Abstract
The many different mechanisms that fungi use to transmit and share genetic material are mediated by a broad range of chromosome and nuclear dynamics. The mechanics underlying nuclear migration are well integrated into detailed models, in which the forces supplied by plus- and minus-end-directed microtubule motors position and move the nucleus in a cell. Although we know much about how cells move nuclei, we know much less about why the cell invests in so many different nuclear ‘dances’. Here, we briefly survey the available models for the mechanics of nuclear migration in fungi and then focus on examples of how fungal cells use these nuclear dances — the movement of intact nuclei in and between cells — to control the integrity, ploidy and assortment of specific genomes or individual chromosomes.
Different fungal species propagate as haploids, diploids, dikaryons, multinucleates, polyploids or aneuploids, and some fungal life cycles include not just one or two but several of these different states. Each of these genome conditions has different challenges and requirements for moving nuclei around the cell, and this movement must be developmentally regulated. Molecular analysis of a range of fungal organisms (TABLE 1) has revealed both similarities to and important differences from the paradigms that have been established in the model yeast Saccharomyces cerevisiae. Here, we discuss how whole nuclei move during normal cell division, how nuclei maintain or alter the ploidy of one or all chromosomes and how different nuclei interact in multinucleate cells. We also discuss some of the additional functions of nuclear movement and our emerging understanding of how nuclear positioning might contribute to the preservation of an ‘immortal’ template chromosome in stem cell-like nuclei. The specific nuclear dynamics discussed here are examples from fungi, but many of them resemble processes that occur in mammalian cells, including cancer cells and stem cells. Thus, in this Review, we survey diverse fungal systems and many different areas of cell biology in order to highlight open questions about the interplay between nuclear content and nuclear migration.
Table 1. Selected model fungi.
| Organism | Phylum | Environment | Utility and areas of investigation | Genome sequence |
|---|---|---|---|---|
| Saccharomyces cerevisiae | Ascomycete | Ubiquitous | Baker's and brewer's yeast; premier eukaryotic model organism used to study a broad range of molecular, cell-biological and genetic questions | 1996 (REF. 96) |
| Candida albicans | Ascomycete | Ubiquitous | Human commensal and opportunistic pathogen with budding yeast and hyphal forms; studies focus on drug resistance, biofilm formation and morphogenesis, as the ability to switch to the hyphal form seems to be important for virulence | 2004 (REF. 97) |
| Schizosaccharomyces pombe | Ascomycete | Ubiquitous | Fission yeast with three chromosomes that are detectable by light microscopy; used to study many aspects of cell cycle progression and the general biology of eukaryotes | 2002 (REF. 98) |
| Neurospora crassa | Ascomycete | Tropical and subtropical; found on dead plant matter following fires | Classic haploid genetic model; used to study circadian rhythms, epigenetics, gene silencing, polarized growth and cell development | 2003 (REF. 99) |
| Ashbya gossypii | Ascomycete | Associated with insects and plants; potential symbiote with both | Filamentous, multinucleate growth with haploid nuclei that divide asynchronously in a common cytoplasm; high genome similarity to S. cerevisiae and highly tractable for molecular genetics; used as a model system for the cell cycle and morphogenesis and in industry to produce riboflavin (vitamin B2) | 2004 (REF. 100) |
| Ustilago maydis | Basidiomycete | Corn smut pathogen | Model organism readily amenable to gene replacement; used to study DNA recombination and repair, and cytoskeleton dynamics; biotrophic organism requiring plant tissue for growth and development | 2006 (REF. 101) |
| Podospora anserina | Ascomycete | Herbivore dung | Classic genetic model with no asexual spores; used to study cytoplasmic inheritance, differentiation and cell death, ageing, multicellularity and epigenetics; close relative of N. crassa | 2008 (REF. 102) |
| Schizophyllum commune | Basidiomycete | Widespread | Split-gill fungus that causes white rot; >20,000 different compatible mating types found worldwide | Incomplete* |
| Coprinopsis cinerea | Basidiomycete | Widespread | Inky-cap mushroom with a sexual cycle that is completed in ∼2 weeks in the laboratory | Draft sequence annotated‡ |
| Heterobasidion parviporum | Basidiomycete | North American and European conifer forests | Root rot fungus and important pathogen of conifers; model system for evolutionary genetics | NA |
| Magnaporthe grisea | Ascomycete | Widespread | Rice blast fungus that also infects grasses; model system for understanding plant–fungal-pathogen interactions | 2005 (REF. 103) |
| Armillaria gallica | Basidiomycete | Hardwood forests | A ‘honey mushroom’, one of the oldest and largest living organisms; important model system for evolutionary genetics | NA |
Entrez Genome Project S. commune entry (see Online links box).
Broad Institute C. cinerea genome project (see Further Information).
Nuclear dances: moving intact nuclei
The paradigms for nuclear migration have emerged from studies in both the budding yeast S. cerevisiae and the multinucleate filamentous fungus Aspergillus nidulans. These have been exhaustively reviewed elsewhere, and we do not attempt to review this vast literature here. Instead, we provide a brief overview of the general mechanics that underlie nuclear migration. Summaries of mitotic cell cycle progression in yeast and dimorphic yeast cells and in filamentous fungi are provided in BOXES 1,2,3.
Box 1. Mitotic cell cycle progression in budding yeast.
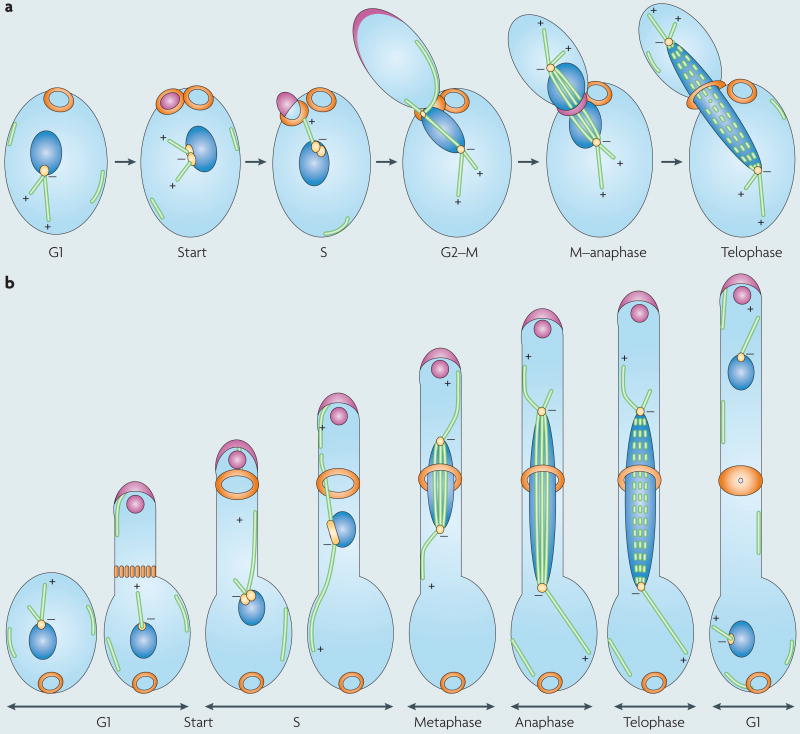
Unicellular yeast
Saccharomyces cerevisiae and Candida albicans yeast cells undergo a well-characterized mitotic cell cycle (see the figure, part a). In G1, a new septin ring forms adjacent to the birth scar (orange rings) or at the opposite pole of the cell (not illustrated). At the G1–S boundary (Start), the spindle pole body (SPB; yellow) duplicates, and the bud (pink) emerges through the septin ring by the action of polarized secretory vesicles in the bud. All microtubules (MTs; green) seem to nucleate at the SPBs, and mitosis is closed (the nuclear envelope does not break down). In C. albicans, free microtubules are evident along the cell cortex. The duplicated SPB forms a short spindle, and astral MTs communicate with the septin ring to orientate the spindle with the mother–bud axis. Spindle oscillations across the bud neck are mediated by dynein acting on the astral MTs. Growth of the bud is mediated by secretory vesicles, which are organized into a crescent-shaped polarisome (pink). During anaphase, spindle elongation forces are sufficient to deliver the sister chromatids into the two daughter nuclei (blue). Polarized growth is re-orientated to the septin ring, the ring contracts, and it is filled with chitin-rich cell wall material before cell separation. Yeast and pseudohyphal cells have similar dynamics, with the main difference being that pseudohyphae spend more time in G2 and, thus, the buds become longer than buds in yeast cells before the onset of anaphase, and cells often do not separate completely, remaining attached in chains of elongated budded cells.
Unicellular hyphae in multimorphic budding yeast
When grown under conditions that induce hyphal growth (see the figure, part b), a C. albicans germ tube evaginates through a band of septin proteins (orange). Polarized growth is driven by a polarisome (pink cresent) in addition to a Spitzenkörper (pink circle), which is a vesicle-organizing centre that drives the highly polarized growth of hyphal tips. Septin ring formation occurs ∼10–20 μm from the basal cell at the time of SPB duplication88, and the ring is laid down just as the tip passes this future site of septation (the presumptum). Nuclei with short spindles migrate to the presumptum by oscillatory movements that are mediated by contacts between astral MTs and the cell cortex and that are primarily dynein dependent. Anaphase occurs across the septin ring, and spindle elongation forces deliver the daughter nuclei into the two compartments (the basal cell and the hyphal tip cell). Other, slower forces move the mother nucleus back into the basal cell. Dissolution of the spindle is followed by contraction of the septin ring and accumulation of cell wall material at the septum, but the cells remain attached to one another.
