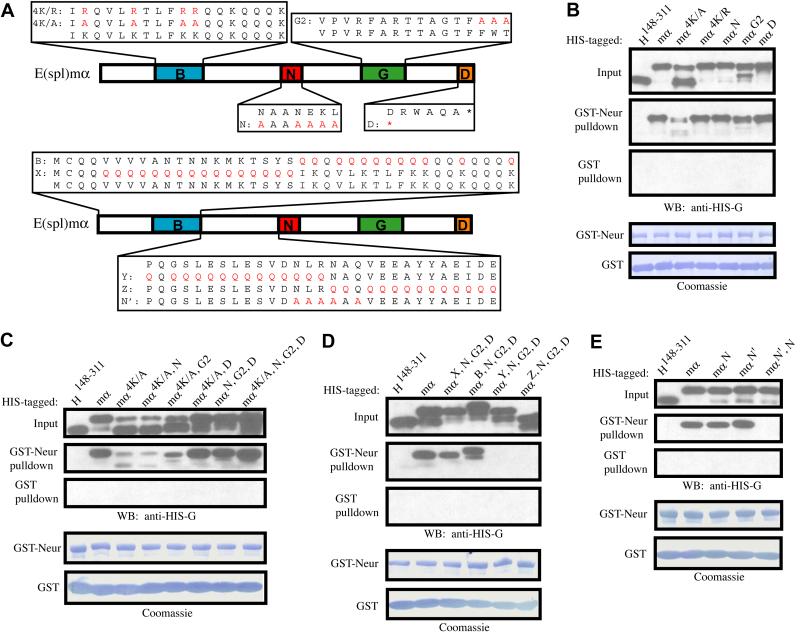
To assess the role of each of its conserved domains/motifs in direct protein-protein interaction with Neur, His-tagged variants of E(spl)mα were generated, similar to those described above for E(spl)m4 (Fig. 2A, upper image). Bacterial cell lysates containing these His-tagged proteins were used in a pulldown assay with bacterially expressed and purified GST-Neur, or GST, bound to Glutathione Sepharose beads. The His-tagged negative control, His-Hairless148-311 (His-H148-311) was chosen because it is of comparable size to His-mα and is not expected to have affinity for GST-Neur. Indeed, His-H148-311 does not bind to GST-Neur in this assay, whereas His-mα shows a strong interaction (Fig. 2B). The E(spl)mα variants His-mα4K/R, His-mαN, His-mαG2, and His-mαD also show efficient binding to GST-Neur, whereas His-mα4K/A shows a substantial decrease in binding (Fig. 2B). These data indicate that none of the three identified motifs in E(spl)mα (N, G, or D) is required for strong in vitro interaction with Neur, while loss of the basic amphipathic character of the B domain impairs, but does not eliminate, binding to Neur.
Fig. 2.
E(spl)mα contains two N motifs capable of interacting with Neur. (A) Variants of E(spl)mα. The B domain and the N, G, and D motifs are indicated. Substituted residues are depicted in red. (B-E) Western blots of pulldown assays of the interaction between Neur and E(spl)mα. Also shown are Coomassie-stained blots depicting amounts of bead-bound GST (control) and GST-Neur proteins in each assay. (B) The variant mα4K/A is the only single mutant that weakens E(spl)mα’s interaction with Neur; this variant also shows an atypical gel migration pattern. (C) Normal gel migration and strong interaction with Neur are both restored in the mα4K/A, D double mutant, and mutation of all four conserved domains/motifs (mα4K/A, N, G2, D) does not abolish the E(spl)mα-Neur interaction. (D) In an mαN, G2, D background, mutation of regions X and B have no affect on E(spl)mα’s binding to Neur, while mutation of either region Y or region Z eliminates the interaction completely. (E) The N’ motif is found in the zone of overlap between regions Y and Z (see A). While the mαN and mαN’ variants interact normally with Neur, the mαN’, N variant has lost this capacity.
We note that while all His-mα variants display more than one band when electrophoresed on SDS-polyacrylamide gels, all but His-mα4K/A exhibit an upper band containing the great majority of the protein, with a minor lower band of variable intensity depending on the preparation. His-mα4K/A instead consistently displays a gel pattern with the majority of the protein in the lower band. While this major lower band of His-mα4K/A binds poorly to GST-Neur, the minor upper band binds relatively more strongly (Fig. 2B). We suggest that this exceptional behavior of His-mα4K/A may be due to abnormal folding of at least a majority of the protein. In any case, we reasoned that the residual Neur interaction observed with His-mα4K/A might be mediated by a second motif that cooperates with the B domain. Based on our misexpression data (above) and a previous report (Bardin and Schweisguth, 2006), it seemed likely that the N motif fills this role.
To test our hypothesis that multiple elements of E(spl)mα are important for its interaction with Neur, additional variants of the protein were generated, each containing mutations in two or more domains/motifs. We find that the double mutants His-mα4K/A, N and His-mα4K/A, G2 behave like His-mα4K/A, in that they both display a major-lower-band gel pattern and interact poorly, though clearly detectably, with GST-Neur (Fig. 2C). By contrast, we were surprised to observe that elimination of the D motif in the double mutant His-mα4K/A, D restores both the protein’s wild-type gel migration pattern and its ability to bind efficiently to GST-Neur. Moreover, both the triple mutant His-mαN, G2, D and the quadruple mutant His-mα4K/A, N, G2, D likewise migrate quite normally and interact strongly with GST-Neur (Fig. 2C). From these data we conclude that none of the recognized conserved domains/motifs of E(spl)mα are required for a strong interaction with Neur. The implication is that E(spl)mα contains one or more uncharacterized motifs capable of interacting with Neur.
The aberrant migration displayed by the His-mα4K/A variant seems to result from disrupting the amphipathicity of the B domain [Fig. 2B; recall that mα4K/R displays a normal migration pattern (Fig. 2B)]. We find that mutating the lysines of the B domain to uncharged, polar glutamines (mα4K/Q) produces this same effect, as does mutating only the five non-polar residues of E(spl)mα’s B domain to glutamine (mα5np/Q; data not shown; see Fig. 2B). In all cases, deletion of the D motif in combination with the B domain mutation restores a wild-type gel migration pattern, as well as strong binding of the protein to Neur (data not shown).
