Abstract
The cancer stem cell paradigm postulates that dysregulated tissue-specific stem cells or progenitor cells are precursors for cancer biogenesis. Consequently, identifying cancer stem cells is crucial to our understanding of cancer progression and for the development of novel therapeutic agents. In this study, we demonstrate that the overexpression of Twist in breast cells can promote the generation of a breast cancer stem cell phenotype characterized by the high expression of CD44, little or no expression of CD24, and increased aldehyde dehydrogenase 1 activity, independent of the epithelial-mesenchymal transition. In addition, Twist-overexpressing cells exhibit high efflux of Hoechst 33342 and Rhodamine 123 as a result of increased expression of ABCC1 (MRP1) transporters, a property of cancer stem cells. Moreover, we show that transient expression of Twist can induce the stem cell phenotype in multiple breast cell lines and that decreasing Twist expression by short hairpin RNA in Twist-overexpressing transgenic cell lines MCF-10A/Twist and MCF-7/Twist as well as in MDA-MB-231 partially reverses the stem cell molecular signature. Importantly, we show that inoculums of only 20 cells of the Twist-overexpressing CD44+/CD24-/low subpopulation are capable of forming tumors in the mammary fat pad of severe combined immunodeficient mice. Finally, with respect to mechanism, we provide data to indicate that Twist transcriptionally regulates CD24 expression in breast cancer cells. Taken together, our data demonstrate the direct involvement of Twist in generating a breast cancer stem cell phenotype through down-regulation of CD24 expression and independent of an epithelial-mesenchymal transition.
Introduction
Tumor biogenesis and metastasis have classically been considered a result of random but sequential genetic alterations that lead to the selection and generation of clonal populations with uncontrollable growth potential [1]. The cancer stem cell theory postulates that carcinogenesis originates in tumor stem or progenitor cells possibly due to dysregulation of normal stem cell self-renewal pathways [2]. Cancer stem cells are capable of self-renewal and can differentiate into multiple cell types in the tumor [3,4]. In addition, they possess high membrane transporter [5] and telomerase activity [6] and have the ability to migrate and metastasize [7]. Evidence supporting the cancer stem cell theory has been demonstrated in hematological malignancies as well as in solid tumors such as those of the prostate [8], colon [9], pancreas [10], brain [11], and breast [12].
A subpopulation of breast cancer stem cells exhibits the phenotype of high CD44, low CD24, and Lin- [12] and the ability to form tumors in xenograft models with very few cell numbers [13]. However, the identity of breast cancer stem cells is not limited to these molecular signatures alone. The expression of aldehyde dehydrogenase 1 (ALDH), which is also observed in hematopoietic progenitor cells, has been proposed as a breast cancer stem cell marker [14,15]. Similarly, CD10, a marker of myoepithelial precursors or bilinear progenitors that generates both luminal and myoepithelial lineages, was identified as a marker of potential breast cancer cell progenitors [16]. Thus, there is accumulating evidence indicating that markers of normal breast stem cells could be additional markers of breast cancer stem cells.
Over the years, compelling evidence has revealed the oncogenic role of Twist in breast cancer biogenesis [17–19]. Twist is a transcription factor normally expressed only in mesodermal tissue in adults and is a crucial master regulator during normal development [20]. However, Twist up-regulation is observed in many cancers such as melanoma [21], T-cell lymphoma [22], prostate cancer [23], gastric carcinomas [24], rhabdomyosarcomas [25], and breast cancer [17]. Mechanistically, overexpression of Twist has been shown to inhibit apoptosis and interfere with p53 tumor suppressor functions [25,26].
Twist has also been associated with stem cell compartments during embryogenesis [27]. Overexpression of mouse Twist in an in vitro mouse embryonic stem cell model system prevents premature muscle cell differentiation, indicating its functions within the stem cell context [28]. In addition, Twist has been described as a component of neural crest stem cells [29]. In development, Twist has been shown to be upregulated through the Sonic hedgehog/Patch pathway and in turn upregulates Gli-1 [30]. This pathway is known to function in the self-renewal of normal stem cells, in part through Gli-protein interactions [30].
Earlier work in our laboratory has demonstrated that overexpressing Twist in breast cells increased invasiveness, motility, angiogenesis, and resistance to radiation [26]. We also demonstrated that Twist is overexpressed in more than 60%of breast tumors [17] and is associated with loss of E-cadherin expression [18] and increased genetic instability [19].
In the present study, we set out to identify and characterize the role of Twist in generating breast cancer stem cells. To test this idea, we overexpressed Twist in the breast adenocarcinoma cell line MCF-7 as well as in the transformed but nonmalignant breast cell line MCF-10A and analyzed the resultant phenotypes for stem cell characteristics. We found that overexpression of Twist significantly increased the stem cell population, as identified by an increase in the number of CD44+/CD24-/low and ALDH+ cells, increased exclusion of Hoechst 33342 and Rhodamine 123 dyes through increased expression of ABCC1 transporters, and initiated the formation of tumors from low-cell inoculums. Mechanistically, we found that Twist was a direct transcriptional repressor of CD24 promoter activity. Taken together, these data strongly support the role of Twist in generating breast cancer stem cells and subsequent resistance to chemotherapy through increased drug efflux.
