Abstract
Telomeric DNA terminates with a single-stranded 3′ G-overhang that in vertebrates and fission yeast is bound by POT1 (Protection Of Telomeres). However, no in vitro telomeric DNA binding is associated with Arabidopsis POT1 paralogs. To further investigate POT1–DNA interaction in plants, we cloned POT1 genes from 11 plant species representing major branches of plant kingdom. Telomeric DNA binding was associated with POT1 proteins from the green alga Ostreococcus lucimarinus and two flowering plants, maize and Asparagus. Site-directed mutagenesis revealed that several residues critical for telomeric DNA recognition in vertebrates are functionally conserved in plant POT1 proteins. However, the plant proteins varied in their minimal DNA-binding sites and nucleotide recognition properties. Green alga POT1 exhibited a strong preference for the canonical plant telomere repeat sequence TTTAGGG with no detectable binding to hexanucleotide telomere repeat TTAGGG found in vertebrates and some plants, including Asparagus. In contrast, POT1 proteins from maize and Asparagus bound TTAGGG repeats with only slightly reduced affinity relative to the TTTAGGG sequence. We conclude that the nucleic acid binding site in plant POT1 proteins is evolving rapidly, and that the recent acquisition of TTAGGG telomere repeats in Asparagus appears to have co-evolved with changes in POT1 DNA sequence recognition.
INTRODUCTION
Telomeres are the ancient nucleoprotein structures that define the physical ends of eukaryotic chromosomes, protecting them from deleterious activities, such as recombination and nucleolytic attack, and providing a means to replenish telomeric DNA lost during replication (1). Defects in telomere structure or length maintenance result in cell proliferation and genome maintenance abnormalities, senescence or apoptosis (2). Telomere structure and composition is conserved across different eukaryotic lineages. Telomeric DNA typically consists of tandem arrays of short G-rich repeats that can reach thousands of nucleotides in length. The extreme 3′-ends of the chromosomes terminate in a single-stranded protrusion termed the G-overhang. Several evolutionarily conserved proteins bind directly to the double-stranded region of the telomeric DNA or to the single-strand G-overhang to form the first layer of telomere-associated protein factors. Together with bridging proteins, these DNA-binding factors constitute a telomere-specific protein complex termed shelterin (2,3).
The green plant lineage represents a monophyletic group of photosynthetic organisms that evolved near the base of eukaryotic life and shared the last common ancestor with fungi and animals ∼1.5 billion years ago (bya) (4). Despite such long divergence time, many aspects of telomere biology are well conserved between plants and animals. The telomere repeat sequence in the vast majority of plants is TTTAGGG, one nucleotide longer than the 6-base sequence TTAGGG found in vertebrates (5). Interestingly, a few outliers exist in the plant kingdom, that harbor atypical or unknown telomere sequence. For example, while many green algae exhibit the canonical plant TTTAGGG repeats (6) (E. Shakirov and D. Shippen, unpublished results), telomeres in the model fresh water alga Chlamydomonas reinhardtii are composed of the eight nucleotide repeat TTTTAGGG (7). Onions (Allium cepa) and related species lack simple telomere repeats and instead appear to harbor terminally located satellite DNA (8). Most intriguing is the situation in two phylogenetically unrelated groups of flowering plants (angiosperms), in which canonical TTTAGGG repeats have been replaced by the vertebrate-type hexanucleotide TTAGGG sequence. One of these plant groups is comprised of a handful of obscure Solanaceae (tomato family) species native to South America (9). The second group consists of a large number of families in the Asparagales order, and represents one of the most successful lineages of extant flowering plants, with 22 000–25 000 currently known species, or nearly 10% of all angiosperms, including Irises, Hyacinths, Agaves and Amaryllis (10). The switch in telomeric DNA sequence from TTTAGGG to TTAGGG is thought to have occurred approximately 90 million years ago (mya) (11) and likely corresponds to a single nucleotide deletion in the template region of the telomerase RNA subunit. This sequence change may have posed a challenge for the plant shelterin complex to maintain chromosome end protection, although the successful diversification of Asparagales argues that these plants accommodated the mutation in telomeric DNA sequence in a short evolutionary time frame. Such compensatory changes likely involved co-evolution of telomere-binding proteins to allow binding to the new telomere repeat sequence, but the molecular mechanisms of these changes are currently unknown.
In vertebrates and fission yeast, POT1 (Protection Of Telomeres) binds single-strand G-rich telomeric DNA with high affinity and plays a pivotal role in mediating telomere length regulation as well as chromosome end protection and cell viability (2,3,12). POT1 proteins are defined by the presence of two N-terminal oligosaccharide/oligonucleotide binding folds (OB-folds), which are responsible for the specific interaction with single-stranded telomeric DNA. OB1 and OB2 contacts with telomeric DNA are primarily mediated by aromatic amino acids, which form stacking interactions with DNA nucleotides (13). Most POT1 proteins studied to date exhibit a minimum binding site (MBS) of 10–12 nucleotides, roughly corresponding to two telomeric repeats, though the most preferred repeat permutation in each case appears to be species-specific (14–16). Interestingly, only a subset of MBS nucleotides is specifically recognized by POT1 proteins, with nucleotides crucial for protein binding scattered throughout the MBS. For many POT1 proteins, the most 3′-terminal MBS nucleotide is buried deep inside the OB-folds (14,17), suggesting a mechanism for how POT1 can protect the G-overhang from nucleases or telomerase action. In addition, while some POT1 proteins clearly prefer 3′-terminal telomeric repeats (12), others can associate with telomeric sequences in the middle or on the 5′-terminus of oligonucleotide substrate (16,18), suggesting that in vivo they can also bind to the displaced G-rich strand in the context of the T-loop. While specific interaction with telomeric DNA in vivo is an essential feature of all POT1 proteins studied to date, human POT1 is delivered to the telomere via protein–protein interactions with its binding partner, TPP1/ACD. The interaction with TPP1 is achieved through a structurally undefined C-terminal domain (19,20). The POT1-TPP1 heterodimer is required for proper shelterin assembly and to regulate telomerase access and processivity (20–23).
POT1-like proteins have also been identified in plants (24–28). Similar to the situation in most other eukaryotes, only a single POT1 gene has been detected in most plants surveyed (Shakirov et al., manuscript in preparation). However, Arabidopsis is an exception as it encodes three highly divergent POT1-like proteins (29; Nelson et al., manuscript in preparation). All three Arabidopsis POT1 proteins are involved in telomere biology, but their functions differ. AtPOT1a is a positive regulator of telomerase activity that physically interacts with the telomerase RNP (30,31), while AtPOT1b and AtPOT1c negatively regulate telomerase activity and participate in chromosome end protection (29; Nelson et al., manuscript in preparation). Strikingly, although AtPOT1 proteins have an architecture similar to yeast and vertebrate POT1 with two N-terminal OB-folds and a C-terminal domain, no in vitro telomeric DNA binding has been demonstrated for POT1 proteins from Arabidopsis or two other closely related plants (26,30). Thus, it is unclear whether telomeric DNA binding is a conserved function of POT1 proteins from the plant kingdom.
Plant systematics has witnessed a remarkable influx of new data revealing evolutionary relationship of various lineages of the green plants. We took advantage of this detailed phylogenetic map to clone POT1 genes from eleven representative members of major plant evolutionary branches. Here we report the initial characterization of the DNA-binding activities of three plant POT1 proteins and provide evidence for significant biochemical differences in POT1 proteins across the plant kingdom. We also demonstrate that POT1 proteins from angiosperms have strong affinity for both TTAGGG and TTTAGGG telomeric repeats, providing a possible explanation for how Asparagales adapted to the recent switch in its telomeric DNA repeat sequence.
