Abstract
Background and Aims
The study of rapid evolution in invasive species has highlighted the fundamental role played by founder events, emergence of genetic novelties through recombination and rapid response to new selective pressures. However, whether rapid adaptation of introduced species can be driven by punctual changes in genome organization has received little attention. In plants, variation in genome size, i.e. variation in the amount of DNA per monoploid set of chromosomes through loss or gain of repeated DNA sequences, is known to influence a number of physiological, phenological and life-history features. The present study investigated whether change in genome size has contributed to the evolution of greater potential of vegetative growth in invasive populations of an introduced grass.
Methods
The study was based on the recent demonstration that invasive genotypes of reed canarygrass (Phalaris arundinacea) occurring in North America have emerged from recombination between introduced European strains. The genome sizes of more than 200 invasive and native genotypes were measured and their genome size was related to their phenotypic traits measured in a common glasshouse environment. Population genetics data were used to infer phylogeographical relationships between study populations, and the evolutionary history of genome size within the study species was inferred.
Key Results
Invasive genotypes had a smaller genome than European native genotypes from which they are derived. This smaller genome size had phenotypic effects that increased the species' invasive potential, including a higher early growth rate, due to a negative relationship between genome size and rate of stem elongation. Based on inferred phylogeographical relationships of invasive and native populations, evolutionary models were consistent with a scenario of genome reduction by natural selection during the invasion process, rather than a scenario of stochastic change.
Conclusions
Punctual reduction in genome size could cause rapid changes in key phenotypic traits that enhance invasive ability. Although the generality of genome size variation leading to phenotypic evolution and the specific genomic mechanisms involved are not known, change in genome size may constitute an important but previously under-appreciated mechanism of rapid evolutionary change that may promote evolutionary novelties over short time scales.
Key words: Biological invasion, evolutionary models, genome size, Phalaris arundinacea, quantile regression, relative growth rate, rapid evolution
INTRODUCTION
Over the past decade, the study of invasive species has provided major insights into the evolutionary mechanisms that allow species range expansions (Lee, 2002; Huey et al., 2005; Facon et al., 2006; Sax et al., 2007). A number of studies have demonstrated that the interactive effects of species immigration history, genetic bottlenecks and emergence of genetic novelties through recombination have a primary influence on how quickly a species will adapt to a new environment and ultimately invade a new region (Huey et al., 2000; Lindholm et al., 2005; Novak and Mack, 2005; Lavergne and Molofsky, 2007; Le Roux et al., 2007; Zayed et al., 2007). These mechanisms only imply a traditional model of evolution through successive allelic substitutions accumulating over time and leading to gradual phenotypic change, but it is becoming recognized that rapid evolution in introduced species may also be driven by punctual changes in genome structure and organization (Lee, 2002; Prentis et al., 2008).
In plants, variation in genome size, i.e. the amount of DNA per monoploid set of chromosomes (Greilhuber et al., 2005), is mainly due to variation in the amount of repeated DNA sequences (Kubis et al., 1998; Kidwell, 2002; Weider et al., 2005) and is thought to play an important role in plant phenotypic evolution (Gregory and Hebert, 1999; Knight et al., 2005; Meagher and Vassiliadis, 2005). Changes in genome size have repeatedly occurred during the evolution of higher plants (Leitch et al., 1998; Weiss-Schneeweiss et al., 2005; Oliver et al., 2007), with periods of genome inflation through retrotransposon activity (Bennetzen, 2002; Bennetzen et al., 2005) and events of punctual genome contraction following unequal homologous or illegitimate recombinations (Devos et al., 2002; Bennetzen et al., 2005). As smaller plant genomes generally result in smaller cells and faster cell division (Bennett, 1998; Francis et al., 2008; Knight and Beaulieu, 2008), genome size ultimately influences a number of plant physiological, phenological and life-history features (Grime and Mowforth, 1982; Grime et al., 1985; Grime, 1998; Knight and Ackerly, 2002; Meagher and Vassiliadis, 2005; Beaulieu et al., 2006, 2007). Indeed, comparative studies have demonstrated that plant species with smaller genomes are more likely to become invasive than their relatives, due to the effect of genome size on key life-history traits that enhance colonization potential, such as seed mass, growth-related traits and generation time (Bennett et al., 1998; Grotkopp et al., 2004). As genome size can rapidly decrease by random loss of repetitive DNA sequences, genome size reduction has the potential to contribute to rapid evolution of invasive ability in introduced plant species, and thus could be a significant mechanism of contemporary phenotypic evolution in plants.
The present study investigated whether genome size reduction can trigger rapid phenotypic evolution in invasive plants using the invasive reed canarygrass (Phalaris arundinacea) as a biological model. This species is invasive in North America where it is currently considered a major threat to native wetland vegetation and bird breeding habitat (Lavergne and Molofsky, 2006). It was recently demonstrated that the invasive ability of this species in North America resulted from the introduction and subsequent recombination of multiple European strains, which allowed the rapid selection of novel genotypes with higher potential of vegetative colonization (Lavergne and Molofsky, 2004, 2007). First, the study tested whether, apart from exhibiting new allelic combinations, novel genotypes of reed canarygrass also have a smaller genome size than European native genotypes from which they are derived. Second, it tested whether potential variation in genome size had phenotypic effects that influence the growth rate and invasive potential of the species. Finally, based on inferred phylogeographical relationships between study populations, different evolutionary models of genome size were fitted to test whether inter-population variation in genome size was more likely to be explained by a scenario of random change or a scenario of natural selection during species invasion.