E(spl)mα and E(spl)m4 each contain multiple N motifs capable of mediating interaction with Neur
Because we have observed that a truncated version of E(spl)mα, extending from the N terminus to a point just N-terminal to the N motif, is capable of binding to Neur (data not shown), we focused on this region in our search for a possible uncharacterized motif capable of interacting with Neur. Using the triple mutant His-mαN, G2, D as a backbone, we substituted large stretches of amino acids in the N-terminal portion of the protein with glutamine residues. The variant mαX, N, G2, D contains a poly-Q stretch covering amino acids 5-20, mαB, N, G2, D a.a. 21-38 (the entire B domain), mαY, N, G2, D a.a. 39-53, and mαZ, N, G2, D a.a. 54-67 (just N-terminal to the N motif; Fig. 2A, lower image).
Region X and the B domain are not required for interaction with Neur, as mutations in these blocks of amino acids did not affect the apparent affinity of the corresponding His-mα variants for GST-Neur (Fig. 2D). However, binding to GST-Neur is completely eliminated for both His-mαY, N, G2, D and His-mαZ, N, G2, D (Fig. 2D), indicating that there exists at least one functional element in the section of E(spl)mα between the B domain and the N motif, possibly near the junction of the Y and Z regions (Fig. 2A, lower image). Repeating the pulldown assay using E(spl)mα variants bearing fewer substituted amino acids in these two regions led to the discovery of an N-like motif [NLRNAQV, termed N’ (“N-prime”)] that spans the junction between Y and Z and is required for interaction with Neur in an mαN, G2, D background (data not shown). To test the possibility that E(spl)mα contains two N motifs that are independently capable of mediating interaction with Neur, the single-motif mutant His-mαN’ and the double mutant His-mαN’, N were generated (Fig. 2A). His-mαN’ behaves like His-mαN and interacts strongly with Neur, while the His-mαN’, N variant lacks affinity for GST-Neur (Fig. 2E). From these data we conclude that E(spl)mα contains two N motifs that are each individually capable of mediating a robust interaction with Neur.
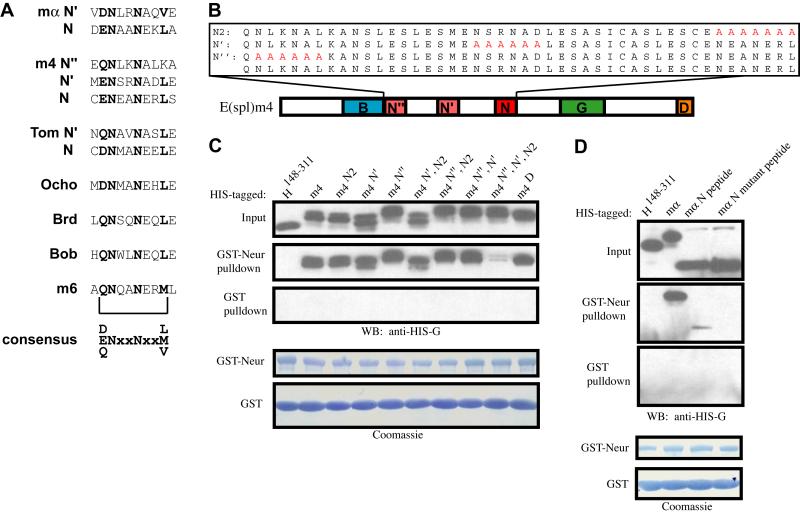
The finding of a second N motif in E(spl)m〈 raised the possibility that other Brd family proteins may also contain additional N motifs. The previously recognized consensus sequence for the N motif was NXANE(K/R)L (Lai et al., 2000b). The N’ element in E(spl)mα shares only the core NXXN with this consensus. Using this simplified motif definition (with X≠N), we find that among the canonical Brd family proteins, E(spl)m4 contains three potential N motifs, while Tom and Ocho include two and one potential N motifs, respectively (Fig. 3A). [We suggest that the existence of a second (N’) motif in Tom is likely to account for the failure of mutations affecting its original N motif to fully eliminate Tom-Neur co-immunoprecipitation (Bardin and Schweisguth, 2006).] The non-canonical family members Brd, Bob, and E(spl)m6 appear to contain only the single previously identified N motif, while E(spl)m2 has no N motifs, even with this looser definition. Aligning all of these N motifs yields the new consensus (D/E/Q)NXXNXX(L/M/V) (Fig. 3A).
Fig. 3.
A refined consensus identifies multiple N motifs in Brd family proteins, each sufficient to mediate binding to Neur. (A) Canonical BFMs E(spl)mα, E(spl)m4, and Tom contain two or more N motifs defined by the new consensus (D/E/Q)NXXNXX(non-polar). Conserved residues within each N motif are shown in bold; note that the N” motif of E(spl)m4 violates the consensus at the non-polar residue position. (B) N-motif variants of E(spl)m4. The B domain and the N, G, and D motifs are indicated. Substituted residues are depicted in red. (C,D) Western blots of pulldown assays. Also shown are Coomassie-stained blots depicting amounts of bead-bound GST (control) and GST-Neur proteins in each assay. (C) The integrity of any of the three N motifs in E(spl)m4 is sufficient for interaction with Neur. Only mutation of all three of these motifs (m4N”, N’, N2) results in the disruption of the E(spl)m4-Neur interaction. (D) An 18-a.a. segment containing the E(spl)mα N motif (AEIDENAANEKLAQLAHS) is sufficient to mediate a weak interaction with Neur. This interaction is dependent on the two asparagine residues of the core NXXN sequence, as a mutant peptide (AEIDEAAAAEKLAQLAHS) fails to bind Neur.
We sought to determine whether all three potential N motifs in E(spl)m4 are capable of mediating interaction with Neur by assaying the single-motif mutants His-m4N2, His-m4N’, and His-m4N”, as well as the triple N-motif mutant and every combination of double N-motif mutant (Fig. 3B,C). Consistent with the results obtained with E(spl)mα, E(spl)m4 is capable of interacting strongly with Neur as long as it contains any of the three N motifs. Binding to Neur is severely reduced only when all three N motifs are mutant (Fig. 3C). Finally, in contrast to a previous finding concerning the N motif of Tom (Bardin and Schweisguth, 2006), we observe that a short peptide containing the N motif of E(spl)mα is sufficient to mediate a weak interaction with Neur in the pulldown assay, in a manner dependent on the two asparagine residues of the NXXN core (Fig. 3D).