MATERIALS AND METHODS
Asparagus telomere length analysis
DNA from Asparagus (Asparagus officinalis) shoots was extracted as described (32). Terminal restriction fragment (TRF) analysis was performed as described (33) with DNA digested with either Tru1I or AluI restriction enzymes (Fermentas, Hanover, MD). 32P 5′-end-labeled (T3AG3)4 and (T2AG3)4TTAG oligonucleotides were used as heptanucleotide and hexanucleotide probes, respectively. Radioactive signals were scanned by a Pharos FX Plus Molecular Imager (Bio-Rad Laboratories, Hercules, CA), and the data were analyzed by Quantity One v.4.6.5 software (Bio-Rad).
In vitro translation and EMSA assays
Expression of plant POT1 proteins in rabbit reticulocyte lysate (RRL) was performed as described (24). EMSA assays were conducted as described (26) with slight modifications. Briefly, each reaction (15 μl total volume) contained equal amounts (4 μl) of RRL-translated plant POT1 protein, 0.5 pmol of the corresponding 32P-labeled telomeric oligonucleotide, 3 μl of 5 × DNA-binding buffer (100 mM Tris–HCl pH 8.0, 250 mM NaCl, 50 mM MgCl, 5 mM EDTA, 5 mM DTT, 25% glycerol) and 1 μl each of non-specific RNA, single-stranded and double-stranded DNA competitors as described in (24). Reactions were incubated at RT for 15 min. For competition assays, 2.5 pmol of cold competitor oligonucleotide was added and the reactions were incubated for an additional 15 min. The complexes were separated on 5% polyacrylamide gel (acrylamide: bisacrylamide 29:1) for 2 h at 150 volts in 0.8 × TBE at RT, dried and exposed to PhosphorImager screens. Screens were scanned by a Pharos FX Plus Molecular Imager and signal intensity was quantified by Quantity One v.4.6.5 software. Each EMSA result was reproduced several times, but due to variations in protein expression levels in RRL, only one representative gel and the corresponding quantification result are shown for each experiment.
Site-directed mutagenesis
Mutagenesis reactions were performed with Pfu Turbo DNA polymerase (Stratagene) according to the manufacturer’s instructions using the following PCR conditions: 94°C 5 min; 18 cycles of 94°C 30 s, 55°C 1 min, 68°C 40 min; followed by 10 min at 68°C. After DpnI (Promega) treatment at 37°C for 4 h, the reaction product was transformed into TOP10F’ competent cells (Invitrogen). Plasmids were purified and mutations verified by sequencing.
Nucleotide sequence accession numbers
cDNAs encoding the following plant POT1 proteins were deposited into the GenBank: HvPOT1 (EU880295), PtrPOT1 (EU880297), HaPOT1 (EU880298), SmPOT1 (EU880301), ZmPOT1a (EU880303), ZmPOT1b (EU880304), GhPOT1 (EU880305), PtaPOT1 (EU880306), StPOT1 (EU883536) and AoPOT1 (FJ516399). The nucleotide sequence of cloned OlPOT1 cDNA was found to correspond to the previously deposited sequence ABO96101.
RESULTS
Telomeric DNA binding by plant POT1 proteins
As part of a larger study on the molecular evolution and functional divergence of POT1 proteins in plants, we cloned POT1 cDNAs from eleven plant species representing major branches on the plant evolutionary tree. These include POT1 sequences from the green alga Ostreococcus lucimarinus, a spikemoss (Selaginella moellendorffii), a pine, three monocotyledonous (barley, maize and Asparagus) and four dicotyledonous (potato, sunflower, poplar and cotton) flowering plants (Supplementary Figure S1). Among the plant species analyzed in our study, three genomes have been sequenced: O. lucimarinus, S. moellendorffii and Populus trichocarpa (poplar). Ostreococcus POT1 is a single-exon gene. POT1 genes in both Selaginella and poplar, as well as the two previously characterized Arabidopsis thaliana POT1 genes, harbor 10 exons with conserved intron positions (data not shown). This evolutionarily conserved gene structure supports the conclusion that the plant POT1 genes are indeed orthologs. Among all plant species analyzed in this study, only maize appears to encode more than one POT1 protein. The two maize POT1 genes were most likely retained after whole-genome duplication in the ancestor of maize (34). ZmPOT1a and ZmPOT1b encode 54.5 and 54.6 kDa proteins, respectively, with 75% overall amino acid similarity to each other. Like the POT1 proteins from the Brassicaceae plants (26), all eleven of the new POT1 proteins display significant sequence conservation and are predicted to harbor two N-terminal DNA-binding OB-folds with secondary structures similar to the human and fission yeast POT1 proteins (J. Croy and D. Wuttke, personal communication).
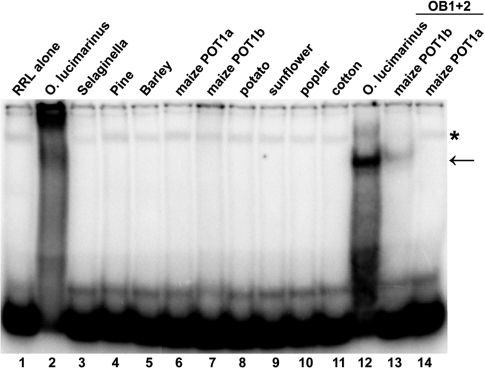
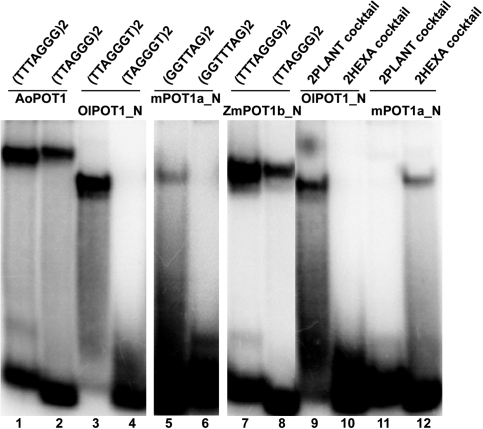
To assess the DNA-binding properties of the new POT1 proteins, we attempted to obtain soluble recombinant proteins using standard expression protocols in Escherichia coli. However, as was previously shown for POT1 proteins from Arabidopsis and related species (26,30), we were unable to generate a sufficient amount of soluble protein from any clone for DNA-binding studies. Therefore, we turned to an in vitro rabbit reticulocyte lysate (RRL) expression system, previously shown to produce soluble vertebrate and plant POT1 proteins (24,26,35). While such approach will not allow us to define DNA-binding constants, we could perform qualitative binding experiments that in previous studies with the mammalian POT1 proteins have provided important comparative insights into POT1 interaction with telomeric DNA (24,35). RRL-expressed plant POT1 proteins were obtained in a soluble form (Supplementary Figure S2) and were subjected to electrophoretic mobility shift assays (EMSA) (Figure 1).
Figure 1.
Analysis of DNA-binding capacity of recombinant plant POT1 proteins. EMSA was performed with a cocktail of seven 32P-labeled 2PLANT oligonucleotides. POT1 proteins from the corresponding plant species are shown above each lane. Binding assays were performed with either full-length POT1 (lanes 2–11) or with truncated proteins bearing only two N-terminal OB-fold domains (lanes 12–14). Asterisk designates a non-specific band often present in the negative RRL-only control (lane 1). Protein–DNA complexes specific to maize and Ostreococcus lucimarinus POT1 proteins are indicated by an arrow.