MATERIAL AND METHODS
Study species and genotypic data
Reed canarygrass (Phalaris arundinacea L., Poaceae) is a long-lived perennial grass that reproduces both sexually and asexually through profuse rhizome propagation (Lavergne and Molofsky, 2004). A hierarchical sampling design (see Table 1) was used and 350 rhizome pieces of reed canarygrass were collected from populations located at the centre and the margin of its native range in Europe (the Czech Republic and southern France, respectively) and of its invasive range in North America (Vermont and North Carolina, respectively).
Table 1.
Climatic description of the four study regions of Phalaris arundinacea, with the Czech Republic and France in its native range, and Vermont and North Carolina in its invasive range
| Lat/Long | No. of sampled populations | Mean genome size (pg)/s.e. | Average temperature (°C) | Maximum temperature of warmest month (°C) | Minimum temperature of coldest month (°C) | Average precipitation (mm) | Precipitation seasonality (coefficient of variation) | |
|---|---|---|---|---|---|---|---|---|
| Czech Republic | 49·00000/14·766667 | 4 | 9·34/0·048 | 8 | 23·6 | −5·2 | 717 | 36 |
| France | 43·616667/3·866667 | 2 | 9·31/0·078 | 13·9 | 28·5 | 1·4 | 743 | 31 |
| Vermont | 44·466667/ − 73·15000 | 3 | 9·14/0·062 | 6·9 | 27·3 | −13·5 | 885 | 23 |
| North Carolina | 35·316667/ − 83·63333 | 3 | 9·21/0·045 | 12 | 28 | −5·3 | 1584 | 12 |
The geographical position of each study region is given in latitude/longitude (Lat/Long). The number of study populations within each region, mean (and standard error) estimated genome size and a set of descriptive climatic variables are also presented.
Based on 12 allozyme markers (Gifford et al., 2002; Lavergne and Molofsky, 2007), it has previously been shown that invasive genotypes of reed canarygrass arose from recombination between introduced European genotypes and that these novel American genotypes have a greater potential for vegetative establishment and propagation than European native genotypes (Lavergne and Molofsky, 2007). The great majority (98·5 %) of invasive genotypes of reed canarygrass that have been identified in the eastern United States (Vermont and North Carolina) have arisen from recombination of genotypes native to disparate European regions such as the Czech Republic and France (Lavergne and Molofsky, 2004, 2007). Thus, only 1·5 % of the study invasive genotypes are non-recombinant genotypes also found in European populations. All genotypes have the same ploidy, corresponding to a diploidized tetraploid cytotype that is originally native to temperate European regions (Lavergne and Molofsky, 2004, 2007).
The study used the same 210 multilocus genotypes that were identified across the 12 populations of reed canarygrass, sampled in Vermont and North Carolina for the North American invasive range, and in the Czech Republic and southern France for the European native range.
Variation in genome size
A solution of nuclei of every genotype was obtained using the following protocol. We chopped 70–90 mg of leaf material with razor blades in 3 mL of ice-cold chopping buffer (Galbraith et al., 1997). The solution obtained was filtered with 40- and 20-μm nylon meshes (Small Parts Inc., Miami Flakes, FL, USA) and then purified by centrifugation (2000 r.p.m., 7 min). The supernatant was carefully removed using a disposable pipette, and the pellet was suspended in 1 mL of staining solution (chopping buffer with 100 µg mL−1 of propidium iodide and 50 µg mL−1 of RNAase A, Sigma-Aldrich, St Louis, MO, USA). An internal standard of known DNA content (chicken erythrocyte nuclei, DNA content 2·50 pg, obtained from Biosure, Grass Valley, CA, USA) was added to the solution and the solution was allowed to stain in the dark for 1 h.
One millilitre of nuclei solution (nuclei from each genotype and the internal standard) was processed using an Epics XL-MCI laser flow cytometer (Beckman Coulter, Miami, FL, USA). The median value of each fluorescence peak (one for the study genotype and one for the standard) was used to compute the genome size of each genotype. Samples were run several times over a 1-h period (to improve stain saturation) and on two different days (with a new nuclei solution). When one of the two measurements differed substantially (more than 5 % difference), the same genotype was processed again and final estimation of the genome size of the genotype was computed by averaging different-day measurements.