The N motifs of E(spl)mα and E(spl)m4 are required to disrupt the Neur-Dl interaction
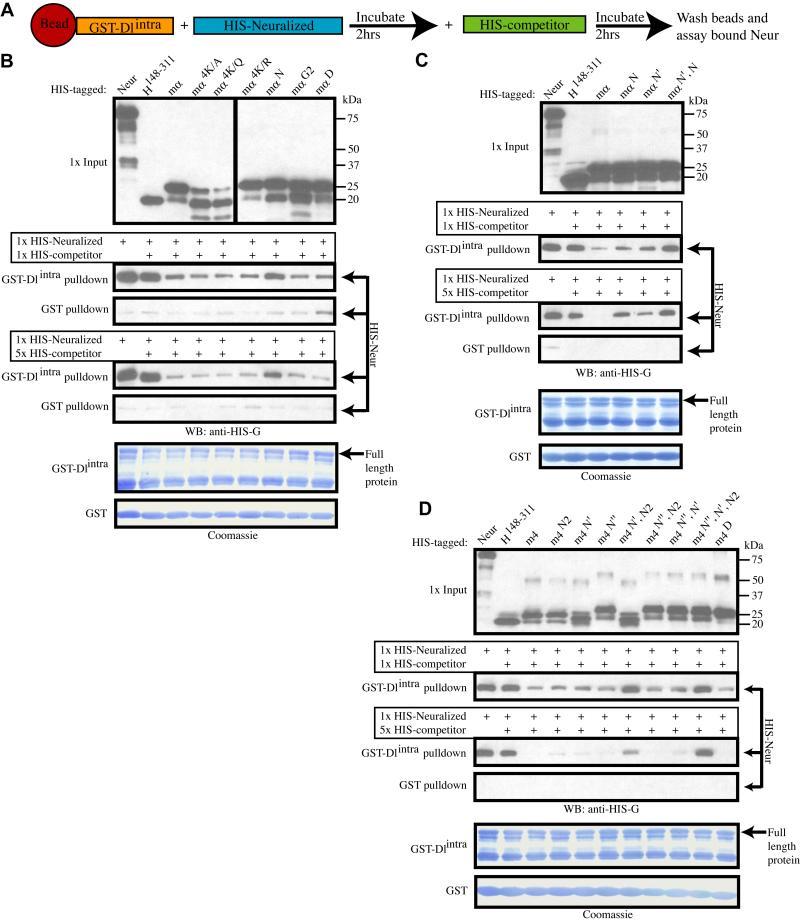
We have seen that misexpression of BFMs in the SOP of a PNC prevents the SOP from sending an effective inhibitory signal to the Notch receptor on non-SOPs, thus disrupting lateral inhibition. Given their interaction with Neur (Giot et al., 2003; Bardin and Schweisguth, 2006), BFMs might do this either by interfering with the E3 ligase activity of Neur, thus preventing the conversion of Dl into an active ligand for Notch, or by interfering with the binding of Neur to its substrate Dl. To test this latter possibility, we employed an in vitro binding inhibition assay (Fig. 4A). Bacterially expressed, purified, GST-tagged Dl intracellular domain (GST-Dlintra), bound to Glutathione Sepharose beads, was incubated with bacterial cell lysate containing His-tagged Neur (His-Neur) to permit binding between the two proteins to occur. Following incubation, a His-tagged competitor, either His-H148-311 (negative control) or a His-mα variant, was added. After a second incubation period, the amount of His-Neur still bound to GST-Dlintra was assayed (Fig. 4A).
Fig. 4.
Brd family proteins compete with Dl for binding to Neur. (A) Schematic of the competition pulldown assay. (B-D) Western blots of competition pulldown assays. Also shown are Coomassie-stained blots depicting amounts of bead-bound GST (control) and GST-Dlintra proteins in each assay. Neur is efficiently pulled down by GST-Dlintra, and this interaction is not strongly affected by the presence of the control competitor H148-311. (B) Addition of wild-type mα, or any variant except mαN, disrupts the Neur-Dlintra interaction in a dose-dependent manner. (B, C) Mutation of either the N or N’ motif weakens E(spl)mα’s ability to disrupt this interaction, while the double mutant mαN’, N lacks this ability (C). (D) Similarly, addition of E(spl)m4 also disrupts the Neur-Dlintra interaction, and the triple mutant m4N”, N’, N2 loses this ability. The N” motif of E(spl)m4 is weaker than the N or N’ motifs at disrupting the Neur-Dlintra interaction, as seen with variant m4N’, N2.
His-Neur binds efficiently to GST-Dlintra, an interaction that is not significantly affected by the addition of the control competitor His-H148-311 (Fig. 4B). The addition of wild-type His-m〈 as a competitor severely reduces the amount of His-Neur that is pulled down with GST-Dlintra, consistent with the interpretation that the binding of Neur to E(spl)mα is able to disrupt and prevent the binding of Neur to Dlintra. Additionally, we find that this competition is dose-dependent: a 5X concentration of His-mα is more effective than a 1X concentration at disrupting the Neur-Dlintra interaction. The E(spl)mα variants His-mα4K/A, His-mα4K/Q, His-mα4K/R, His-mαG2, and His-mαD are all equally capable of disrupting the Neur-Dlintra interaction, excluding a requirement for the B domain and the G and D motifs for this activity. The variant His-mαN also retains some ability to disrupt the Neur-Dlintra interaction; however, its efficiency is reduced compared to that of wild-type mα (Fig. 4B).
The intact N’ motif seemed the most likely source of the residual activity of His-mαN in this assay. We tested this inference by comparing the activities of His-mαN, His-mαN’, and His-mαN’, N. While His-mαN and His-mαN’ both show a weakened ability to disrupt the Neur-Dlintra interaction when compared with His-mα, the double mutant His-mαN’, N has lost this ability completely (Fig. 4C). These data indicate that the N and N’ motifs of E(spl)mα each contribute independently to the protein’s capacity to disrupt binding between Neur and Dlintra.
Since E(spl)m4 contains three functional N motifs that mediate binding to Neur (see Fig. 3B,C), we asked if all three likewise contribute to the protein’s ability to disrupt the Neur-Dl interaction. The single-motif mutants His-m4N, His-m4N’, and His-m4N” are each able to compete for binding to Neur nearly as efficiently as wild-type His-m4, suggesting that the presence of two intact N motifs in these variants is sufficient to disrupt Neur-Dlintra binding (Fig. 4D). The double-motif mutants His-m4N”, N and His-m4N”, N’ are also very effective at disrupting the Neur-Dlintra interaction, indicating that the presence of either the N or N’ motif is largely sufficient to confer this capacity. The variant His-m4N’, N shows a significant decrease in competitive ability, suggesting that the N” motif is functionally weaker than the N and N’ motifs. Finally, as expected, the triple-motif mutant His-m4N”, N’, N is completely impaired in its ability to disrupt the Neur-Dlintra interaction, consistent with its near lack of binding affinity for Neur (see Fig. 3C).
