Abstract
The high-throughput accurate mass and time (AMT) tag proteomic approach was utilized to characterize the proteomes for cytoplasm, cytoplasmic membrane, periplasm, and outer membrane fractions from aerobic and photosynthetic cultures of the gram-nagtive bacterium Rhodobacter sphaeroides 2.4.1. In addition, we analyzed the proteins within purified chromatophore fractions that house the photosynthetic apparatus from photosynthetically grown cells. In total, 8300 peptides were identified with high confidence from at least one subcellular fraction from either cell culture. These peptides were derived from 1514 genes or 35% percent of proteins predicted to be encoded by the genome. A significant number of these proteins were detected within a single subcellular fraction and their localization was compared to in silico predictions. However, the majority of proteins were observed in multiple subcellular fractions, and the most likely subcellular localization for these proteins was investigated using a Z-score analysis of estimated protein abundance along with clustering techniques. Good (81%) agreement was observed between the experimental results and in silico predictions. The AMT tag approach provides localization evidence for those proteins that have no predicted localization information, those annotated as putative proteins, and/or for those proteins annotated as hypothetical and conserved hypothetical.
Keywords: Rhodobacter sphaeroides, comparative proteomics, Fourier transform ion cyclotron resonance mass spectrometry (FTICR MS), localization
Introduction
Developing a systems-level understanding of the physiology of a microorganism such as Rhodobacter sphaeroides, a α-3-purple nonsulfur eubacterium 1,2 of significant environmental importance,3–6 requires not only knowledge about the identity and relative abundance of proteins in the bacterium, but also knowledge about the subcellular localization of these proteins. Traditional methods for protein localization are often based on generating hybrid proteins, such as green fluorescent protein fusions,7 Western blots and two-dimensional polyacrylamide gel electrophoresis (2-D PAGE) on individual subcellular fractions, as well as predictions from in silico analyses of annotated gene products. With the exception of the in silico methods, these technologies are often limited to the analysis of proteins present in relatively high abundances, and often require either specific hypotheses or some prior knowledge of localization. Additionally, protein extracts from individual subcellular fractions are often difficult to obtain due to sample quantity limitations and purity issues, which ultimately impacts the localization information that can be obtained from these methods.
Herein, we report the results of a localization study that utilized the accurate mass and time (AMT) tag approach8–10 to investigate 1514 proteins associated with subcellular fractions obtained from aerobic and photosynthetic cell cultures of R. sphaeroides strain 2.4.1. The demonstrated efficiency of the AMT tag approach in terms of sample utilization, high-throughput capability, sensitivity, and large number of identified proteins make it well-suited for subcellular proteome studies of bacteria and other cells.10 We identified peptides in cytoplasm, cytoplasmic membrane, periplasm, and outer membrane subcellular fractions from aerobic and photosynthetic cells, as well as in the chromatophore fraction that houses the photosynthetic apparatus of this facultative photosynthetic bacterium. Localization for many of these proteins had not been observed beyond that implied by prediction algorithms and functional annotation.
Experimental Section
Culture Conditions
Isolated batch cultures of R. sphaeroides 2.4.1 (ATCC# BAA-808) were grown in Sistrom’s minimal medium that contained succinate prepared according to established protocols.11 Batch cultures were sparged with 30% O2, 69% N2, and 1% CO2 using a Linde RM4575 Mass Flowmeter/Flowcontroller (ASGE, Middlesex, NJ) to create an environment suitable for aerobic growth, and with 95% N2 and 5% CO2 under white light (3 W/m2) to create an environment suitable for photosynthetic cell development. Cell growth was monitored turbidimetrically with a Klett–Summerson colorimeter (Klett Manufacturing Co., New York, NY) that was equipped with a No. 66 filter. Cells were harvested when cell densities reached ~2 × 108 cfu/mL to prevent O2 limitation in aerobic cultures and to minimize shading of the photosynthetic cultures. Harvested cells were placed on ice and then pelleted by centrifugation at 5000 rpm for 20 min at 4 °C. After decanting, cell pellets were immediately frozen in liquid nitrogen and stored at −80 °C.
Cell Fractionation
Aerobic and photosynthetic cell pellets collected from 2 to 3 biological replicates were suspended in warm (37 °C) Tris (100 mM, pH 8.0) buffer that contained 20% sucrose and gently agitated in a water bath set at 37 °C. The lysozyme-EDTA method12 as adapted for R. sphaeroides13 was used to obtain subcellular fractionations of the cytoplasm, cytoplasmic membrane, periplasm, and outer membrane. Purity of subcellular fractions was determined by measuring glutathione dependent formaldehyde dehydrogenase (GSH–FDH) activity and pyridine nucleotide transhydrogenase activity, which have been previously reported as marker proteins for the cytoplasm14 and cytoplasmic membrane,15 respectively. Antibody detection of cytochrome c554 was monitored as a marker protein for the periplasm,16 while the major outer membrane protein was monitored as a marker for the outer membrane.17
Protein Extraction and Digestion
Proteins from the cytoplasm and periplasm subcellular fractions were denatured and reduced by adding urea and thiourea to final concentrations of 7 and 2 M, respectively. Dithiothreitol (DTT, 50 mM prepared fresh) was added to a final concentration of 5 mM, and the samples were incubated at 60 °C for 30 min. Following incubation, each fraction was diluted 10-fold with 100 mM NH4-HCO3 (pH 8.4) to reduce the salt concentration. A volume of 1 M CaCl2 was added to each diluted fraction to a final concentration of 1 mM, and the protein samples were digested at 37 °C for 4 h by using sequencing grade trypsin (Roche, Indianapolis, IN) at a ratio of 1 unit per 50 units of protein (1 unit = ~1 µg of protein). Following incubation, the digested cytoplasm and periplasm fractions were desalted by loading each sample onto an appropriately sized C-18 SPE column (Supelco, St. Louis, MO). Three column volumes of methanol were passed through the column, followed by 2 column volumes of Nanopure water to remove the salt. After the sample passed through the column, the column was washed by 4 column volumes of a 95% acetonitrile (ACN), 0.1% trifluoro-acetic acid (TFA) solution. Peptides were eluted by passing 1 column volume of 80% ACN, 0.1% TFA solution through the column. Peptides were collected and concentrated to a final volume that ranged from 50 to 100 µL. Peptide concentrations were measured by using the BCA assay (Pierce Chemical Co., Rockfort, IL) according to the manufacturer’s instructions.