This protocol allowed us to overcome the methodological caveats associated with the quantification of intraspecific variation in genome size pointed by Greilhuber (2005). Whether genome size varied consistently between invasive and native genotypes was then tested by fitting a linear mixed effects model (Pinheiro and Bates, 2000), with populations as random effects and range of genotypes (invasive vs. native) as a fixed effect. Finally, the observed mean genome size (and the extent of intraspecific variation in genome size) was compared with the range of values generally observed in angiosperms, particularly in monocot species. To do so, we used the Kew Gardens database of genome sizes (Bennett and Leitch, 2004) to download the genome size of 4427 species. Comparison with the reed canarygrass genome size was done using histograms and estimation of genome size quantiles among these 4427 species.
Phenotypic measurements and relation to genome size
A sub-sample of 90 genotypes (49 invasive and 41 native genotypes) were randomly selected, and four clones of each were grown (from equal sized rhizome pieces) for 78 d, in a common glasshouse environment (22–25 °C day temperature). Morphological measurements were taken until final harvest when an estimation of dry biomass production was obtained. As genome size should influence rates of cell division and morphological growth, the following developmental traits were computed: stem elongation rate (stem length increment between 15 and 78 d of growth), leaf production rate (leaf number increment between 15 and 41 d of growth) and relative growth rate (dry biomass production at 78 d).
The relationship between developmental traits and genome size was investigated using robust linear regressions and linear quantile regressions. Robust regressions were used instead of classic linear regressions in order to reduce the influence of potential outliers in the data (Hosmer and Lemeshow, 1989). Quantile regression models, which are increasingly used in ecological research (Cade and Noon, 2003; Cade et al., 2005), allow one to model the entire probabilistic distribution of a dependent variable (instead of only the expected value as in a classic regression). A quantile regression quantifies the effect of an independent variable on the probabilistic distribution of a dependent variable around a given quantile τ (i.e. the probability that the value of a dependent variable is less than τ). Fitting quantile regression for the full range of τ (usually from 0·05 to 0·95) provides a more complete view of the relationship between a dependent variable and putative independent variables. The overall effect of each independent variable is thus non-parametric and can take any form. Note that in the case of a symmetric distribution, the 0·5-quantile regression is equivalent to a classic linear regression. The use of quantile regression has been repeatedly advocated to depict complex functional relationships between variables, especially in the case of heterogeneous responses to limiting factors, i.e. where changes in the dependent variable differ between different parts of its distribution (Cade et al., 2005). Quantile regression has been used several times to investigate the effect of genome size on plant phenotypic traits (Knight and Ackerly, 2002; Beaulieu et al., 2006).
Models of genome size evolution
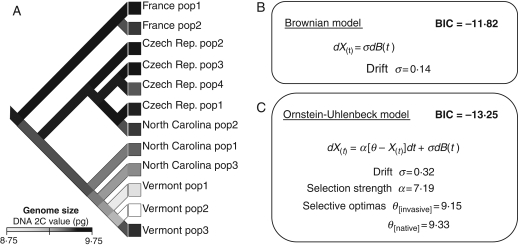
Based on allele frequency data of 34 distinct allozyme alleles, a UPGMA tree of the 12 study populations was constructed, and this tree was used to reconstruct the phylogeographical history of genome size variation in reed canarygrass. First, ancestral states of genome size were calculated using a square parsimony reconstruction (Maddison and Maddison, 2006). Second, alternative evolutionary models were fitted following the method implemented by Butler and King (2004) to assess whether changes in genome size along the branches of the phylogeographical tree were better explained by a scenario of stochastic change (Brownian motion) or scenarios of adaptive evolution (Ornstein–Uhlenbeck process). Three models were fitted.
Model 1 (Brownian model): genome size evolves stochastically along branches of the tree, conforming to evolution by random drift or selection in a rapidly and randomly changing environment.
Model 2 (Ornstein–Uhlenbeck with one selective optimum): genome size evolves under drift and natural selection towards a unique optimum common to all study populations.
Model 3 (Ornstein–Uhlenbeck with two selective optima): genome size evolves under drift and natural selection towards two distinct selective optima for invasive and native populations.
For more details on the structure of different models, see Fig. 4. The results obtained were robust to the type of phylogeographical reconstruction and to randomization in branch lengths of the tree (analysis not shown).
Fig. 4.
Squared-parsimony reconstruction of ancestral genome size (A) and fit of models of genome size evolution (B, C) based on phylogeographical structure of study populations of Phalaris arundinacea. The phylogeographical hypothesis was estimated through a UPGMA tree of study populations (12 polymorphic allozyme markers, 34 alleles overall), with France and Czech populations for native range of the species and North Carolina and Vermont populations in its invasive range. Change in genome size along the branches of the tree (defined as X(t)) was modelled using two distinct evolutionary models: a Brownian model, based on a single drift term (B), and an Ornstein–Uhlenbeck model, based on a drift term and a selection term with two different selective optima for invasive and native populations (C). Model parameters, namely drift (σ), selection intensity (α) and selective optima (θ[invasive], θ[native]) were estimated by likelihood maximization. In both models, the function B(t) is a random normal function of branch length used to simulate stochastic population processes. Goodness of fit of different models was compared using the Bayesian Information Criterion (BIC) and likelihood ratio test (see Results). An additional Ornstein–Uhlenbeck model with only one selective optimum (thus assuming θ[invasive] = θ[native]) was also fitted but is not shown here since it had the lowest fit (BIC = −9·12).
