Abstract
ETS-domain transcription factors play important roles in controlling gene expression in a variety of different contexts; however, these proteins bind to very similar sites and it is unclear how in vivo specificity is achieved. In silico analysis is unlikely to reveal specific targets for individual family members and direct experimental approaches are therefore required. Here, we take advantage of an inducible dominant-negative expression system to identify a group of novel target genes for the ETS-domain transcription factor Elk-1. Elk-1 is thought to mainly function through cooperation with a second transcription factor SRF, but the targets we identify are largely SRF-independent. Furthermore, we demonstrate that there is a high degree of overlapping, cell type-specific, target gene binding by Elk-1 and other ETS-domain transcription factors. Our results are therefore consistent with the notion that there is a high degree of functional redundancy in target gene regulation by ETS-domain transcription factors in addition to the specific target gene regulation that can be dictated through heterotypic interactions exemplified by the Elk-1-SRF complex.
INTRODUCTION
Eukaryotic transcription factors are classified into families based on the identity of their DNA-binding domains. In many cases, the shared structure of the DNA binding domain offers limited opportunity for providing unique DNA binding specificities to individual family members, and hence a substantial overlap in DNA sequences recognised is apparent. It is currently not fully clear how this lack of sequence selectivity impacts on target gene selection in vivo, but interactions with other transcription factors in defined modules are thought to provide the driving force for generating specific promoter recognition. The ETS-domain transcription factor family provides a good example of these phenomena (1,2).
There are 27 known members of the ETS-domain transcription factor family known in human cells (1,2). A large number of these are actually co-expressed in any given cell type (3). All of these proteins bind to a sequence containing a core GGAA/T motif that is embedded in a longer 10 bp region (1,2). Subtle differences in in vitro site selectivity are observed for individual family members, but it is not known how this impacts on DNA binding in vivo. In some cases, heterologous transcription factor partner proteins have been identified that, along with an ETS-domain protein, form a module that binds to composite elements and hence provides further binding specificity. The ternary complex factor (TCF) subfamily of ETS-domain proteins represent important exemplars of this type of action and are commonly found to bind promoters in a complex with SRF (4–6). Elk-1, SAP-1 and SAP-2/Net, comprise the TCF subfamily of ETS-domain transcription factors and they are characterized by the presence of a short protein interaction motif known as the B-box, which enables them to interact cooperatively with a second transcription factor SRF (7–9). This protein–protein interaction with SRF apparently permits less stringent DNA binding requirements on the TCF partner (10). However, despite this emerging paradigm, there are several instances where TCF binding occurs in the absence of obvious SRF binding to the same promoter (e.g. PAI-1; 11,12). While interaction with SRF appears to be a property unique to the TCF subclass, it is unclear how the specificity of promoter recognition is achieved in the absence of SRF, and whether other ETS-domain proteins can potentially substitute for the TCFs in this context. Indeed, a recent ChIP-chip study revealed an apparent high degree of redundancy in promoter binding amongst the ETS-domain transcription factors, Ets-1, GABPα and Elf-1 in Jurkat cells (13). However, in the same study, it was concluded that no redundancy of promoter binding occurs with the TCF protein Elk-1, thereby suggesting that Elk-1 has a unique repertoire of target genes, potentially dictated by its interaction with SRF.
To begin to investigate the target specificity of Elk-1, we set out to identify more target genes due to the small number of known targets. Elk-1 exhibits overlapping in vitro DNA binding specificities with other ETS-domain proteins (14) and moreover, also shares the ability to interact with SRF with other members of the TCF subfamily. Thus, to circumvent the possibility of redundancy of function with other TCFs in particular, we used a dominant-negative approach to identify new genes regulated by Elk-1. Microarray analysis revealed a number of potential Elk-1 target genes and we focussed on one particular group that was consistently down-regulated by a constitutively repressive form of Elk-1, Elk-1-En, under a variety of conditions. These were verified as direct Elk-1 targets and shown largely not to be targets of its partner protein SRF. Furthermore, by knockdown approaches, we show latent redundancy of ETS-domain protein binding. This is further emphasised by the observation that other ETS-domain transcription factors can bind the same sites in different cell types. Our data therefore reveal that Elk-1 can function more widely in an SRF-independent manner, but that this function is highly redundant with other ETS-domain transcription factors.
MATERIALS AND METHODS
Plasmid constructs
The following plasmids were used in mammalian cell transfections. pSRE-Luc (pAS821) contains two copies of the c-fos SRE (nucleotides −357 to −275, containing both an SRF binding site and an adjacent ets motif) upstream of a minimal tk promoter and the luciferase gene (15). The MCL-1, BCL10, FTSJ2, SASS6, FLJ14803, PSMD8 and MYC-driven luciferase reporter vectors were generated by SwitchGear Genomics, and contain ∼1 kb of promoter fragment cloned into the pGL3 vector (Promega). The luciferase reporter vector (pAS2351) containing the SAS10 promoter was produced by ligating a PCR product into pGL3 vector using SacI/HindIII sites. The following primers were used for PCR: (ADS1615) GCCGAGCTCAGCAACGTATCAAAAGTTCAG, (ADS1616) CTCAAGCTTGGCTCACAATCTCAGGTTTTAC. pAS1407 is a pcDNA3.1-derived plasmid encoding full length Elk-1 fused to the Engrailed repression domain and Flag epitope {Elk-En} (16); pAS348 is a Rous sarcoma virus (RSV) promoter-driven vector, encoding full-length wild-type human Elk-1 fused to residues 410–490 of VP16 {Elk-VP16} (17); pMLV-SRF-VP16 encodes full-length SRF fused to the VP16 activation domain (kindly provided by Richard Treisman), and pRL encoding Renilla luciferase (Promega) was used to monitor transfection efficiency.
Tissue culture, cell transfection, reporter gene assays, RT–PCR and RNA interference
EcR293(Elk-En)#1.3 (16,18), HEK 293T and HeLa cells were grown in DMEM supplemented with 10% fetal bovine serum, SH-Sy5y cells were grown in DMEM/F12 (1:1) medium supplemented with 10% FBS, and U937 cells were grown in RPMI 1640, with 10% serum. Transfections were performed with polyethylenimine (PEI) (Polysciences) for HEK 293T cells or Amaxa Nucleofector system for U937 cells according to the manufacturers’ instructions.
To induce, differentiation, U937 cells were treated with 50 nM PMA for 3 h and then grown in DMEM and 10% FBS for up to 72 h to allow differentiation.
For reporter gene assays, typically 0.2 µg of reporter plasmid and 50 ng of pRL were co-transfected with 0.005–0.1 µg of expression plasmids. Cell extracts were prepared and luciferase activity was measured 24 h after transfection using the Dual-Luciferase Reporter Assay System (Promega) according to the supplier’s protocol.