Proteins from the outer and cytoplasmic membrane fractions were centrifuged at 4 °C and 100 000 rpm for 10 min. The resulting supernatant was decanted and each pellet was washed by using mild sonication to suspend the pellet in 100 mM NH4-HCO3 (pH 7.8) and then centrifuged at 100 000 rpm for 5 min at 4 °C. Following centrifugation, pellets were suspended in a solubilization solution of 7 M urea, 2 M thiourea, and 1% CHAPS in 50 mM NH4HCO3 (pH 7.8), and a volume of 194 mM DTT solution (prepared fresh) was added to a final sample concentration of 9.7 mM DTT. Pellets from the outer membrane and cytoplasmic membrane fractions were treated with trypsin as described above, with the exception that a 50 mM NH4HCO3 (pH 7.8) buffer was used for the 10-fold dilution. Following digestion, the pH of each digest was slowly lowered to just below 4.0 by adding small volumes (1 to 2 µL) of 20% formic acid. Removal of salts and detergent was performed with a 1 mL SCX SPE column (Supelco, St. Louis, MO) and vacuum manifold by first passing three column volumes of methanol through the column. The column was conditioned by a series of rinses (6 column volumes each) of the following three solutions: 10 mM ammonium formate in 25% ACN, pH 3.0 (solution 1), 500 mM ammonium formate in 25% ACN, pH 6.8 (solution 2), and Nanopure water. After passing the acidified sample through the column, the column was washed with 10 column volumes of solution 1, and peptides were eluted by using 1 column volume of solution 2. Peptides were concentrated, and their concentration measured as described above.
Sample Analysis by LC–MS
The reference database for R. sphaeroides was generated as previously described.10,18,19 Briefly, tryptic digestions of readily obtained R. sphaeroides samples were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Peptides were identified from the resulting spectra by using the SEQUEST algorithm20 and filtered using criteria established by Washburn et al.21 The database contained ~24 437 unique peptide sequences along with the theoretical mass and observed normalized elution time (NET) associated with each mass measurement. These mass and time tags served as two-dimensional markers for identifying peptides in subsequent high throughput LC–MS analyses of R. sphaeroides.
Triplicate samples from each subcellular fraction were consecutively analyzed by capillary LC coupled to a 9.4 T (Bruker Daltonics) Fourier transform ion cyclotron resonance (FTICR) mass spectrometer.8,22 Separations on the LC system were achieved by using 5000 psi reversed-phase packed capillaries (150 µm i.d. × 360 µm o.d.; Polymicro Technologies, AZ) and the following two mobile phase solvents: 0.2% acetic acid and 0.05% TFA in water (A) and 0.1% TFA in 90% acetonitrile/10% water (B). Flow through the capillary high-pressure LC column was ~1.8 µL/min when equilibrated to 100% mobile phase A. The eluent from the LC column was infused into the FTICR by an electrospray ionization source. This source was interfaced to an electrodynamic ion funnel assembly that was coupled to a radio frequency quadrupole for collisional ion focusing and highly efficient ion accumulation prior to ion transport to the cylindrical ICR cell for analysis. Mass spectra were acquired with approximately 105 resolution and analyzed by using ICR-2LS software.10
Data Analysis
The reference peptide database was filtered to remove all peptides that had discriminant scores (i.e., identification confidence scores)23 <0.9 prior to their use to match the mass and elution time features detected by LC–FTICR to the mass and elution time information on these peptides within the database. During the matching stage, a spatially localized confidence score was calculated24,25 for each peptide detected by LC-FTICR, and peptides with spatially localized confidence scores <0.7 were also excluded. Measured arbitrary abundances of peptides were determined by integrating the areas under each LC–FTICR peak for the detected peptide.10 Protein abundances were estimated by averaging the abundances of multiple peptides for a given protein.19,26 Z-scores were first calculated for each protein19 to compare these estimated abundances across subcellular fractions within the aerobic and photosynthetic cell cultures, and then were clustered by using hierarchical clustering algorithms available in OmniViz analysis software. Because bias can be introduced when protein abundances are estimated by averaging multiple peptides for a protein detected in multiple subcellular fractions (e.g., more than one peptide in one fraction and only one peptide in another fraction), the minimum calculated Z-score for a given subcellular fraction was assigned to proteins identified by only peptide within that fraction.