RESULTS
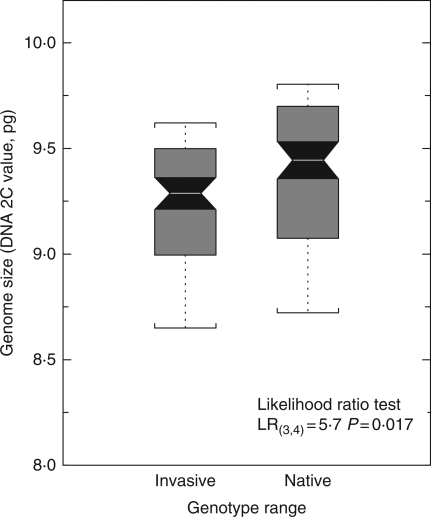
Based on the entire sample of 210 distinct multilocus genotypes identified across the study populations, flow cytometry procedures yielded a mean genome size of 9·26 pg across all study populations of reed canarygrass. This places reed canarygrass around the 40th quantile of monocot genome sizes, and around the 62th quantile of angiosperm genome sizes (Supplementary Data Fig. S1, available online). The replication of flow cytometry procedures on each study genotype demonstrated a significant intraspecific variation in genome size within reed canarygrass. Linear mixed models revealed that North American genotypes have a significantly smaller genome size (2C-value) than European genotypes (Fig. 1). This result was robust to the exclusion of invasive genotypes that were non-recombinant native genotypes.
Fig. 1.
Variation of diploid genome size between invasive and native genotypes of Phalaris arundinacea (sample size n = 210). Difference was tested using a likelihood-ratio test based on a linear mixed effect model with populations as random effects, and origin of genotypes (invasive vs. native) as a fixed effect. Black shaded notches indicate 95 % confidence intervals of the mean genome size of each range.
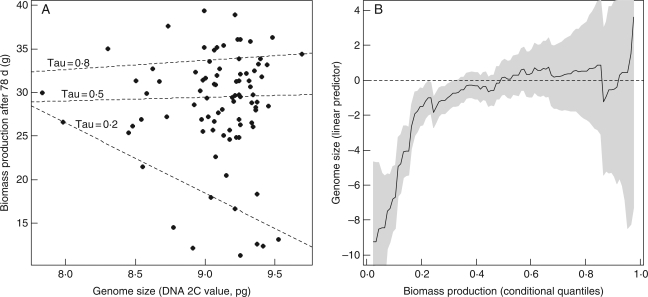
Based on the morphological data obtained in the common glasshouse experiment, it was found that the early growth rate of the 90 study genotypes (measured as biomass production over 78 d) and their genome size exhibited a triangular relationship, with no genotypes having both a slow biomass production and a low genome size (Fig. 2A). Quantile regressions showed that biomass production of a genotype was negatively correlated with its genome size for lower quantiles of biomass production (Fig. 2A). When quantile regressions were repeated over the entire range of quantiles of biomass production, the probability for biomass production of a genotype to be less than lower quantiles (lower than the 5th–40th quantiles) was negatively correlated with its genome size (Fig. 2B); hence, the linear predictor of genome size becomes negative for lower quantiles of growth rates (Fig. 2B).
Fig. 2.
(A) Relationship between early growth rate (measured as biomass production after 78 d) and genome size among the 90 study genotypes of reed canarygrass, with regression lines (dashed lines) fitted by quantile regression of biomass production as a function of genome size, for three different quantiles (tau = 0·8, 0·5 and 0·2). (B) Results of quantile regressions of biomass production as a function of genome size obtained for the entire range of biomass production quantiles. The solid line depicts the effect of genome size of genotypes (linear predictor, y-axis) on the probabilistic distribution of the biomass production of genotypes, for the 5th–95th quantiles of the data. The shaded area represents the 95 % confidence intervals of the linear predictor of genome size for each quantile of growth rate. For more details on quantile regression methodology, see Materials and Methods.
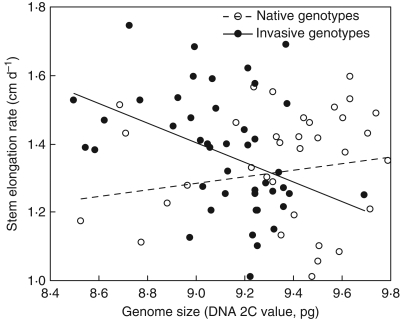
Based on robust regression procedures, there was also evidence for a negative effect of genome size on stem elongation rate (Fig. 3): this effect was significant for invasive genotypes (χ21 = 8·50, P = 0·0036) but not for native genotypes (χ2 = 1·27, d.f. = 1, P = 0·26). As a result, invasive genotypes had a higher rate of leaf canopy expansion (measured as leaf production rate, 10 000-permutation test, P = 0·0024) and a higher growth rate (measured as biomass production, 10 000-permutation test, P = 0·0013) relative to native European genotypes. Thus, larger genomes may ultimately limit plant growth rate in reed canarygrass, and the genome shrinkage that occurred in introduced populations enhanced early growth rate of novel genotypes relative to native European genotypes by modifying some developmental traits, including faster stem elongation rate and a greater potential of leaf canopy expansion.