For the yeast and vertebrate POT1 proteins, two telomeric repeats are sufficient for in vitro-binding (13). Therefore, we used a cocktail of 32P 5′-labeled oligonucleotides corresponding to two repeats of the seven possible permutations of the plant telomere repeat (2PLANT cocktail probe) for EMSA. Under standard gel-shift conditions (35), stable telomeric DNA binding was observed for two full-length plant POT1 proteins: OlPOT1 from the green alga O. lucimarinus (Figure 1, lane 2) and AoPOT1 from A. officinalis (garden asparagus) (see below). In addition, a band with intensity slightly above background was observed in the well for the maize (Zea mays) ZmPOT1b protein (Figure 1, lane 7). The intensity of this band increased when a specific oligonucleotide was used instead of the oligonucleotide cocktail (data not shown).
Telomeric DNA binding by mammalian and yeast POT1 proteins is enhanced with constructs containing only the N-terminal OB-folds (12,35,36), possibly due to modulation of DNA binding by the protein C-terminus. The OB-fold domains of all 11 plant POT1 proteins were expressed in RRL (Supplementary Figure S2 and data not shown) and tested in EMSA. Deletion of the C-terminus improved the binding of OlPOT1_N (amino acids 1–363) to the plant telomeric DNA cocktail, resulting in the formation of a single well-defined protein-DNA complex (Figure 1, lane 12). In addition, a truncated version of maize POT1b, ZmPOT1b_N (amino acids 1–326) also formed single weak protein-DNA complex (Figure 1, lane 13). However, the second maize POT1 protein, ZmPOT1a, failed to bind telomeric DNA as either a full-length protein or an OB-fold truncation (Figure 1, lanes 6 and 14). With the exception of AoPOT1_N (see below), we detected no telomeric DNA binding by any other OB-fold truncated plant POT1 protein (data not shown). We also asked whether plant POT1 proteins could bind an oligonucleotide corresponding to the C-rich strand of telomeric DNA, as is the case for one of the POT1-like proteins from Caenorhabditis elegans (37). No C-strand binding was observed for any of the plant POT1 proteins (data not shown). Nevertheless, our data imply that the ability of POT1 to bind G-rich telomeric DNA has not been completely lost throughout plant kingdom.
DNA-binding properties of OlPOT1_N from green alga
Ostreococcus lucimarinus is a species of Prasinophytes, a clade of green algae that belongs to the oldest diverging (over 1 bya) branch of the photosynthetic eukaryotic lineage, and is a sister clade to all land plants (4). Consequently, analysis of the DNA-binding characteristics of OlPOT1_N may provide insight into the mechanisms of telomeric DNA recognition by the ancestral plant POT1 protein and how these properties have evolved in land plants.
To determine which of the seven permutations of the plant telomeric repeat OlPOT1_N binds best, we performed competition experiments with 32P-labeled 2PLANT cocktail probe and a 5-fold excess of individual cold 2PLANT oligonucleotides. A representative gel and corresponding quantification are shown in Supplementary Figure S3A. The intensity of the shifted band (fraction bound) in the absence of competitors was measured and set as 1.0, and the remaining signal intensity after the addition of competitors was expressed as a fraction of 1. We found that all individual 2PLANT oligonucleotides competed efficiently for binding (Supplementary Figure S3A).
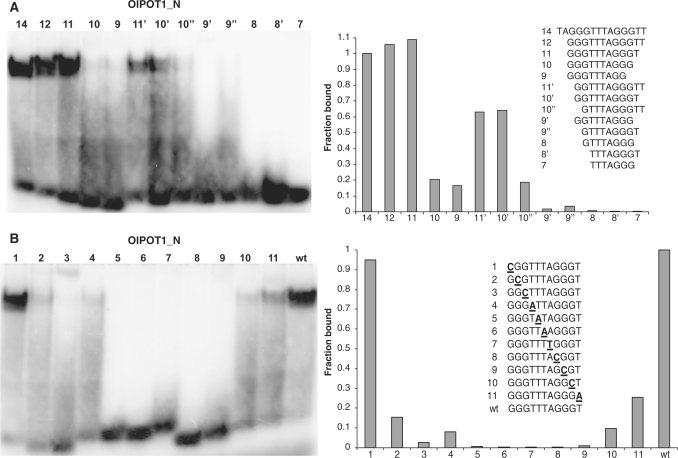
Next we determined the minimum DNA sequence required for OlPOT1_N binding. EMSA was performed using TAGGGTTTAGGGTT and a series of single nucleotide truncations from either the 5′- or 3′-end of this substrate (Figure 2A). Deletion of the first two nucleotides from the 5′-end (oligonucleotide 12) did not decrease binding. However, removal of three nucleotides decreased binding by over 30% (oligonucleotide 11′) and deletion of four abolished nearly all binding (oligonucleotide 10″). In both cases, a smear trailing down to free probe was observed, suggesting partial dissociation of the complex during electrophoresis. Only one nucleotide could be removed from the 3′-end without detectable loss of DNA binding (oligonucleotide 11). Deletion of two nucleotides (oligonucleotide 10) decreased binding to only 15% of the full-length oligonucleotide. Therefore, the minimum tight-binding sequence (core MBS) for OlPOT1_N appears to be GGTTTAGGGT (oligonucleotide 10′). Addition of one G residue at the oligonucleotide 5′-end improved binding almost 2-fold (oligonucleotide 11). Thus, as little as 10 nucleotides are necessary for OlPOT1_N binding and may comprise its MBS, while an 11-nt GGGTTTAGGGT oligonucleotide represents the best tight-binding substrate.
Figure 2.
Characterization of DNA-binding activity of recombinant POT1_N protein from Ostreococcus lucimarinus. (A) EMSA identifies the minimum binding site of OlPOT1_N. Equal amounts of OlPOT1_N were incubated with the indicated oligonucleotides and the protein-DNA complexes were separated by native PAGE. (B) Identification of nucleotides recognized by OlPOT1_N in GGGTTTAGGGT. Numbers indicate nucleotide positions that were substituted with complementary nucleotides (bold and underlined). Representative EMSA scans are shown in left panels. For each scan, the signal intensity (fraction of protein bound) is plotted on the right with binding to TAGGGTTTAGGGTT (A) or GGGTTTAGGGT (wt) (B) set at 1.0.
Studies with non-plant POT1 proteins reveal that only a subset of nucleotides within the MBS are specifically recognized (13). To identify nucleotides in the OlPOT1_N MBS critical for protein interaction, a series of oligonucleotides with single complementary nucleotide substitutions were tested for OlPOT1_N binding (Figure 2B). Unexpectedly, we discovered that all core MBS nucleotides are crucial for OlPOT1_N binding (Figure 2B, lanes 2–11). The only nucleotide change that did not affect OlPOT1_N binding was the most 5′-terminal G (Figure 2B, lane 1), which is not a part of the core MBS. A nucleotide in this position may be required for improved protein-DNA complex stability, but may not contribute to specific interactions with OlPOT1_N protein. Overall, we conclude that OlPOT1_N requires at least 10 telomeric nucleotides for efficient binding. However, unlike the vertebrate and yeast POT1 proteins, all nucleotides in the core MBS appear to make specific and crucial contacts with OlPOT1_N.
DNA-binding properties of POT1b_N from maize
Maize is an angiosperm species that harbors canonical plant TTTAGGG telomere repeats (38). We tested ZmPOT1b_N binding to the seven permutations of the plant telomere repeat sequence using competition assays (Supplementary Figure S3B). Although incubation with several 2PLANT competitors leads to slightly decreased signal intensity (Supplementary Figure S3B, lanes 6–8), only the addition of (TTTAGGG)2 and (GTTTAGG)2 resulted in significant competition (Supplementary Figure S3B, lanes 2 and 3). These data indicate that ZmPOT1b_N recognizes two different permutations of the plant telomere repeat.