+, microtubule plus end; −, microtubule minus end.
Box 2. Mitotic cell cycle progression in fission yeast.
In Schizosaccharomyces pombe cells (see the figure), G1 is very short-lived, and thus cytokinesis and cell separation are accomplished during S phase of the next cell cycle. Microtubule-organizing centres (MTOCs; yellow), primarily at the nuclear periphery, nucleate microtubules (MTs; green) in interphase, in addition to the spindle pole bodies, which nucleate the mitotic spindle. Before cell division, equatorial MTOCs nucleate MTs from the future site of septation. In general, MT minus ends (−) are located towards the centre of the cell and the plus ends (+) are located towards the tips.
Box 3. Mitosis in filamentous fungi.
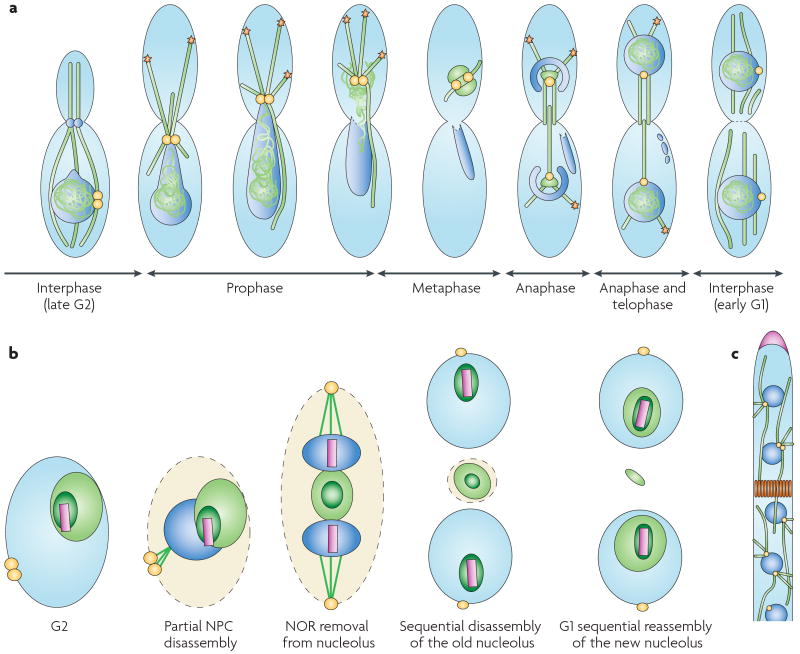
Ustilago maydis
In the basidomycete corn smut fungus, Ustilago maydis, yeast growth (see the figure, part a) is accompanied by microtubule organizing centre (MTOC)-mediated nucleation of microtubules (MTs; green) from the bud neck region (blue circles). The plus-end localization of dynein (orange stars) mediates spindle elongation and is essential for retrograde endosome motility. In hyphae, MTs nucleate near the central nucleus (dark blue), or the central pair of nuclei in dikaryotic cells89. Nuclear envelope breakdown begins with extension of the nuclear envelope. In prophase, dynein in the bud cortex pulls on MTs and spindle pole bodies (SPBs; yellow) that are attached to condensed chromosomes. The nuclear envelope eventually breaks at the SPB, and the chromosomes congregate with the SPB outside the old nuclear envelope. Once the chromosomes are separated in anaphase, a new nuclear envelope forms around the daughter chromosomes.
Aspergillus nidulans
In the most apical hyphal compartment, Aspergillus nidulans nuclei divide in a parasynchronous wave, and SPBs of post-mitotic nuclei nucleate cytosolic MTs90,91. The basal compartments, which are separated from the apical compartment by a septum that is established by septin rings, contain nuclei that are arrested in interphase until a side branch emerges, at which time the nuclei re-enter the division cycle. Cytosolic MTs are present at interphase and generally disappear during mitosis, except in the hyphal tip92. A. nidulans nucleolar proteins are separated from the ribosomal DNA that makes up the nucleolar organizing region (NOR) and are expelled from daughter nuclei to the cytoplasm between them (see the figure, part b). Unlike most nuclear reorganizations, this process can occur even when spindle attachment is abrogated93. Nuclear DNA in hyphae undergoes a partially open mitosis, in which nuclear pore complexes (NPCs) disassemble, making the nucleus more permeable during mitosis. In addition, on the initiation of mitosis, DNA (dark blue) condenses and nucleolar proteins (dark green) are exported from the nucleus, whereas the NOR (pink) is retained. Following mitosis, nucleolar proteins are imported through reassembled NPCs in the nuclear membrane to form new nuclei.
Ashbya gossypii
This ascomycete is thought to nucleate MTs exclusively from nuclear-membrane-associated SPBs. Short astral MTs associate with the cortex, and longer cytosolic microtubules extend for tens of microns through the cytosol. The nuclei divide asynchronously, and therefore in the figure (part c), individual nuclei are depicted in different states of SPB duplication and separation94,95.
Microtubules (MTs) and microtubule-organizing centres (MTOCs) have a central and conserved role in moving nuclei. Kinetochore MTs in the nucleus segregate chromosomes by pulling them towards the poles. Interpolar MTs facilitate nuclear pole separation by pushing against each other. Outside the nucleus, astral MTs emanate from the MTOC to orientate the mitotic spindle across the mother–daughter cell junction through interactions with the cell cortex and to promote pole separation. In some organisms, such as S. cerevisiae, all MTs originate from spindle pole bodies (SPBs) (FIG. 1). In other organisms (for example, the fission yeast Schizosaccharomyces pombe (BOX 2) and the basidiomycete Ustilago maydis (BOX 3)), MTOCs arise at extra-SPB positions during interphase1 or during mating, when mating-specific MTOCs form at the tips of mating projections2. In S. pombe, MT dynamics, along with plus-end tracking (+TIP) proteins, which localize to the plus ends of MTs, have central roles in nuclear positioning. The cylindrical shape, small size and symmetrical division plane of fission yeast all facilitate a simple mechanism of nuclear positioning that relies on MT dynamics. In this system, parallel arrays of MTs interact with the cortex and exert pushing forces on the nucleus1,3. However, in most systems, particularly in large filamentous fungal hyphae, MT dynamics alone are not sufficient for nuclear positioning during interphase, and nuclear migration is also powered by molecular motors.
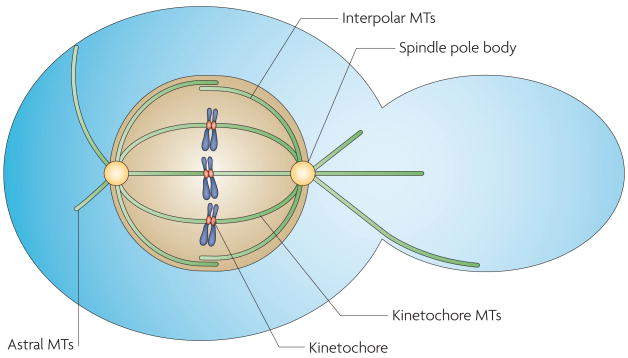
Figure 1. Microtubules and nuclear movement.
In Saccharomyces cerevisiae, all microtubules (MTs) originate from the spindle pole body. Kinetochore MTs are required for chromosome segregation, and interpolar MTs are required for nuclear pole separation. Astral MTs interact with the cell cortex and septin ring at the bud neck to orientate the mitotic spindle across the mother–daughter cell junction.
S. cerevisiae
Work from a number of laboratories has shown that the dynein heavy chain (Dyn1) has a key role in S. cerevisiae nuclear positioning, as it provides the main force that pulls the nucleus, through astral MTs. For example, S. cerevisiae dyn1 mutants exhibit abnormal nuclear localization and elevated levels of binucleate cells4. During G2 and early M phase in the cell cycle, the entire mitotic spindle, including the duplicated chromosomes, undergoes a series of oscillations across the mother–bud neck. This requires Dyn1 and interactions between astral MTs and actin cables, mediated by the MT-associated proteins karyogamy protein 9 (Kar9) and Bim1, which tether the S. cerevisiae type v myosin Myo2 to the plus ends of MTs4. In the absence of Kar9 or the spindle polarity determinants Bni1 and_bud site selection protein 6 (Bud6), dynein-mediated nuclear movement occurs earlier and more rapidly than in wild-type cells, indicating that Bud6 and Bni1 normally attenuate dynein activity5, although they probably do so through independent mechanisms6. Similarly, in response to double-strand breaks, the mitosis entry checkpoint protein 1 (Mec1)-dependent checkpoint signals through Rad52 and checkpoint kinase 1 (Chk1) to repress dynein-dependent nuclear migration through the bud neck7. This balance of forces ensures that, in S. cerevisiae, the mitotic spindle is properly orientated to span the mother–bud neck and that replicated chromosomes are accurately delivered to both mother and daughter cells. One key question that remains to be resolved is how mitotic checkpoints help cells to determine when both nuclei have been properly positioned. This process involves the asymmetrical association of proteins such as Kar9 and Dyn1 with the SPB that localizes to daughter cells8,9.