Materials and Methods
Cell Culture
Cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Generation of the MCF-7/Twist cell line has been described [17]. The MCF-10A/Twist cell line was prepared in a similar fashion using the pCMV-Twist plasmid. For transient expression, cells were transfected using either retroviral constructs (generated in-house) or lentiviral constructs (Open Biosystems, Huntsville, AL).
Flow Cytometry
Cells were washed in Hank's balanced salt solution (HBSS) and harvested using 0.25% trypsin-EDTA (Invitrogen, Carlsbad, CA), counted, and resuspended in 1% fetal calf serum (FCS)-phosphate-buffered saline (PBS; 1 x 106 cells/400 µl). Cells were incubated with CD44-fluorescein isothiocyanate (FITC) and CD24-phycoerythrin (PE; BD Pharmingen, San Diego, CA), along with respective controls for 45 minutes at 4°C in the dark. After incubation, cells were centrifuged and resuspended in 500 µl of 1% FCS-PBS and acquired on a FACScan II Cytometer (BD Immunocytometry Systems, San Jose, CA). No-antibody and single-antibody controls were used to compensate the sample readings and for designating quadrants. Analysis was done using Cell Quest software (BD Immunocytometry Systems) on a Macintosh platform.
Efflux Assays
Semiconfluent cells were washed once with HBSS and resuspended in growth medium at 1 x 106 cells/ml and 5 µg/ml Hoechst 33342 (Sigma-Aldrich, St. Louis, MO) and incubated for 45 minutes at 37°C in a constant-temperature water bath. Subsequently, the cells were washed once in cold HBSS/2% BSA and resuspended in warm medium and incubated for 45 minutes at 37°C for efflux. Cells were centrifuged and mounted on a slide before photomicroscopy.
For Rhodamine 123 (Sigma-Aldrich) staining, cells were trypsinized and washed twice in PBS/1% FCS. One million cells were resuspended in 100 µl of PBS/1% FCS and stained with 100 to 500 ng/ml Rhodamine 123 for 15 to 30 minutes at 37°C. Cells were washed twice at room temperature, resuspended, and incubated without Rhodamine 123 for 15 to 30 minutes at 37°C to allow for efflux to occur. Rhodamine efflux was subsequently determined by flow cytometry on a FACScan II flow cytometer.
ALDEFLUOR Assay
The ALDEFLUOR assay was performed as described by the manufacturer (Stem Cell Technologies, Vancouver, British Columbia, Canada). Briefly, cells were harvested and resuspended in assay buffer at 1 x 106 cells/ml and added to a tube containing 5 µl/ml of activated ALDEFLUOR substrate BAAA (BODIPY-aminoacetaldehyde). Half the sample was transferred to a tube containing the ALDH inhibitor diethylaminobenzaldehyde. Both samples were incubated at 37°C for 30 minutes. The cells were resuspended in assay buffer and assayed on a FACScan II flow cytometer. The ALDH bright region was based on the control diethylaminobenzaldehyde sample that was gated to have less than five events.
Mammosphere Generation
Culture cells were harvested in trypsin-EDTA and carefully resuspended in mammary epithelial growth medium (Lonza, Walkersville, MD). Cells were filtered through sterile filters (BD Discovery Labware, Bedford, MA) to obtain a single cell suspension and plated at 5000 cells per well of a six-well ultra low-attachment plate (Corning, Lowell, MA). Mammospheres were assayed 3 to 10 days after cell plating. An average of five fields/well was counted using ImagePro software (Media Cybernetics, Bethesda, MD), and experiments were done in quadruplicates.
Promoter Assays
The CD24 promoter (1600 bp) was polymerase chain reaction (PCR)-amplified from human genomic DNA and cloned into the pGL4 vector upstream of the luciferase reporter (Promega, Madison, WI). Promoter assays were carried out in 24-well plates containing 50,000 cells. MCF-7 cells were transiently cotransfected with pGL4-CD24 reporter construct, pEF1a-Twist plasmid, phRL-TK Renilla control plasmid, and pCR3.1 nonspecific plasmid. After transfection, the cells were incubated for varying times (12, 24, and 48 hours), assayed using the dual luciferase kit (Promega), and quantified on a luminometer (Berthold Sirius, Oak Ridge, TN).
Chromatin Immunoprecipitation
MCF-7/Twist cells were cross-linked with formaldehyde and quenched with glycine. The cells were lysed to release chromatin, which was sheared by sonication to an average size of 500 to 1000 bp. The chromatin was precleared with preblocked, formalin-fixed, heat-killed Staphylococcus aureus cells. The positive control used was an anti-acetyl histone H3 antibody (Millipore, Billerica, MA), whereas the negative controls consisted of no-antibody and no-chromatin samples. The antibody-chromatin-Staph A complex was washed in stringent-buffered conditions and eluted from the complex, reverse cross-linked at high temperature, and phenol-chloroform extracted. The resulting purified DNA was PCR-amplified using primers that flank the putative Twist binding sites within the CD24 promoter.