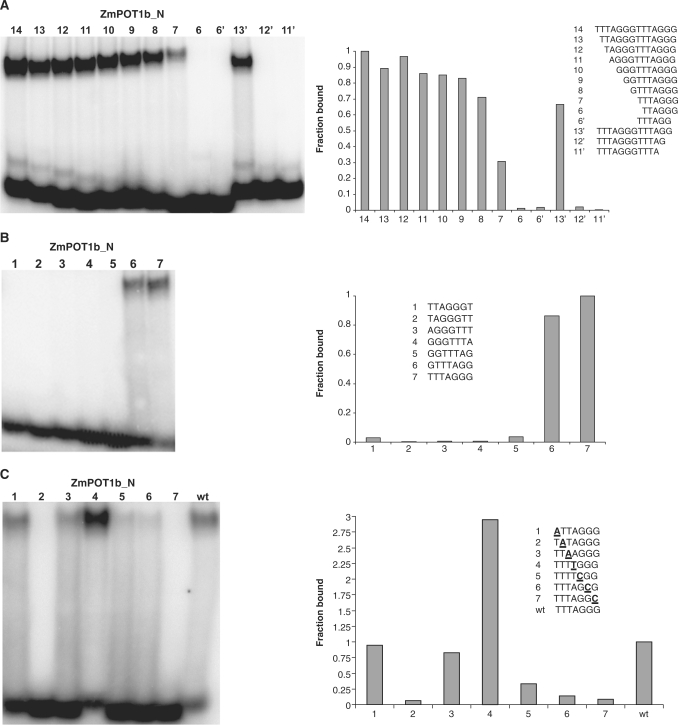
Next we determined the minimum number of nucleotides required for ZmPOT1b_N interaction with telomeric DNA (Figure 3A). In contrast to the situation with OlPOT1_N, removal of as many as seven nucleotides from the 5′-end of the oligonucleotide (a full plant TTTAGGG repeat) did not abolish binding (Figure 3A, oligonucleotide 7). On the other hand, only a single nucleotide could be removed from the 3′-end of (TTTAGGG)2 oligonucleotide (Figure 3A, compare oligonucleotides 13′ and 12′). Since two different probes containing 6 nucleotides each failed to bind ZmPOT1b_N (Figure 3A, lanes 6 and 6′), these data indicate that the MBS necessary for efficient ZmPOT1b_N binding consists of seven nucleotides, one full plant repeat.
Figure 3.
Characterization of ZmPOT1b_N interaction with telomeric DNA. (A) Identification of the MBS for ZmPOT1b_N. Equal amounts of ZmPOT1b_N were incubated with the indicated oligonucleotides and protein-DNA complexes were separated by native PAGE. (B) Analysis of ZmPOT1b_N binding to different permutations of a single plant telomere repeat. Numbers indicate seven possible repeat permutations. (C) Analysis of nucleotides specifically recognized by ZmPOT1b_N in TTTAGGG. Numbers indicate nucleotide positions that were substituted with complementary nucleotides (bold and underlined). For all panels, EMSA scans are shown on the left, and radioactive signal intensity is plotted on the right with ZmPOT1b_N binding to TTTAGGGTTTAGGG (A) or TTTAGGG (B, C) set at 1.0.
To further evaluate the MBS of ZmPOT1b_N, we tested ZmPOT1b_N binding to oligonucleotides representing the seven permutations of the plant telomere repeat (1PLANT). Similar to the permutation analysis of 14-nt 2PLANT probes (Supplementary Figure S3B), ZmPOT1b_N stably associated with two 7-nt 1PLANT probes, TTTAGGG and GTTTAGG (Figure 3B, lanes 6 and 7). Among the two, TTTAGGG appeared to be a slightly better substrate, suggesting that this sequence represents the preferred MBS for ZmPOT1b_N.
We tested the relative importance of each MBS nucleotide for efficient ZmPOT1b_N binding using complementary nucleotide substitutions (Figure 3C). ZmPOT1b_N binding was significantly reduced or abolished with substitutions in positions 2 (TATAGGG), 5 (TTTACGG), 6 (TTTAGCG) and 7 (TTTAGGC) (Figure 3C, lanes 2, 5–7). Nucleotide changes in position 1 (ATTAGGG) and position 3 (TTAAGGG) did not lead to a substantial decrease in ZmPOT1b_N binding, while a change in position 4 (TTTTGGG) actually improved binding 3-fold over the wild-type (Figure 3C, compare lane 4 with wt). These data suggest that unlike the situation with OlPOT1_N, not all nucleotides in ZmPOT1b_N MBS make crucial contributions to binding.
Interestingly, only three out of seven nucleotide positions (1, 4 and 5) differ in the two acceptable ZmPOT1b_N permutations, T1T2T3A4G5G6G7 and G1T2T3T4A5G6G7. Since changes in positions 1 and 4 do not lead to decreased protein binding, the increased ZmPOT1b_N binding to TTTAGGG versus GTTTAGG may reflect a difference at position 5, with G being preferred over A. Overall, although TTTAGGG is the natural telomere repeat sequence in both O. lucimarinus and maize, the MBS and the relative importance of individual nucleotides within it vary dramatically between OlPOT1_N and ZmPOT1b_N.
DNA-binding properties of Asparagus POT1 protein
The plant lineages leading to maize and Asparagus diverged only 100–110 mya (11), a relatively recent event in the evolutionary history of land plants. To study Asparagus POT1, we first confirmed that A. officinalis possesses hexanucleotide TTAGGG repeats using the TRF assay. As expected, a consensus plant telomere probe (T3AG3)4 hybridized to the control Arabidopsis telomeric DNA (Supplementary Figure S4A, lanes 1 and 3), but not to Asparagus DNA (lanes 2 and 4), while a TTAGGG-specific telomere probe recognized Asparagus DNA, but not Arabidopsis (Supplemental Figure 4A, lanes 5–8). Strikingly, Asparagus telomere tracts appear to be at least an order of magnitude longer than in Arabidopsis (Supplemental Figure 4A, compare lanes 1 and 3 with 6 and 8).
To evaluate Asparagus POT1 binding to the hexanucleotide telomere repeat sequences, we examined the affinity of full-length AoPOT1 for an oligonucleotide cocktail containing equal amounts of all six possible permutations of two TTAGGG telomere repeats (2HEXA cocktail probe). A single shifted band was formed (Supplementary Figure 3C, lane 1). To determine which permutation(s) of the human telomere repeat were recognized, competition assays were performed with a 5-fold excess of individual cold 2HEXA oligonucleotides (Supplementary Figure S3C, lanes 2–7). Among all 2HEXA oligonucleotides, (TTAGGG)2 was the most efficient competitor (Supplementary Figure S3C, lane 2). The specificity of AoPOT1 for (TTAGGG)2 was confirmed in a direct binding assay. AoPOT1 efficiently bound (TTAGGG)2, while the mouse mPOT1a_N protein was unable to bind this repeat permutation (Supplementary Figure S4B, compare lanes 1 and 3). Thus, although the telomere repeat sequence in Asparagus and vertebrates is the same, AoPOT1 prefers the permutation terminating in TTAGGG, while mPOT1a_N and other vertebrate POT1 proteins prefer the permutation terminating in GGTTAG (14,16,18,35).