Fig. 3.
Relationship between stem elongation rate and genome size of genotypes of Phalaris arundinacea. The relationship was assessed separately for native genotypes and invasive genotypes by fitting a robust regression. This relationship was significantly negative for invasive genotypes (solid line) but not significant for native genotypes (dashed line). See Results for statistical tests.
Squared-parsimony reconstruction of ancestral genome sizes mapped on the phylogeographical tree showed a consistent decrease of genome size along branches leading to invasive populations (Fig. 4A). The evolutionary model that best fitted the phylogeographical patterns of genome size accounted for drift and selection with two distinct selective optima for invasive and native populations, according to Bayesian information criteria (Fig. 4B, C). Accordingly, likelihood ratio tests showed that the drift-selection model with two selective optima explained the data better than with a single optimum (LR = 6·61, d.f. = 3, P = 0·010) and better than a pure drift model (LR = 8·88, d.f. = 3, P = 0·031). While accounting for phylogeographical relationships between study populations, genome size differences observed between invasive and native genotypes best fitted an evolutionary scenario in which reduction in genome size has been maintained by natural selection during the invasion process.
DISCUSSION
Recent evidence suggests that many invasive species have been introduced multiple times into a new region and that this situation may favour the emergence of novel genotypes with high invasive potential through genetic recombination (Bossdorf et al., 2005; Lavergne and Molofsky, 2007; Rosenthal et al., 2008). Here we suggest that, in addition to exhibiting new allelic combinations, these novel genotypes may also present genomic re-arrangements that may have immediate phenotypic consequences and enhance their invasive ability. The results presented here are based on the previous evidence that invasive genotypes of reed canarygrass occurring in North America are for the most part (here 98·5 %) recombinants of multiple European genotypes that were introduced in the last 200 years, and that this history of multiple introductions and subsequent recombinations sparked post-immigration evolution of invasive ability (Lavergne and Molofsky, 2007; Novak, 2007). It is also shown herein that novel genotypes of reed canarygrass occurring in its invasive range have a smaller genome than European genotypes from which they emerged through recombination. This observed smaller genome size had direct phenotypic effects that increased early growth rate of the species in its introduced range, a pattern that proved to be robust to bootstrapping procedures. As the greater potential for vegetative growth of novel genotypes relative to native European genotypes contributes to the invasive potential of the species under natural conditions (Lavergne and Molofsky, 2007), the present results suggest that the observed genome size reduction partly promoted the evolution of higher colonization potential and invasive ability in introduced populations of reed canarygrass. Based on phylogeographical relationships between study populations, evolutionary models indicate that the most likely scenario involved natural selection for smaller genome size during invasion, rather than a scenario of stochastic changes in genome size.
The results here are consistent with earlier studies that have shown that a change in genome size can be a product of natural selection (Meagher and Costich, 1996; Meagher and Vassiliadis, 2005; Nilsson et al., 2005) and that artificial selection for more rapid plant development can result in smaller genome size (Rayburn et al., 1994). Through its documented effects on cell size and cell division rate or on patterns of gene expression (Zuckerkandl, 2002; Cavalier-Smith, 2005; Francis et al., 2008; Knight and Beaulieu, 2008), genome size may impose a developmental constraint on plant growth (Knight et al., 2005). Thus, genome size reduction may have conferred a selective advantage to novel genotypes of reed canarygrass by allowing faster rates of plant canopy expansion, due to a negative relationship between genome size and stem elongation. The smaller genomes that characterize introduced populations may have had immediate consequences on early developmental traits enhancing the potential of competition for light that have been later exposed to natural selection.
A significant intraspecific variation in plant genome size was documented here. As it is commonly established that variation in genome size is mostly due to repetitive DNA elements (reviewed by Meagher and Vassiliadis, 2005), and as the vast majority of invasive genotypes of reed canarygrass are recombinants of genotypes drawn from across Europe, it is likely that this genome downsizing has occurred by unequal or non-homologous recombination between different European ecotypes, which purged repeated DNA sequences from the genome. Such mechanisms have been shown to be responsible for deletion of repeated DNA sequences in model plant species (Bennetzen, 2002; Devos et al., 2002; Bennetzen et al., 2005). However, we cannot completely rule out that aneuploidy may also have caused genome downsizing in reed canarygrass, although chromosome counts performed on a small subset of genotypes confirmed that chromosome number may be constant between study genotypes and that both invasive and native genotypes are tetraploids (2n = 42). There is now need for further work to determine which categories of repeated DNA elements (transposable elements, retro-elements, micro/mini satellites and ribosomal genes) have been lost to drive rapid genome shrinkage and which specific genomic mechanisms cause this association between smaller genome and accelerated plant growth rate.