Results
Peptide Quality and Confidence of Protein Identifications
Approximately 8300 high quality peptides were detected from 54 LC–FTICR–MS analyses; each of these analyses were comprised of 2 to 3 biological replicates per subcellular fraction from both aerobic and photosynthetic cell cultures. All peptide identifications were within a mass measurement accuracy of 6 ppm and a normalized LC elution time of 0.025 (less than ±2.5% difference between previously observed elution time and measured elution time in an LC–FTICR analysis). Additionally, all identified peptides had discriminant scores of at least 0.9 (maximum of 1.0), and spatially localized confidence scores (SliC) of at least 0.7 (maximum of 0.99). This latter quality score represents a measure of uniqueness among the peptides detected by LC–FTICR (within a mass measurement accuracy and normalized LC elution time windows24,25) that match multiple peptide sequences in the reference database. The discriminant score,23 which is derived from 7 measurements determined by LC-MS/MS and SEQUEST analyses, represents a measure of quality for a peptide sequence in the reference database.23,27 By filtering the database on a discriminant threshold of 0.9, the number of false positive identifications included in the reference database (used for matching LC–FTICR features) is reduced.19,23,28
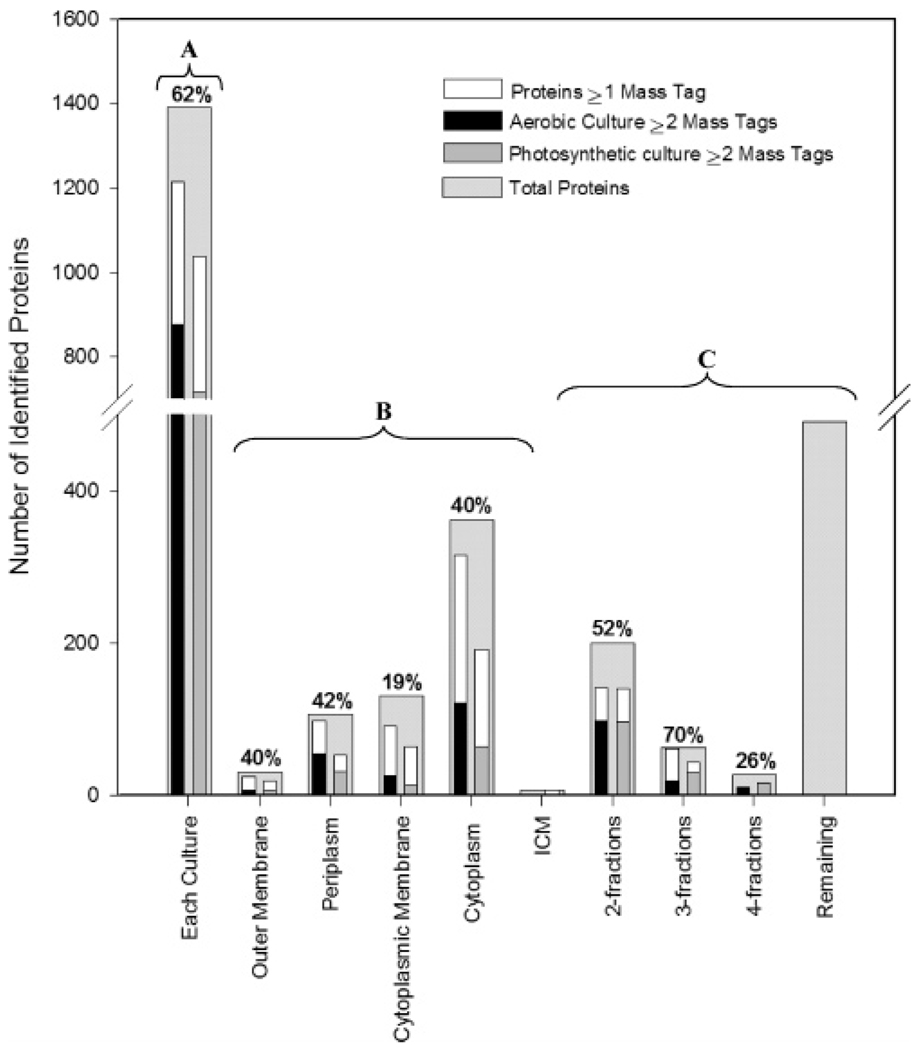
The ~8300 high quality peptide identifications corresponded to 1748 proteins and represented 41% coverage of the 4269 proteins predicted to be encoded by the genome (http://genome.jgi-psf.org/draft_microbes/rhosp/rhosp.home.html) and 57% of the genes for which RNA is detected by whole genome expression analysis in these (or similar) cultures.29 When reproducibility was factored in (i.e., the protein had to be observed in at least 2 of the 3 biological replicates per cell culture and in at least 4 of the 6 instrument analyses), the total number of observed proteins was reduced to 1514, which corresponded to 35% coverage of the predicted proteins. Additionally, proteins identified by more than a single peptide within a subcellular fraction were viewed with higher confidence than proteins identified by a single peptide, even though the peptide was detected in at least 4 of 6 instrument analyses (Figure 1).
Figure 1.
Number of proteins identified from application of the AMT approach for (A) all proteins detected in the aerobic and photosynthetic cell cultures, (B) proteins solely detected within a subcellular fraction, and (C) proteins detected in multiple subcellular fractions. Percentages at the top of each bar correspond to proteins identified in at least 4 of 6 instrument analyses by at least one high quality peptide and observed in both the aerobic and photosynthetic cell cultures.
We observed a large difference in the number of proteins detected in each subcellular fraction and between cell cultures. In Figure 1, the 1514 proteins are subdivided by the following: (A) culture condition, (B) subcellular fraction, i.e., proteins that were observed solely within a single subcellular fraction, and (C) multiple subcellular fractions, i.e., proteins were consistently observed across more than one subcellular fraction. Additionally, a number of proteins (~460) present in both cell cultures were detected in 2 or more subcellular fractions in either one or both cultures, possibly due to sample contamination, to association of soluble proteins with membrane samples, and/or to entrapment of periplasmic proteins in vesicles when cytoplasmic membrane or chromatophore samples were generated. Sixty-two percent (62%) of the total number of identified proteins were common among the two cell cultures (Figure 1A), of which 57% of these were identified by 2 or more peptides. This finding suggests that a large number of proteins were expressed at detectable levels under both respiratory and photosynthetic conditions. This overlap varied with each subcellular fraction.