By contrast, the nuclear migration that is involved in karyogamy (the fusion of haploid nuclei following conjugation between gametes) seems to be powered primarily by interactions between cytoplasmic MTs that extend from the SPBs to interconnect the fusing nuclei10. Depolymerization of SPB-bound MTs pulls the nuclei together, and these nuclei then fuse near one edge of the SPBs. a careful study of this nuclear congression found that karyogamy is mediated by the interaction of MT plus ends rather than by MT sliding and extensive overlap11. The prevailing model is that during karyogamy MT plus-end interactions from oppositely orientated MTOCs provide the force to move nuclei through the cytoplasm11. Mutations causing defects in karyogamy also provide powerful genetic tools for performing cytoduction12.
Filamentous fungi
Most filamentous fungi have multinucleate hyphal compartments. Thus, in expanding myce lia, daughter nuclei remain in the same compartment after mitosis (which is not spatially and temporally coordinated with cell division; see REF. 13 for an exception). However, the nuclei are evenly distributed along the hyphal compartments, which balances the cytoplasmic volume and contents that are sampled by each nucleus. MTs and their motors have central roles in positioning nuclei in hyphae, and this has been reviewed in detail elsewhere14. The role of kinesins in nuclear positioning has been elusive, potentially because of functional redundancy among the conventional kinesins; for example, in A. nidulans there are at least 11 different forms. There is some, limited, evidence in A. nidulans for a role for kinesins in destabilizing MTs and localizing dynein to promote nuclear migration, but on the whole much remains to be discovered about the role of kinesins in nuclear migration in filamentous fungi15,16.
By contrast, dynein is central to nuclear migration in many filamentous fungi. Mutations in dynein components, such as the classic nud alleles of A. nidulans and the ropy alleles of Neurospora crassa, have revealed that dynein has a broad range of functions. In Ashbya gossypii, N. crassa and A. nidulans dynein mutants, the nuclei are no longer evenly spaced, leading to clumping of nuclei in regions of the mycelia17–19. In A. gossypii, the loss of dynein leads to the clustering of all the nuclei at hyphal tips, leaving vast areas (tens of microns) of cytosol devoid of nuclei17 (BOX 3). By contrast, in A. nidulans dynein mutants, all of the nuclei clump at the germ bubble by the ascospore (the area of the hypha that is farthest from the hyphal tip)17,19. In these systems, dynein ensures the balanced distribution and even spacing of nuclei throughout the hyphal tube. However, it is not known whether this is accomplished directly, by dynein acting as a conventional motor, or whether it is an indirect effect of dynein on MT dynamics14. In U. maydis, between three and six small, motile, γ-tubulin-containing MTOCs nucleate MTs at the boundary of mother and budding daughter cells, and dynein-based transport of MTs and MTOCs is required to polarize the MT cytoskeleton20.
Candida albicans
The human pathogen Candida albicans21 can form yeast, pseudohyphae or true hyphae, depending on the environmental conditions22,23. The nuclear dynamics in C. albicans yeast and pseudohyphae resemble those in S. cerevisiae: the nuclei cross the mother–bud neck, and the elongated mitotic spindle oscillates across the neck in a dynein-dependent manner24,25 (FIG. 2). Spindle elongation is sufficient to deliver a single nucleus to each daughter cell. The nuclear dynamics in C. albicans true hyphae are different: the nucleus migrates ∼10 μm into the growing germ tube to the presumptum, which is the site of germ tube division and septum formation24. The spindles undergo dynein-dependent oscillatory movements that cross the presumptum (Supplementary information S1 (movie)). In the absence of dynein in both hyphae and yeast (FIG. 2c), the velocity of nuclear movement is reduced such that mitosis ensues in the mother cell. However, a spindle checkpoint mediated by Bub2 prevents premature cytokinesis, such that a post-mitotic daughter nucleus often succeeds in eventually reaching the bud. The slower movement of nuclei in dynein mutants is probably mediated by MT plus-end dynamics25.
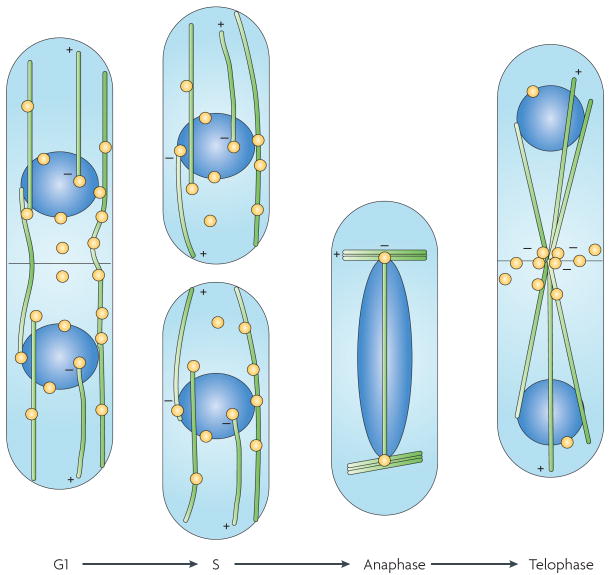
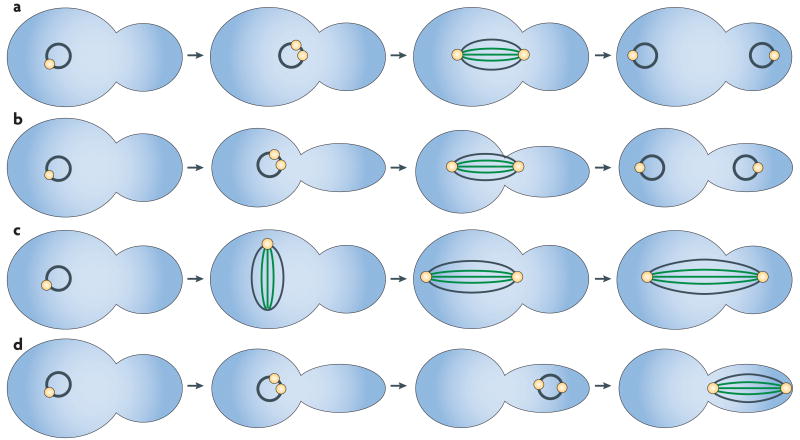
Figure 2. Mitotic dynamics in Candida albicans.
a,b | In wild-type yeast (part a) and pseudohyphal (part b) cells, the nuclei are aligned by the mitotic spindle (spindle pole bodies are in yellow, kinetochore microtubules are in green and interpolar microtubules are in black) and divide across the mother–bud neck. c | In yeast cells lacking the dynein heavy chain, the nuclei divide in the mother cell and a checkpoint delays cell cycle progression until the nucleus enters the daughter cell. d | In pseudohyphal cells lacking dynein, nuclei migrate into the daughter cell and divide there. There is no checkpoint to ensure that the mother nucleus returns to the mother cell. In this schematic, the details of microtubule attachment to chromosomes and of inter-microtubule interactions are not shown.
C. albicans pseudohyphal cells that lack dynein exhibit a distinctive nuclear dance: the nuclei move into the daughter cell before mitosis and divide there (FIG. 2d). In the absence of dynein, there is no oscillation across the mother–bud neck. Importantly, in pseudohyphae there is no evident checkpoint-mediated delay of cytokinesis when the entire mitotic spindle is in the bud; the result is the formation of a dikaryotic daughter cell and an anucleate mother cell. In subsequent divisions, the binucleate cells continue to grow and divide with multiple nuclei (K. Finley, personal communication). a similar multinuclear phenotype is seen in a subpopulation of mutants with cell cycle defects, such as those lacking forkhead transcription factor 2 (Fkh2)26 or the cell cycle kinase Dbf227. Interestingly, in some of the cells that lack Fkh2 the spindles seem to be in synchronous stages of the cell cycle, whereas in other multinucleate cells mitotic spindles coexist with duplicated SPBs, suggesting that different stages of the cell cycle can coexist in the same cytosol (E. Bensen, personal communication). Similarly, in S. pombe, nuclear alignment and the plane of division is determined by the nuclear oscillations that occur just before mitosis, and defects in MT attachment to the nucleus lead to misaligned spindles and the mis-segregation of nuclei28,29. Thus, defects in nuclear movement, like defects in cell cycle progression, can result in multinucleate cells.
Dancing partners: genome integrity
MTs and their motors are not simply a generic ‘mass transit system’ in which astral MTs move nuclei. With each turn of the cell cycle there is another layer of choreography within each migrating nucleus, as the chromosomes move in the process of attaching to kinetochore MTs. Recent work indicates that there is an intimate association between the molecular machines that move the DNA and the amount and content of the DNA that is transported. Controlled MT dynamics combined with specific MT motors ensure that each individual chromosome is correctly tethered to, and later appropriately released by, its kinetochore MTs.