A number of studies suggest that intraspecific variation in genome size may also be correlated with climatic factors (Knight and Ackerly, 2002; Knight et al., 2005), suggesting that the results here could be a product of local adaptation of reed canarygrass populations along climatic gradients. However, invasive and native study regions were approximately matched in terms of latitude, thus limiting the effect of this confounding factor. A closer look at geographical position and mean climatic variables of each study region indeed shows that there is no relationship between average genome size within study regions and their position along temperature and precipitation gradients (Table 1). Thus, the smaller genome size of North American genotypes of reed canarygrass is more likely to be a byproduct of the history of repeated introduction of European strains and further illegitimate recombination between them, which created the opportunity for novel genomic organization to emerge.
The observed greater growth rate in genotypes of reed canarygrass that have smaller genomes may have important implications in the search for biological features leading to species invasiveness and the understanding of adaptive evolution in introduced plants. Comparative studies have previously demonstrated that plant species with smaller genomes are more likely to become invasive than their close relatives, because smaller genomes enhance colonization ability through higher early growth rate and production of propagules with greater dispersal potential (Bennett et al., 1998; Grotkopp et al., 2004). Thus, the capacity of species with smaller genomes to become invasive when transported into a new region has been mainly regarded as a ‘pre-adaptation’. However, the present results suggest that genome size can rapidly change following the introduction of a species into a new region and has immediate phenotypic consequences that enhance its invasive ability. This may posit a potential new mechanism by which key life-history traits may evolve rapidly following the introduction of a species into a new region, involving a punctual change in genome organization. More comparative data of genome size between invasive and native conspecific plant populations are needed to determine whether genome size reduction is a general evolutionary mechanism whereby introduced plants rapidly become more aggressive.
This study provides preliminary evidence that a sudden reduction in genome size, probably occurring over very few generations, may result in rapid phenotypic evolution. Change in genome size might constitute an important but previously under-appreciated mechanism of rapid evolutionary change promoting evolutionary novelties over short time scales, which could have important implications from an evolutionary point of view. The underlying mechanisms of rapid adaptive evolution may not only involve successive allelic substitutions as traditionally acknowledged for microevolution, but also regulatory or epigenetic effects resulting from punctual changes in genome size or genome organization (Zuckerkandl, 2002; Cavalier-Smith, 2005). A large number of genetic and genomic mechanisms may allow rapid adaptive change and influence species contemporary response to climate change, making predictions based on species current characteristics largely insufficient.
SUPPLEMENTARY DATA
ACKOWLEDGEMENTS
This research was supported by USDA hatch and USDA CSREES award 2003-35320-13503. We thank Michael Fay, Charles Knight and an anonymous referee for the significant improvements they suggested on our manuscript. Jean-Baptiste Ferdy, Joël Mathez, Claire Lagaye, Max Debussche, Stepan Husak, Jitka Klimesova, Jan Kvet and Gary Kauffmann helped with sampling. Flow cytometry was realized with the help of Scott Tighe (Flow Cytometry Facility, UVM College of Medicine). We also thank Dave Barrington, Heather Driscoll and Phil Lintilhac (Department of Plant Biology, UVM) for their valuable guidance in the genetic study, as well as Colleen Armstrong and Nina Mosle from the UVM greenhouse facility. Experiments were realized with the help of M. Bower, N. Muenke, C. Brodersen, R. Winkelstein, B. Williams, M. Fitzgerald, R. Collins, M. Patel and M. Garibaldi.
Supplementary Material
REFERENCES
- Beaulieu J, Moles AT, Leitch IJ, Bennett MD, Dickie JB, Knight CA. Correlated evolution of genome size and seed mass. New Phytologist. 2006;173:422–437. doi: 10.1111/j.1469-8137.2006.01919.x. [DOI] [PubMed] [Google Scholar]
- Beaulieu J, Leitch IJ, Knight CA. Genome size evolution in relation to leaf strategy and metabolic rates revisited. Annals of Botany. 2007;99:495–505. doi: 10.1093/aob/mcl271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD. Plant genome values: how much do we know? Proceedings of the National Academy of Sciences of USA. 1998;95:2011–2016. doi: 10.1073/pnas.95.5.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ. Angiosperm DNA C-values database. Kew: Royal Botanic Gardens; 2004. (release 5·0) [Google Scholar]
- Bennett MD, Leitch IJ, Hanson L. DNA amounts in two samples of angiosperm weeds. Annals of Botany. 1998;82:121–134. [Google Scholar]
- Bennetzen JL. Mechanisms and rates of genome expansion and contraction in flowering plants. Genetica. 2002;115:29–36. doi: 10.1023/a:1016015913350. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Ma J, Devos KM. Mechanisms of recent genome size variation in flowering plants. Annals of Botany. 2005;95:127–132. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossdorf O, Auge H, Lafuma L, Rogers WE, Siemann E, Prati D. Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia. 2005;144:1–11. doi: 10.1007/s00442-005-0070-z. [DOI] [PubMed] [Google Scholar]
- Butler MA, King AA. Phylogenetic comparative analysis: a modeling approach for adaptive evolution. The American Naturalist. 2004;164:683–695. doi: 10.1086/426002. [DOI] [PubMed] [Google Scholar]
- Cade BS, Noon BR. A gentle introduction to quantile regression for ecologists. Frontiers in Ecology and the Environment. 2003;1:412–420. [Google Scholar]