For the cytoplasmic membrane, only 19% of proteins were present in both cell cultures (12% when only proteins identified by 2 or more peptides were considered). Such a small overlap for this fraction reflects the different proteins utilized by R. sphaeroides for energy gathering and conversion purposes related to aerobic respiration and photosynthesis. For example, 2 annotated H+-transporting ATPase subunits (RSP3929 and RSP7012) encoded on plasmid a were uniquely detected in the photosynthetic cell culture. RSP3929 is part of a gene cluster (RSP3929–2936) that encodes a F0F1 type ATP synthase that is upregulated (transcriptome) under anoxic dark conditions.30 Many energy-related proteins, particularly those involved in light gathering and electron transport, are contained within the intracytoplasmic membrane (chromatophore fraction) that develops under low oxygen tension conditions in both the presence and absence of light.31 Of the 214 total proteins detected in the chromatophore fraction, 101 had more than one identifying peptide, and all 214 proteins were observed in additional subcellular fractions, with the highest number of proteins common to the cytoplasmic (96 proteins) and outer membrane (80-proteins) fractions.
Proteins Identified Solely within a Single Subcellular Fraction
Overall, the predicted localization (PSORTB algorithms32) of proteins identified solely within a subcellular fraction was in 81% agreement with the localization determined experimentally by proteome analysis. However, agreement varied according to subcellular fractions. For the outer membrane, agreement was 100% for proteins identified by 2 or more peptides (10 proteins) and was 71% for the periplasm. Agreement generally improved for proteins that were detected in both cell cultures versus proteins detected in only a single culture. We observed 98% agreement between predicted and observed localizations for proteins detected solely in the cytoplasm in both cell cultures; whereas, the agreement was 94% for proteins detected in the cytoplasm and in only one cell culture, i.e., either the aerobic or the photosynthetic. No agreement between experimentally determined and predicted localizations was observed for proteins within the chromatophore since this subcellular fraction is not in the prediction algorithms.
While PSORTB32 was unable to predict localizations for 241 detected proteins, proteome analysis was able to detect most of these proteins in the cytoplasm (124 proteins), the periplasm (54 proteins), cytoplasmic membrane (42 proteins), and outer membrane (21 proteins). An implied localization can be inferred for many of these proteins from predicted annotations or functional assignments. However, 89 of the 241 proteins had no functional assignment, had only a general function prediction,33 or were annotated as hypothetical/conserved hypothetical. Table 1 provides a subset of the 241 proteins that are either annotated as conserved hypothetical/hypothetical or assigned an unknown or general function of R. sphaeroides. The cell culture in which the protein was detected also is provided in this table, along with the observed subcellular fraction and the number of unique identifying peptides per protein. Note, these proteins were identified by at least 2 high quality peptides per analysis in at least 4 of the 6 instrument analyses.
Table 1.
Subset of 241 Proteins Observed within a Single Sub-cellular Fraction, Where Predicted Localization Could Not Be Assigneda
| ORF ID | cell culture |
unique peptides |
percent coverage |
description |
|---|---|---|---|---|
| Cytoplasm | ||||
| RSP0205 | A&P | 3 | 3 | hypothetical protein |
| RSP0380 | A&P | 6 | 49 | conserved hypothetical protein |
| RSP0924 | A&P | 3 | 27 | predicted nucleotide-utilizing enzyme/ competence-damage associated protein |
| RSP1940 | A | 4 | 31 | conserved hypothetical protein |
| RSP2350 | A | 3 | 39 | hypothetical protein |
| RSP2728 | A&P | 3 | 34 | phospholipase/carboxylesterase |
| RSP3069 | A&P | 3 | 30 | putative oxidoreductase protein/putative flavoprotein or flavodoxin/FMN reductase |
| RSP3419 | A | 3 | 24 | conserved hypothetical protein |
| RSP3919 | A&P | 3 | 17 | conserved hypothetical protein |
| Cytoplasmic Membrane | ||||
| RSP3425 | A&P | 3 | 24 | conserved hypothetical protein |
| Periplasm | ||||
| RSP0086 | A&P | 7 | 52 | conserved hypothetical protein |
| RSP0345 | A | 6 | 30 | possible ABC transporter, periplasmic/outer membrane lipoprotein binding protein |
| RSP0600 | A&P | 4 | 33 | hypothetical protein |
| RSP1107 | A&P | 3 | 22 | hypothetical protein |
| RSP1544 | A&P | 6 | 26 | hypothetical signal peptide protein |
| RSP1807 | A&P | 5 | 37 | conserved hypothetical protein |
| RSP1844 | A&P | 3 | 42 | hypothetical protein |
| RSP2309 | A | 4 | 26 | conserved hypothetical protein |
| RSP2583 | A | 3 | 10 | conserved hypothetical protein |
| RSP3306 | P | 3 | 19 | hypothetical protein |
| RSP3923 | A&P | 10 | 34 | conserved hypothetical protein |
| Outer Membrane | ||||
| RSP0334 | A&P | 7 | 50 | hypothetical protein |
| RSP1496 | A&P | 2 | 16 | conserved hypothetical protein |
| RSP1697 | A | 2 | 18 | ypothetical protein |
| RSP2271 | A&P | 3 | 16 | hypothetical protein |
| RSP3594 | P | 3 | 22 | antifreeze protein, type I |
| RSP6127 | A | 2 | 13 | hypothetical protein |
These proteins were identified by a minimum of 2 high-quality peptides and are annotated as putative, or hypothetical. The subcellular fraction at the head of each group indicates the observed subcellular fraction. A = Aerobic cell culture; P = Photosynthetic cell culture, A&P = Both the aerobic and photosynthetic cell cultures.