Animal Experiments
Five to ten million MCF-7/Twist cells were labeled with CD44-FITC and CD24-PE antibodies as described earlier. Subsequently, the labeled cells were sorted into CD44+/CD24-/low and CD44+/CD24+ subpopulations using a FACS Diva flow cytometer. The cells were cultured for one to two splittings before harvesting. The cells were trypsinized and resuspended with media/Matrigel (1:1) before orthotopically injecting various cell numbers in the mammary fat pad of 4- to 6-week-old female severe combined immunodeficient (SCID) mice (National Cancer Institute, Fredrick, MD). All animal experiments were conducted in compliance with protocols instituted by the Institutional Animal Care and Use Committee of the Johns Hopkins University School of Medicine.
Immunohistochemistry
Immunohistochemistry was performed as described by us earlier [17]. Briefly, tumors were fixed overnight in 4% formalin and embedded in paraffin for sectioning. Antigen retrieval was carried out by boiling in sodium citrate for 10 minutes. Blocking was performed with normal goat serum. Sections were incubated overnight with 1:50 dilution of Twist primary antibody (in-house generated) and for 30 minutes with secondary antirabbit biotinylated antibody (Vector Laboratories, Burlingame, CA). Avidin-biotin mixture was added on the slides, followed by detection with DAB followed by counterstaining with hematoxylin. Sections were mounted using Permount (Thermo-Fisher Scientific, Waltham, MA) and photographed on a Nikon Eclipse 80i fluorescence microscope using a CoolSnap ES camera (Nikon Instruments).
Results
A CD44+/CD24-/low Subpopulation Is Evident in Twist-Overexpressing Breast Cells
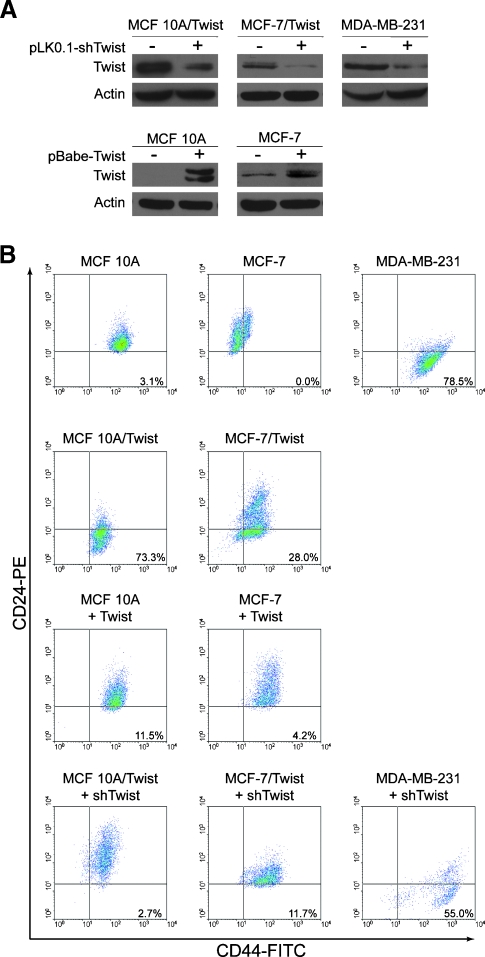
Recent studies have demonstrated that the cell surface expression pattern of CD44 and CD24 constitutes a major identifying marker of breast cancer stem cells. To determine the role of Twist in inducing the breast cancer stem cell subpopulation, we overexpressed and knocked down Twist in a panel of normal immortalized mammary cells and breast cancer cell lines. MCF-10A/Twist, MCF-7/Twist, and MDA-MB-231 cell lines were used as models for stable Twist overexpression as well as for transiently knocking down Twist expression using short hairpin RNA (shRNA) lentiviral constructs. Parental MCF-10A and MCF-7 cell lines were used as Twist-nonexpressing models into which Twist was transiently expressed using retroviral constructs. Immunoblot analysis was used to determine the levels of Twist expression in the transgenic cell lines as well as in Twist knockdown cell lines. We obtained a significant decrease in Twist expression in all three cell lines using shRNA lentiviral constructs (Figure 1A). Use of empty vector controls did not alter the Twist expression levels in these cell lines (data not shown).
Figure 1.
Cell surface expression of CD44 and CD24 in Twist-dysregulated breast cells. (A) Immunoblots showing the downregulation of Twist in MCF-10A/Twist, MCF-7/Twist, and MDA-MB-231 breast cancer cell lines by short hairpin RNA against Twist delivered using lentiviral vectors. Lower panels show Twist transient expression levels in MCF-10A and MCF-7 cell lines expressing Twist by retroviral delivery. The antibody against Twist was generated in-house and was validated. Actin was scored as loading control. (B) Flow cytometry analysis of various cell lines for CD44 and CD24 expression. Unstained and single-antibody stained cells were used as controls for setting the quadrants. The events in the lower right quadrant represent the CD44+/CD24-/low subpopulation. The analyzed data are represented as dot plots of (first row) immortalized normal mammary epithelial cell line MCF-10A and breast cancer cell lines MCF-7 and MDA-MB-231, (second row) stable Twistoverexpressing cell lines MCF-10A/Twist and MCF-7/Twist, (third row) transiently transduced Twist-expressing MCF-10A and MCF-7, and (fourth row) Twist knockdown in MCF-10A/Twist, MCF-7/Twist, and MDA-MB-231 breast cancer cells. Ten thousand viable cells were gated for each dot plot acquisition. Results are representative of five separate experiments.