Real-time RT–PCR was carried out as described previously (19). The following primer-pairs were used for RT–PCR experiments. ELK1, ADS2113 (5′-GGTGGTGAATTCAAGCTGGT-3′) and ADS2114 (5′-ATTTGGCATGGTGGAGGTAA-3′), MCL1, ADS2221 (5′-AGACCTTACGACGGGTTGG-3′) and ADS2222 (5′-ATGGTTCGATGCAGCTTTCT-3′); FOS, ADS4029 (5′-AGAATCCGAAGGGAAAGGAA-3′) and ADS4030 (5′-CTTCTCCTTCAGCAGGTTGG-3′); CHERP, ADS1609 (5′-TCGGAGGCGAATTCTACAGT-3′) and ADS1610 (5′-AGGTTCCACTGGCTCTGCT-3′); SAS10 ADS1611 (5′-GTCAACAGGAGGCAGAGGAG-3′) and ADS1612 (5′-GGTGATTCCTTTCGCAACAT-3′); ZNF410, ADS1613 (5′-CCGGAGTTTTTGTCCACTTC-3′) and ADS1614 (5′-AAACTGTCATGGGCCAACTC-3′); BCL10, ADS1903 (5′-CCTCACTGAAGTGAAGAAGG-3′) and ADS1904 (5′-ATAGATTCAACAAGGGTGTCCA-3′); SAP-1, ADS2276 (5′-TCGCAAGAACAAGCCTAACA-3′) and ADS2277 (5′-CTTTCCCTCCATTCTCCACA-3′) and 18S internal control, ADS4005 (5′-TCAAGAACGAAAGTCGGAGGTT-3′) and ADS4006, (5′-GGACATCTAAGGGCATCACAG-3′).
siRNAs against Elk-1, SAP-1, FLI-1 and matched GAPDH control, were constructed by the SilencerTM siRNA construction kit (Ambion). Human Elk-1 target sequences were: 5′-AAGGCAAUGGCCACAUCAUCU-3′ (ADS 1926/1927) and 5′-AAUUCAAGCUGGUGGAUGCAG-3′ (ADS 1928/1929), the SAP-1 target sequence was 5′-AAGUAAAUAAUUCAUCAAGAU-3′ (ADS 1934/1935), and FLI-1 target sequences were 5′-AAGUUCACUGCUGGCCUAUAA (ADS1930/1931) and 5′-AACGUCAAGCGGGAGUAUGAC-3′ (ADS1932/1933). The siRNAs against SRF and matched control siRNA (Santa Cruz) were made synthetically. To carry out RNA interference (RNAi) for HeLa cells, a two-step transfection protocol was performed in 12-well plates as described previously (19). U937 cells were transfected using an Amaxa Nucleofector system with 2.25 μM (3 µg) of siRNA. For transfections with siRNA constructs against Elk-1 or FLI-1, a mixture of two different targeting constructs was used.
Western blot analysis
Western blotting was carried out with the primary antibodies; Elk-1 (Epitomics), phospho-Elk-1(Ser383) (Cell Signalling), phospho-ERK-1 and -2 (Cell Signalling), SRF (Santa Cruz), Fli-1 (Santa Cruz), Actin (Abcam), GAPDH (Abcam) essentially as described previously (19).
For PMA stimulation, HeLa cells or SH-Sy5y cells were serum starved for 24 h and then treated with 10 nM PMA for indicated times. Cells were collected, lysed in cell lysis buffer (Tropix) with phosphatase inhibitors (10 mM NaF, 10 mM β-glycerol phosphate, 1 mM Na3VO4) and subjected to immunoblotting.
ChIP assays
Chromatin immunoprecipitation (ChIP) assays using control IgG (Upstate) or antisera specific to Elk-1 (Epitomics), SAP-1 (Santa Cruz), SRF (Santa Cruz) and Fli-1 (Santa Cruz) were performed as described previously (20) except that cross-linking for U937 cells was performed for 15 rather than the standard 10 min. Bound promoters were detected by PCR using primers for the human CHERP, FTSJ2, ZNF410, SAS10, LSR68, MOAP-1, promoters (primers listed in Supplementary Table S8), respectively or for the c-FOS promoter or SRF intron 3 as described previously (21).
Quantitative PCR was performed at least in duplicate, from at least two independent experiments, using Quantitect SYBR green PCR reagent (Qiagen) and a Rotorgene 3000 machine (Corbett Research). Results were analysed with Rotorgene 6.0 software (Corbett Research) relative to input using the standard curve method.
Microarray and bioinformatics analysis
The dominant-negative expression experiments were performed in four independent batches each containing the six samples (starved, EGF 15 min and EGF 30 mins, each with or without ponasterone A [PonA]). RNA quality was checked using the RNA 6000 Nano Assay, and analysed on an Agilent 2100 Bioanalyser (Agilent Technologies). RNA was quantified using a Nanodrop ultra-low-volume spectrophotometer (Nanodrop Technologies). Affymetrix human genome U133A microarrays were used as described previously (16). The technical quality control of array data was performed with dChip (V2005) using the default settings (www.dchip.org; 22). Background correction and quantile normalization were performed using RMA in Bioconductor (23) using an Affymetrix Chip Definition (CDF) from AffyProbeMiner gene consistent ccds 1.8.0 distribution (AffyProbeMiner: a web resource for computing or retrieving accurately redefined Affymetrix probe sets; 24). Since a Principal Components Analysis showed a strong batch effect, a batch removal step was performed with Partek Genomics Solution (version 6.3, Copyright 2005, Partek Inc., St Charles, MO, USA). Expression values were then analysed for differential expression due to ponA exposure in three separate two-way analyses (starved, EGF 15 min and EGF 30 min) using Cyber-T (25). The mean of expression changes for each probeset under each condition was then calculated and a gene list of 877 differentially expressed genes was created by filtering for probesets with (i) a P < 0.05 in any of the three tests of starved, EGF 15 min or EGF 30 min (ii) fold change less than 1 (natural number scale, since we were only interested in genes down-regulated by dominant negative Elk-1). This gene list was segregated into eight clusters based on similarity of expression profile across the dataset using a K-means clustering algorithm. Clustering was performed on the means of each sample group (log 2) that had been z-transformed [for each probeset across all six conditions, the mean was set to zero, standard deviation to 1). K-means clustering was done on the basis of similarity of profiles across the dataset using the ‘Super Grouper’ plugin of maxdView software (available from http://bioinf.man.ac.uk/microarray/maxd/)]. The similarity metric used to quantify the similarity between two profiles was ‘Slope’.
In each of the eight gene clusters, Affymetrix probe identifiers were converted into Ensembl (version 49; http://www.ensembl.org; 26) gene identifiers, using the Clone/GeneID converter (http://idconverter.bioinfo.cnio.es; 27). Affymetrix probe sets associated with more than one Ensembl identifier were rejected, resulting in an unambiguous set of genes corresponding to each expression result from the microarray experiment. The Ensembl data mining tool Biomart was used to extract the genome coordinates of the Ensembl genes identified above. The promoter regions were then identified as −1000 bp and +1000 bp of the most 5′ transcription start site, bearing in mind the strand on which the gene resides. Genomic sequence corresponding to the human genome (hg18/NCBI36) was obtained using Galaxy (http://g2.trac.bx.psu.edu; 28). The resulting sequences were repeat masked. A set of 300 random Ensembl gene promoters were also prepared.
Novel motif discovery within the promoter sequences was performed using the stand-alone version of Weeder 1.3.1 (29). Searches were performed using the parameter ‘small’ (motifs of length 6 and 8), using both sequence strands and allowing the expectation that motif may appear more than once in a given sequence. The identification of known Elk-1 and SRF motif consensus sequences within the promoter sequences was performed using a PERL script. The script identified matches to supplied IUPAC consensus sequence in the forward and reverse strands of repeat masked sequences.
The intersect between lists of genes associated with the eight gene clusters and the dataset containing promoters identified as bound by Elk-1, SRF (32) or GABPα (30) was performed using an online tool ‘Comparison of two lists’ (http://jura.wi.mit.edu/bioc/tools/compare.html).
The Z-score statistic was calculated by first identifying the overlap of gene symbols associated with each cluster to a second gene list; 1000 lists of random gene symbols (each containing an equal number of symbols to the gene cluster that was tested) was then compared against the second gene list. The mean and standard deviation of the percentage overlap of gene symbols of the 1000 comparisons was calculated and compared to the observed mean percentage overlap. A two-tailed P-value of the Z-score was calculated using the Microsoft Excel NORMSDIST function P-value = 2*(1−(NORMSDIST(Z-score))).