Among the proteins listed in Table 1, several were observed to have a large percent coverage of amino acid sequences (that ranged from 42% to 52%), including RSP0334 that was detected in the outer membrane (50% coverage of 223 residues) and annotated as hypothetical, and RSP0086 that was observed in the periplasm (52% coverage of 175 residues) and annotated as conserved hypothetical. The large coverage is due in part to the number of residues, but also is due to their detection in both cell cultures. We observed several proteins where the percent coverage of amino acid sequence was significant (>20%), but were only detected within one cell culture. Further evaluation as to whether these proteins are indeed unique to the observed cell culture needs to be undertaken. Nevertheless, detection of these proteins, which were previously annotated as hypothetical/conserved hypothetical or categorized as having an unknown function or unknown predicted localization, confirms their presence from a functional proteomic standpoint and adds significant insight into their localization within the cellular matrix.
Proteins Identified in Two or More Subcellular Fractions
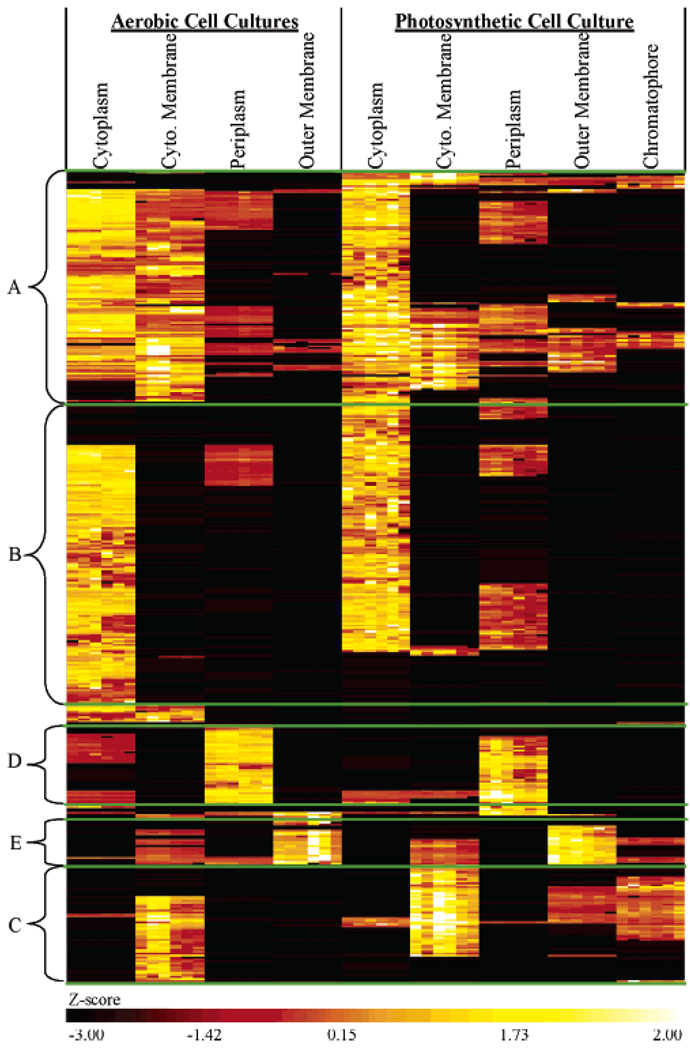
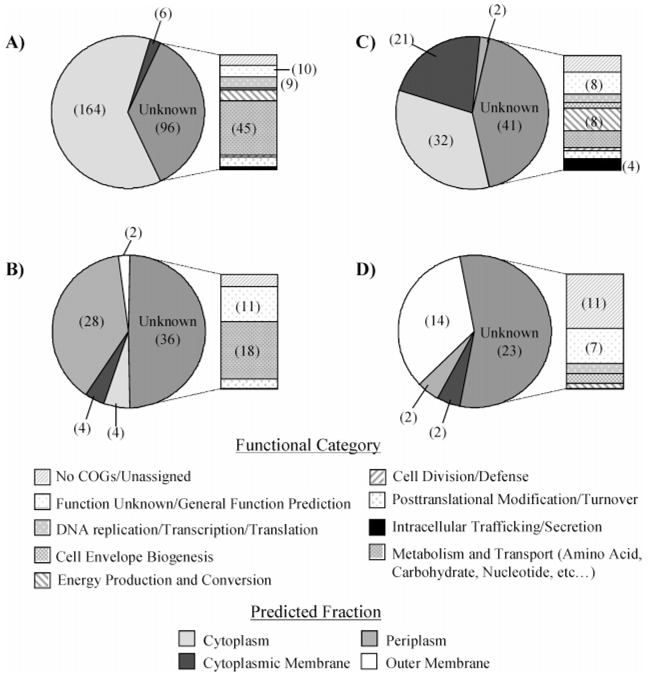
Almost half of the proteins identified by at least 2 peptides in one subcellular fraction were identified in more than one fraction (Figure 1). For these proteins, we wanted to know whether the primary fraction of localization could be inferred from a comparison of calculated Z-scores19,26 that were obtained from protein abundances measured by LC-FTICR. Clustering on the basis of Z-scores grouped 476 out of 672 proteins into a specific fraction (Figure 2, B–E); the remaining proteins could not be resolved to any particular fraction (Figure 2A). The differences observed in Z-scores for the largest group of proteins (266) suggested the cytoplasm fraction as the most likely localization (Figure 2B). Most of these 266 proteins were observed in the cytoplasm of both aerobic and photosynthetic cell cultures. An in silico analysis (PSORTB) of proteins observed within the cytoplasm grouping showed 96% agreement. Nevertheless, a significant number of these 266 proteins (96) had an unknown predicted localization (Figure 3A). Categorization of these proteins according to Clusters of Orthologous Groups (COGS)33 revealed a majority were placed into categories associated with activities that take place in the cytoplasm (Figure 3A), such as metabolism of substrates (amino acids, carbohydrates, nucleic acids, etc.), DNA replication, transcription, translation, cell division, and post-translational modification.