S. cerevisiae
In S. cerevisiae, cells are normally maintained as haploids or diploids, although crosses can be manipulated to yield triploid and tetraploid strains. Although deviations in normal ploidy can occur through endoreplication, non-disjunction or mating of non-haploid strains (reviewed in REF. 30), most tetraploidy is assumed to arise unintentionally, as a by-product of defects such as a failure of cytokinesis. Cytokinesis can fail as a consequence of subtle defects in the segregation of a single pair of chromosomes or because of wholesale defects in mitotic processes (reviewed in REFS 31,32). In mammalian cells, such ‘4n’ cells usually undergo apoptosis. However, when they do divide, these cells exhibit high levels of genome instability, resulting in the extensive aneuploidy that is typical of cancer cells (reviewed in REF. 30).
In S. cerevisiae, mitotically dividing tetraploids exhibit high levels of genome instability, including the accumulation of point mutations during S phase at a twofold higher rate than that in diploid cells33. A genome-wide screen for ploidy-specific lethality revealed that only a small number of genes are necessary to maintain tetraploidy33. These genes fall into three main functional groups: genes encoding components of the mitotic spindle, including the SPB; genes that are necessary for the establishment of chromosome cohesion, including several kinetochore components; and genes that are required for homologous recombination33. This suggests that the presence of extra chromosome copies causes cells to expend more energy on whole-chromosome dynamics and maintaining the integrity of the DNA. It has been proposed that extra stress is placed on tetraploid cells, because MTs do not increase in length despite the higher number of chromosomes on a tetraploid mitotic spindle33.
When unstressed and salt-stressed haploid, diploid and tetraploid S. cerevisiae strains were followed for ∼1,800 doublings, all cultures converged to diploidy, suggesting that the diploid state has a selective advantage in these conditions34. Furthermore, diploids had an advantage over haploids when serial cultures of mutator (msh2Δ) strains were propagated, presumably owing to the ability of the diploid state to buffer deleterious mutations35. Despite the instability of cells with increased ploidy, many industrial S. cerevisiae strains are polyploid36. It is likely that the fitness of a strain with increased ploidy depends on the specific chromosomes that are amplified.
Aneuploidy is assumed to arise from defects in the associations between individual kinetochores and their kinetochore MTs. aneuploidy was studied in haploid yeast strains that each carried an extra copy of one or more chromosomes37. These disomic (haploid + 1) aneuploids shared several phenotypes, including sensitivity to conditions that interfere with protein folding, protein synthesis and cell cycle progression. In addition, the transcription level of ∼400 genes was altered in almost all of the aneuploid strains, suggesting that, irrespective of the chromosome involved, disomic haploids exhibit growth defects and changes in transcription due to the presence of extra chromosomes37. Similarly, trisomic mouse cell lines (diploid + 1) exhibited reduced proliferation and cellular fitness38. an important open question is: why do aneuploid cancer cells exhibit increased proliferative capacity39, but aneuploid S. cerevisiae and mouse cells exhibit reduced fitness37? It is likely that specific aneuploidies provide fitness advantages under specific stress conditions40,41.
C. albicans
In contrast to S. cerevisiae, C. albicans is much more tolerant of aneuploidies and translocations. Whole-chromosome aneuploidies arise frequently following transformation of laboratory strains42–44, with trisomy being much more prevalent than monosomy. aneuploidy, including whole-chromosome and segmental amplifications, has been detected in 50% of C. albicans strains that have elevated resistance to fluconazole41, the most commonly used antifungal drug. One specific segmental aneuploidy, isochromosome 5l, in which 2 copies of the left arm of chromosome 5 flank the centromere, accounts for at least 20% of these aneuploidies41. The elevated copy number of two genes on isochromosome 5l, one near the telomere and one near the centromere, is responsible for most of the resistance that is seen in strains carrying this isochromosome45.
The high level of tolerance for aneuploidy and translocations could be because C. albicans does not undergo the conventional meiotic pairing that requires alignment of homologous chromosomes. alternatively, this tolerance may stem from the flexibility in spindle length between yeast, pseudohyphal and hyphal C. albicans cells24. This flexibility would allow C. albicans to accommodate extra chromosomes by adjusting its spindle geometry to the changing needs of a dynamic genome in a manner that does not occur in S. cerevisiae. Such genome plasticity is far from unique and has been seen in many other fungi. among the human pathogens, Candida glabrata undergoes rapid changes in karyotype during bloodstream infections46,47, and Cryptococcus neoformans, a basidiomycete, also exhibits aneuploidy48. In addition, supernumerary chromosomes (or dispensable ‘B’ chromosomes) have arisen in specific fungal isolates and seem to have been acquired by horizontal transmission (for examples, see REFS 49–51). In some cases, supernumerary chromosomes are associated with virulence factors and confer pathogenic properties on an otherwise benign fungus.
The ability of cancer cells and certain fungal pathogens to adapt to the aneuploid state is striking. On the basis of work in budding yeast, it is likely that this occurs at least partially through adaptive pressure on the MTs and MT motors that move chromosomes and nuclei. Thus, multimorphic fungi hold promise for studying the basic mechanisms that facilitate aneuploidy and, by implication, tumorigenesis and pathogenesis. It is becoming clear that MTs and MT motors are not passive shuttles of chromosomes and nuclei but instead respond to changes in the environment and to the genotypic alterations that arise as a consequence of changes in ploidy or, as we discuss below, when two different genotypes share a cell.
Dikaryons: a nuclear two-step
Although we have a clear mechanistic understanding of nuclear migration in mating and vegetative cells for a range of model ascomycetes and a few basidiomycetes with yeast forms, there is a paucity of information about the regulation of nuclear dynamics in many basidiomycete mushrooms. Dikaryons are cells in which two haploid nuclei, one from each parent cell, share a single cytoplasm for a period of time without undergoing nuclear fusion or meiosis52. The dikaryon stage dominates the life cycles of many basidiomycetes, such as mushrooms. By contrast, filamentous ascomycetes, such as aspergilli or Podospora anserina, produce transient dikaryons as part of their sexual cycle. Dikaryons also form following mating in S. cerevisiae karyogamy (kar) mutants53,54. The receipt of an unknown environmental signal ends the dikaryon stage and triggers rapid karyogamy, meiosis and fruiting-body development. In many systems, dikaryons emerge from a heterogeneous mixture of nuclei in the large syncytia that form when the parental mycelia fuse. The process of dikaryon formation involves nuclear migration and sorting of the nuclei by genotype to ensure that each dikaryon contains a balance of each parental genome (FIG. 3).
Figure 3. Overview of dikaryon formation.
a | Hyphae of many types of filamentous fungi can fuse regardless of mating type. b | If fusion occurs between mates with compatible mating loci (blue and pink nuclei), rapid nuclear migration and exchange between the two parental mycelia is initiated. It is unknown how microtubules (MTs) and MT-based motors are regulated during this process, but there is no migration if noncompatible mates fuse. c | The nuclei migrate until they reach the distal tip of a hypha and then pair with a nucleus from the other parent. In some cells, a specialized polarized cell structure forms, called a clamp, crozier or hook cell (depending on the species). Some dikaryons form without a clamp. The clamp cell is a side projection of the hypha, and one nucleus migrates up into this projection while the opposite mating-type nucleus remains in the hypha. Both nuclei divide synchronously. Owing to the placement of the septa and the subsequent fusion of the clamp cell back to the main hypha, one daughter nucleus from each mitosis ends up in each cell of the hyphal tube. d | This intricate process produces a dikaryotic hypha: it is compartmentalized such that two parental nuclei share the same cytosol. The dikaryon state can persist for extended periods and there can be exchange of genetic material between nuclei.
When hyphae of compatible mating types fuse, the nuclei rapidly migrate (for example, up to 2–3 mm per hour in Schizophyllum commune55 and a remarkable 4 cm per hour in Coprinellus congregatus56) to the distal reaches of the partner mycelium56,57. unlike yeast, filamentous fungi that form dikaryons do not use pheromones to recognize mates extracellularly, as the hyphae will fuse (a process termed anastomosis) regardless of mating type52. nuclei do not initiate migration when cell fusion is with the same mating type, so the rapid increase in motility is triggered by the coexistence of compatible nuclei, as defined by their mating-type loci52. Presumably, there are regulators of MT motors and MT organization that are controlled (directly or indirectly) by the mating-type loci and that lead to the induction of nuclear migration, but these regulators have not been identified.
On arrival at the hyphal tips, nuclei of opposite mating types pair through MT associations, but the basis for detecting the opposite mate for such pairing is unknown. One possibility is that the pheromones and pheromone receptors that are encoded by the mating-type locus are spatially limited to either the nuclear membrane58 or to the cell cortex surrounding the nuclei. In the filamentous ascomycete P. anserina, genetic evidence points to autonomous nuclear signals, such as nuclear-membrane markers, that sort the nuclei so that each dikaryon contains two different parental nuclei59. Remarkably, in multiparental matings, in which there are multiple competing genotypes in a single cell, the most genetically diverse nuclei will form dikaryon pairs60.