The functional profiling of genes that belong to the eight Elk-En-regulated clusters was performed using the DAVID bioinformatics resource 2008 (31; http://david.abcc.ncifcrf.gov/). Gene symbol gene lists for each of the eight clusters were uploaded into DAVID and the ‘functional annotation tool’ was used to identify over-represented GO terms (‘molecular function’ levels 3 and 5, plus ‘biological property’ Level 5), reporting uncorrected P-values.
RESULTS
Identification of genes regulated by dominant-negative version of Elk-1
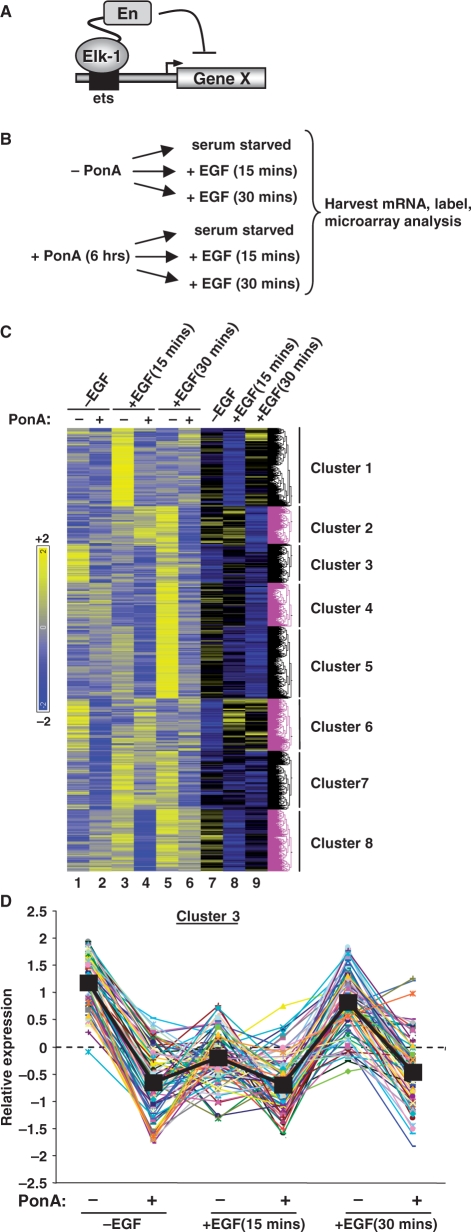
To identify genes regulated by Elk-1, we used a stable EcR293 cell line containing a ponasterone A (PonA)-inducible gene encoding full-length Elk-1 fused to the engrailed repression domain (Elk-En) (Figure 1A; 16,18). By retaining all the functional domains of Elk-1, we predicted that we would increase our chances of identifying bona fide target genes; additional domains such as the SRF-binding B-box would potentially contribute to the target selectivity of Elk-1. Indeed, our previous studies demonstrated that Elk-En functioned as expected to repress the activity of known Elk-1 target genes and mutation of the B-box blocked the repressive activity of Elk-En against promoters regulated by the Elk-1-SRF module (18). The level of Elk-En expression was roughly equivalent to that of endogenous Elk-1 found in HeLa cells at both the mRNA and protein levels (Supplementary Figure S1).
Figure 1.
Identification of Elk-1-regulated genes. (A) Diagrammatic representation of the mode of action of Elk-En. Elk-En binds to ETS motifs within promoters and represses promoter activity. (B) Illustration of the induction regime used for sample preparation. The expression of Elk-En is stimulated by adding ponasterone A (PonA) to the EcR-293 (Elk-En) cells for 6 h. Cells were grown in the presence or absence of PonA, and either left serum starved or treated with EGF for 15 or 30 min before harvesting. (C) Cluster analysis of 877 genes, which show consistent downregulation upon PonA addition under at least one of the three experimental conditions (serum starved, 15 min EGF and 30 min EGF). Columns 1–6 show the relative expression of the groups of probesets representing each gene. Columns 7–9 represent the same data but show fold change in response due to PonA treatment. Hierarchical clustering was performed on columns 1–6. (D) Expression profiles of individual genes in cluster 3. Each gene is represented by a different coloured line. The black squares and line represents the average expression value of genes in this cluster under each of the six experimental conditions. For each gene, the mean of the probe sets across all six conditions was set as zero, and expression levels (log2) are presented relative to this value.
We compared the mRNA expression profiles of EcR293(Elk-En) cells in the absence and presence of induction of Elk-En expression by PonA treatment, under three different conditions; (i) in serum starved cells (ii) in cells stimulated with EGF for 15 min and (iii) in cells stimulated with EGF for 30 min (Figure 1B). The latter two conditions result in ERK MAP kinase pathway activation, which is a known upstream regulator of Elk-1 activity (3,4,11). Elk-En was induced with PonA addition for 3 h prior to EGF stimulation and mRNA isolation. Three experimental replicates were performed under each condition. A total of 877 genes displayed significantly decreased expression (Supplementary Table S1; P < 0.05) in response to Elk-En expression, under at least one of these conditions. These genes were then clustered according to their responses to EGF treatment and to PonA treatment. This gave rise to eight clusters of genes with different response patterns (Figure 1C; Supplementary Figure S2; Supplementary Table S1). Genes from cluster 7 appeared to follow the expected pattern for regulation by Elk-1: their expression is induced in response to EGF stimulation at both 15 and 30 min and down-regulated in response to induction of Elk-En expression. Clusters 5 and 8 also show up-regulation by EGF at both time-points but down-regulation by Elk-En only following EGF treatment. These clusters contain known Elk-1 target genes such as FOS (cluster 8), EGR1 (cluster 7) and IER2 (cluster 5). In contrast, the expression of the 77 genes in cluster 3 was barely affected by EGF treatment, although they were consistently down-regulated by Elk-En under all conditions analysed (Figure 1C and D). The patterns of gene expression changes in the other clusters are not readily explicable, and might have arisen due to indirect effects through Elk-En controlling the expression of genes encoding other transcription factors or regulatory molecules; for example, the early increases in gene expression due to PonA treatment in cluster 2 could arise due to Elk-En repressing a repressor protein that controls this set of genes. We therefore decided to focus on cluster 3 genes as these appear to be a novel class of Elk-1 target gene: they are consistently down-regulated by Elk-En under all conditions and, unlike known Elk-1 target genes, do not respond to MAPK pathway activation by EGF.
We also investigated whether any of the expression clusters were enriched for genes belonging to a common functional category by using the DAVID bioinformatics resource (31). Under the stringent criteria that we set, no GO terms were significantly enriched in clusters 1, 2, 6 and 7. In contrast, either when combined or analysed individually, clusters 2, 4, 5 and 8 showed enrichment of a number of functional categories (Supplementary Tables S2 and S3). For example, cluster 3 was enriched for genes involved in nucleotide processing/metabolism and DNA-dependent transcription, whereas cluster 5 genes were involved in more general biosynthetic processes, sugar metabolism, and translation. Cluster 4 was enriched for genes involved in cell structure, intracellular transport and nucleotide binding. Cluster 8 genes were associated with nucleotide biosynthesis. When considered together, genes within clusters 2, 4, 5 and 8 showed enrichment for a combination of these functional categories but also revealed enrichment of genes associated with apoptosis. Together, these results therefore reveal that the unique expression patterns of clusters 3, 4, 5 and 8 in response to EGF treatment and Elk-En expression also reflect potential co-regulation of distinct categories of target genes.