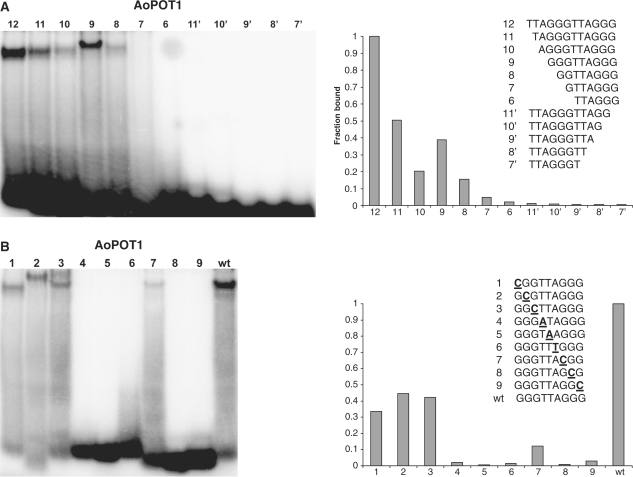
AoPOT1 binding was assessed with a series of single-nucleotide truncations of the (TTAGGG)2 oligonucleotide to define the MBS. Removal of even a single nucleotide from the 3′-end of the oligonucleotide was not tolerated (Figure 4A, oligonucleotides 11′-7′). On the other hand, removal of as many as 4 nucleotides from the 5′-end did not completely abolish binding, though it led to a substantial reduction in signal intensity (Figure 4A, oligonucleotide 8). While these data suggest that the MBS is the 8-nt GGTTAGGG, we noticed that AoPOT1 binding to a 9-nt oligonucleotide GGGTTAGGG is improved 2-fold (oligonucleotide 9), suggesting that GGGTTAGGG represents the best tight-binding substrate.
Figure 4.
Analysis of Asparagus POT1 interaction with telomeric DNA. (A) Identification of the MBS for AoPOT1. Equal amounts of AoPOT1 were incubated with the indicated oligonucleotides and protein-DNA complexes were separated by native PAGE. (B) Analysis of nucleotides specifically recognized by AoPOT1 in GGGTTAGGG. Numbers indicate nucleotide positions that were substituted with complementary nucleotides (bold and underlined). For all panels, EMSA scans are shown on the left, and radioactive signal intensity is plotted on the right with AoPOT1 binding to TTAGGGTTAGGG (A) or GGGTTAGGG (B) set at 1.0.
We next examined the relative contribution of each MBS nucleotide to AoPOT1 binding using a series of complementary nucleotide substitutions in GGGTTAGGG (Figure 4B). Although mutations in the first three 5′-terminal positions decreased signal intensity to only 34–45% of the original oligonucleotide (Figure 4B, oligonucleotides 1–3), mutations in all other positions abolished binding almost completely (Figure 4B, oligonucleotides 4–9). Thus, nucleotides 4–9 (a complete TTAGGG telomere repeat) make the most important contributions to AoPOT1 binding, while guanines 1–3 may be important for the stability of AoPOT1-DNA interaction. This situation is different from OlPOT1_N, where all the nucleotides in the MBS are important for binding, and ZmPOT1b_N, where several nucleotide positions in the MBS exhibit more relaxed specificity.
POT1 proteins from Asparagus and maize bind both hexa- and heptanucleotide telomere repeats
Our TRF and EMSA data indicate that AoPOT1 may bind TTAGGG repeats in vivo, however it was unclear whether this protein retained the ability to interact with the ancestral TTTAGGG sequence. We found that AoPOT1 binding to two heptanucleotide repeats is 2-fold better than to two hexanucleotide repeats (Figure 5, lanes 1 and 2), suggesting that the ancestral TTTAGGG sequence is still a preferred substrate for AoPOT1. We next tested whether the ability to bind both types of telomere repeats is a conserved feature of POT1 proteins. As with AoPOT1, ZmPOT1b_N bound both types of repeats (Figure 5, lanes 7 and 8), displaying 2.5-fold better binding to (TTTAGGG)2 than to (TTAGGG)2. In striking contrast, we could not detect binding by O. lucimarinus POT1_N to any permutation of the hexanucleotide repeat in a direct binding assay (Figure 5, lanes 4 and 10). Similarly, mouse POT1a_N failed to bind any permutation of the plant telomere repeat (Figure 5, lanes 6 and 11). Thus, recognition of both hexanucleotide and heptanucleotide telomere repeats is an evolutionarily conserved feature of POT1 proteins from maize and Asparagus, which dates back to at least 100–110 mya. The inability of OlPOT1_N to bind hexanucleotide repeats suggests that the algal POT1 protein either lost the ability to bind such repeats after the divergence of land plants and green algae, or this property evolved independently in angiosperms.
Figure 5.
AoPOT1 and ZmPOT1b_N bind both hepta- and hexanucleotide telomere repeats. EMSA results are shown for AoPOT1, OlPOT1_N, mPOT1a_N and ZmPOT1b_N. POT1 proteins were incubated with the indicated oligonucleotides consisting of two full hepta- or hexanucleotide telomere repeats, or with an oligonucleotide cocktail containing all possible permutations of the heptanucleotide or hexanucleotide telomere repeat.
We noted that the first six 3′-terminal positions in the MBS of POT1 proteins from angiosperms (GGGTTAGGG for AoPOT1 and TTTAGGG for ZmPOT1b_N) are identical and constitute one full hexanucleotide repeat. The remaining 5′-terminal nucleotides in each MBS do not appear to make crucial contacts with the corresponding POT1 proteins (Figures 4B, lanes 1–3 and 3C, lane 1). Thus, the ability of AoPOT1 and ZmPOT1b_N to bind both hexa- and heptanucleotide telomere repeats can be explained if both proteins fail to discriminate between different nucleotides in the 5′-terminal positions of their respective MBS. In support of this model, we found that AoPOT1 and ZmPOT1b_N can bind to all variations of GGNTTAGGG and NTTAGGG, respectively (Supplementary Figure S5).
End-binding specificity of plant POT1 proteins
Vertebrate and yeast POT1 proteins differ in their preference for telomeric repeats at the 3′-end of the DNA substrate (13). Therefore, we asked if plant POT1 proteins also exhibit such preference by performing competition assays with oligonucleotides containing two telomere repeats at the 5′- or 3′-end or in the middle of a longer DNA oligonucleotide (Figure 6). Interestingly, the plant POT1 proteins behaved differently in these competition assays. Complex formation of OlPOT1_N with labeled (TTAGGGT)2 was significantly reduced or abolished with all competitors, suggesting that this protein binds telomeric repeats regardless of their position in the substrate (Figure 6A). On the other hand, increasing competition was observed for ZmPOT1b_N with oligonucleotides carrying the telomere repeats on the 5′-end, middle and on the 3′-end (Figure 6B), respectively. Finally, AoPOT1 binding to (TTAGGG)2 could only be competed with a substrate harboring two hexanucleotide telomere repeats on the 3′-end of the oligonucleotide (Figure 6C). We conclude that POT1 preference for the position of telomeric repeats on the DNA substrate is species-specific and not evolutionarily conserved.
Figure 6.
End-binding preference of plant POT1 proteins. (A) EMSA results are shown for reactions with OlPOT1_N using radioactively labeled (TTAGGGT)2 oligonucleotide as a probe in the absence (lane 1) or presence of 5X excess cold competitors containing the same telomeric sequence located either 5′-terminally (lane 2) or 3′-terminally (lane 4) to the 10-nt non-telomeric sequence NS10 (CTCTACCAAA), or flanked by 5-nt non-telomeric NS5 sequences (CTCTA and CCAAA) on both ends (lane 3). The fraction of complex bound to the labeled oligonucleotide is plotted on the right with binding in the absence of competitor set at 1.0. (B) and (C) Competition assays for ZmPOT1b_N bound to radioactively labeled (TTTAGGG)2 oligonucleotide (B) and AoPOT1 bound to radioactively labeled (TTAGGG)2 oligonucleotide (C). Lane designation and quantification as in (A).