The N motifs of E(spl)mα and E(spl)m4 are required to confer a misexpression phenotype
Knowing that E(spl)mα and E(spl)m4 contain multiple functional N motifs, we hypothesized that the remaining intact N motifs (N’ and N”) are responsible for the substantial residual ability of FLAG-m4N1 to disrupt lateral inhibition and generate an extra-bristle phenotype (see Fig. 1E). To test this proposition, we generated and misexpressed a transgene construct encoding the triple mutant FLAG-m4N”, N’, N2. As predicted, FLAG-m4N”, N’, N2 fails to confer the misexpression phenotype (0.03 extra bristles per fly; Fig. 5A).
Fig. 5.
The N motifs of E(spl)m4 and E(spl)mα are required for the gain-of-function phenotype. (A) Misexpression of FLAG-m4 using the sca-GAL4 driver produces an extra-bristle phenotype that is weakened with the FLAG-m4N1 variant and completely absent in the FLAG-m4N”, N’, N2 triple mutant variant. (B) Misexpression of mα with sca-GAL4 also produces an extra-bristle phenotype that is dependent on the presence of the N motifs. The variant mα4K/A is unable to generate a gain-of-function phenotype, as was observed with FLAG-m44K/A (see Fig. 1E). Error bars indicate standard errors; asterisks denote statistical significance of differences from the wild-type (A, FLAG-m4; B, mα) results (P<0.04; Mann-Whitney U test).
Misexpressing E(spl)mα variants in this same manner produces results comparable to those for E(spl)m4. Wild-type mα misexpression produces a mean of 9.6 extra bristles per fly while mαN’, N lacks a significant capacity to disrupt lateral inhibition, producing only 0.85 extra bristles per fly (Fig. 5B). Interestingly, mαN is only slightly less efficient than wild-type mα in conferring the misexpression phenotype (6.7 extra bristles per fly), while mαN’ is severely impaired in this ability (0.97 extra bristles per fly). This may suggest that the in vivo affinity of Neur for specific N motifs may vary by more than what is observed using in vitro binding assays (see Fig. 2E). Also comparable to the E(spl)m4 results, mα4K/R is capable of strongly disrupting lateral inhibition, generating 8.1 extra bristles per fly, while mα4K/A nearly lacks this ability, yielding 0.11 extra bristles per fly (Fig. 5B). Finally, mαD produces 6.5 extra bristles per fly, indicating that, like FLAG-m4D, this variant retains the capacity to disrupt lateral inhibition (Fig. 5B; see Fig. 1E).
The differing phenotypic effects of the various E(spl)mα variants in the gain-of-function assay could potentially be attributable to differences in protein accumulation in vivo. However, visualization of misexpressed V5-tagged versions of these variants by immunofluorescence shows comparable levels of accumulation (see Supplementary Fig. S2A-E).
Dlintra contains an N motif that is required for binding to Neur
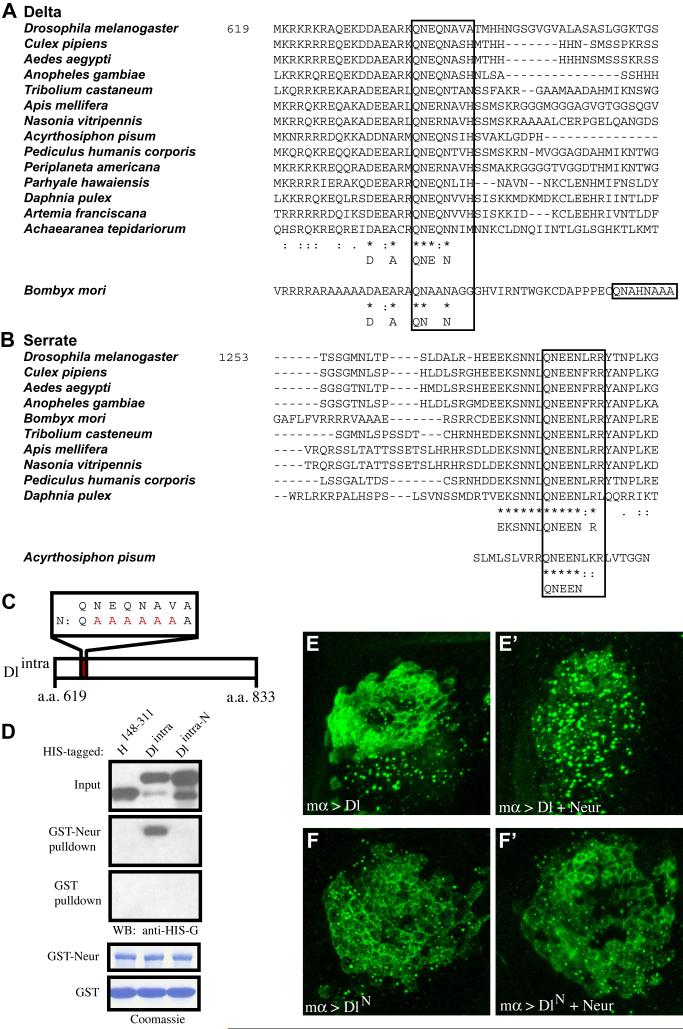
Our findings that N motifs are required both for the binding of Brd family proteins to Neur, and for their ability to compete with Dlintra for binding to Neur, raised the possibility that Dlintra and Brd family proteins compete in vivo for the same binding site(s) on Neur. This model would suggest that Dlintra contains a motif similar to the N motif of Brd family proteins. Indeed, a survey of the amino acid sequence of the intracellular domain of Dl (a.a. 619-833) uncovered a motif near the transmembrane domain that strongly resembles an N motif (QNEQNAVA) and shows a high level of conservation in other arthropods (Fig. 6A). The presence of this conserved N motif in Dl is consistent with a similar mode of interaction for Dl and Brd family proteins with Neur. We have also found a putative N motif in the intracellular domain of the other Drosophila Notch ligand, Ser (Fig. 6B), and we note that the region containing it has previously been found to be important for the activation of Notch signaling and for Ser-Neur co-immunoprecipitation (Glittenberg et al., 2006). The conserved N motif [QNEEN(L/F)RR] we have identified in arthropod Ser proteins contrasts very significantly in its proposed critical residues with the (E/D)(E/D)X2-3NNX5NX3-5NP(L/I) motif suggested by Glittenberg et al. (2006) to be shared between insect Ser and vertebrate Jagged proteins; for example, the first asparagine residue of the N motif’s critical NXXN core is unconstrained (“X”) in the latter consensus.