Figure 2.
Heat map of Z-scores clustered using hierarchical algorithms used to identify the likely fraction of localization for proteins detected in multiple fractions. (A) the primary fraction of localization could not be resolved, (B) cytoplasm, (C) cytoplasmic membrane, (D) periplasm, and (E) outer membrane.
Figure 3.
Number of proteins detected in multiple subcellular fractions where the most likely fraction of localization was observed for (A) cytoplasm, (B) cytoplasmic membrane, (C) periplasm, and (D) outer membrane. Pie charts breakdown these proteins by predicted localization utilizing PSORTB algorithms. Bars represent COGs functional categories for proteins with an unknown predicted localization.
The next largest cluster of proteins (96) corresponded to the cytoplasmic membrane (Figure 2C). Here, in silico observations were only in 38% agreement; a larger number of proteins were predicted to localize to the cytoplasm than to the cytoplasmic membrane (Figure 3B). However, almost half of the 96 proteins were assigned an unknown localization that spanned a number of COGs functional categories. Many of the proteins within these categories are known to localize to the cytoplasmic membrane. For example, RSP0693 (annotated as a CcoP subunit of cbb3-type c cytochrome oxidase), RSP1035, and RSP1036 (both annotated as FoF1 ATP synthase, subunit B) are involved in energy production and conversion, and localize to the cytoplasmic membrane. The four proteins assigned to functions related to intracellular trafficking and secretion localize to the cytoplasmic membrane (based on assigned annotations), including 2 twin-arginine translocation proteins, RSP2540 (TatA) and RSP6059 (TatB),34 and a putative secindependent translocation protein RSP3102 (TatE). Although agreement with in silico results were poor, the annotations associated with proteins that have an unknown predicted localization suggested that our observed localization based on the AMT approach and Z-score analysis was in better agreement than those suggested by localization algorithms for this subcellular fraction.
We observed a cluster of 73 proteins whose primary subcellular localization was the periplasm (Figure 2D) in both the aerobic and photosynthetic cell cultures. An in silico analysis of these proteins was in 76% agreement, with only 10 proteins predicted to localize elsewhere (Figure 3C). However, as with the other fractions, almost 50% of these proteins had an unknown predicted localization. COGs analysis of these unknown proteins revealed a large number (18 proteins) categorized as proteins involved in metabolism and transport functions. Of these proteins, 13 were annotated as substrate binding periplasm proteins that are involved in transport, which indicates that their grouping with the other proteins that were predicted to localize to the periplasm was correct. An additional 17 proteins with no COGs information or unknown function were annotated as putative, hypothetical/conserved hypothetical.
Observed and in silico results for the 41 proteins grouped in the outer membrane (Figure 2E) were in 78% agreement for 18 of the 41 proteins; the remaining proteins had no predicted (unknown) localization (Figure 3D). The majority of these unknown proteins either could not be placed into a COGs functional category or were categorized as having an unknown function, which suggests that the proteome of the R. sphaeroides outer membrane is relatively uncharacterized. Those proteins that could be categorized were placed into the categories of energy production and conversion (a possible membrane bound Class I monoheme cytochrome c, RSP0775); transcription/translation (2 proteins); and cell envelope biogenesis (2 proteins). The 2 proteins in the transcription/translation category are annotated as bacterial regulatory (RSP1550) and conserved hypothetical (RSP2616) proteins; their relationship to the outer membrane is unclear. The 2 proteins in the cell envelope biogenesis category are annotated as a peptidoglycan binding protein (RSP2543) and putative lipoprotein (RSP0891). Observation of this latter protein in the outer membrane subcellular fraction provides evidence that this protein should be considered as nonputative. Thirty additional proteins observed in the outer membrane group also are annotated as either putative, hypothetical, or conserved hypothetical. They have been placed in Supporting Information Table 1, along with information regarding the number of unique peptides detected by LC –FTICR and percent coverage, to provide helpful information for future characterization.
Discussion
Proteomic analyses of prokaryotes (including our previous work using the AMT tag approach) typically focus on obtaining information from soluble, insoluble, and global protein extracts.8,19 Such a practice does not add biological information that pertains to the specific localization of proteins within the cellular matrix, particularly those proteins assigned putative functions, proteins annotated as hypothetical/conserved hypothetical, and those proteins with assigned functions that have no associated localization information. The objective of this study was to evaluate the utility of the AMT tag approach for identifying proteins associated with the cytoplasm, cytoplasmic membrane, periplasm, and outer membrane subcellular fractions for R. sphaeroides aerobic and photosynthetic cell cultures. An additional subcellular fraction, the chromatophore that houses the specialized photosynthetic apparatus was also analyzed for the photosynthetic cell culture (details to be reported in a future publication). In terms of R. sphaeroides, obtaining protein localization information is important for enhancing our understanding of the biology associated with its various metabolic lifestyles and the transitions between the aerobic and photosynthetic cell.