Elk-1 directly regulates cluster 3 genes
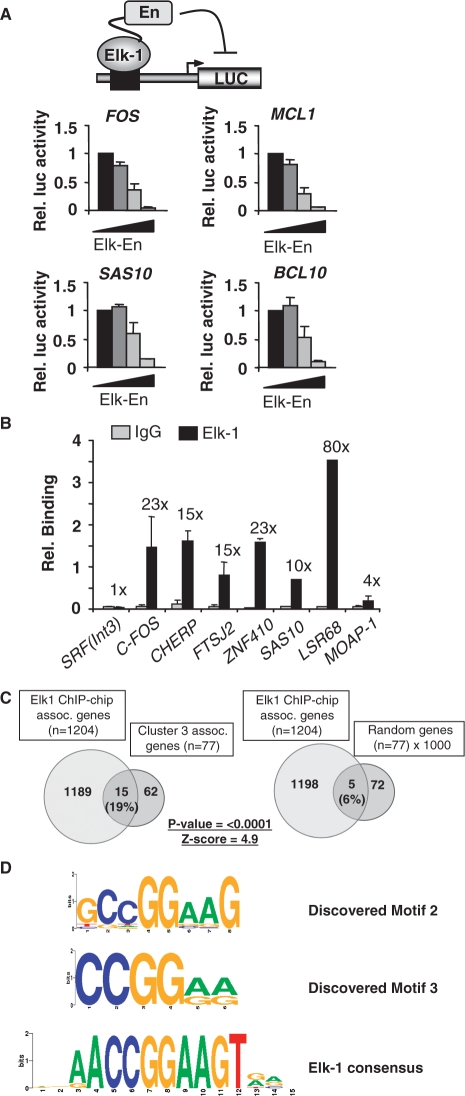
To establish whether genes in cluster 3 are directly targeted by Elk-1, we first carried out reporter gene analysis using promoters derived from genes in this cluster. Increasing amounts of Elk-En inhibited the expression of the known target genes c-FOS and MCL-1, as observed previously (Figure 2A; 16). Similar dose-dependent Elk-En-mediated repression of the promoters from the cluster 3 genes SAS10 and BCL10 was observed (Figure 2A). Importantly, this effect is specific as the SASS6, FLJ14803 and PSMD8 promoters do not respond to Elk-En to the same extent (Supplementary Figure S3A). Thus, Elk-En acts on cluster 3 gene promoters to alter their transcription rates.
Figure 2.
Elk-1 binds to cluster 3 gene promoters. (A) Elk-En down-regulates cluster 3 gene promoters. HEK293T cells were transiently transfected with a luciferase construct containing the indicated promoters and with increasing concentrations of Elk-En(WT) (0, 0.5, 5, 50 ng; increasing concentrations indicated by a triangle). The results were normalized to renilla luciferase activity, and the activity in the absence of Elk-En given the value of one. Error bars represent standard deviation calculated from three biologically independent replicates, each representing the average of two samples. (B) ChIP validation of Elk-1 binding to cluster 3 genes promoters. ChIP assay from HeLa cells using Elk-1-specific antibodies or a non-specific rabbit IgG, followed by real-time PCR. An SRF intronic region [SRF(Int3)] and the c-FOS promoter region were used as negative and positive controls respectively. Relative binding indicates percentage bound relative to input. Fold enrichment of Elk-1 over IgG control precipitation is shown above the bars. Results shown are representative of at least two independent experiments and are the average of two samples, and standard deviation. (C) Venn diagrams illustrating the overlap between cluster 3 genes and Elk-1 target genes identified by ChIP-chip analysis. As a comparison, the average overlap between the 1204 target genes associated with regions identified by ChIP-chip analysis and 1000 randomly selected groups of 77 genes is shown. (D) Motif logos of two of the top three sequence motifs discovered by Weeder in cluster 3 gene promoters. The Elk-1 binding motif identified from site selection studies (14) is shown at the bottom.
To determine whether Elk-1 binds to cluster 3 promoters directly at endogenous levels of Elk-1 expression we performed ChIP analysis in HeLa cells. We included ZNF410 in this analysis as although it does not appear in the final cluster 3 list (due to narrowly missing the cut-off thresholds), earlier analyses indicated that it was consistently down-regulated by Elk-En. Direct binding of Elk-1 was clearly observed to 5 of 6 genes analysed (Figure 2B). The other promoter tested from MOAP1, exhibited lower levels of binding, which might reflect that either: (i) steric hindrance occurs at this promoter; (ii) it is an indirect target; or (iii) we have tested the wrong part of the promoter.
Further insights into direct Elk-1 target genes amongst the genes in cluster 3 could be discerned from comparisons to a recent ChIP-chip study in Hela cells (32). In total, 77/877 (9%) genes showing down-regulation by Elk-En under at least one condition, were also identified by ChIP-chip analysis as direct Elk-1 targets (Supplementary Table 4). Importantly though, another 15 promoters were revealed as direct Elk-1 targets by comparing the genes in cluster 3 with promoter regions shown to be bound by Elk-1 by ChIP-chip analysis. This overlap was highly significant when compared to a series of randomly generated equivalent sized datasets (Figure 2C; Supplementary Tables S1 and S4). These random datasets represent the average of 1000 lists of gene symbols that contain the same number of gene symbols as found in each cluster that was tested. Indeed, this value represented nearly 20% of all cluster 3 genes, whereas the overlap was not as large for any of the other clusters, with the next nearest being clusters 5 and 8 (12% overlap), and the lowest being cluster 7 (4% overlap) (Supplementary Table S4). Importantly, although lower in percentage terms, the overlap with clusters 5 and 8 was still significant (Z-scores 2.8 and 2.1, respectively).
To further investigate the nature of the promoters controlling cluster 3 genes, we searched for motifs that resembled the consensus binding motif AACCGGAAGT that was identified for Elk-1 by in vitro site-selection analysis (14). We searched within the proximal promoter regions defined as ±1 kb from the most 5′ transcriptional start site of each gene. Using three different overlapping hexameric submotifs derived from the last 8 nucleotides in this decamer, we could find significant over-representation in cluster 3 gene promoters: 79–85% of all promoters contained either CCGGAA, CGGAAG or GGAAGT (Supplementary Table S5). Moreover, if we relaxed the stringency to CCGGAW, CGGAWG or GGAWGT, the incidence of these motifs rose to 85–90%. Lower incidences of octameric representations of this binding motif were found: either AACCGGAA, ACCGGAAG or CCGGAAGT were found in 35% of all cluster 3 promoters, but these were substantially enriched compared to only 11% incidence in a random dataset. Thus, Elk-1 binding motifs are vastly over-represented in cluster 3 gene promoters. Cluster 5, cluster 8 and to a lesser extent cluster 4, also exhibited over-representation of Elk-1 binding motifs (Supplementary Table S5). This is consistent with the observation that these clusters also contain significant numbers of Elk-1 targets identified by ChIP-chip analysis. As an alternative approach we used Weeder (28) to look for over-represented motifs in each of the clusters. Two of the three highest ranking motifs closely resembled the Elk-1 binding motif identified by in vitro site-selection analysis (Figure 2D) (14). Similarly high scoring motifs that resembled ETS transcription factor binding sites could be discerned in clusters 4, 5 and 8 (Supplementary Figure S2).
Together, these results demonstrate that a large proportion of cluster 3 gene promoters represent direct Elk-1 targets and this is reflected by the over-representation of Elk-1 binding motifs within these promoters. Similarly, clusters 4, 5 and 8 exhibit some of these characteristics and hence also likely represent direct Elk-1 targets.