Fig. 6.
The intracellular domain of Dl contains an N motif that is required for in vitro binding and in vivo responsiveness to Neur. (A,B) Alignments of segments of the intracellular domains of Dl (A) and Ser (B) show strong conservation of an N motif in the arthropods. (C) Cartoon illustrating the mutant N motif variant of Dlintra. Substituted residues are depicted in red. (D) Western blot of a pulldown assay shows that the N motif of Dlintra is required for its interaction with Neur. Also shown are Coomassie-stained blots depicting amounts of bead-bound GST (control) and GST-Neur proteins in each assay. (E,F) 30-40-μm confocal stack images of anti-Dl antibody stains of wing imaginal disc cells expressing Dl (E), DlN (F), Dl + Neur (E’), or DlN + Neur (F’) under the control of the mα-GAL4 driver. Both apical and basal regions of the tissue are included in the image stack. In contrast to wild-type Dl, the intracellular localization of DlN is not responsive to the presence of Neur.
To investigate the possible requirement for its putative N motif in Dl’s interaction with Neur, we created versions of His-tagged Dlintra with and without an N-motif mutation (His-Dlintra-N and His-Dlintra; Fig. 6C). Using a pulldown assay with GST-Neur or GST, we find that His-Dlintra interacts strongly with GST-Neur, while the mutant His-Dlintra-N fails to interact (Fig. 6D). This tells us that Dl and Brd family proteins interact with Neur via similar motifs, and are most likely competing for the same binding site(s) in Neur.
The N motif of Delta is required for its Neur-dependent endocytosis
Coexpression of Neur and Dl in vivo leads to the endocytosis of Dl at the cell surface into intracellular vesicles (Lai et al., 2001; Pavlopoulos et al., 2001). Having identified an N motif in the intracellular domain of Dl that is required to mediate its interaction with Neur in vitro, we proceeded to test whether a form of Dl mutant for this motif (DlN) would be impaired in its ability to undergo Neur-dependent endocytosis. When expressed alone under the control of either the mα-GAL4 or the dpp-GAL4 driver, both Dl and DlN are found associated with the plasma membrane in the apical region of the cell and in intracellular vesicles basally (Fig. 6E,F; data not shown; see Fig. S2F,G). This indicates that DlN, like wild-type Dl, can localize properly to the cell cortex and is capable of being trafficked into vesicles. When coexpressed with Neur, wild-type Dl is depleted from the apical cell surface and appears primarily in intracellular vesicles that are more numerous and larger in size than when Neur is not present (Fig. 6E,E’). However, when Neur and DlN are coexpressed, the intracellular localization of DlN is left unchanged (Fig. 6F,F’). This result supports the conclusion that the N motif in its intracellular domain is required in vivo for endocytosis of Dl in a Neur-dependent manner.
Identification of a Brd family gene in the crustacean Daphnia pulex
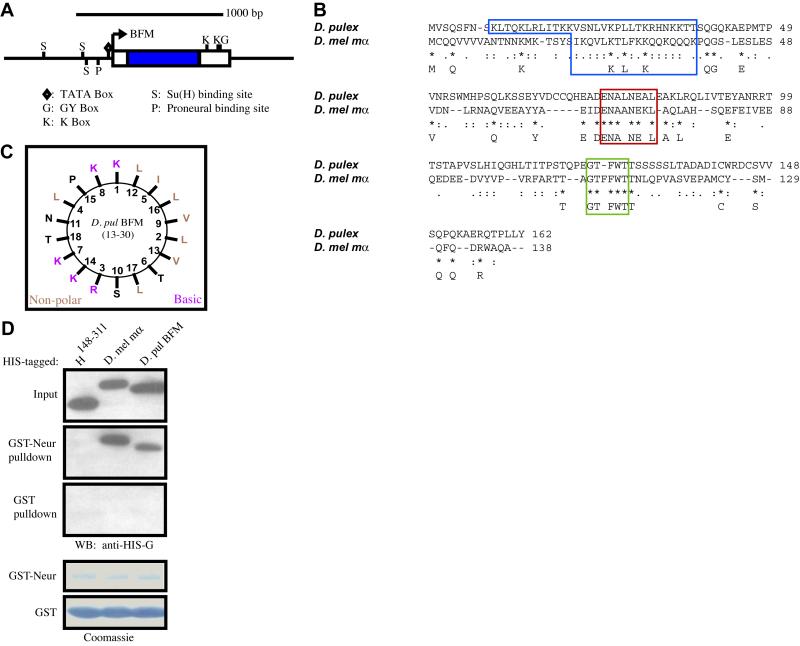
The recent availability of a genome sequence assembly for the waterflea, Daphnia pulex, presented us with the opportunity to search for Brd family genes in a crustacean. Unsurprisingly, standard BLAST searches yield no significant matches to any known BFMs. Using the knowledge that Brd family genes in insects are typically found in the vicinity of conserved bHLH-R genes of the Hairy/Enhancer of split (Hes) class, we first identified scaffolds containing Hes genes, and then looked nearby for possible BFMs. Using this approach, we successfully identified a Daphnia BFM approximately 6 kb upstream of the bHLH-R gene that encodes the Hes protein Dp15 (Simionato et al., 2007) (Fig. 7A). This crustacean BFM appears to be regulated in a manner consistent with BFM regulation in Drosophila (Singson et al., 1994; Bailey and Posakony, 1995; Nellesen et al., 1999), as its immediate upstream region contains a proneural protein binding site, a “lone” binding site for Su(H), and a Su(H) paired site (SPS) (Bailey and Posakony, 1995) within 200 bp of the TATA element. Moreover, two K boxes and a single GY box are found in the predicted 3′ UTR of the gene, indicating that the transcript is likely subject to the same miRNA-mediated negative regulation as Drosophila BFMs (Lai and Posakony, 1997; Leviten et al., 1997; Lai et al., 1998; Lai, 2002; Stark et al., 2003; Lai et al., 2005) (Fig. 7A). The Daphnia BFM gene is predicted to encode a 162-aa protein (Fig. 7B), containing a basic amphipathic alpha-helix (Fig. 7C) as well as an N motif (ENALNEAL) and a variation of the G motif (GTFWT vs. GTFFWT typically found in Drosophila BFMs). It does not include the C-terminal D motif (Fig. 7B).