Mutational analysis of plant POT1 proteins
Structurally characterized POT1 proteins from humans and Schizosaccharomyces pombe share a number of conserved primary sequence and secondary structure elements, which are crucial for specific interaction with telomeric DNA (14,39). The availability of Asparagus POT1 provides an opportunity to analyze the importance of these amino acids and protein regions for TTAGGG repeat recognition in the context of a full-length plant POT1 protein. As expected from studies in yeast and humans (12,35,36), the C-terminal region of AoPOT1 (amino acids 322–504) was dispensable for DNA binding (Figure 7A, lane 2). Moreover, full-length AoPOT1 and AoPOT1_N have similar binding properties, although the latter showed slightly reduced binding to the 8-nt GGTTAGGG substrate (Supplementary Figure S6). Further truncation of AoPOT1 to eliminate the second OB-fold (amino acids 1–167) completely abolished DNA-binding activity (Figure 7A, lane 3). Thus, two N-terminal OB-folds are sufficient for telomeric DNA binding by AoPOT1.
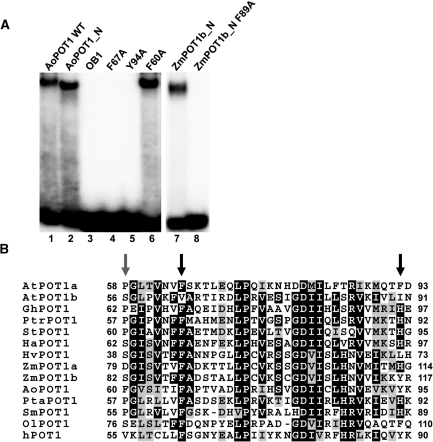
Figure 7.
Mutational analysis of plant POT1 proteins. (A) EMSA assays of AoPOT1 and ZmPOT1b_N truncation and point mutants. DNA-binding reactions were performed with wild-type AoPOT1 (lane 1), AoPOT1 truncation constructs (lanes 2 and 3) and point mutants (lanes 4–6) as well as with wild-type ZmPOT1b_N (lane 7) and its point mutant F89A (lane 8). The labeled probes are GGGTTAGGG for AoPOT1 and TTTAGGG for ZmPOT1b_N. (B) Partial alignment of plant POT1 proteins with human POT1. An OB1 region with a high degree of inter-kingdom amino acid similarity is shown. Black arrows indicate the positions of two catalytically important human POT1 residues and the corresponding aromatic amino acids F67 and Y94 in AoPOT1. Grey arrow designates the location of F60 in AoPOT1, which has no effect on telomeric DNA binding. Numbers indicate amino acid positions relative to the start codon. Abbreviations: At, Arabidopsis thaliana; Gh, Gossypium hirsutum; Ptr, Populus trichocarpa; St, Solanum tuberosum; Ha, Helianthus argophyllus; Hv, Hordeum vulgare; Zm, Zea mays; Ao, Asparagus officinalis; Pta, Pinus taeda; Sm, Selaginella moellendorffii and Ol, Ostreococcus lucimarinus. Alignment was generated with MEGA 3 software (52) and visualized in the BOXSHADE format.
The crystal structure of human POT1 protein identified conserved F62 and Y89 residues in OB1, which are critical for POT1 interaction with telomeric DNA (Figure 7B) (14). Most plant POT1 proteins have a nearly invariant phenylalanine in the first position (F67 in AoPOT1) and a large amino acid with a bulky side chain (mostly Y, H or F) in the second position (Y94 in AoPOT1). Consistent with previous reports for mammalian POT1 proteins (14,35), alanine substitutions of the corresponding Asparagus POT1 amino acids F67 and Y94 completely abolished DNA binding (Figure 7A, lanes 4 and 5). A similar result was obtained in ZmPOT1b_N with a F89A mutation (corresponding to F62 in human POT1) (Figure 7A, compare lanes 7 and 8). As a control, a F60A mutation in AoPOT1, which affects a non-conserved amino acid, had no effect on AoPOT1 binding (Figure 7A, lane 6). Altogether, these results argue that evolutionarily conserved aromatic amino acids in OB1 are important for telomeric DNA binding across kingdoms, and imply that the overall architecture of OB1 in plant POT1 proteins is similar to that of its mammalian and yeast counterparts.
DISCUSSION
The POT1 protein family represents an evolutionarily conserved group of telomeric DNA-binding factors with essential functions in chromosome end protection and telomere length regulation. Although the major POT1 functions appear to be conserved in most branches of eukaryotic life, previous data in plants indicated that Arabidopsis POT1 proteins evolved unusual functions in regulating telomerase, a property not dependent on physical contact with telomeric DNA (30). This study addresses this phenomenon further and provides a reconciling view that in some plant species POT1 may indeed have functions similar to those described for non-plant POT1 proteins.
DNA-binding properties of plant POT1 proteins
A well-established phylogenetic hierarchy for the major groups within the green plant lineage allows us an opportunity to examine changes in telomere-related genes in an evolutionary context. Here we compare the DNA-binding properties of three plant POT1 proteins: OlPOT1_N, from the earliest branching lineage analyzed in our study, and AoPOT1 and ZmPOT1b_N from angiosperms. Our biochemical analysis reveals fundamental differences in the nucleic acid binding activity of POT1 proteins across the plant kingdom (Supplemental Table S1). Several of the DNA-binding properties of OlPOT1 are reminiscent of non-plant POT1 proteins and contrast sharply with AoPOT1 and ZmPOT1b. Specifically, the minimum tight-binding sequence of OlPOT1_N consists of 10 nucleotides, a number similar to MBS of POT1 proteins from yeast and animals (13). In contrast, POT1 proteins from angiosperms require fewer nucleotides for efficient binding, with Asparagus POT1 (8–9 nucleotides) being on the lower end of the spectrum and ZmPOT1b_N displaying a short MBS of only 7 nucleotides. Interestingly, two polypeptides harboring a single OB-fold, S. pombe Pot1pN and C. elegans CeOB2, recognize a shorter, 6-nucleotide MBS (37,40). Thus, ZmPOT1b_N has the smallest MBS among all currently characterized POT1 proteins bearing at least two OB-folds.
Second, plant POT1 proteins show significant variation in the way they interact with cognate DNA. For other POT1 proteins, only a subset of MBS nucleotides is specifically recognized and makes important contributions to binding (13). A similar situation is observed for ZmPOT1b_N, where four out of seven nucleotides in the MBS are required for binding. Asparagus POT1 follows the same trend, with six of the nine MBS nucleotides needed for binding, although all these crucial AoPOT1 MBS nucleotides are localized at the 3′-end of the oligonucleotide. In contrast, all 10 MBS nucleotides are required for OlPOT1 binding, a phenomenon not previously observed for other POT1 proteins. These data suggest that the mechanism responsible for specific recognition of single-stranded telomeric DNA may significantly differ between POT1 proteins from green algae and land plants. Overall, we conclude that the telomeric DNA-binding properties of plant POT1 proteins are evolving rapidly.
Evidence for co-evolution of telomeric DNA and POT1 proteins in plants
The biochemical similarities and differences between ZmPOT1b_N and AoPOT1 may provide clues to the apparent co-evolution of Asparagus POT1 and the telomere repeat sequence in this species. ZmPOT1b_N requires only 7 nt (one full TTTAGGG repeat) for efficient binding, while AoPOT1 requires a very similar, but longer nine-nucleotide substrate GGGTTAGGG. Since the three 5′-terminal Gs do not appear to make significant contributions to AoPOT1 binding, our data suggest that these additional nucleotides in Asparagus POT1 MBS may stabilize this protein’s interaction with shorter human-type telomere repeats. Notably, the remaining six nucleotides in AoPOT1 MBS, TTAGGG, are identical to the 3′-end nucleotides present in ZmPOT1b_N MBS. Complementary substitutions in most of these nucleotides completely abolish protein binding, suggesting that TTAGGG sequence is crucial for specific interaction with POT1 proteins. These data may also explain the requirement of AoPOT1 and, to a lesser extent, ZmPOT1b_N for the presence of telomeric repeats on the 3′-end of DNA oligonucleotides. Such 3′-end positioning is likely necessary to improve or stabilize Asparagus POT1 interaction with the G-overhang, which, in turn, may result in better regulation of G-overhang length or interaction with telomerase.