Comparison of our results with previously published results obtained by analyzing the soluble, insoluble, and global fractions of the global R. sphaeroides proteome revealed similar coverage in terms of predicted proteins for this organism (34% vs 32%; a minimum of 2 unique peptides per protein). However, in this study, we applied a more stringent discriminant score threshold to increase the reliability of proteins contained in the reference database.23 We were also able to add localization evidence for 241 proteins detected solely within a single subcellular fraction, as well as localization evidence for 196 proteins detected across multiple subcellular fractions that have either an unknown predicted localization or implied localization from annotation. The detection of 30 putative, hypothetical/conserved hypothetical proteins within the outer membrane not only confirmed their identity as actual proteins, but also predicted/assigned a subcellular fraction to aid in future functional predictions.
Confidence in these localization assignments was evaluated by comparing the assignments with in silico results and annotated descriptions. The in silico results were in good agreement with our observations, with the exception of the cytoplasmic membrane fraction. A possible reason for this finding is that the prediction algorithms may not be as robust for this subcellular fraction as they are for the other subcellular fractions, or possibly the proteins that interact with cytoplasmic membrane proteins may have been carried into this fraction during subcellular fractionation. This possibility is evidenced by the large overlap of proteins observed between the cytoplasm and cytoplasmic membrane, particularly with the aerobic cell culture, where Z-scores could not differentiate whether the cytoplasm or cytoplasmic membrane was the most likely fraction of localization (Figure 2A). Of future interest is an investigation into the biological significance, if any, of these proteins; especially, those that exhibited similar Z-scores in adjacent fractions or appeared to change localization dependent on the cell culture.
A large number of proteins observed in multiple subcellular fractions did not have any in silico localization assignment, which was potentially due to the training set of data used by the learning machines for prediction and to the existence of “unconventional” targeting signal peptides.35 The observation of a protein across multiple fractions is in part due to the difficulty of obtaining highly pure subcellular fractions. Additionally, the potential for detecting this contamination is increased because of the high sensitivity of the analyses.8,10,18 As evidence, proteins assigned to the cytoplasm by way of Z-score analysis and clustering were also commonly observed in the periplasm subcellular fraction and vice versa for proteins assigned to the periplasm (Figure 2, parts B and D). A parallel observation was made for the more insoluble membrane proteins (Figure 2, parts C and E), which indicates the difficulty of separating soluble proteins into cytoplasm and periplasm fractions, and insoluble proteins into cytoplasmic membrane and outer membrane fractions. Indeed, general purity of subcellular fractionation obtained by utilizing the lysozyme-EDTA method12 adapted for R. sphaeroides13 is reported to range from 80% to >90%.36 In addition, proteins that exhibited differences in Z-scores that were consistent across the aerobic and photosynthetic cell cultures resulted in the most discernible clusters for a given subcellular fraction. For example, the 476 proteins that were resolved into a particular subcellular fraction corresponded to ~10 clusters (in some instances adjacent clusters targeted the same fraction). The 196 proteins (Figure 2A) that could not be resolved into any particular fraction corresponded to ~11 clusters (based on hierarchical clustering).
The analyses in the current study represent the first steps toward probing the biology of the R. sphaeroides photosynthetic cell state with large scale proteomic measurements. We have demonstrated good agreement with predicted and observed localization information, and have reported on the localization of proteins with no previous localization information. The next step is to establish our ability to confidently distinguish proteins within different subcellular fractions before proceeding to an evaluation of the differential abundance of the proteins present in both the aerobic and photosynthetic cell states. We are currently assessing different strategies and tools for identifying the differential abundance of the proteins observed in multiple subcellular fractions and in both cell cultures. By adding this capability to our localization evaluation, we will be able to provide a more complete picture of the R. sphaeroides proteome.
Supplementary Material
Acknowledgment
Portions of this work were supported by the U. S. Department of Energy (DOE) Office of Biological and Environmental Research GtL:Genomes to Life program at Pacific Northwest National Laboratory. Grant (ER63232-1018220-0007203), and United States Public Health Service grant (USPHS GM15590). We thank J. N. Adkins and N. G. Colton for critical and editorial reviews of this work, and N. Tolić and M. E. Monroe for development of the data analysis tools. Pacific Northwest National Laboratory is a multiprogram national laboratory operated by Battelle for the DOE under Contract No. DE-AC05-76RL01830.