Fig. 7.
The genome of the crustacean Daphnia pulex encodes a Brd family protein capable of binding to Drosophila Neur. (A) GenePalette illustration of the genomic region containing the Daphnia Brd family gene. Blue rectangle indicates the protein-coding region of this intronless gene; white rectangles represent untranslated regions. Transcriptional and post-transcriptional regulatory sequence motifs shared with other arthropod Brd family genes (see Materials and methods) are shown. (B) ClustalW alignment of the predicted sequence of the Daphnia BFM with D. mel E(spl)mα (blue box = B domain, longer in the Daphnia sequence to include an extended region of high amphipathicity; red box = N motif; green box = G motif). (C) Helical wheel plot of the Daphnia BFM’s B domain. (D) Western blot of a pulldown assay, showing the conserved ability of the Daphnia BFM to interact specifically with Drosophila Neur. Also shown are Coomassie-stained blots depicting amounts of bead-bound GST (control) and GST-Neur proteins in each assay.
To test whether the D. pulex Brd family protein retains the property of binding to Neur, we generated a His-tagged version and performed a pulldown assay with D. melanogaster GST-Neur. Drosophila Neur is indeed able to interact with the Daphnia BFM in this assay, suggesting a conserved function of Brd family proteins in insects and crustaceans (Fig. 7D).
Discussion
Multiple Neur-binding motifs in Brd family proteins
We have presented evidence here that the two canonical Brd family proteins encoded in the Drosophila E(spl)-C, E(spl)mα and E(spl)m4, contain previously unidentified sequence motifs that are structurally and functionally similar to the recognized N motif common to nearly all BFMs (Lai et al., 2000b). It appears that each of these motifs is independently capable of mediating binding to the E3 ubiquitin ligase Neur, and we show that the presence of at least one such motif is necessary for this interaction. Henceforth, we will refer to all of these sequence elements as NXXN motifs, both because of their characteristic pattern of asparagine residues and to avoid confusion with the “N” symbol for Notch.
The availability of whole-genome and EST sequence data for a broad range of insects has permitted the identification of BFMs in 12 Drosophila species and in other Dipterans (Anopheles gambiae, Aedes aegypti, Culex pipiens, Ceratitis capitata, and Haematobia irritans), several Lepidopterans (Bombyx mori, Manduca sexta, Heliconius erato, Antheraea assama, Samia cynthia ricini, and Plodia interpunctella), a Hymenopteran (Apis mellifera), a Coleopteran (Tribolium castaneum), a Hemipteran (Acyrthosiphon pisum), and a Phthirapteran (Pediculus humanus corporis); we also report here the recognition of BFMs in three more distantly related arthropods, the Crustaceans Daphnia pulex, Artemia franciscana, and Callinectes sapidus (Fig. 7; see Fig. S3; J. R. Fontana and J. W. Posakony, unpublished). This in turn affords us the opportunity to refine our definition of the NXXN motif consensus [(D/E/Q)NXXNXX(I/L/M/V); see Fig. S3].
The overall picture that emerges from our examination of arthropod BFM NXXN motifs is that both the appearance of secondary (generally non-canonical) Brd family genes in the genome, and the appearance of additional NXXN motifs within a given Brd family protein, permit much greater variability in NXXN motif sequence to arise. It is tempting to interpret this as a form of subfunctionalization (Lynch and Force, 2000), even at the level of individual duplicated motifs within one protein. It is also reasonable to suggest that multiple NXXN motifs arise within even canonical BFMs such as Drosophila E(spl)mα, E(spl)m4, and Tom because this has the effect of lowering the dissociation constant between these proteins and Neur, making them more efficient inhibitors of Notch signaling. Indeed, we see that mutation of just one of the NXXN motifs in E(spl)mα or E(spl)m4 is sufficient to decrease the efficacy of these proteins in disrupting lateral inhibition (see Fig. 5).
Lastly, we note that the short, relatively loose, consensus for the NXXN motif defined here is not unprecedented for target sequences bound by E3 ubiquitin ligases. For instance, the WW domain of Nedd4 proteins, a family of HECT-domain E3 ubiquitin ligases, binds the small PY motif consensus (L/P)PXY found in targets such as the sodium channel ENaC (Kasanov et al., 2001).
Conserved NXXN motifs in the intracellular domains of Notch ligands
The definition of a looser consensus for the Neur interaction motifs in Brd family proteins permitted the immediate recognition of potential NXXN motifs in the intracellular domains of the Notch ligands Dl and Ser (Fig. 6A,B). The high level of conservation of these motifs in otherwise divergent sequence strongly suggests their functional importance. Indeed, we find NXXN motifs at comparable positions in the intracellular domains of Dl and Ser ligands from non-arthropod protostomes as well, including the nematode Xiphinema index, the polychaete annelid Capitella sp. I, the cephalopod mollusc Euprymna scolopes, and the gastropod mollusc Lottia gigantea (see Fig. S4A). Equally striking is the presence of similar conserved NXXN motifs in both the Delta-like1 (Fig. S4B) and Jagged1 (Fig. S4C) proteins of vertebrates, which suggests strongly that in these species, too, the NXXN motif mediates the interaction between Notch ligands and Neur orthologs. Moreover, the finding that both BFMs and Notch ligands make use of a similar conserved motif to bind to Neur suggests the feasibility of identifying other Neur substrates computationally.
NXXN motif-dependent regulation of Notch signaling
We have demonstrated here for the first time that Brd family NXXN motifs are required for the inhibitory activities of these proteins in two assays, in vitro inhibition of Neur-Dl interaction, and antagonism of Notch signaling activity in vivo. Likewise, we have shown that the NXXN motif of Dl is required both for its interaction with Neur in vitro, and for the Neur-dependent endocytosis of Dl in vivo.
Our results support a specific model for how Brd family proteins function as antagonists of Notch pathway signaling activity; namely, that BFMs and the intracellular domains of Notch ligands compete directly, via their respective NXXN motifs, for binding to Neur. Thus, NXXN motifs are essential mediators both of the activation of the Notch pathway by the ligands Dl and Ser [which require Neur-dependent ubiquitination to be fully functional (Pavlopoulos et al., 2001; Wang and Struhl, 2004)], and of its inhibition by Brd family proteins (which act to prevent this modification as competitive antagonists of the Neur-substrate interaction).