The CD44+/CD24-/low subpopulation was evaluated by flow cytometry in various parental cell lines (MCF-10A, MCF-7, MDA-MB-231), transgenic cell lines (MCF-10A/Twist, MCF-7/Twist), Twist-overexpressing cell lines (MCF-10A-Twist and MCF-7-Twist), and in Twist knockdown breast cancer cell lines (MCF-10A/Twist-shTwist, MCF-7/Twist-shTwist, MDA-MB-231-shTwist). As seen in Figure 1B, we observed the nontumorigenic cell lines MCF-10A (3.1%) and MCF-7 (0.0%) to have a low degree of the CD44+/CD24-/low subpopulation. This is in comparison to the higher percentage of CD44+/CD24-/low subpopulation in the metastatic MDA-MB-231 cell line (78.5%) (top row). To evaluate if Twist-overexpressing breast cancer cells have altered CD44 and CD24 expression, we analyzed multiple MCF-10A/Twist and MCF-7/Twist clones for CD44 and CD24 expression. As seen in the second row, MCF-10A/Twist cells demonstrated increased CD44+/CD24-/low (73.3%) compared with parental MCF-10A cells (3.1%). In addition, the MCF-7/Twist cell line exhibited a higher CD44+/CD24-/low subpopulation (28.0%) compared with parental MCF-7 cells (0.0%). To confirm that the observed changes were caused by the effector functions of Twist and not caused by clonal selection, we determined the CD44 and CD24 levels in transient Twist-overexpressing breast cancer cell lines MCF-10A and MCF-7. As seen in the third row, MCF-10A and MCF-7 transduced with Twist showed an increase in the CD44+/CD24-/low subpopulation (from 3.1% to 11.5% and from 0.0% to 4.2%, respectively).
Knockdown of Twist Reverses the Stem Cell Phenotype
Because Twist overexpression altered CD44 and CD24 expression levels, we wanted to determine if decreasing Twist expression could reverse the cancer stem cell phenotype back to parental status. Toward this goal, we knocked down Twist expression in MCF-10A/Twist and MCF-7/Twist cell lines, using lentiviral-mediated shRNA knockdowns (Figure 1A). These cell lines showed lowered Twist expression when compared with parental MCF-10A/Twist and MCF-7/Twist cells. Subsequent analysis of the CD44+/CD24-/low subpopulation by flow cytometry exhibited a loss in the CD44+/CD24- phenotype in both Twist knockdown cell lines—MCF-10A/Twist (reduced from 73.3% to 2.7%) and MCF-7/Twist (reduced from 28.0% to 11.7%; Figure 1B, fourth row). Knockdown of Twist expression in MDA-MB-231 cells decreased the CD44+/CD24-/low subpopulation from 78.5% in the parental cells to 55% in the knockdown cells (fourth row). Collectively, these data demonstrate that Twist is a major factor modulating the CD44+/CD24-/low subpopulation in breast cancer cells.
With respect to epithelial-mesenchymal transition (EMT) markers in Twist knockdown MCF-10A/Twist and MCF-7/Twist cells, we observed an increase in E-cadherin levels but little or no change in vimentin levels (Figure 2A). This further supports our hypothesis that Twist expression potentiates EMT and down-regulating Twist can partially reverse this phenotype. In addition, we show that transient expression of Twist in MCF-7 cells altered CD44 and CD24 levels but did not induce any change in protein expression of the EMT markers E-cadherin and vimentin (Figure 2B). This is in contrast to high vimentin and low E-cadherin expression observed in stable Twist-expressing MCF-7 cells (Figure 2B).
Figure 2.
Epithelial-mesenchymal marker expression in Twist cells. (A) Immunoblots showing epithelial marker E-cadherin and mesenchymal marker vimentin in transient knockdown of Twist in stable Twist-expressing cell lines MCF-10A/Twist and MCF-7/ Twist. (B) Immunoblots showing E-cadherin and vimentin in MCF-7 and MCF-7/Twist cells transiently expressing Twist. Transduced cells were lysed and immunoblotted with antibodies against E-cadherin, vimentin (Santa Cruz Biotechnology, Santa Cruz, CA), and Twist (in-house generated). Actin (Sigma-Aldrich) was used as loading control.
MCF-7/Twist Cells Show Increased Efflux of Hoechst 33342 and Rhodamine 123 Dyes
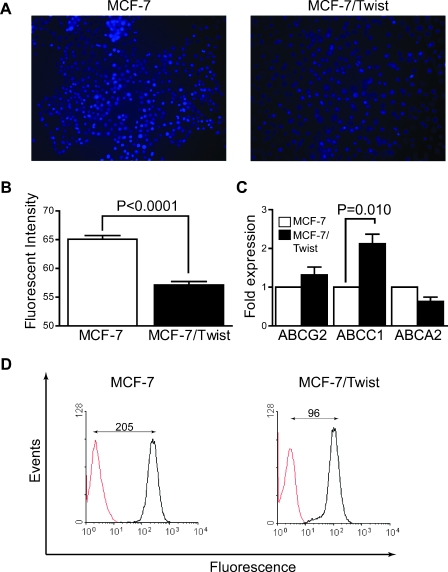
A characteristic of stem cells is increased drug resistance brought about by elevated expression of ATP-binding cassette (ABC) transporters on the cell surface [31]. The functionality of these transporters can be characterized by studying the efflux of vital dyes such as Hoechst 33342 and Rhodamine 123 from treated cells [32]. Because Twist expression also induces chemoresistance [33], we initially evaluated the ability of MCF-7/Twist cells to extrude Hoechst 33342 dye microscopically [31]. As shown in Figure 3 (A and B), MCF-7/Twist cells retained significantly less dye compared with parental MCF-7 cells (relative fluorescence intensity per cell of 57 vs 65). To determine factors responsible for increased efflux, we carried out quantitative reverse transcription-PCR (qRT-PCR) for the expression of various stem cell factors such as ABC transporters ABCG2 [31] and ABCC1 (MRP1) [34]. The results obtained revealed a significant increase in ABCC1 (MRP1) and a lesser increase in ABCG2 transcript levels in Twist-overexpressing breast cancer cells (Figure 3C).