- Cade BS, Noon BR, Flather CH. Quantile regression reveals hidden bias and uncertainty in habitat models. Ecology. 2005;86:786–800. [Google Scholar]
- Cavalier-Smith T. Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Annals of Botany. 2005;95:147–175. doi: 10.1093/aob/mci010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Brown JKM, Bennetzen JL. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Research. 2002;12:1075–1079. doi: 10.1101/gr.132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facon B, Genton BJ, Shykoff J, Jarne P, Estoup A, David P. A general eco-evolutionary framework for understanding bioinvasions. Trends in Ecology & Evolution. 2006;21:130–135. doi: 10.1016/j.tree.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Francis D, Davies MS, Barlow PW. A strong nucleotypic effect on the cell cycle regardless of ploidy level. Annals of Botany. 2008;101:747–757. doi: 10.1093/aob/mcn038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith DW, Lambert GM, Macas J, Dolezel J. Analysis of nuclear DNA content and ploidy in higher plants. In: Robinson JP, Darzynkiewicz Z, Dean PN, et al., editors. Current protocols in cytometry. New York: John Wiley & Sons, Inc.; 1997. pp. 7.6.1–7.6.22. [DOI] [PubMed] [Google Scholar]
- Gifford ALS, Ferdy J-B, Molofsky J. Genetic composition and morphological variation among populations of the invasive grass, Phalaris arundinacea. Canadian Journal of Botany. 2002;80:779–785. [Google Scholar]
- Gregory TR, Hebert PDN. The modulation of DNA content: proximate causes and ultimate consequences. Genome Research. 1999;9:317–324. [PubMed] [Google Scholar]
- Greilhuber J. Intraspecific variation in genome size in angiosperms: identifying its existence. Annals of Botany. 2005;95:91–98. doi: 10.1093/aob/mci004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greilhuber J, Doležel J, Lysák MA, Bennett MD. The origin, evolution and proposed stabilization of the terms ‘genome size’ and ‘C-value’ to describe nuclear DNA contents. Annals of Botany. 2005;95:255–260. doi: 10.1093/aob/mci019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grime JP. Plant classification for ecological purposes: is there a role for genome size? Annals of Botany. 1998;82(suppl. A):117–120. [Google Scholar]
- Grime JP, Mowforth MA. Variation in genome size – an ecological interpretation. Nature. 1982;299:151–153. [Google Scholar]
- Grime JP, Shacklock JML, Brand SR. Nuclear DNA contents, shoot phenology and species co-existence in a limestone grassland community. New Phytologist. 1985;100:435–445. [Google Scholar]
- Grotkopp E, Rejmanek M, Sanderson MJ, Rost TL. Evolution of genome size in pines (Pinus) and its life-history correlates: supertree analyses. Evolution. 2004;58:1705–1729. doi: 10.1111/j.0014-3820.2004.tb00456.x. [DOI] [PubMed] [Google Scholar]
- Hosmer DW, Lemeshow S. Applied logistic regression. New York: Wiley; 1989. [Google Scholar]
- Huey RB, Gilchrist GW, Carlson ML, Berrigan D, Serra L. Rapid evolution of a geographic cline in size in an introduced fly. Science. 2000;287:308–309. doi: 10.1126/science.287.5451.308. [DOI] [PubMed] [Google Scholar]
- Huey RB, Gilchrist GW, Hendry AP. Using invasive species to study evolution: case studies with Drosophila and salmon. In: Sax DF, Stachowicz JJ, Gaines SD, editors. Species invasions. Insights into ecology, evolution, and biogeography. Sunderland, MA: Sinauer Associates, Inc.; 2005. pp. 139–164. [Google Scholar]
- Kidwell MG. Transposable elements and the evolution of genome size in eukaryotes. Genetica. 2002;115:49–63. doi: 10.1023/a:1016072014259. [DOI] [PubMed] [Google Scholar]
- Knight CA, Ackerly DD. Variation in nuclear DNA content across environmental gradients: a quantile regression analysis. Ecology Letters. 2002;5:66–76. [Google Scholar]
- Knight CA, Beaulieu J. Genome size scaling through phenotype space. Annals of Botany. 2008;101:759–766. doi: 10.1093/aob/mcm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight CA, Molinari NA, Petrov DA. The large genome constraint hypothesis: evolution, ecology and phenotype. Annals of Botany. 2005;95:177–190. doi: 10.1093/aob/mci011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis S, Schmidt T, Heslop-Harrison JS. Repetitive DNA elements as a major component of plant genomes. Annals of Botany. 1998;82(suppl. A):45–55. [Google Scholar]
- Lavergne S, Molofsky J. Reed canary grass (Phalaris arundinacea L.), as a biological model in the study of plant invasions. Critical Reviews in Plant Sciences. 2004;23:415–429. [Google Scholar]
- Lavergne S, Molofsky J. Control strategies for the invasive reed canarygrass (Phalaris arundinacea L.) in North American wetlands: the need for an integrated management plan. Natural Areas Journal. 2006;26:208–214. [Google Scholar]
- Lavergne S, Molofsky J. Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proceedings of the National Academy of Sciences of the USA. 2007;104:3883–3888. doi: 10.1073/pnas.0607324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux JJ, Wieczorek AM, Wright MG, Tran CT. Super-genotype: global monoclonality defies the odds of nature. PLoS One. 2007;2:e570. doi: 10.1371/journal.pone.0000590. doi:10.1371/journal.pone.0000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CE. Evolutionary genetics of invasive species. Trends in Ecology and Evolution. 2002;17:386–391. [Google Scholar]
- Leitch IJ, Chase MW, Bennett MD. Phylogenetic analysis of DNA C-values provide evidence for a small ancestral genome size in flowering plants. Annals of Botany. 1998;82(suppl. A):85–94. [Google Scholar]
- Lindholm AK, Breden F, Alexander HJ, Chan W-K, Thakurta SG, Brooks R. Invasion success and genetic diversity of introduced populations of guppies Poecilia reticulata in Australia. Molecular Ecology. 2005;14:3671–3682. doi: 10.1111/j.1365-294X.2005.02697.x. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. 2006. Version 1.11. http://mesquiteproject.org .