In some fungal species, nuclear pairing initiates the formation of a clamp or crozier cell; this is a specialized projection that connects two adjacent hyphal cells and facilitates the segregation of two daughter nuclei, one of each mating type, into distinct cellular compartments (FIG. 3). In species that form clamp cells, one nucleus divides in the clamp cell and the other divides in the main hypha, but the nuclei have spindles of different lengths, which helps to ensure sorting of the daughter nuclei into different cells. In Coprinopsis cinerea, the genotypes alternate positions along the hypha so that in one cell a nucleus with one mating type enters the clamp cell and in the adjacent cell the nucleus with the other mating type enters the clamp cell (FIG. 3). This produces a perfect alternating pattern of parental genotypes along the dikaryotic hypha61. However, clamp cell formation is not essential for stable and accurate dikaryon formation and is not seen in all species that can form dikaryons62. In species that do not form clamp cells, different spindle lengths, different spindle elongation rates or simply a small enough starting distance between the two nuclei ensure that the spindles overlap in anaphase, enabling a ‘two-step’ swap of different sister nuclei. This is seen in S. pombe dikaryons, which can be forced to form using conditional cell cycle mutants such that, after mitosis, the sister nuclei (that is, nuclei originating from the same mother nucleus) are systematically segregated to different cells63. Thus, spindle elongation-based sorting mechanisms occur in the absence of a clamp cell and during otherwise normal vegetative growth, even in a yeast63.
Dikaryons are neither true haploids nor true diploids, and this unique state of ploidy seems to lend them some adaptive power. Mutations can be more readily expressed phenotypically in dikaryons than in homokaryotic, haploid mycelia64. In laboratory selection experiments with S. commune64, the biparental nuclei in the dikaryon underwent co-adaptation as a result of compensatory changes. Some basidiomycetes form stable heterokaryons, in which multiple nuclei from each mating partner are maintained in a multinuclear compartment. Intriguingly, the balance of parental genotypes in the heterokaryons deviates from a strict 1/1 ratio depending on environmental conditions65. Work in Heterobasidion parviporum suggests that individual nuclei are under selection65. In addition, nuclei with different genotypes or different epigenetic states also differ in their rates of replication and migration, further adding to the intriguing connections between genotype, ploidy and nuclear migration that are seen in filamentous fungal cells65,66.
It has long been known that the nuclei in a dikaryon can communicate: exchange of genetic material and somatic recombination occurs between genotypes60,64. Interestingly, the exact position of the two nuclei in a dikaryon influences the specific genes that are expressed in these nuclei. For example, in S. commune dikaryons, when the nuclei are close together (<2 microns apart) a different set of hydrophobin-encoding genes is expressed than when the nuclei are further apart67. Thus, the precise positions of the biparental nuclei in the dikaryon can specify the transcriptional programme of the cell. Intriguingly, a similar proximity effect has been seen in S. pombe cells. The nuclei in induced multinucleate fission yeast are different sizes, depending on their proximity to other nuclei. This indicates that nuclei sharing a single cell may be able to locally sense and respond to the presence of the other nuclei in the same cell68.
The dikaryotic state ensures that an organism is poised for meiosis whenever the proper environmental cues are sensed and also confers functional and fitness benefits on the cell. Dikaryon formation requires the regulation of nuclear migration when there are multiple cohabiting genotypes. The variation in the length and alignment of spindles may be the basis of alternating genotypes, but it does not explain how opposite mates find each other and pair in large syncytia (for example, in mushrooms). There are still many open questions to be answered, including how nuclei recognize one another as ‘different’ in a common cytoplasm, and how this information is converted to the signals that regulate the migration, positioning and sorting of the genotypes into dikaryons. Studying these questions in model filamentous fungi will probably reveal new lines of communication between the genome and MT motor machinery.
Reasons to dance: harnessing nuclear migration
Nuclear movement in fungi is not only used in specialized sexual cells or to position nuclei relative to the plane of cell division, but it also contributes to other basic nuclear functions in vegetative cells. In N. crassa hyphae, chromatin structure responds to the direction of nuclear migration, thereby establishing intranuclear polarity and often mirroring the hyphal axis of polarity69. This suggests that chromatin organization either responds to the external MT-based forces that are encountered by a migrating nucleus or, conversely, influences the direction of nuclear migration. notably, polarization of histone localization and modification is also seen in migrating uninucleate mammalian cells in tissue culture, suggesting that signals coordinating nuclear or cell migration with chromatin organization may be evolutionarily conserved70.
U. maydis makes use of dynein-dependent nuclear migration for the mechanical process of tearing open the nuclear envelope71 (BOX 3). This basidiomycete undergoes ‘open’ mitosis71,72, meaning that the nuclear envelope breaks down with each cell cycle. This is unlike the closed mitosis in most ascomycetes, such as budding yeast, in which the nuclear envelope remains intact and mitosis occurs in the nucleus73, or the partially open mitosis in A. nidulans, in which the envelope remains intact but the nuclear pores open to allow non-selective transport74 (BOX 3). In animal cells, open mitosis is achieved by fragmenting the nuclear membrane and disassembling the nuclear lamina. By contrast, U. maydis uses the energy from MT motors to rip apart the nuclear membrane and enable cytoplasmic MTs to form the kinetochore attachments that are necessary for chromosome segregation71. Presumably, this is an evolutionary intermediate or at least an alternative solution to mixing the nucleoplasm and cytosol.
Nuclear oscillations might also protect genome integrity in budding and fission yeast. a dicentric S. cerevisiae strain undergoing chromosome breaks due to the attempted segregation of the two centromeres can be rescued by DNA repair mechanisms. These repair processes are accompanied by MT-dependent, dynein-independent nuclear oscillations75. Similarly, the prophase zygote in S. pombe, which exists for a short period following conjugation and before meiosis, undergoes dramatic chromosomal DNA oscillations, termed ‘horse tail’ movements76. These occur through dynein-dependent movement of astral MTs that are bound to SPBs which are, in turn, attached to telomeres from the time of conjugation to the end of meiosis I76. The movement has been proposed to assist in the alignment of homologous chromosomes before meiotic recombination77. Recent work presented a mechanism for these oscillations that may be applicable to nuclear migration in many systems. In an elegant marriage of in vivo microscopy and mathematical modelling, vogel et al. provide evidence for the collective self-organization of dynein motors, during horse tail movements, into an asymmetrical distribution on MTs. The distribution changes as a consequence of mechanical forces on the MTs and thus generates the oscillations78. Therefore, nuclear movements seem to facilitate other DNA transactions as well as chromosome segregation in these model yeasts.
Nuclear migration is also required for the assembly of an appressorium, which is a specialized extension of a germ tube that uses turgor pressure to penetrate rigid plant cell walls. In the filamentous ascomycete Magnaporthe grisea (rice blast fungus), the appressorium ultimately builds up to 8 MPa of turgor to cross the plant cuticle79. This process requires nuclear migration and the completion of one cycle of mitosis in a germ tube. Intriguingly, after the single mitosis, one nucleus migrates back into the spore body and, from there, triggers an autophagy programme that is essential for appressorium development. It has been proposed that checkpoints monitor the completion of mitosis and couple it to the autophagy program. This checkpoint signal may be ‘carried’ back to the spore by the migrating nucleus to trigger autophagy and thereby enable appressorium formation. Thus, nuclear migration might be used as a messenger system to ensure linear progression through a developmental programme that is necessary for plant infection.
The perpetual dance: the immortal strand
As early as 1968, evidence existed for intriguing positional differences between nuclei of different ages that shared the cytoplasm of A. nidulans vegetative hyphae. Rosenberger and Kessel incubated germinating conidia in radioactive adenine to label the DNA of the ‘founding’ nucleus in the spore80. They then analysed where the labelled nuclei were found along the hypha. after multiple rounds of mitosis with random segregation of chromatids, the radioactivity should have been dispersed between the nuclei. Instead, these early pulse–chase studies indicated that the two nuclei proximal to the hyphal tip contained the labelled ‘old’ DNA, and the more basal nuclei contained the ‘new’ DNA. To establish this gradient of genome age along a hypha, non-random sister chromatid segregation must be coupled with nuclear segregation that can position nuclei on the basis of the presence of the template strand.
A few years later, John Cairns proposed his ‘immortal strand’ hypothesis, in which non-random segregation of old and new DNA occurs to protect the ‘original’ genome in the asymmetric division of stem cells81. In self-renewing division cycles, the asymmetric distribution of chromosomes on the basis of DNA age enables the original or template genome (and, perhaps, any imprinting) to be preserved without the risks of replication errors. Interestingly, in Armillaria gallica, a wood rot honey mushroom that is clonal and can live for more than 1,000 years, there is extremely low sequence variation between nuclei that are separated by hundreds of metres in individuals found in nature82,83. Such stability in the sequence is puzzling and unexpected in nuclei that are physically far apart and presumably separated by many rounds of nuclear division. One mechanism to preserve the original genotype throughout the extensive mycelia is through a repository of stem-cell-like nuclei, which mitigate variation over time and space.