Figure 3.
Efflux studies in MCF-7 and MCF-7/Twist cells. (A) Representative photomicrographs of Hoechst efflux staining in MCF-7/Twist cells compared with parental MCF-7 cells. The cells, after efflux, were photographed using a Nikon Eclipse 80i fluorescence microscope. (B) Histogram showing quantification of fluorescence intensity per cell. Cell fluorescence (n = 6) in the blue channel was analyzed using dedicated software developed in IDL programming environment that provides an operator-free segmentation of images and determines the average relative fluorescence per cell. Three images were analyzed per sample. (C) Histogram showing expression of drug transporters ABCG2, ABCC1, and ABCA1 in MCF-7/Twist and parental MCF-7 cells. (D) Histogram showing efflux of Rhodamine 123 stain in MCF-7/Twist and in MCF-7 cells. The amount of Rhodamine 123 dye in the cells was determined by flow cytometry. Results are representative of three separate experiments.
Subsequently, we confirmed the ability of MCF-7/Twist cells to extrude Rhodamine 123 dye by analyzing mean fluorescence intensity as determined by flow cytometry. As seen in Figure 3D, Rhodamine 123 was excluded more from MCF-7/Twist than the MCF-7 cells (96 vs 205). These results confirm that the chemoresistance of MCF-7/Twist cells is partially conferred by the overexpression of at least one of the transporter genes, ABCC1.
MCF-7/Twist Cells Have Increased Aldehyde Dehydrogenase Activity
Expression of ALDH within the presumptive stem cell phenotype identifies and enriches self-renewing populations within the breast milieu [15]. Given our initial finding that the expression of Twist in breast cells promotes the stem cell phenotype, we analyzed for ALDH expression using the ALDEFLUOR assay (Stem Cell Technologies) in a panel of breast cancer cell lines as well as in MCF-10A/Twist and MCF-7/Twist transgenic cells. As shown in Figure 4 (first row), the percentage of ALDH-positive cells was generally low in the breast cancer cell lines analyzed. However, stable expression of Twist increased the ALDH-positive cells from 0.66% to 1.71% in MCF-10A/Twist cells and from 0.04% to 2.81% in MCF-7/Twist cells (Figure 4, second row). An increase in the ALDH-positive cells was also observed in MCF-10A (from 0.66% up to 0.93%) and MCF-7 (from 0.04% up to 1.2%) transduced with Twist-overexpressing retroviral constructs (Figure 4, third row).
Figure 4.
ALDH activity in Twist-dysregulated breast cells. Dot plots of various cell lines analyzed by flow cytometry for ALDH activity. Cells were treated with ALDEFLUOR in the presence or absence of ALDH inhibitor DEAB. After treatment, the samples were analyzed by flow cytometry for the presence of ALDH bright cells. The ALDH bright region was based on the control DEAB sample that was gated to have less than five events. The cell percentage numbers for ALDH positivity are indicated in the bottom right of each histogram. The values presented are the averages of three independent experiments. The analyzed data are represented as dot plots of (first row) parental MCF-10A and MCF-7 breast cancer cell lines, (second row) stable Twist-overexpressing MCF-10A/Twist and MCF-7/Twist breast cancer cell lines, (third row) breast cell lines MCF-10A and MCF-7 transiently transduced with Twist-expressing retroviral constructs, and (fourth row) Twist knockdown breast cancer cell lines MCF-10A/Twist and MCF-7/Twist. Results are representative of five separate experiments.
In further support of the role of Twist in altering ALDH activity, we estimated ALDH in Twist knockdown cell lines. As shown in Figure 4 (fourth row), loss of Twist decreased the number of ALDH-positive cells in MCF-10A/Twist (from 1.71% down to 0.13%) and MCF-7/Twist (1.2% down to 0.86%). Interestingly, MDA-MB-231 has a very low subpopulation of ALDH+ cells (0.01%), which is not affected by Twist knockdown (data not shown). Overall, these results strengthen our hypothesis that Twist is a regulator of the breast cancer stem cell phenotype.