SRF binding is not detectable at the majority of cluster 3 gene promoters
The established paradigm for Elk-1 function is that it works through forming complexes with a second transcription factor, SRF. However, there are a growing number of instances where SRF-independent promoter regulation has been documented. Since cluster 3 genes do not respond to MAP kinase signalling in a manner expected of an SRF-Elk-1 module, we checked whether SRF was important in controlling these genes.
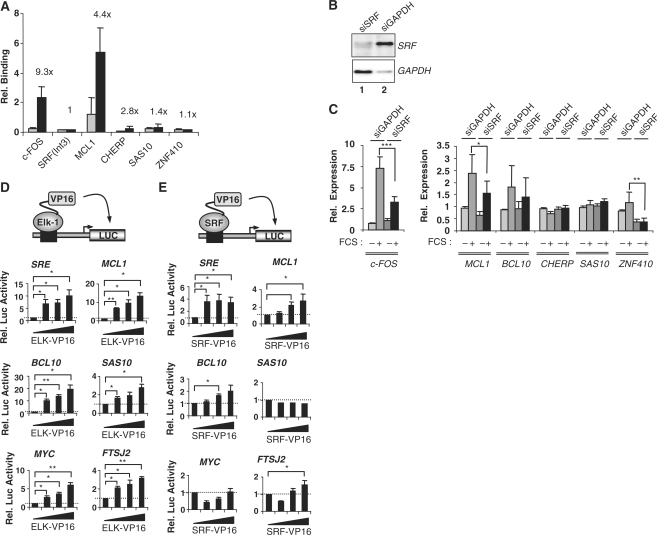
First, we analysed SRF binding by ChIP analysis. In contrast to the high occupancy observed with Elk-1, only one of the four genes tested in this cluster (MCL1) had significant SRF binding (Figure 3A). MCL1 is a known target of the Elk-1-SRF complex (16,33). In addition, we compared the genes identified as high confidence direct SRF targets in both HeLa (32) and Jurkat (30) cells by ChIP-chip and ChIP-Seq analysis respectively, to the Elk-En-regulated genes in each cluster. A statistically significant overlap was seen with genes in cluster 3, but the total number of genes involved was low (Supplementary Table S6), further emphasising that many genes in this cluster seem to be regulated independently from SRF. Consistent with this small overlap, there was no evidence for over-representation of the SRF binding motif CCW6GG in the promoters of cluster 3 genes (Supplementary Table S5). The ChIP-derived SRF dataset also exhibited a small but statistically significant overlap with genes in clusters 5 and 8, but not with any of the other clusters (Supplementary Table S6).
Figure 3.
Role of SRF in cluster 3 gene regulation. (A) SRF binding to selected cluster 3 gene promoters. HeLa cells were serum starved for 24 h, followed by ChIP with SRF or non-specific rabbit IgG antibodies. ChIP DNA was subjected to qPCR. Relative binding indicates percentage binding relative to input sample. The ratio between SRF and non-specific IgG is indicated above the bars. An SRF intronic region [SRF(Int3)] and the c-FOS promoter region were used as negative and positive controls, respectively. Error bars represent standard deviation calculated from three biologically independent replicates and the average of two samples. (B) Western blot analysis of siRNA-mediated knockdown of SRF and GAPDH. (C) Quantitative RT–PCR analysis of cluster 3 genes upon SRF knockdown. HeLa cells were transfected with siRNA against SRF or GAPDH. Cells were either left in serum free medium for 48 h or treated with 20% FCS for 30 min and mRNA levels were measured. Error bars represent standard deviations calculated from three biologically independent replicates, each representing the average of two samples. Asterisks denote differences with statistical significances (*P < 0.05, **P < 0.01, ***P < 0.001) as determined by the Student t-test. (D and E) Reporter gene assays in HEK293T cells with the indicated promoter–reporter constructs and with either increasing concentrations of Elk-VP16 (0, 5, 10, 50 ng) (D) or SRF-VP16 (0, 10, 50, 100 ng) (E). Luciferase activity was measured 24 h after transfection. The results were normalized to renilla luciferase activity, and the activity in the absence of cotransfected VP16 fusion protein taken as 1. Error bars represent standard deviation calculated from three biologically independent replicates, each being the average of duplicate samples. Statistical differences between the bracketed samples were determined by Student’s t-test (* P < 0.05; **P < 0.01).
To probe potential functional regulatory links between SRF and cluster 3 genes, we reduced SRF levels by siRNA treatment. Knockdown of SRF resulted in little change in the expression of cluster 3 genes, with the exception of MCL1 and ZNF410 (Figure 3B and C). However, in the latter case, this regulation is likely indirect or through a site located outside the region tested by ChIP. We also compared the response of several of the cluster 3 promoters to the expression of constitutively active Elk-1 (Elk-VP16) or SRF (SRF-VP16) fusion proteins. Elk-VP16 activated all the tested promoters in a dose-dependent manner (Figure 3D). This effect was specific as a number of control promoters were unresponsive to Elk-VP16 (Supplementary Figure S3B). In contrast, SRF-VP16 activated an SRE-driven reporter, the MCL1 promoter and the BCL10 promoter in a dose-dependent manner but failed to promote significant activation of the SAS10 and MYC promoters (Figure 3E). Similarly, little activation of the FTSJ2 promoter was observed, with only slight increases detected at the highest levels of transfected SRF.
Collectively, these various lines of evidence demonstrate that the Elk-1 target genes in cluster 3 are largely regulated in an SRF-independent manner.
Redundant binding of ETS-domain proteins to cluster 3 gene promoters
To establish a potential functional role for Elk-1 in controlling the activity of the genes in cluster 3, we determined the expression levels of several of these target genes in HeLa cells upon knockdown of Elk-1 levels by siRNA treatment. However, depletion of Elk-1 had little effect on the activity of these target genes either in the absence or presence of PMA (data not shown). Indeed, while the Elk-1 target gene FOS was rapidly and potently activated in both HeLa and SH-Sy5y cells in response to ERK pathway activation by PMA treatment, only small changes in the expression of the cluster 3 genes CHERP, SAS10 and ZNF410 were observed (Supplementary Figure S4C). Elk-1 was efficiently activated as revealed by increased phosphorylation in response to PMA treatment in both cell types (Supplementary Figure S4C).
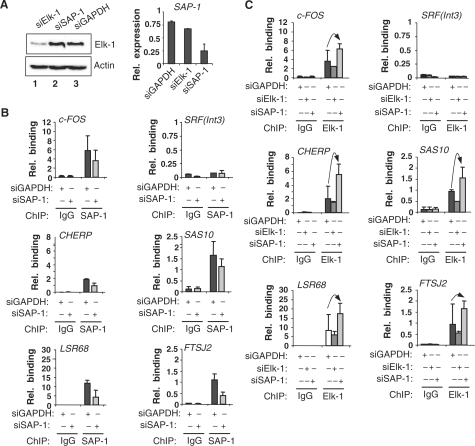
One possible reason for this lack of response to Elk-1 depletion is functional redundancy, where alternative ETS-domain transcription factors might substitute for Elk-1. In particular, the closely related TCF subfamily member SAP-1 is a likely candidate. SAP-1 binding was detected at the promoters of all the cluster 3 genes analysed (Figure 4B). To establish the specificity of the SAP-1 ChIP, we depleted SAP-1 levels in HeLa cells by siRNA treatment, and on each target promoter, reductions in SAP-1 binding were observed (Figure 4B). The specific depletion of SAP-1 mRNA levels was confirmed by RT–PCR (Figure 4A). Next, we examined the effect of reducing SAP-1 levels on Elk-1 binding to promoters of cluster 3 genes. As expected, siRNA-mediated depletion of Elk-1 levels caused a reduction in the association of Elk-1 to the tested promoters (Figure 4C). In contrast, depletion of SAP-1 resulted in a reciprocal increase in Elk-1 binding which was particularly significant at the CHERP and SAS10 promoters (Figure 4C; Supplementary Figure S5). Similar effects were observed in 293T cells (data not shown). Western analysis confirmed the effectiveness and specificity of Elk-1 depletion (Figure 4A). Thus, these findings are consistent with redundant binding of the TCFs Elk-1 and SAP-1 at cluster 3 gene promoters.