Role of Brd family proteins during lateral inhibition
Taken together, the results presented here and in previous reports (Pavlopoulos et al., 2001; Li and Baker, 2004; Wang and Struhl, 2004; Bardin and Schweisguth, 2006; De Renzis et al., 2006) support the following relatively simple model for BFM function during lateral inhibition. In response to Notch signaling, Brd family genes are transcriptionally activated specifically in the non-SOP cells of the PNC (Nellesen et al., 1999; Castro et al., 2005). There the encoded BFM proteins act to inhibit Neur-dependent ubiquitination of Dl, by the mechanism of directly competing with Dl for binding to Neur via their respective NXXN motifs. Inhibiting the endocytosis-dependent activation of the Dl ligand in non-SOPs would in turn have the effect of preventing these cells from becoming “strong signalers” that otherwise might laterally inhibit the SOP itself, or might themselves be resistant to signaling.
A critical question prompted by the simple model described above for Brd family protein function during lateral inhibition is, Why is inhibition of Neur activity by BFMs in non-SOPs necessary if Neur is highly expressed only in SOPs? Work currently in progress in our laboratory (S. W. Miller and J. W. Posakony, unpublished observations) has established that neur is indeed actively transcribed in multiple cells early in the development of the PNC, potentially necessitating the deployment of BFMs as Neur inhibitors, in order to eliminate any threat to correct cell fate specification this may pose.
Evolutionary history of the Brd protein family
Our identification of a Brd family gene in the Crustacean Daphnia pulex pushes back the origin of this family to perhaps the Silurian era, more than 400 Mya. The presence of a B domain and both N and G motifs in the predicted protein product (see Fig. 7B,C) suggests that the ancestral BFM must have contained at least these three elements. The D motif may either have been lost in the crustaceans, or have appeared sometime later in the hexapod lineage. It also seems clear that both transcriptional and post-transcriptional modes of Brd family gene regulation have been conserved from a deep ancestor. The presence of high-affinity binding sites for proneural proteins and Su(H) in the immediate upstream region of the Daphnia BFM strongly suggests that it uses the “S+P” transcriptional regulatory code first uncovered in studies of Drosophila Brd family and bHLH repressor genes (Singson et al., 1994; Bailey and Posakony, 1995; Nellesen et al., 1999; Castro et al., 2005). Likewise, the presence of K and GY boxes in the gene’s 3′ UTR makes it equally likely that it is subject to the same miRNA-mediated negative regulation that applies to most Drosophila BFMs (Lai and Posakony, 1997; Lai et al., 1998; Stark et al., 2003; Lai et al., 2005). Our demonstration that the Daphnia Brd family protein binds efficiently to Drosophila Neur in vitro (see Fig. 7D) indicates that this functionality, too, is an ancient property of the family. Finally, the close genomic association of the Daphnia BFM with a Hes-type bHLH repressor gene, just as in insect E(spl)-C’s (Schlatter and Maier, 2005), suggests both that this proximity dates from the common ancestor, and that the association is maintained by selection, for an as-yet unknown reason. It is striking that so many features of the structure, function, and regulation of the Brd gene family have survived for such an extraordinarily long time.
Supplementary Material
Acknowledgments
We would like to thank Brian Castro, Feng Liu, Mariano Loza Coll, Steven Miller, Mark Rebeiz, Nick Reeves, and Sui Zhang for both material and mental support, and Gentry Patrick and Jeff Keil for helpful discussions. We also thank Tammie Stone for generating various in vitro reagents and for help with formulating pulldown assay conditions. We are grateful to Dr. Matthias Westphal at the Center for Genomics and Bioinformatics, Indiana University, Bloomington, for Daphnia pulex (Log50 strain) and protocol suggestions. F. Liu, M. Loza Coll, and S. Miller provided very useful critical comments during the preparation of the manuscript. J.R.F. was supported by a National Institutes of Health (NIH) pre-doctoral training grant in Developmental Biology. This work was supported by NIH grant GM075270 to J.W.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Apidianakis Y, Nagel AC, Chalkiadaki A, Preiss A, Delidakis C. Overexpression of the m4 and mα genes of the E(spl)-Complex antagonizes Notch mediated lateral inhibition. Mech. Dev. 1999;86:39–50. doi: 10.1016/s0925-4773(99)00099-4. [DOI] [PubMed] [Google Scholar]
- Bailey AM, Posakony JW. Suppressor of Hairless directly activates transcription of Enhancer of split Complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- Bardin A, Schweisguth F. Bearded family members inhibit Neuralized-mediated endocytosis and signaling activity of Delta in Drosophila. Dev. Cell. 2006;10:245–255. doi: 10.1016/j.devcel.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Barolo S, Carver LA, Posakony JW. GFP and β-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques. 2000;29:726–732. doi: 10.2144/00294bm10. [DOI] [PubMed] [Google Scholar]
- Bellaiche Y, Gho M, Kaltschmidt J, Brand A, Schweisguth F. Frizzled regulates localization of cell-fate determinants and mitotic spindle rotation during asymmetric cell division. Nat. Cell Biol. 2001;3:50–57. doi: 10.1038/35050558. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Castro B, Barolo S, Bailey AM, Posakony JW. Lateral inhibition in proneural clusters: Cis-regulatory logic and default repression by Suppressor of Hairless. Development. 2005;132:3333–3344. doi: 10.1242/dev.01920. [DOI] [PubMed] [Google Scholar]