Ability of Self-renewal of the CD44+/CD24-/low Subpopulation in MCF-7/Twist Cells
One of the defining characteristics of stem cells is the property of self-renewal or asymmetric cell division. To ascertain whether the ability of asymmetric division is an inherent characteristic of Twist+ cells, we flow sorted the CD44+/CD24-/low subpopulation from MCF-7/Twist cells using antibodies against CD44 and CD24. After purification, the enriched cells were allowed to divide in vitro, and the CD44+/CD24-/low subpopulation was estimated at regular time intervals. The initial percentage of the purified subpopulation of CD44+/CD24-/low cells was greater than 98% (data not shown). As shown in Figure 5A, continuous culture of the purified cells decreased the CD44+/CD24-/low subpopulation from 72% (generation 2) to 31% at generation 13, which is an indication of self-renewal that characterizes the stem cell phenotype. After passage 15, the CD44+/CD24-/low subpopulation stabilized at 20% to 30% for the next 20 generations (data not shown).
Figure 5.
Self-renewal and mammosphere formation of Twist-expressing MCF-7 cells. MCF-7/Twist cells were stained with CD44 and CD24 antibodies and flow sorted to purify the CD44+/CD24-/low subpopulation. The purified MCF-7/Twist CD44+/CD24-/low subpopulation was propagated in culture, and the percentage of CD44 and CD24 cells was estimated by flow cytometry. (A) Histograms of chronological changes in the CD44+/CD24-/low subpopulation during a period of 13 generations. (B) qRT-PCR analysis of Twist, CD44, and CD24 transcript levels in the purified CD44+/CD24-/low versus CD44+/CD24+ subpopulations of cells. (C and D) Mammosphere formation by stable Twist-overexpressing cell lines, MCF-7/Twist, and MCF-10A/Twist. Top rows show phase-contrast photomicrographs of mammospheres at a magnification of x20. The lower rows show viability of the cells as indicated by the fluorescence of Calcein-AM stain. Data are represented as histograms on the right. Experiments were performed in quadruplicates and an average of five fields per well were counted.
As a further confirmation of the cancer cell stem phenotype, we carried out qRT-PCR for the expression of Twist, CD44, and CD24 transcript levels in the CD44+/CD24-/low and CD44+/CD24+ subpopulations. CD24 expression was four-fold lower (P = .002) in the CD44+/CD24-/low subpopulation compared with the CD44+/CD24+ subpopulation (Figure 5B). In addition, Twist expression was 15% higher (P = .003) in the CD44+/CD24-/low subpopulation compared with the CD44+/CD24+ subpopulation. No significant difference was observed in the CD44 expression between these two subpopulations.
Increased Mammosphere Formation in Twist-Overexpressing Cells
Because the ability to form mammospheres is a characteristic of cancer stem cells, we evaluated the ability of MCF-10A/Twist and MCF-7/Twist cells to form mammospheres in culture. MCF-7/Twist cells formed mammospheres that were disaggregated and unlike the classic rounded mammosphere phenotype observed in culture (Figure 5C). Similar results were reported using the highly aggressive breast cancer cell line, MDA-MB-231, which also exhibits a large subpopulation of CD44+/CD24-/low cells [35]. Nonetheless, these cells were alive and present in significantly higher numbers (55 vs 30, P < .0001) compared with parental MCF-7 cells when analyzed by the Calcein-AM green staining (lower rows). With respect to MCF-10A/Twist cells, significantly larger numbers of mammospheres (53 vs 32, P = .002) were generated compared with the parental MCF-10A cells (Figure 5D). This was observed in two independent MCF-10A/Twist clones (data not shown). In addition, the mammospheres generated using MCF-10A/Twist cells were significantly larger than the parental MCF-10A cells.
Twist Downregulates CD24 Expression in Breast Cancer Cells
After the observation that Twist decreased CD24 expression, we sought to characterize if this repression was transcriptionally regulated. We analyzed the 1.6-kb CD24 promoter region for E-box sequences (CANNTG) to which Twist can potentially bind. As shown in Figure 6A, the putative CD24 promoter contains eight E-box sequences. This region was PCR-amplified and cloned into the pGL4 luciferase reporter plasmid (Promega). Transient transfection assays using the CD24 promoter-reporter construct and a Twist expression plasmid in MCF-7 cells showed a significant down-regulation of the reporter gene, 24 and 48 hours after transfection (Figure 6B). A further confirmation of this regulation was demonstrated using protein extracts from MCF-7/Twist cells and scored for CD24 protein levels. As shown in Figure 6C, CD24 protein expression was significantly reduced in MCF-7/Twist compared with parental MCF-7 cells.
Figure 6.
Twist transcriptional regulation of CD24 expression. (A) Schematic representation of the CD24 promoter sequence showing the location of putative TWIST binding sites relative to the transcription start site (+1). Tick marks denote the canonical E-box sequences (CANNTG) to which Twist can potentially bind. The numbers above the tick marks indicate the relative position of E-box sequences within the promoter region. (B) Reporter assays were performed using CD24 promoter-reporter constructs in MCF-7 with exogenous Twist expression plasmid added and estimated for 2 days. The results of the promoter assays are represented as a histogram showing normalized luciferase readings in CD24 promoter activity. (C) Immunoblot analysis of cell extracts from MCF-7 and MCF-7/Twist cells scored for CD24 (Santa Cruz Biotechnology) and Twist protein expression. Actin was used as a loading control. (D and E) In vivo binding of Twist protein to the CD24 promoter sequence. ChIP was carried out using MCF-7/Twist cells and analyzed using CD24 promoter-specific primers by PCR. Identical volumes from the final precipitate were used for the PCRs except for the input, which was diluted 10-fold.