Figure 4.
Redundancy of Elk-1 and SAP-1 binding to cluster 3 promoters. (A) HeLa cells were transfected with the indicated siRNAs and Elk-1 expression monitored by western analysis (left panel) and SAP-1 expression by qRT–PCR (right panel). (B and C) qPCR-ChIP analysis of SAP-1 (B) and Elk-1 (C) binding to the indicated promoters in HeLa cells. Where indicated, cells were transfected with siRNA against Elk-1, SAP-1 or GAPDH. Samples from serum-starved HeLa cells taken 48 h after transfection with the indicated siRNAs. ChIP was performed using antibodies against SAP-1 (B) or Elk-1 (C) or non-specific rabbit IgG. Note that the same IgG data are presented in (B and C) as these experiments were performed at the same time. The relative binding is the percentage binding relative to input sample. Error bars represent standard deviations calculated from two biologically independent replicates each being the average of duplicate samples. Arrows indicate the increase of Elk-1 binding to the promoters upon SAP-1 knock-down.
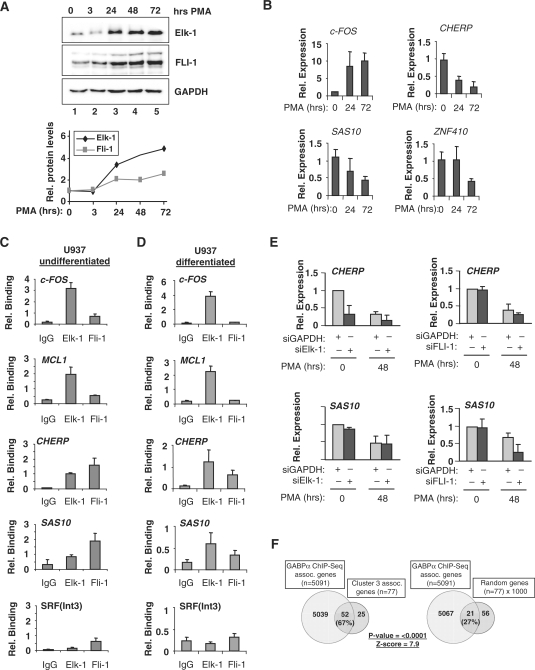
Further functional redundancy is also possible in other cell types and where other ETS-domain transcription factors are expressed. Indeed, this has been shown to be the case for the ETS-domain proteins, Ets-1, Elf-1 and GABPα: ChIP-chip analysis has suggested a high degree of redundant promoter occupancy in Jurkat cells (13). We therefore tested whether promoter occupancy by different, more divergent, ETS-proteins could be detected in undifferentiated and differentiated monocytic-like U937 cells. U937 cells express both Elk-1 and FLI-1, but the ratio of these two ETS-domain proteins varies depending on differentiation status. This is largely due to the larger PMA-induced increases in Elk-1 levels that are seen as U937 cells are stimulated to differentiate (≈5-fold increase for Elk-1 compared to ≈3-fold for FLI-1; Figure 5A). Next, we examined the expression of several cluster 3 target genes in response to stimulation of U937 cells with PMA (Figure 5B). FOS expression increased following stimulation by PMA, which is known to activate the ERK pathway and hence activate Elk-1 on the FOS promoter. However, in contrast, the expression of the cluster 3 genes CHERP, SAS10 and ZNF410 decreased substantially after PMA stimulation (Figure 5B) suggesting a non-linear relationship between Elk-1 levels and target gene activity.
Figure 5.
Cluster 3 gene regulation upon monocytic U937 cell differentiation. (A) Western analysis of changes in Fli-1 and Elk-1 levels upon U937 cell differentiation following PMA treatment for the indicated times. Quantification of the expression of each protein relative to the amount in undifferentiated cells (taken as 1) is shown below. (B) Reduced expression of cluster 3 genes upon monocyte differentiation. U937 cells were taken either before PMA-induced differentiation or 24 or 72 h after PMA addition. Expression levels of indicated genes were measured by quantitative RT–PCR. Error bars represent standard deviations calculated from two biologically independent replicates which are the average of two samples. (C and D) Dynamic changes in Elk-1 and Fli-1 binding to cluster 3 gene promoters upon monocyte differentiation. qPCR of ChIP DNA from undifferentiated (C) and differentiated (D) U937 cells using Elk-1 or Fli-1 specific antibodies or a non-specific total rabbit IgG (IgG). Differentiated cells were analysed 48 hrs after PMA stimulation. The relative binding is the percentage binding relative to input sample. Data are the average of at least two independent experiments and duplicate samples, ± standard deviation. (E) siRNA-mediated Elk-1 and FLI-1 depletion and cluster 3 gene expression in U937 cells. U937 cells were transfected with 3 µg of siRNA targeted against Elk-1 or FLI-1 (dark grey bars) or GAPDH (light grey bars). Cells were left for 24 h after transfection, then undifferentiated cells (0 h) were collected, and differentiated cells were created by treatment with 50 nM PMA for 3 h and left to differentiate for another 48 h. mRNA was isolated and the expression levels of indicated genes in the presence of reduced levels of Elk-1 (left) or FLI-1 (right) was analysed using qRT–PCR. Error bars represent standard deviations calculated from three biologically independent replicates and the average of two samples. (F) Venn diagrams illustrating the overlap between cluster 3 genes and GABPα target genes identified by ChIP-seq analysis (30). As a comparison, the average overlap between the 5091 target genes identified by ChIP-seq analysis and 1000 randomly selected groups of 77 genes is shown.
Next, we tested the promoter occupancy of several cluster 3 genes by FLI-1 and Elk-1. ChIP analysis demonstrated that both Elk-1 and FLI-1 bind the promoters from CHERP and SAS10 in undifferentiated (Figure 5C) and differentiated (Figure 5D) U937 cells. However, the relative ratios of Elk-1 and FLI-1 bound differed depending on differentiation status, with relatively more binding seen in the differentiated cells, which is commensurate with the increasing Elk-1 levels in these cells. In contrast, Elk-1 was the major binding protein on the FOS and MCL1 promoters in U937 cells irrespective of differentiation status (Figure 5C and D). Together these results demonstrate redundancy of ETS-domain protein binding to cluster 3 gene promoters where dynamic changes in promoter occupancy are observed during U937 cell differentiation. Moreover, they underline the distinctive role for Elk-1 on promoters such as FOS and MCL1 where SRF promotes selectivity in ETS-domain protein binding.
To probe functional links between promoter occupancy, and transcription factor binding and promoter activation, we examined the expression of the cluster 3 genes CHERP and SAS10 following siRNA-mediated depletion of Elk-1 or FLI-1 in U937 cells (Figure 5E). CHERP levels were reduced by depletion of Elk-1, which was most apparent in undifferentiated cells. In contrast depletion of FLI-1 had little effect on CHERP levels. Reciprocal effects were seen on SAS10 expression, where Elk-1 depletion had little effect but FLI-1 depletion caused a marked reduction in expression in differentiated cells. Thus, despite the binding of both transcription factors being detectable on both promoters in both differentiated and undifferentiated cells, there is a clear delineation in function that can be attributed to each transcription factor in regulating a distinct target gene under different cellular differentiation states.