What is the molecular basis for the stable association of POT1 proteins from angiosperms with hexanucleotide telomere repeats? Since both ZmPOT1b_N and AoPOT1 are capable of specifically binding to TTAGGG repeats, this biochemical feature must have evolved in the common ancestor of maize and Asparagus. We note that AoPOT1 and ZmPOT1b have a similar tolerance to nucleotide substitutions in certain MBS positions. Specifically, two different nucleotides, T (the extra T nucleotide present only in the plant TTTAGGG repeat, but not in the hexanucleotide TTAGGG sequence) and G (which replaces this T in the context of AoPOT1 MBS GGGTTAGGG) are equally tolerated by ZmPOT1b and AoPOT1 in the seventh MBS position (counting from the 3′-end of each oligonucleotide). This property may have originally evolved as a response to the known ability of many plant telomerases to naturally generate mutant telomere repeats containing one less or one more T (25,41,42). These so-called T-slippage events represent the most commonly detected type of telomerase error in plants in vitro and in vivo. The ability to tolerate T-slippage could have potentially allowed the ancestral POT1 protein to remain bound to the mutant human-type TTAGGG sequences. Another possibility is that decreased affinity to the cognate telomere repeats may help to dislodge POT1 from the G-overhang by other DNA-binding proteins, such as RPA or the CST complex during telomere replication (43).
Recognition of non-cognate telomere repeats is not unique to higher plant POT1 proteins. Oxytricha nova telomere end-binding protein (OnTEBP) stably associates with non-cognate telomeric sequences by facilitating significant conformational changes in DNA oligonucleotides via a phenomenon termed nucleotide shuffling, during which DNA sequence register shifts and entire nucleotides are excluded from the protein-DNA complex (44). Chicken and human POT1 proteins are also capable of interacting with non-cognate DNA oligonucleotides in competition assays (12,16). Similarly, in direct EMSA assays, S. pombe POT1 specifically binds DNA sequences resembling telomere repeats present in Tetrahymena thermophyla, O. nova and even Saccharomyces cerevisiae (45). Taken together, these observations suggest that relaxed DNA sequence specificity may be a common characteristic of POT1 proteins. This property could be especially beneficial in organisms such as Paramecium, where telomerase synthesizes an unusually high number of mutant telomere repeats (46). Likewise, in Asparagus plant lineage relaxed POT1 telomeric DNA sequence specificity would be beneficial in an evolutionary context and may have contributed to the survival of this entire plant order.
Evolutionary changes in plant POT1 functions
Interestingly, we failed to detect in vitro telomeric DNA binding for eight POT1 proteins from the evolutionarily diverse group of plant organisms analyzed in this study and for six previously characterized POT1 proteins from the Brassicaceae family of plants, which includes Arabidopsis (26). We can not rule out the possibility that some RRL-expressed plant POT1 proteins lack proper post-translational modifications or other requisites for efficient binding to telomeric DNA in vitro. However, we note that RRL-expressed POT1 proteins from yeast (12), mammals (36) and three different plants (this study) can efficiently bind telomeric DNA under the same conditions. Moreover, several lines of evidence suggest that telomeric DNA binding may not be the major in vivo function of POT1 proteins in Arabidopsis and, perhaps, in other plants. In striking contrast to yeast and mammalian POT1 proteins, Arabidopsis POT1a acts as a positive regulator of telomerase activity and is only enriched at the telomeres in S-phase, when telomerase is thought to act (30). Moreover, Arabidopsis POT1b appears to be a negative regulator of telomerase activity (E. Shakirov, A. Nelson and D. Shippen, in preparation). Recent data indicate that AtPOT1a and AtPOT1b associate directly with Arabidopsis telomerase RNA in regions outside the telomere template domain (C. Cifuentes-Rojas et al., manuscript in preparation). While RNA binding is associated with other OB-fold containing proteins, for example, translation factors (47), POT1 proteins have not been previously reported to bind RNA. Thus, the interaction of Arabidopsis POT1a and POT1b with telomerase RNA appears to represent a major evolutionary shift in plant POT1 functions from DNA to RNA binding.
Arabidopsis and maize are currently the only plants known to harbor more than one POT1 orthologue. In both cases, POT1 genes were likely duplicated around 30 mya, when the lineages leading to Arabidopsis and maize experienced independent whole-genome duplication events (34,48,49). Despite a similar evolutionary timeframe, the fate of these duplicated POT1 genes appears to be distinct. First, like the two mouse POT1 proteins, which evolved partially non-overlapping functions (36,50,51), the maize POT1 paralogs share ∼75% amino acid similarity. In contrast, Arabidopsis POT1a and POT1b display much lower sequence conservation, retaining only ∼50% amino acid similarity overall. Second, while both Arabidopsis POT1 proteins bind telomerase RNA instead of telomeric DNA, only one of the maize POT1 proteins, ZmPOT1a, lost the ability to bind telomeric DNA, raising the interesting possibility that ZmPOT1a evolved to bind the maize telomerase RNA. Although further analysis of the telomere complex in maize will be required to test this model, the comparative biochemical analysis of plant POT1 proteins described here underscores the remarkably rapid evolution of the OB-fold nucleic acid binding interface.
ACCESSION NUMBERS
EU880295, EU880297, EU880298, EU880301, EU880303–EU880306, EU883536, FJ516399.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM065383 to D.E.S.]. Funding for open access charge: National Institutes of Health [GM065383 to D.E.S.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Sandy Chang for providing the mouse POT1a_N construct. They are also grateful to Thomas D. McKnight and to members of the Shippen laboratory for insightful discussions and critical reading of the manuscript.