Twist Binds In Vivo to E-box Sequences in the CD24 Promoter
To establish that the down-regulation of CD24 by Twist was due to the direct binding of Twist to the CD24 promoter sequence, we performed chromatin immunoprecipitation (ChIP) assays using MCF-7 cells overexpressing Twist (Figure 6D). A set of PCR primers (5′-TGCCCCTTAGAATTGCTGTT-3′ and 5′-TCATTGAACCTGGAAGTGG-3′) was designed for specific amplification of the Twist binding sites (E-boxes) from -1256 and -1107 bp within the CD24 promoter. As shown in Figure 6D, the use of this primer set in PCR of ChIP DNA generated from Twist immunoprecipitations resulted in a specific amplified product of 149 bp, whereas no amplification was seen in the samples that were processed in the absence of precipitating antibody or no input chromatin. We confirmed the PCR by qRT-PCR amplification (Figure 6E). These results indicate that Twist binds directly or as part of a complex to the endogenous CD24 promoter in vivo.
Twist+/CD44+/CD24-/low Subpopulation Exhibits Increased Tumorigenic Potential In Vivo
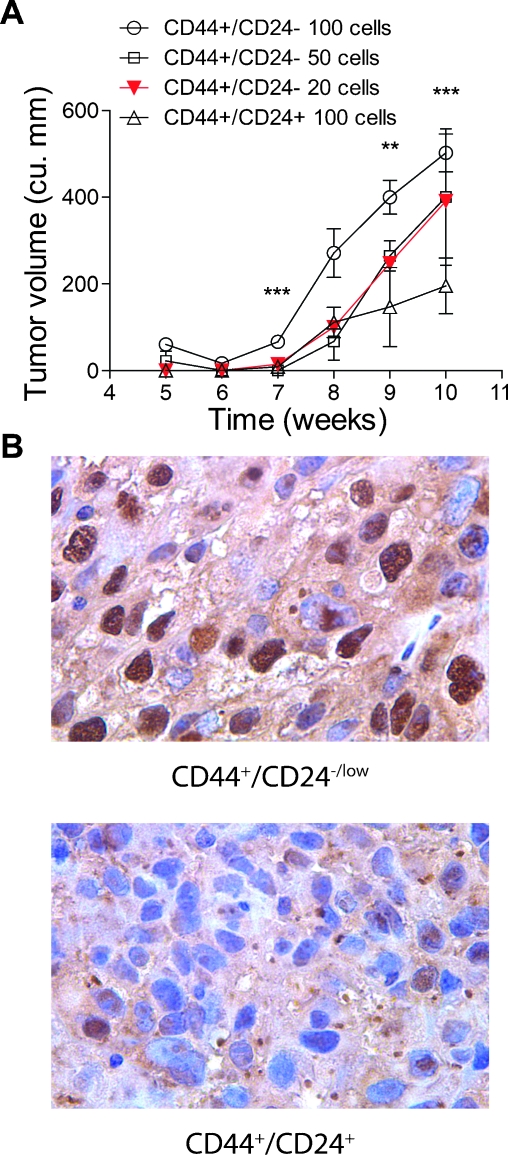
A hallmark of cancer stem cells is the ability to form tumors with limited number of cells. To determine the minimum number of cells of the Twist+/CD44+/CD24-/low subpopulation required to initiate and establish tumor growth within the mammary fat pad, we injected 100, 50, and 20 purified cells into the orthotopic site in SCID mice. We also injected 100 cells of the Twist+/CD44+/CD24+ purified subpopulation. After an incubation period of 7 weeks, we observed tumor growth in mice injected with 20 cells (Figure 7A). There was a trend toward increased latency as the cell inoculums became smaller with tumor uptake time being approximately 2 to 3 weeks less in the 100-cell inoculums of the Twist+/CD44+/CD24-/low subpopulation compared with that of the Twist+/CD44+/CD24+ subpopulation.
Figure 7.
Growth of low inoculums of purified CD44+/CD24-/low and CD44+/CD24+ subpopulations in SCID mice. (A) Graphical representation of growth rates of orthotopic xenograft tumors using flow-sorted cells from the Twist+/CD44+/CD24-/low and CD44+/CD24+ subpopulations. We transplanted 100, 50, and 20 cells of the Twist+/CD44+/CD24-/low subpopulation, and 100 cells from the Twist+/CD44+/CD24+ subpopulation. Four mice per inoculum were used for this study. Significance was analyzed by a two-sided t-test (**P<.05, ***P <.005). (B) Twist immunohistochemical staining of representative tumors from the CD44+/CD24-/low and CD44+/CD24+ subpopulations. Photomicrographs were taken in bright field settings at a magnification of x40 using Nikon Eclipse 80i fluorescence microscope.
To study the expression levels of Twist in both the CD44+/CD24-/low- and the CD44+/CD24+-derived tumors, we performed immunohistochemistry on tumor sections. We found that tumors generated from the Twist+/CD44+/CD24+ subpopulation had lower Twist expression compared with tumors derived from the Twist+/CD44+/CD24-/low subpopulation (Figure 7B).