As there is apparently a large overlap in promoter binding by Elk-1 and FLI-1, we wondered whether other ETS transcription factors might also occupy similar sites. As a further test for this potential redundancy in promoter binding, we compared the occupancy of promoters in each of the eight gene expression clusters derived from Elk-En induction, with the promoter binding for the divergent ETS-domain transcription factor GABPα determined by ChIP-Seq analysis in Jurkat cells (30) (Supplementary Table S7). Cluster 3 gene promoters exhibited substantial overlap with promoters occupied by GABPα (67% of genes; P < 0.0001) (Figure 5F). Clusters 4, 5 and 8 also showed substantial overlap with GABPα binding events (Supplementary Table S7; 43, 57 and 52%, respectively) but the other four clusters showed essentially random overlap with this dataset. Thus, we have identified a number of genes that are direct Elk-1 targets but the majority of which can also be redundantly targeted by different ETS-domain transcription factors.
Collectively, these results demonstrate that Elk-1 plays a role in regulating the expression of at least a subset of cluster 3 genes; however, a particularly characteristic feature of cluster 3 genes is redundant promoter binding by Elk-1 and other ETS-domain transcription factors. This redundant binding is also exhibited amongst other gene clusters identified as potential direct Elk-1 targets, illustrating a more widespread redundancy in promoter recognition by ETS transcription factors and potentially redundancy in gene regulation.
DISCUSSION
The overlapping in vitro DNA binding specificities of ETS-domain transcription factors suggests that either additional mechanisms are needed for in vivo selectivity or that they can bind to largely overlapping sets of promoters. Previous studies on the TCF subfamily of ETS-domain transcription factors have provided a model whereby in vivo selectivity is largely dictated through interaction with a second transcription factor SRF (4–6). However, here we have identified a group of Elk-1 target genes that are occupied by Elk-1 in an SRF-independent manner. Moreover, we demonstrate that many of these new targets can also be bound by other ETS-domain proteins under different conditions, suggesting a high degree of functional redundancy in promoter binding amongst ETS-domain transcription factors. Thus, while Elk-1 is specifically directed to some target genes by an SRF-Elk-1 transcription factor core module, many other targets bind Elk-1 independently of SRF.
Our results also extend recent observations that demonstrated highly overlapping promoter binding patterns between the ETS-domain transcription factors Ets-1, Elf-1 and GABPα (13), suggesting a high degree of functional redundancy amongst ETS-domain proteins. Here, we add further weight to this conclusion as we show that redundant promoter occupancy can be observed for Elk-1 and the closely related transcription factor SAP-1, and also between Elk-1 and the more distantly related FLI-1. Such redundant promoter binding makes functional interrogation of ETS-domain transcription factor action difficult to achieve as knockdown of one member is likely compensated for by a different member. This was apparent in our study where depletion of Elk-1 levels in HeLa cells had little effect on target gene expression (data not shown). However, in an alternative cell type (monocytic U937 cells), we were able to see an effect of Elk-1 depletion on CHERP gene expression, indicating that the cellular environment has a key role in dictating the functional specificity of individual ETS-domain proteins. Indeed, although both Elk-1 and FLI-1 were shown to bind the SAS10 promoter, only depletion of FLI-1 caused a reduction in SAS10 gene expression, and this effect was only observed in differentiated cells. It is currently unclear what contributes to the ability of Elk-1 and FLI-1 to specifically regulate CHERP and SAS10 expression respectively, but different signalling conditions or the availability of different partner proteins, most likely dictates these different functional outcomes.
The main conclusions of this study are also supported by comparative analysis of a much larger set of Elk-1 targets identified in a recent ChIP-chip study (32). 77/877 (9 %) targets from the current study were also identified in the highest confidence data set from the ChIP-chip analysis; within cluster 3 a higher percentage (15/77, 19%) was identified. Moreover, when a more comprehensive ChIP-Seq dataset for GABPα binding was analysed, the overlap was even more striking: 333/877 (38 %) targets from the current study were also identified in the ChIP-Seq analysis. In several clusters, the overlap was even higher (e.g. 52/77, 68% in cluster 3). This emphasises the quality of our dataset from the Elk-En experiment and also further underlines the likely redundancy of ETS protein binding to the promoters from these genes.
Comparison of ChIP-chip datasets for Elk-1 and SRF demonstrated that although co-occupancy of promoters by SRF and Elk-1 accounted for around 22% of the Elk-1 targets, overall co-occupancy was a rather infrequent event with 78% of promoters not co-bound by SRF (32). Thus the paradigm derived from studies of the FOS promoter, where SRF is an obligate binding partner, appears to apply to a subset of genes rather than, the majority of Elk-1 target genes. This finding is further emphasized here, as we find little evidence for a substantial overlap in Elk-1 and SRF occupancy at target genes. Interestingly, there are a small number of promoters in the Elk-En target gene clusters that overlap with a high confidence SRF binding dataset created by identifying common targets in both ChIP-Chip (32) and ChIP-Seq (30) studies. These promoters are predominantly in clusters 3, 4, 5 and 8 (15/16 genes), which are the highest confidence Elk-1 targets. Overall, therefore there is an important subset of Elk-1 targets that are also bound by SRF, further emphasizing the importance of this transcription factor module, yet the major mode of function of Elk-1 appears to be independent of SRF, which then obliges Elk-1 to compete with other ETS transcription factors for promoter occupancy.
In cases where highly redundant binding of transcription factors occurs, such as that observed amongst the ETS-domain family, elucidating function is difficult by conventional single protein knockdown studies. However, the current study demonstrates a possible way forward through using a dominant-negative type of overexpression approach. Alternative strategies such as introducing promoter mutations in the ETS factor binding site are also informative but not amenable to interrogating large numbers of potential target genes. The use of an inducible dominant-negative type Elk-1 construct (Elk-En) coupled to expression array analysis, enabled us to identify a high confidence dataset that represented potential targets for ETS-domain transcription factors and subsequent clustering further filtered this dataset. Four of the clusters (3, 4, 5 and 8 comprising 459/877 {53%} Elk-En-regulated genes) represented potential direct targets by several criteria: (i) qPCR-ChIP demonstrated that a large number of cluster 3 genes are direct Elk-1 targets; (ii) there is a significant overlap in these clusters with Elk-1 direct targets identified by ChIP-chip (Supplementary Table S4); (iii) there is an overrepresentation of ETS transcription factor-like binding motifs in these clusters; and (iv) there is a significant overlap in these clusters with direct targets of GABPα identified by ChIP-Seq (Supplementary Table S4). Thus, while we focussed on cluster 3 for validation purposes, clusters 4, 5 and 8 also represent high confidence targets for binding by Elk-1 and other ETS transcription factors, and provide a valuable dataset for others investigating promoter regulation. Furthermore, genes in clusters, 3, 4, 5 and 8 showed over-representation of several functional categories and when considered together, revealed apoptosis regulation as a new functional category (Supplementary Tables S2 and S3). This is in keeping with our previous observation that Elk-En expression promotes apoptosis, in part through repressing MCL-1 expression (16). The association with apoptosis regulation appears more specific to Elk-1 than other ETS transcription factors as this category was not identified previously for genes commonly bound by Ets-1 and Elf-1, although the other functional categories identified in the current study overlap with subcategories of the housekeeping genes identified as commonly bound by other ETS proteins (13).