Footnotes
Supporting Information Available: Thirty additional proteins observed in the outer membrane group also are annotated as either putative, hypothetical, or conserved hypothetical. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Woese CR. Microbiol. Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woese CR, Weisburg WG, Paster BJ, Hahn CM, Tanner RS, Krieg NR, Koops HP, Harms H, Stackebrandt E. Syst. Appl. Microbiol. 1984;5:327–336. doi: 10.1016/s0723-2020(84)80034-x. [DOI] [PubMed] [Google Scholar]
- 3.Fang HHP, Liu H, Zhang T. Int. J. Hydrogen Energ. 2005;30:785–793. [Google Scholar]
- 4.Martinezluque M, Dobao MM, Castillo F. FEMS Microbiol. Lett. 1991;83:329–334. [Google Scholar]
- 5.Nepple BB, Kessi J, Bachofen R. J. Ind. Microbiol. Biotechnol. 2000;25:198–203. [Google Scholar]
- 6.Joshi HM, Tabita FR. Proc. Natl. Acad. Sci. U.S.A. 1996;93:14515–14520. doi: 10.1073/pnas.93.25.14515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Rourke NA, Meyer T, Chandy G. Curr. Opin. Chem. Biol. 2005;9:82–87. doi: 10.1016/j.cbpa.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Lipton MS, Pasa-Tolic L, Anderson GA, Anderson DJ, Auberry DL, Battista KR, Daly MJ, Fredrickson J, Hixson KK, Kostandarithes H, Masselon C, Markillie LM, Moore RJ, Romine MF, Shen YF, Stritmatter E, Tolic N, Udseth HR, Venkateswaran A, Wong LK, Zhao R, Smith RD. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11049–11054. doi: 10.1073/pnas.172170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romine MF, Elias DA, Monroe ME, Auberry K, Fang RH, Fredrickson JK, Anderson GA, Smith RD, Lipton MS. OMICS. 2004;8:239–254. doi: 10.1089/omi.2004.8.239. [DOI] [PubMed] [Google Scholar]
- 10.Smith RD, Anderson GA, Lipton MS, Pasa-Tolic L, Shen YF, Conrads TP, Veenstra TD, Udseth HR. Proteomics. 2002;2:513–523. doi: 10.1002/1615-9861(200205)2:5<513::AID-PROT513>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 11.Cohen-Bazire G, Sistrom WR, Stanier RY. J. Cell. Comput. Physiol. 1957;49:25–68. doi: 10.1002/jcp.1030490104. [DOI] [PubMed] [Google Scholar]
- 12.Weiss RL. J. Bacteriol. 1976;128:668–670. doi: 10.1128/jb.128.2.668-670.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tai SP, Kaplan S. J. Bacteriol. 1985;164:181–186. doi: 10.1128/jb.164.1.181-186.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barber RD, Rott MA, Donohue TJ. J. Bacteriol. 1996;178:1386–1393. doi: 10.1128/jb.178.5.1386-1393.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson JB. J. Bioenerg. Biomembr. 1991;23:715–741. doi: 10.1007/BF00785998. [DOI] [PubMed] [Google Scholar]
- 16.Flory JE, Donohue TJ. J. Bacteriol. 1995;177:4311–4320. doi: 10.1128/jb.177.15.4311-4320.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deal CD, Kaplan S. J. Bacteriol. 1983;154:1015–1020. doi: 10.1128/jb.154.2.1015-1020.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasa-Tolic L, Masselon C, Barry RC, Shen YF, Smith RD. Biotechniques. 2004;37:621. doi: 10.2144/04374RV01. [DOI] [PubMed] [Google Scholar]
- 19.Callister SJ, Goddard CD, Zeng X, Roh JH, Dominguez MA, Tavano CL, Kaplan S, Donohue TJ, Smith RD, Lipton MS. J. Microbiol. Methods. doi: 10.1016/j.mimet.2006.04.021. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eng JK, McCormack AL, Yates JR. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 21.Washburn MP, Wolters D, Yates JR. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 22.Belov ME, Anderson GA, Wingerd MA, Udseth HR, Tang KQ, Prior DC, Swanson KR, Buschbach MA, Strittmatter EF, Moore RJ, Smith RD. J. Am. Soc. Mass Spectrom. 2004;15:212–232. doi: 10.1016/j.jasms.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Strittmatter EF, Kangas LJ, Petritis K, Mottaz HM, Anderson GA, Shen YF, Jacobs JM, Camp DG, Smith RD. J. Proteome Res. 2004;3:760–769. doi: 10.1021/pr049965y. [DOI] [PubMed] [Google Scholar]
- 24.Anderson KK, Monroe ME, Daly DS. Proc. Intern. Conf. METMBS; 2004. pp. 151–156. [Google Scholar]
- 25.Norbeck AD, Monroe ME, Adkins JN, Anderson KK, Daly DS, Smith RD. J. Am. Soc. Mass Spectrom. 2005;16:1239–1249. doi: 10.1016/j.jasms.2005.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adkins JN, Monroe ME, Auberry KJ, Shen Y, Jacobs JM, Camp DG, II, Moore RJ, Rodland KD, Smith RD, Pounds JG. Proteomics. 2005;5:3454–3466. doi: 10.1002/pmic.200401333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eng JK, McCormack AL, Yates JR. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 28.Qian WJ, Liu T, Monroe ME, Strittmatter EF, Jacobs JM, Kangas LJ, Petritis K, Camp DG, Smith RD. J. Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 29.Roh JH, Smith WE, Kaplan S. J. Biol. Chem. 2004;279:9146–9155. doi: 10.1074/jbc.M311608200. [DOI] [PubMed] [Google Scholar]
- 30.Pappas CT, Sram J, Moskvin OV, Ivanov PS, Mackenzie RC, Choudhary M, Land ML, Larimer FW, Kaplan S, Gomelsky M. J. Bacteriol. 2004;186:4748–4758. doi: 10.1128/JB.186.14.4748-4758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chory J, Donohue TJ, Varga AR, Staehelin LA, Kaplan S. J. Bacteriol. 1984;159:540–554. doi: 10.1128/jb.159.2.540-554.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gardy JL, Spencer C, Wang K, Ester M, Tusnady GE, Simon I, Hua S, deFays K, Lambert C, Nakai K, Brinkman FSL. Nucleic Acids Res. 2003;31:3613–3617. doi: 10.1093/nar/gkg602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatusov RL, Galperin MY, Natale DA, Koonin EV. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berks BC, Palmer T, Sargent F. Curr. Opin. Microbiol. 2005;8:174–181. doi: 10.1016/j.mib.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 35.Schneider G, Fechner U. Proteomics. 2004;4:1571–1580. doi: 10.1002/pmic.200300786. [DOI] [PubMed] [Google Scholar]
- 36.Rott MA, Witthuhn VC, Schilke BA, Soranno M, Ali A, Donohue TJ. J. Bacteriol. 1993;175:358–366. doi: 10.1128/jb.175.2.358-366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.