- Meagher TR, Costich DE. Nuclear DNA content and floral evolution in Silene latifolia. Proceedings of the Royal Society of London Series B – Biological Sciences. 1996;263:1455–1460. [Google Scholar]
- Meagher TR, Vassiliadis C. Phenotypic impacts of repetitive DNA in flowering plants. New Phytologist. 2005;168:71–80. doi: 10.1111/j.1469-8137.2005.01527.x. [DOI] [PubMed] [Google Scholar]
- Nilsson AI, Koskiniemi S, Eriksson S, Kugelberg E, Hinton JCD, Andersson DI. Bacterial genome size reduction by experimental evolution. Proceedings of the National Academy of Sciences of the USA. 2005;102:12112–12116. doi: 10.1073/pnas.0503654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak SJ. The role of evolution in the invasion process (Commentary) Proceedings of National Academy of Sciences of the USA. 2007;104:3671–3672. doi: 10.1073/pnas.0700224104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak SJ, Mack RN. Genetic bottlenecks in alien plant species. Influence of mating systems and introduction dynamics. In: Sax DF, Stachowicz JJ, Gaines SD, editors. Species Invasions. Insights into ecology, evolution, and biogeography. Sunderland, MA: Sinauer Associates, Inc; 2005. pp. 201–228. [Google Scholar]
- Oliver MJ, Petrov D, Ackerly D, Falkowski P, Schofield OM. The mode and tempo of genome size evolution in eukaryotes. Genome Research. 2007;17:594–601. doi: 10.1101/gr.6096207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro JC, Bates DM. Mixed-effects models in S and S-PLUS. New York: Springer; 2000. [Google Scholar]
- Prentis PJ, Wilson JRU, Dormontt EE, Richardson DM, Lowe AJ. Adaptive evolution in invasive species. Trends in Plant Science. 2008;13:288–294. doi: 10.1016/j.tplants.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Rayburn AL, Dudley JW, Biradar DP. Selection for early flowering results in simultaneous selection for reduced nuclear DNA content in maize. Plant Breeding. 1994;112:318–322. [Google Scholar]
- Rosenthal DM, Ramakrishnan AP, Cruzan MB. Evidence for multiple sources of invasion and intraspecific hybridization in Brachypodium sylvaticum (Hudson) Beauv. in North America. Molecular Ecology. 2008;17:4657–4669. doi: 10.1111/j.1365-294X.2008.03844.x. [DOI] [PubMed] [Google Scholar]
- Sax DF, Stachowicz JJ, Brown JH, et al. Ecological and evolutionary insights from species invasions. Trends in Ecology & Evolution. 2007;22:465–471. doi: 10.1016/j.tree.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Weider LJ, Elster JJ, Crease TJ, Mateos M, Cotner JB, Markow TA. The functional significance of ribosomal (r)DNA variation: impacts on the evolutionary ecology of organisms. Annual Review of Ecology, Evolution and Systematics. 2005;2005:219–242. [Google Scholar]
- Weiss-Schneeweiss H, Greilhuber J, Schneeweiss GM. Genome size evolution in holoparasitic Orobanche (Orobanchaceae) and related genera. American Journal of Botany. 2005;93:148–156. doi: 10.3732/ajb.91.3.439. [DOI] [PubMed] [Google Scholar]
- Zayed A, Constantin SA, Packer L. Successful biological invasion despite a severe genetic load. PLoS One. 2007;2:e868. doi: 10.1371/journal.pone.0000868. doi:10.1371/journal.pone.0000868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E. Why so many noncoding nucleotides? The eukaryote genome as an epigenetic machine. Genetica. 2002;115:105–129. doi: 10.1023/a:1016080316076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.