It is not clear why we also identified the genes in clusters 1, 2, 6 and 7 as potentially regulated by Elk-En using this approach as they seem to not represent direct Elk-1 targets. Indeed, these genes do not contain any over-represented subcategories. One likely explanation for their occurrence is that these genes represent indirect effects whereby Elk-En causes the repression of a gene that encodes an activator(s) of the genes in these clusters. However, it is likely that at least some of the genes in these clusters might well represent direct targets of ETS transcription factors as clustering analysis is not definitive, although clearly further verification would be needed. Indeed, the reciprocal is also true for clusters 3, 4, 5 and 8, which although of high confidence, probably contain a number of false positives that are not supported by additional evidence from ChIP-chip data or the presence of potential binding motifs. Thus in general, the target genes that are supported by multiple lines of evidence provide a higher confidence dataset.
In summary, our findings provide further insights into target gene selection by members of the ETS-domain transcription factors, highlighting redundancy of binding at some promoters but highly selective function at others, dictated by heterotypic interactions with other transcription factors. In a broader context, these results have an impact on framing our understanding of how other multi-gene families of transcription factors might function through specific and redundant promoter recognition.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Wellcome Trust; the AICR, Cancer Research UK; a Royal Society-Wolfson award (to A.D.S.). Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Anne Clancy, Karren Palmer and Leanne Wardleworth for excellent technical assistance; Andy Hayes in the core microarray facility; Stefan Roberts and members of our laboratory for comments on the article and stimulating discussions; and Richard Treisman for reagents.
REFERENCES
- 1.Graves BJ, Petersen JM. In: Advances in Cancer Research. Woude GV, Klein G, editors. Vol. 75. San Diego: Academic Press; 1998. pp. 1–55. [Google Scholar]
- 2.Sharrocks AD. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 3.Hollenhorst PC, Jones DA, Graves BJ. Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res. 2004;32:5693–5702. doi: 10.1093/nar/gkh906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharrocks AD. Complexities in ETS-domain transcription factor function and regulation: lessons from the TCF (ternary complex factor) subfamily. The Colworth Medal Lecture. Biochem. Soc. Trans. 2002;30:1–9. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 5.Shaw PE, Saxton J. Ternary complex factors: prime nuclear targets for mitogen-activated protein kinases. Int. J. Biochem. Cell Biol. 2003;35:1210–1226. doi: 10.1016/s1357-2725(03)00031-1. [DOI] [PubMed] [Google Scholar]
- 6.Buchwalter G, Gross C, Wasylyk B. Ets ternary complex transcription factors. Gene. 2004;324:1–14. doi: 10.1016/j.gene.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 7.Dalton S, Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68:597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- 8.Shore P, Sharrocks AD. The transcription factors Elk-1 and serum response factor interact by direct protein-protein contacts mediated by a short region of Elk-1. Mol. Cell Biol. 1994;14:3283–3291. doi: 10.1128/mcb.14.5.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hassler M, Richmond TJ. The B-box dominates SAP-1-SRF interactions in the structure of the ternary complex. EMBO J. 2001;20:3018–3028. doi: 10.1093/emboj/20.12.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Treisman R, Marais R, Wynne J. Spatial flexibility in ternary complexes between SRF and its accessory proteins. EMBO J. 1992;11:4631–4640. doi: 10.1002/j.1460-2075.1992.tb05565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchwalter G, Gross C, Wasylyk B. The ternary complex factor Net regulates cell migration through inhibition of PAI-1 expression. Mol. Cell Biol. 2005;25:10853–10862. doi: 10.1128/MCB.25.24.10853-10862.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhong S, Fromm J, Johnson DL. TBP is differentially regulated by c-Jun N-terminal kinase 1 (JNK1) and JNK2 through Elk-1, controlling c-Jun expression and cell proliferation. Mol. Cell Biol. 2007;27:54–64. doi: 10.1128/MCB.01365-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shore P, Sharrocks AD. The ETS-domain transcription factors Elk-1 and SAP-1 exhibit differential DNA binding specificities. Nucleic Acids Res. 1995;23:4698–4706. doi: 10.1093/nar/23.22.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seth A, Gonzalez FA, Gupta S, Raden DL, Davis RJ. Signal transduction within the nucleus by mitogen-activated protein kinase. J. Biol. Chem. 1992;267:24796–24804. [PubMed] [Google Scholar]
- 16.Vickers ER, Kasza A, Kurnaz IA, Seifert A, Zeef LA, O'D;onnell A, Hayes A, Sharrocks AD. Ternary complex factor-serum response factor complex-regulated gene activity is required for cellular proliferation and inhibition of apoptotic cell death. Mol. Cell Biol. 2004;24:10340–10351. doi: 10.1128/MCB.24.23.10340-10351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price MA, Rogers AE, Treisman R. Comparative analysis of the ternary complex factors Elk-1, SAP-1a and SAP-2 (ERP/NET) EMBO J. 1995;14:2589–2601. doi: 10.1002/j.1460-2075.1995.tb07257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vickers ER, Sharrocks AD. The use of inducible engrailed fusion proteins to study the cellular functions of eukaryotic transcription factors. Methods. 2002;26:270–280. doi: 10.1016/S1046-2023(02)00031-2. [DOI] [PubMed] [Google Scholar]
- 19.Yang SH, Sharrocks AD. PIASx acts as an Elk-1 coactivator by facilitating derepression. EMBO J. 2005;24:2161–2171. doi: 10.1038/sj.emboj.7600690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'D;onnell A, Yang SH, Sharrocks AD. MAP kinase-mediated c-fos regulation relies on a histone acetylation relay switch. Mol. Cell. 2008;29:780–785. doi: 10.1016/j.molcel.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasza A, O'D;onnell A, Gascoigne K, Zeef LA, Hayes A, Sharrocks AD. The ETS domain transcription factor Elk-1 regulates the expression of its partner protein, SRF. J. Biol. Chem. 2005;280:1149–1155. doi: 10.1074/jbc.M411161200. [DOI] [PubMed] [Google Scholar]
- 22.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl Acad. Sci. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 24.Liu H, Zeeberg BR, Qu G, Koru AG, Ferrucci A, Kahn A, Ryan MC, Nuhanovic A, Munson PJ, Reinhold WC, et al. AffyProbeMiner: a web resource for computing or retrieving accurately redefined Affymetrix probe sets. Bioinformatics. 2007;23:2385–2390. doi: 10.1093/bioinformatics/btm360. [DOI] [PubMed] [Google Scholar]
- 25.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 26.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36(Database Issue):D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alibés A, Yankilevich P, Cañada A, Díaz-Uriarte R. ID converter and IDC light: conversion and annotation of gene and protein IDs. BMC Bioinformatics. 2007;8:9. doi: 10.1186/1471-2105-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pavesi G, Mereghetti P, Mauri G, Pesole G. Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32:W199–W203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 32.Boros J, Donaldson IJ, O'D;onnell A, Odrowaz ZA, Zeef L, Lupien M, Meyer CA, Liu XS, Brown M, Sharrocks AD. Elucidation of the ELK1 target gene network reveals a role in the coordinate regulation of core components of the gene regulation machinery. Genome Res. 2009 doi: 10.1101/gr.093047.109. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Townsend KJ, Zhou P, Qian L, Bieszczad CK, Lowrey CH, Yen A, Craig RW. Regulation of MCL1 through a serum response factor/Elk-1-mediated mechanism links expression of a viability-promoting member of the BCL2 family to the induction of hematopoietic cell differentiation. J. Biol. Chem. 1999;274:1801–1813. doi: 10.1074/jbc.274.3.1801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.