Recent evidence from mammalian stem cells supports the idea that self-renewing stem cells can retain their ‘template’ DNA. Experiments in both adult muscle stem cells (‘satellite’ cells) and mouse embryonic stem cells showed that non-random chromosome segregation occurs84,85. Thus, the most apical tip zone of some fungal hyphae potentially contains a self-renewing pool of nuclei that is selectively retained in this position. alternatively, in A. gallica there might be repositories of ‘stem nuclei’ throughout the mycelia. An exciting possibility is that the molecular mechanisms underlying this process may be conserved from fungi to mammalian stem cells. Although asymmetrical segregation is not widespread in eukaryotic cells (for example, it is not seen in S. cerevisiae86), many potential mechanisms resulting in asymmetrical chromatid segregation87 can be envisioned. Accordingly, this might be an ancient molecular process that is used in hyphal development. A. nidulans, A. gallica and perhaps other filamentous fungi can be powerful, genetically tractable models for identifying possible conserved mechanisms of selective template DNA maintenance and of transmission of template age into spatial coordinates in the cell.
Concluding remarks
The study of nuclear and chromosome dynamics in diverse fungal systems provides ample evidence that there is tremendous variety in how and why cells move nuclei. The nuclear ‘dances’ that are powered by MTs and MT motors ensure the clearance of the division plane and the segregation of euploid genomes. In addition, fungi exploit the MT-based migration of nuclei and individual chromosomes for diverse purposes that range from sorting nuclei by genotype or age to facilitating recombination and generating genome alterations such as aneuploidy. Nuclear movements respond to the genome content, or its expression, as well as to nuclear position. The diverse fungal models that we have discussed are powerful tools for studying the molecular mechanisms that underlie the generation of genetic diversity during mitotic and meiotic divisions, the maintenance of low levels of diversity in stem cells and the generation of polyploidy and aneuploidy in tumour cells.
Supplementary Material
Acknowledgments
We thank K. Finley for discussions and critical reading of the manuscript and S. Forsberg for information regarding Schizosaccharomyces pombe. J.B. is supported by the US National Institutes of Health (R01AI0624273, R01AI075096 and R01 DE14666). A.S.G. is supported by the National Science Foundation (MCB-0719126) and is the recipient of a Basil O'Connor award from the March of Dimes.
- Dikaryon
A hyphal cell in which two compatible nuclei are maintained without karyogamy (nuclear fusion). Ascomycete dikaryons can produce crozier cells, whereas basidiomycete dikaryons can produce clamp connections for the formation of the dikaryotic state.
- Aneuploid
A cell with an abnormal number of chromosomes; for example, in a diploid organism, the lack of one copy of a chromosome (monosomy) or the presence of an extra copy of a chromosome (triploidy).
- Microtubule-organizing centre
(MTOC). A structure that nucleates and often retains a connection to microtubules. MTOCs at the centrosome (or centriole or spindle pole body) organize the mitotic (and meiotic) spindle apparatus.
- Spindle pole body
In yeast cells, the microtubule-organizing centre that functions like a centrosome and is usually associated with the nuclear membrane for part or all of the cell cycle.
- Dynein
A minus-end-directed microtubule motor protein that transports cellular cargo along microtubules. In fungi, dynein is a key motor protein that is responsible for nuclear movement through interactions with astral microtubules.
- Cytoduction
The production of a cell with a mixed cytoplasm but only one of the two parental nuclei. In Saccharomyces cerevisiae, cytoduction is accomplished by mating cells with defects in nuclear fusion (karyogamy).
- Supernumerary chromosomes
Small extra chromosomes that are generally dispensable for normal cell functions but that, in some cases, are required for pathogenicity and thus are ‘conditionally dispensable’.
- Dicentric
A chromosome that has two functional centromeres. When they are tethered to opposite poles of the mitotic spindle the chromosome will break during mitosis.
Footnotes
Databases
Entrez Gene: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene msh2
Entrez Genome Project: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=genomeprj
Ashbya gossypii | Aspergillus nidulans | Candida albicans | Candida glabrata | Coprinopsis cinerea | Cryptococcus neoformans | Magnaporthe grisea | Neurospora crassa | Podospora anserina | Saccharomyces cerevisiae | Schizophyllum commune | Schizosaccharomyces pombe | Ustilago maydis
UniProtKB: http://www.uniprot.org
Bim1 | Bni1 | Bub2 | Bud6 | Chk1 | Dbf2 | Dyn1 | Fkh2 | Kar9 | Mec1 | Myo2 | Rad52
Further Information
Judith Berman's homepage:
http://www.cbs.umn.edu/labs/berman
Amy Gladfelter's homepage:
http://www.dartmouth.edu/∼gladfelterlab
Broad Institute C. cinerea genome project:
http://www.broadinstitute.org/annotation/genome/coprinus cinereus/GenomesIndex.html
See online article: S1 (movie)
References
- 1.Sawin KE, Tran PT. Cytoplasmic microtubule organization in fission yeast. Yeast. 2006;23:1001–1014. doi: 10.1002/yea.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petersen J, Heitz MJ, Hagan IM. Conjugation in S. pombe: identification of a microtubule-organising centre, a requirement for microtubules and a role for Mad2. Curr Biol. 1998;8:963–966. doi: 10.1016/s0960-9822(98)70397-5. [DOI] [PubMed] [Google Scholar]
- 3.Tran PT, Marsh L, Doye V, Inoue S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeh E, Skibbens R, Cheng J, Salmon E, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeh E, et al. Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol Biol Cell. 2000;11:3949–3961. doi: 10.1091/mbc.11.11.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huisman SM, Segal M. Cortical capture of microtubules and spindle polarity in budding yeast - where's the catch? J Cell Sci. 2005;118:463–471. doi: 10.1242/jcs.01650. [DOI] [PubMed] [Google Scholar]
- 7.Dotiwala F, Haase J, Arbel-Eden A, Bloom K, Haber JE. The yeast DNA damage checkpoint proteins control a cytoplasmic response to DNA damage. Proc Natl Acad Sci USA. 2007;104:11358–11363. doi: 10.1073/pnas.0609636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liakopoulos D, Kusch J, Grava S, Vogel J, Barral Y. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell. 2003;112:561–574. doi: 10.1016/s0092-8674(03)00119-3. [DOI] [PubMed] [Google Scholar]
- 9.Grava S, Schaerer F, Faty M, Philippsen P, Barral Y. Asymmetric recruitment of dynein to spindle poles and microtubules promotes proper spindle orientation in yeast. Dev Cell. 2006;10:425–439. doi: 10.1016/j.devcel.2006.02.018. [DOI] [PubMed] [Google Scholar]; Together with reference 8, this work demonstrates the asymmetrical localizations of Kar9 and the cytoplasmic dynein motor protein. Both localize to the SPB or to microtubules that are bound for the bud and serve to move that end of the spindle into the bud. Phosphorylation of these proteins is necessary for their asymmetrical localization.