REFERENCES
- 1.Gilson E, Geli V. How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 2.Xin H, Liu D, Songyang Z. The telosome/shelterin complex and its functions. Genome Biol. 2008;9:232. doi: 10.1186/gb-2008-9-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 4.Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 2004;21:809–818. doi: 10.1093/molbev/msh075. [DOI] [PubMed] [Google Scholar]
- 5.McKnight TD, Shippen DE. Plant telomere biology. Plant Cell. 2004;16:794–803. doi: 10.1105/tpc.HistPersp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higashiyama T, Maki S, Yamada T. Molecular organization of Chlorella vulgaris chromosome I: presence of telomeric repeats that are conserved in higher plants. Mol. Gen. Genet. 1995;246:29–36. doi: 10.1007/BF00290130. [DOI] [PubMed] [Google Scholar]
- 7.Petracek ME, Lefebvre PA, Silflow CD, Berman J. Chlamydomonas telomere sequences are A+T-rich but contain three consecutive G-C base pairs. Proc. Natl Acad. Sci. USA. 1990;87:8222–8226. doi: 10.1073/pnas.87.21.8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pich U, Fuchs J, Schubert I. How do Alliaceae stabilize their chromosome ends in the absence of TTTAGGG sequences? Chromosome Res. 1996;4:207–213. doi: 10.1007/BF02254961. [DOI] [PubMed] [Google Scholar]
- 9.Sykorova E, Lim KY, Chase MW, Knapp S, Leitch IJ, Leitch AR, Fajkus J. The absence of Arabidopsis-type telomeres in Cestrum and closely related genera Vestia and Sessea (Solanaceae): first evidence from eudicots. Plant J. 2003;34:283–291. doi: 10.1046/j.1365-313x.2003.01731.x. [DOI] [PubMed] [Google Scholar]
- 10.Fajkus J, Sykorova E, Leitch AR. Telomeres in evolution and evolution of telomeres. Chromosome Res. 2005;13:469–479. doi: 10.1007/s10577-005-0997-2. [DOI] [PubMed] [Google Scholar]
- 11.Wikstrom N, Savolainen V, Chase MW. Evolution of the angiosperms: calibrating the family tree. Proc. Biol. Sci. 2001;268:2211–2220. doi: 10.1098/rspb.2001.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 13.Croy JE, Wuttke DS. Themes in ssDNA recognition by telomere-end protection proteins. Trends Biochem. Sci. 2006;31:516–525. doi: 10.1016/j.tibs.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 15.Croy JE, Podell ER, Wuttke DS. A new model for Schizosaccharomyces pombe telomere recognition: the telomeric single-stranded DNA-binding activity of Pot11-389. J. Mol. Biol. 2006;361:80–93. doi: 10.1016/j.jmb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Wei C, Price CM. Cell cycle localization, dimerization, and binding domain architecture of the telomere protein cPot1. Mol. Cell Biol. 2004;24:2091–2102. doi: 10.1128/MCB.24.5.2091-2102.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 18.Loayza D, Parsons H, Donigian J, Hoke K, de Lange T. DNA binding features of human POT1: A nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J. Biol. Chem. 2004;279:13241–13248. doi: 10.1074/jbc.M312309200. [DOI] [PubMed] [Google Scholar]
- 19.Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu D, Safari A, O'C;onnor MS, Chan DW, Laegeler A, Qin J, Songyang Z. PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- 21.Loayza D, de Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;424:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 22.Kelleher C, Kurth I, Lingner J. Human protection of telomeres 1 (POT1) is a negative regulator of telomerase activity in vitro. Mol. Cell Biol. 2005;25:808–818. doi: 10.1128/MCB.25.2.808-818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 24.Baumann P, Podell E, Cech TR. Human Pot1 (protection of telomeres) protein: cytolocalization, gene structure, and alternative splicing. Mol. Cell Biol. 2002;22:8079–8087. doi: 10.1128/MCB.22.22.8079-8087.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shakirov EV, Salzberg SL, Alam M, Shippen DE. Analysis of Carica papaya telomeres and telomere-associated proteins: insights into the evolution of telomere maintenance in Brassicales. Tropical Plant Biol. 2008;1:202–215. doi: 10.1007/s12042-008-9018-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shakirov EV, McKnight TD, Shippen DE. POT1-independent single-strand telomeric DNA binding activities in Brassicaceae. Plant J. 2009;58:1004–1015. doi: 10.1111/j.1365-313X.2009.03837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuchar M, Fajkus J. Interactions of putative telomere-binding proteins in Arabidopsis thaliana: identification of functional TRF2 homolog in plants. FEBS Lett. 2004;578:311–315. doi: 10.1016/j.febslet.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 28.Tani A, Murata M. Alternative splicing of Pot1 (Protection of telomere)-like genes in Arabidopsis thaliana. Genes Genet. Syst. 2005;80:41–48. doi: 10.1266/ggs.80.41. [DOI] [PubMed] [Google Scholar]
- 29.Shakirov EV, Surovtseva YV, Osbun N, Shippen DE. The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol. Cell Biol. 2005;25:7725–7733. doi: 10.1128/MCB.25.17.7725-7733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Surovtseva YV, Shakirov EV, Vespa L, Osbun N, Song X, Shippen DE. Arabidopsis POT1 associates with the telomerase RNP and is required for telomere maintenance. EMBO J. 2007;26:3653–3661. doi: 10.1038/sj.emboj.7601792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossignol P, Collier S, Bush M, Shaw P, Doonan JH. Arabidopsis POT1A interacts with TERT-V(I8), an N-terminal splicing variant of telomerase. J. Cell Sci. 2007;120:3678–3687. doi: 10.1242/jcs.004119. [DOI] [PubMed] [Google Scholar]
- 32.Cocciolone SM, Cone KC. Pl-Bh, an anthocyanin regulatory gene of maize that leads to variegated pigmentation. Genetics. 1993;135:575–588. doi: 10.1093/genetics/135.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fitzgerald MS, Riha K, Gao F, Ren S, McKnight TD, Shippen DE. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl Acad. Sci. USA. 1999;96:14813–14818. doi: 10.1073/pnas.96.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl Acad. Sci. USA. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He H, Multani AS, Cosme-Blanco W, Tahara H, Ma J, Pathak S, Deng Y, Chang S. POT1b protects telomeres from end-to-end chromosomal fusions and aberrant homologous recombination. EMBO J. 2006;25:5180–5190. doi: 10.1038/sj.emboj.7601294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 37.Raices M, Verdun RE, Compton SA, Haggblom CI, Griffith JD, Dillin A, Karlseder J. C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell. 2008;132:745–757. doi: 10.1016/j.cell.2007.12.039. [DOI] [PubMed] [Google Scholar]
- 38.Burr B, Burr FA, Matz EC, Romero-Severson J. Pinning down loose ends: mapping telomeres and factors affecting their length. Plant Cell. 1992;4:953–960. doi: 10.1105/tpc.4.8.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- 40.Lei M, Baumann P, Cech TR. Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe. Biochemistry. 2002;41:14560–14568. doi: 10.1021/bi026674z. [DOI] [PubMed] [Google Scholar]
- 41.Fitzgerald MS, Shakirov EV, Hood EE, McKnight TD, Shippen DE. Different modes of de novo telomere formation by plant telomerases. Plant J. 2001;26:77–87. doi: 10.1046/j.1365-313x.2001.01010.x. [DOI] [PubMed] [Google Scholar]
- 42.Mizuno H, Wu J, Katayose Y, Kanamori H, Sasaki T, Matsumoto T. Chromosome-specific distribution of nucleotide substitutions in telomeric repeats of rice (Oryza sativa L.) Mol. Biol. Evol. 2008;25:62–68. doi: 10.1093/molbev/msm227. [DOI] [PubMed] [Google Scholar]
- 43.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 44.Theobald DL, Schultz SC. Nucleotide shuffling and ssDNA recognition in Oxytricha nova telomere end-binding protein complexes. EMBO J. 2003;22:4314–4324. doi: 10.1093/emboj/cdg415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trujillo KM, Bunch JT, Baumann P. Extended DNA binding site in Pot1 broadens sequence specificity to allow recognition of heterogeneous fission yeast telomeres. J. Biol. Chem. 2005;280:9119–9128. doi: 10.1074/jbc.M414511200. [DOI] [PubMed] [Google Scholar]
- 46.McCormick-Graham M, Haynes WJ, Romero DP. Variable telomeric repeat synthesis in Paramecium tetraurelia is consistent with misincorporation by telomerase. EMBO J. 1997;16:3233–3242. doi: 10.1093/emboj/16.11.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kellogg EA. Relationships of cereal crops and other grasses. Proc. Natl Acad. Sci. USA. 1998;95:2005–2010. doi: 10.1073/pnas.95.5.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schranz ME, Mitchell-Olds T. Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. Plant Cell. 2006;18:1152–1165. doi: 10.1105/tpc.106.041111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 51.Palm W, Hockemeyer D, Kibe T, de Lange T. Functional dissection of human and mouse POT1 proteins. Mol. Cell Biol. 2009;29:471–482. doi: 10.1128/MCB.01352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.