- De Renzis S, Yu J, Zinzen R, Wieschaus E. Dorsal-ventral pattern of Delta trafficking is established by a Snail-Tom-Neuralized pathway. Dev. Cell. 2006;10:257–264. doi: 10.1016/j.devcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Deblandre GA, Lai EC, Kintner C. Xenopus neuralized is a ubiquitin ligase that interacts with XDelta1 and regulates Notch signaling. Dev. Cell. 2001;1:795–806. doi: 10.1016/s1534-5807(01)00091-0. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Kobayakawa Y, Tamura K, Kimura K, Kawaichi M, Tanimura T, Honjo T. Suppressor of Hairless, the Drosophila homologue of RBP-Jk, transactivates the neurogenic gene E(spl)m8. Jpn. J. Genet. 1995;70:505–524. doi: 10.1266/jjg.70.505. [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, Mcdaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, Dasilva A, Zhong J, Stanyon CA, Finley RL, Jr., White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, Mckenna MP, Chant J, Rothberg JM. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Glittenberg M, Pitsouli C, Garvey C, Delidakis C, Bray S. Role of conserved intracellular motifs in Serrate signalling, cis-inhibition and endocytosis. EMBO J. 2006;25:4697–4706. doi: 10.1038/sj.emboj.7601337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz U, Giebel B, Campos-Ortega JA. The basic-helix-loop-helix domain of Drosophila lethal of scute protein is sufficient for proneural function and activates neurogenic genes. Cell. 1994;76:77–87. doi: 10.1016/0092-8674(94)90174-0. [DOI] [PubMed] [Google Scholar]
- Kasanov J, Pirozzi G, Uveges A, Kay B. Characterizing Class I WW domains defines key specificity determinants and generates mutant domains with novel specificities. Chem. Biol. 2001;8:231–241. doi: 10.1016/s1074-5521(01)00005-9. [DOI] [PubMed] [Google Scholar]
- Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Lai EC, Bodner R, Kavaler J, Freschi G, Posakony JW. Antagonism of Notch signaling activity by members of a novel protein family encoded by the Bearded and Enhancer of split gene complexes. Development. 2000a;127:291–306. doi: 10.1242/dev.127.2.291. [DOI] [PubMed] [Google Scholar]
- Lai EC, Bodner R, Posakony JW. The Enhancer of split Complex of Drosophila includes four Notch-regulated members of the Bearded gene family. Development. 2000b;127:3441–3455. doi: 10.1242/dev.127.16.3441. [DOI] [PubMed] [Google Scholar]
- Lai EC, Burks C, Posakony JW. The K box, a conserved 3′ UTR sequence motif, negatively regulates accumulation of Enhancer of split Complex transcripts. Development. 1998;125:4077–4088. doi: 10.1242/dev.125.20.4077. [DOI] [PubMed] [Google Scholar]
- Lai EC, Deblandre GA, Kintner C, Rubin GM. Drosophila Neuralized is a ubiquitin ligase that promotes the internalization and degradation of Delta. Dev. Cell. 2001;1:783–794. doi: 10.1016/s1534-5807(01)00092-2. [DOI] [PubMed] [Google Scholar]
- Lai EC, Posakony JW. The Bearded box, a novel 3′ UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development. 1997;124:4847–4856. doi: 10.1242/dev.124.23.4847. [DOI] [PubMed] [Google Scholar]
- Lai EC, Rubin GM. neuralized functions cell-autonomously to regulate a subset of Notch-dependent processes during adult Drosophila development. Dev. Biol. 2001;231:217–233. doi: 10.1006/dbio.2000.0124. [DOI] [PubMed] [Google Scholar]
- Lai EC. Ph.D. Dissertation. Department of Biology, University of California; San Diego: 1999. Regulation of pattern formation during development of the Drosophila peripheral nervous system. [Google Scholar]
- Lai E, Tam B, Rubin G. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourtois M, Schweisguth F. The neurogenic Suppressor of Hairless DNA-binding protein mediates the transcriptional activation of the Enhancer of split Complex genes triggered by Notch signaling. Genes Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- Leviten MW, Lai EC, Posakony JW. The Drosophila gene Bearded encodes a novel small protein and shares 3′ UTR sequence motifs with multiple Enhancer of split Complex genes. Development. 1997;124:4039–4051. doi: 10.1242/dev.124.20.4039. [DOI] [PubMed] [Google Scholar]
- Leviten MW, Posakony JW. Gain-of-function alleles of Bearded interfere with alternative cell fate decisions in Drosophila adult sensory organ development. Dev. Biol. 1996;176:264–283. doi: 10.1006/dbio.1996.0133. [DOI] [PubMed] [Google Scholar]
- Li Y, Baker NE. The roles of cis-inactivation by Notch ligands and of neuralized during eye and bristle patterning in Drosophila. BMC Dev. Biol. 2004;4:5. doi: 10.1186/1471-213X-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado-Pimentel ME, Jordan N, Aisemberg G. Affinity purification of GST fusion proteins for immunohistochemical studies of gene expression. Protein Expr. Purif. 2002;26:260–265. doi: 10.1016/s1046-5928(02)00524-7. [DOI] [PubMed] [Google Scholar]
- Nakao K, Campos-Ortega JA. Persistent expression of genes of the Enhancer of split complex suppresses neural development in Drosophila. Neuron. 1996;16:275–286. doi: 10.1016/s0896-6273(00)80046-x. [DOI] [PubMed] [Google Scholar]
- Nellesen DT, Lai EC, Posakony JW. Discrete enhancer elements mediate selective responsiveness of Enhancer of split Complex genes to common transcriptional activators. Dev. Biol. 1999;213:33–53. doi: 10.1006/dbio.1999.9324. [DOI] [PubMed] [Google Scholar]
- Pavlopoulos E, Pitsouli C, Klueg KM, Muskavitch MA, Moschonas NK, Delidakis C. Neuralized encodes a peripheral membrane protein involved in Delta signaling and endocytosis. Dev. Cell. 2001;1:807–816. doi: 10.1016/s1534-5807(01)00093-4. [DOI] [PubMed] [Google Scholar]
- Rebeiz M, Posakony JW. GenePalette: A universal software tool for genome sequence visualization and analysis. Dev. Biol. 2004;271:431–438. doi: 10.1016/j.ydbio.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Schlatter R, Maier D. The Enhancer of split and Achaete-Scute complexes of Drosophilids derived from simple ur-complexes preserved in mosquito and honeybee. BMC Evol. Biol. 2005;5:67. doi: 10.1186/1471-2148-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, Degnan B, Vervoort M. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol. Biol. 2007;7:33. doi: 10.1186/1471-2148-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singson A, Leviten MW, Bang AG, Hua XH, Posakony JW. Direct downstream targets of proneural activators in the imaginal disc include genes involved in lateral inhibitory signaling. Genes Dev. 1994;8:2058–2071. doi: 10.1101/gad.8.17.2058. [DOI] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell R, Cohen S. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Struhl G. Drosophila Epsin mediates a select endocytic pathway that DSL ligands must enter to activate Notch. Development. 2004;131:5367–5380. doi: 10.1242/dev.01413. [DOI] [PubMed] [Google Scholar]
- Zaffran S, Frasch M. Barbu: an E(spl) m4/ma-related gene that antagonizes Notch signaling and is required for the establishment of ommatidial polarity. Development. 2000;127:1115–1130. doi: 10.1242/dev.127.5.1115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.