- 10.Byers B, Goetsch L. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb Symp Quant Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- 11.Molk JN, Salmon ED, Bloom K. Nuclear congression is driven by cytoplasmic microtubule plus end interactions in S. cerevisiae. J Cell Biol. 2006;172:27–39. doi: 10.1083/jcb.200510032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spencer F, Simchen G. Transfer of YAC clones to new hosts by karyogamy-deficient mating. Curr Protoc Hum Genet. 2001;Chapter 5 doi: 10.1002/0471142905.hg0514s06. Unit 5.14. [DOI] [PubMed] [Google Scholar]
- 13.Wolkow TD, Harris SD, Hamer JE. Cytokinesis in Aspergillus nidulans is controlled by cell size, nuclear positioning and mitosis. J Cell Sci. 1996;109:2179–2188. doi: 10.1242/jcs.109.8.2179. [DOI] [PubMed] [Google Scholar]
- 14.Xiang X, Fischer R. Nuclear migration and positioning in filamentous fungi. Fungal Genet Biol. 2004;41:411–419. doi: 10.1016/j.fgb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 15.Requena N, et al. Genetic evidence for a microtubule-destabilizing effect of conventional kinesin and analysis of its consequences for the control of nuclear distribution in Aspergillus nidulans. Mol Microbiol. 2001;42:121–132. doi: 10.1046/j.1365-2958.2001.02609.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Li S, Fischer R, Xiang X. Accumulation of cytoplasmic dynein and dynactin at microtubule plus ends in Aspergillus nidulans is kinesin dependent. Mol Biol Cell. 2003;14:1479–1488. doi: 10.1091/mbc.E02-08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alberti-Segui C, Dietrich F, Altmann-Johl R, Hoepfner D, Philippsen P. Cytoplasmic dynein is required to oppose the force that moves nuclei towards the hyphal tip in the filamentous ascomycete Ashbya gossypii. J Cell Sci. 2001;114:975–986. doi: 10.1242/jcs.114.5.975. [DOI] [PubMed] [Google Scholar]
- 18.Plamann M, Minke PF, Tinsley JH, Bruno KS. Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J Cell Biol. 1994;127:139–149. doi: 10.1083/jcb.127.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiang X, Beckwith S, Morris N. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc Natl Acad Sci USA. 1994;91:2100–2104. doi: 10.1073/pnas.91.6.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fink G, Steinberg G. Dynein-dependent motility of microtubules and nucleation sites supports polarization of the tubulin array in the fungus Ustilago maydis. Mol Biol Cell. 2006;17:3242–3253. doi: 10.1091/mbc.E05-12-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berman J, Sudbery PE. Candida albicans: a molecular revolution built on lessons from budding yeast Nature Rev Genet. 2002;3:918–932. doi: 10.1038/nrg948. [DOI] [PubMed] [Google Scholar]
- 22.Berman J. Morphogenesis and cell cycle progression in Candida albicans. Curr Opin Microbiol. 2006;9:595–601. doi: 10.1016/j.mib.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Finley KR, Berman J. Microtubules in Candida albicans hyphae drive nuclear dynamics and connect cell cycle progression to morphogenesis. Eukaryot Cell. 2005;4:1697–1711. doi: 10.1128/EC.4.10.1697-1711.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finley KR, Bouchonville KJ, Quick A, Berman J. Dynein-dependent nuclear dynamics affect morphogenesis in Candida albicans by means of the Bub2p spindle checkpoint. J Cell Sci. 2008;121:466–476. doi: 10.1242/jcs.015172. [DOI] [PubMed] [Google Scholar]
- 26.Bensen ES, Filler SG, Berman J. A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot Cell. 2002;1:77–98. doi: 10.1128/EC.1.5.787-798.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez-Novo A, et al. Dbf2 is essential for cytokinesis and correct mitotic spindle formation in Candida albicans. Mol Microbiol. 2009;72:1364–1378. doi: 10.1111/j.1365-2958.2009.06729.x. [DOI] [PubMed] [Google Scholar]
- 28.Vogel SK, Raabe I, Dereli A, Maghelli N, Tolic-Norrelykke I. Interphase microtubules determine the initial alignment of the mitotic spindle. Curr Biol. 2007;17:438–444. doi: 10.1016/j.cub.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 29.Daga RR, Nurse P. Interphase microtubule bundles use global cell shape to guide spindle alignment in fission yeast. J Cell Sci. 2008;121:1973–1980. doi: 10.1242/jcs.011825. [DOI] [PubMed] [Google Scholar]
- 30.Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 31.Rieder CL, Maiato H. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev Cell. 2004;7:637–651. doi: 10.1016/j.devcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Andreassen PR, Lohez OD, Margolis RL. G2 and spindle assembly checkpoint adaptation, and tetraploidy arrest: implications for intrinsic and chemically induced genomic instability. Mutat Res. 2003;532:245–253. doi: 10.1016/j.mrfmmm.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 33.Storchova Z, et al. Genome-wide genetic analysis of polyploidy in yeast. Nature. 2006;443:541–547. doi: 10.1038/nature05178. [DOI] [PubMed] [Google Scholar]; A screen of the yeast deletion collection for triploid and tetraploid mutants that are defective for growth. Almost all of the mutations found are in genes that are involved in homologous recombination, sister chromatid cohesion or mitotic-spindle function. This study also shows that wild-type tetraploids have higher levels of genome instability than diploids and that some of this is due to an increased incidence of defects in the attachment of kinetochores to both spindle poles.
- 34.Gerstein AC, Chun HJ, Grant A, Otto SP. Genomic convergence toward diploidy in Saccharomyces cerevisiae. PLoS Genet. 2006;2:e145. doi: 10.1371/journal.pgen.0020145. [DOI] [PMC free article] [PubMed] [Google Scholar]; During experimental evolution of haploid, diploid and tetraploid cells for ∼1,800 doublings, all strains converged to diploidy regardless of the presence or absence of stress, supporting the idea that genome size evolution is constrained and, in S. cerevisiae, favours a diploid genome size.
- 35.Thompson DA, Desai MM, Murray PW. Ploidy controls the success of mutators and nature of mutations during budding yeast evolution. Curr Biol. 2006;16:1581–1590. doi: 10.1016/j.cub.2006.06.070. [DOI] [PubMed] [Google Scholar]; A comparison of wild-type and mismatch-repair-defective diploid and haploid strains grown under stress or non-stress conditions, which shows that diploid strains acquire mutations that allow them to grow well in several stress conditions. By contrast, haploids acquire more specific mutations for improved growth under only the selected condition. Thus, increased mutagenesis favours the adaptation of diploids relative to haploids.
- 36.Querol A, Bond U. The complex and dynamic genomes of industrial yeasts. FEMS Microbiol Lett. 2009;293:1–10. doi: 10.1111/j.1574-6968.2008.01480.x. [DOI] [PubMed] [Google Scholar]
- 37.Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 38.Williams BR, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 40.Hughes TR, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nature Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 41.Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selmecki A, Bergmann S, Berman J. Comparative genome hybridization reveals extensive aneuploidies in C. albicans laboratory strains. Mol Microbiol. 2005;55:1553–1565. doi: 10.1111/j.1365-2958.2005.04492.x. [DOI] [PubMed] [Google Scholar]
- 43.Chen X, Magee BB, Dawson D, Magee PT, Kumamoto CA. Chromosome 1 trisomy compromises the virulence of Candida albicans. Mol Microbiol. 2003;51:551–565. doi: 10.1046/j.1365-2958.2003.03852.x. [DOI] [PubMed] [Google Scholar]
- 44.Bouchonville K, Forche A, Tang KE, Selmecki A, Berman J. Aneuploid chromosomes are highly unstable during DNA transformation of Candida albicans. Eukaryot Cell. 2009;8:1554–1566. doi: 10.1128/EC.00209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selmecki AM, Gerami-Nejad M, Paulsen C, Forche A, Berman J. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol Microbiol. 2008;68:624–641. doi: 10.1111/j.1365-2958.2008.06176.x. [DOI] [PubMed] [Google Scholar]
- 46.Shin JH, et al. Changes in karyotype and azole susceptibility of sequential bloodstream isolates from patients with Candida glabrata candidemia. J Clin Microbiol. 2007;45:2385–2391. doi: 10.1128/JCM.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polakova S, et al. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc Natl Acad Sci USA. 2009;106:2688–2693. doi: 10.1073/pnas.0809793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu G, et al. Comparative hybridization reveals extensive genome variation in the AIDS-associated pathogen Cryptococcus neoformans. Genome Biol. 2008;9:R41. doi: 10.1186/gb-2008-9-2-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harimoto Y, et al. Expression profiles of genes encoded by the supernumerary chromosome controlling AM-toxin biosynthesis and pathogenicity in the apple pathotype of Alternaria alternata. Mol Plant Microbe Interact. 2007;20:1463–1476. doi: 10.1094/MPMI-20-12-1463. [DOI] [PubMed] [Google Scholar]
- 50.He C, Rusu AG, Poplawski AM, Irwin JAG, Manners JM. Transfer of a supernumerary chromosome between vegetatively incompatible biotypes of the fungus Colletotrichum gloeosporioides. Genetics. 1998;150:1459–1466. doi: 10.1093/genetics/150.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miao VP, Covert SF, VanEtten HD. A fungal gene for antibiotic resistance on a dispensable (“B”) chromosome. Science. 1991;254:1773–1776. doi: 10.1126/science.1763326. [DOI] [PubMed] [Google Scholar]
- 52.Brown AJ, Casselton LA. Mating in mushrooms: increasing the chances but prolonging the affair. Trends Genet. 2001;17:393–400. doi: 10.1016/s0168-9525(01)02343-5. [DOI] [PubMed] [Google Scholar]
- 53.Conde J, Fink GR. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci USA. 1976;73:3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kurihara LJ, Beh CT, Latterich M, Schekman R, Rose MD. Nuclear congression and membrane fusion: two distinct events in the yeast karyogamy pathway. J Cell Biol. 1994;126:911–923. doi: 10.1083/jcb.126.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]; This classic paper analyzes a series of kar mutants in S. cerevisiae and shows that they affect two distinct processes during nuclear fusion following mating. One class is required for the microtubule-mediated movement of nuclei so that they become juxtaposed. The second class mediates membrane interactions, resulting in nuclear fusion. Double-mutant analysis reveals that the nuclear movement step always precedes the membrane fusion step.
- 55.Niederpruem DJ. Direct studies of dikaryotization in Schizophyllum commune. I. Live inter-cellular nuclear migration patterns. Arch Microbiol. 1980;128:162–171. doi: 10.1007/BF00406154. [DOI] [PubMed] [Google Scholar]
- 56.Ross IK. Nuclear migration rates in Coprinus congregatus: a new record? Mycologia. 1976;68:418–422. [Google Scholar]
- 57.Raudaskoski M. The relationship between B-mating-type genes and nuclear migration in Schizophyllum commune. Fungal Genet Biol. 1998;24:207–227. doi: 10.1006/fgbi.1998.1069. [DOI] [PubMed] [Google Scholar]
- 58.Banuett F, Herskowitz I. Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc Natl Acad Sci USA. 1989;86:5878–5882. doi: 10.1073/pnas.86.15.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zickler D, Arnaise S, Coppin E, Debuchy R, Picard M. Altered mating-type identity in the fungus Podospora anserina leads to selfish nuclei, uniparental progeny, and haploid meiosis. Genetics. 1995;140:493–503. doi: 10.1093/genetics/140.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raper JR. Genetics of Sexuality in Higher Fungi. The Ronald Press; New York: 1966. [Google Scholar]
- 61.Iwasa M, Tanabe S, Kamada T. The two nuclei in the dikaryon of the homobasidiomycete Coprinus cinereus change position after each conjugate division. Fungal Genet Biol. 1998;23:110–116. doi: 10.1006/fgbi.1997.1019. [DOI] [PubMed] [Google Scholar]
- 62.Kues U. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol Mol Biol Rev. 2000;64:316–353. doi: 10.1128/mmbr.64.2.316-353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okazaki K, Niwa O. Dikaryotic cell division of the fission yeast Schizosaccharomyces pombe. Biosci Biotechnol Biochem. 2008;72:1531–1538. doi: 10.1271/bbb.80035. [DOI] [PubMed] [Google Scholar]
- 64.Clark TA, Anderson JB. Dikaryons of the basidiomycete fungus Schizophyllum commune: evolution in long-term culture. Genetics. 2004;167:1663–1675. doi: 10.1534/genetics.104.027235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.James TY, Stenlid J, Olson A, Johannesson H. Evolutionary significance of imbalanced nuclear ratios within heterokaryons of the basidiomycete fungus Heterobasidion parviporum. Evolution. 2008;62:2279–2296. doi: 10.1111/j.1558-5646.2008.00462.x. [DOI] [PubMed] [Google Scholar]
- 66.Raper CA. B-mating-type genes influence survival of nuclei separated from heterokaryons of Schizophyllum. Exp Mycol. 1985;9:149–160. [Google Scholar]
- 67.Schuurs TA, Dalstra HJ, Scheer JM, Wessels JG. Positioning of nuclei in the secondary mycelium of Schizophyllum commune in relation to differential gene expression. Fungal Genet Biol. 1998;23:150–161. doi: 10.1006/fgbi.1997.1028. [DOI] [PubMed] [Google Scholar]
- 68.Neumann FR, Nurse P. Nuclear size control in fission yeast. J Cell Biol. 2007;179:593–600. doi: 10.1083/jcb.200708054. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study addresses the mechanistic basis for control of nuclear size in S. pombe, demonstrating that the size of the nucleus is probably under cytosolic control and that the nucleocytoplasmic ratio is not directly controlled by the DNA content, as mutants with widely varying ploidy have a stable ratio.
- 69.Freitag M, Hickey PC, Raju NB, Selker EU, Read ND. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet Biol. 2004;41:897–910. doi: 10.1016/j.fgb.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 70.Gerlitz G, et al. Migration cues induce chromatin alterations. Traffic. 2007;8:1521–1529. doi: 10.1111/j.1600-0854.2007.00638.x. [DOI] [PubMed] [Google Scholar]
- 71.Straube A, Weber I, Steinberg G. A novel mechanism of nuclear envelope break-down in a fungus: nuclear migration strips off the envelope. EMBO J. 2005;24:1674–1685. doi: 10.1038/sj.emboj.7600644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Theisen U, Straube A, Steinberg G. Dynamic rearrangement of nucleoporins during fungal “open” mitosis. Mol Biol Cell. 2008;19:1230–1240. doi: 10.1091/mbc.E07-02-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Byers B, Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.De Souza CP, Osmani AH, Hashmi SB, Osmani SA. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr Biol. 2004;14:1973–1984. doi: 10.1016/j.cub.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 75.Thrower DA, Stemple J, Yeh E, Bloom K. Nuclear oscillations and nuclear filament formation accompany single-strand annealing repair of a dicentric chromosome in Saccharomyces cerevisiae. J Cell Sci. 2003;116:561–569. doi: 10.1242/jcs.00251. [DOI] [PubMed] [Google Scholar]
- 76.Yamamoto A, Tsutsumi C, Kojima H, Oiwa K, Hiraoka Y. Dynamic behavior of microtubules during dynein-dependent nuclear migrations of meiotic prophase in fission yeast. Mol Biol Cell. 2001;12:3933–3946. doi: 10.1091/mbc.12.12.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ding DQ, Chikashige Y, Haraguchi T, Hiraoka Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J Cell Sci. 1998;111:701–712. doi: 10.1242/jcs.111.6.701. [DOI] [PubMed] [Google Scholar]
- 78.Vogel SK, Pavin N, Maghelli N, Julicher F, Tolic-Norrelykke IM. Self-organization of dynein motors generates meiotic nuclear oscillations. PLoS Biol. 2009;7:e1000087. doi: 10.1371/journal.pbio.1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work combines live-cell imaging, laser ablation and mathematical modelling to build a model for the generation of nuclear oscillations during S. pombe meiosis. Dynein is shown to self-organize in a dynamic pattern, alternating between microtubules that radiate in opposite directions from the SPB.
- 79.Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science. 2006;312:580–583. doi: 10.1126/science.1124550. [DOI] [PubMed] [Google Scholar]
- 80.Rosenberger RF, Kessel M. Nonrandom sister chromatid segregation and nuclear migration in hyphae of Aspergillus nidulans. J Bacteriol. 1968;96:1208–1213. doi: 10.1128/jb.96.4.1208-1213.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 82.Smith ML, Bruhn JN, Anderson JB. The fungus Armillaria bulbosa is among the largest and oldest living organisms. Nature. 1992;356:428–431. [Google Scholar]
- 83.Hodnett B, Anderson JB. Genomic stability of two individuals of Armillaria gallica. Mycologia. 2000;92:894–899. [Google Scholar]
- 84.Conboy MJ, Karasov AO, Rando TA. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 2007;5:e102. doi: 10.1371/journal.pbio.0050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shinin V, Gayraud-Morel B, Tajbakhsh S. Template DNA-strand co-segregation and asymmetric cell division in skeletal muscle stem cells. Methods Mol Biol. 2009;482:295–317. doi: 10.1007/978-1-59745-060-7_19. [DOI] [PubMed] [Google Scholar]
- 86.Neff MW, Burke DJ. Random segregation of chromatids at mitosis in Saccharomyces cerevisiae. Genetics. 1991;127:463–473. doi: 10.1093/genetics/127.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lew DJ, Burke DJ, Dutta A. The immortal strand hypothesis: how could it work? Cell. 2008;133:21–23. doi: 10.1016/j.cell.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 88.Hazan I, Sepulveda-Becerra M, Liu H. Hyphal elongation is regulated independently of cell cycle in Candida albicans. Mol Biol Cell. 2002;13:134–145. doi: 10.1091/mbc.01-03-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Steinberg G. Tracks for traffic: microtubules in the plant pathogen Ustilago maydis. New Phytol. 2007;174:721–733. doi: 10.1111/j.1469-8137.2007.02072.x. [DOI] [PubMed] [Google Scholar]
- 90.Su W, Li S, Oakley BR, Xiang X. Dual color imaging of nuclear division and mitotic spindle elongation in live cells of Aspergillus nidulans. Eukaryot Cell. 2004;3:553–556. doi: 10.1128/EC.3.2.553-556.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oakley BR. Tubulins in Aspergillus nidulans. Fungal Genet Biol. 2004;41:420–427. doi: 10.1016/j.fgb.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 92.Horio T, Oakley BR. The role of microtubules in rapid hyphal tip growth of Aspergillus nidulans. Mol Biol Cell. 2005;16:918–926. doi: 10.1091/mbc.E04-09-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ukil L, De Souza CP, Liu HL, Osmani SA. Nucleolar separation from chromosomes during Aspergillus nidulans mitosis can occur without spindle forces. Mol Biol Cell. 2009;20:2132–2145. doi: 10.1091/mbc.E08-10-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper studies the fate of nucleolar components during semi-closed mitosis in A. nidulans, finding that nucleolar proteins that are important for ribosome assembly are extruded from the nucleus into a novel cytoplasmic structure that remains at the site of the metaphase plate. Disruption of spindle assembly blocks this process in wild-type cells but not in checkpoint-deficient mutants, suggesting that the segregation of nuclei and nucleoli is coupled by cell cycle checkpoints.
- 94.Alberti-Segui C, Dietrich F, Altmann-Johl R, Hoepfner D, Philippsen P. Cytoplasmic dynein is required to oppose the force that moves nuclei towards the hyphal tip in the filamentous ascomycete Ashbya gossypii. J Cell Sci. 2001;114:975–986. doi: 10.1242/jcs.114.5.975. [DOI] [PubMed] [Google Scholar]
- 95.Gladfelter AS, Hungerbuehler AK, Philippsen P. Asynchronous nuclear division cycles in multinucleated cells. J Cell Biol. 2006;172:347–362. doi: 10.1083/jcb.200507003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goffeau A, et al. Life with 6000 genes. Science. 1996;274:563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 97.Jones T, et al. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci USA. 2004;101:7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wood V, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 99.Galagan JE, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 100.Dietrich FS, et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- 101.Kamper J, et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature. 2006;444:97–101. doi: 10.1038/nature05248. [DOI] [PubMed] [Google Scholar]
- 102.Espagne E, et al. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 2008;9:R77. doi: 10.1186/gb-2008-9-5-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dean RA, et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434:980–986. doi: 10.1038/nature03449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.