Discussion
Tumor origin has classically been conceived as a result of an accumulation of mutations within the breast epithelium that cause cells to escape cell death and maintain the mutator phenotype [1,36]. The cancer stem cell theory postulates that these mutations or other cellular changes occur in subpopulations of long-lived cancer stem cells, which are the origin of tumorigenesis [2]. Initial identification of breast cancer stem cells was based on a combination of CD44+, CD24-, and Lin- markers [12]. Subpopulations purified based on these markers had a 10- to 50-fold increase in the ability to form tumors in NOD/SCID mice. Subsequently, a number of markers have been defined such as ALDH [15], CD10 [16], and CD133 [37].
On the basis of the need to identify novel breast cancer stem cell markers, investigators have demonstrated that an in vitro EMT can induce stem cell phenotypes [38]. However, the ontogeny of generating the stem cell phenotype, before the onset of EMT, has not been determined. Moreover, the occurrence of EMT in breast cancers is itself extremely rare [39], and the role of Twist and Snail as causal factors has only been demonstrated in vitro [17,38]. This is the first report of Twist mechanistically inducing the stem cell phenotype in breast cancer cells. On the basis of our results using transient Twist-overexpressing cells, we observed dysregulation of CD44 and CD24 expression before any biochemical evidence of EMT. These data indicate that generation of the breast cancer stem cell phenotype by Twist precedes an EMT within our experimental settings. It is possible that Twist regulates other factors that are expressed during stable Twist expression, which may be absent during transient Twist expression. These factors may be one reason why stable Twist expression leads to a higher CD44/CD24 subpopulation compared with transient Twist expression. Another possible explanation is that genetic changes induced early during tumor biogenesis provide a transient state for cellular adaptation resulting in the low expression of unique molecular markers including those of cancer stem cells. Subsequent evolution of these cells and escape from cellular senescence result in the establishment of stem cell markers in a clonal population that is distinct from the bulk of the tumor resulting in a larger percentage of cells with CD44+/CD24-/low expression.
In our present study, we demonstrate that Twist expression in breast cells can augment the CD44+/CD24-/low or breast cancer stem cell. The significance of this finding is that loss of CD24 in breast cancer patients along with increased CD44 expression may promote metastasis [13]. Our results conclusively demonstrate that Twist binds directly or as part of a complex to the endogenous CD24 promoter in vivo causing it to be transcriptionally downregulated. This is supported by flow cytometry data, which demonstrate a loss of CD24 in breast cancer cells with Twist expression. This observation, in combination with the finding that Twist expression is increased in high-grade breast tumors [17], may provide a link between the loss of CD24 expression and the breast cancer stem cell phenotype. Conversely, it is also possible that Twist directly acts on CD44 because there is a distinct increase in CD44 expression in Twist-overexpressing cells as determined by flow cytometry. Overall, this establishes Twist expression as one of the key factors that directly drives cells toward a CD44+/CD24-/low phenotype or at least is necessary for its maintenance.
Twist overexpression has been correlated with drug resistance to microtubule-targeting anticancer drugs taxol and vincristine [33]. This is significant in that ABCC1 (MRP1) is the major drug efflux pump for vincristine [34]. Our results are the first direct evidence demonstrating the increased ABCC1 transporter levels in Twist-overexpressing cells leading to the development of chemoresistance and development of the stem cell phenotype in these cells.
Besides CD44 and CD24, additional markers have recently been identified to classify cancer stem cells. Of these, ALDH1 [14] has gained importance as a potential marker of breast cancer stem cell phenotype [15]. In support of published data, we have also identified an ALDH+ subpopulation in Twist-expressing breast cancer cells. Interestingly, MDA-MB-231, which has very high CD44+/CD24-/low levels, exhibits a negligible amount of ALDH+ cells (data not shown). This could be either due to long-term growth in vitro or to the genetic heterogeneity of the cell type.
Another feature of cancer stem cells is the propensity to form mammospheres in culture. Our results using Twist-overexpressing cells to form mammospheres are in close agreement with recently published work using breast tumors, with the exception of MCF-7/Twist cells, which did not result in the formation of the classic mammosphere phenotype. The formation of such nonclassic mammosphere formation has been reported for the MDA-MB-231 cell line [35,40], which intriguingly exhibits a very high CD44+/CD24-/low subpopulation as well as a high Twist expression. This would indicate that the phenomenon of mammosphere generation is dependent on a number of factors such as the degree of cellular differentiation, genetic makeup, and the biochemical parameters of the transformed cells. Furthermore, it is also possible that Twist-induced breast cancer stem cells will be heterogeneous with varying self-renewal capacity. This is supported by our data, which demonstrate that the purified CD44+/CD24-/low subpopulation was able to initiate tumor growth in the mammary fat pad with extremely low inoculums (20 cells) compared with the CD44+/CD24+ subpopulation. This ability to form tumors from low inoculums conclusively demonstrates that Twist is a primary driver of the breast cancer stem cell phenotype. More importantly, Twist expression was higher in the nucleus of cells from the xenograft tumors generated by the CD44+/CD24-/low subpopulation compared with those formed from the CD44+/CD24+ subpopulation. This indicates that the generation of breast cancer stem cells by Twist expression results in a heterogeneous population of cells with varying degrees of tumorigenicity and may parallel to that observed in breast cancer patient sample.
In conclusion, this is the first report of a single gene, Twist, being able to promote the generation of breast cancer stem cells in a preclinical model through the transcriptional regulation of CD24. This is a mechanistic finding that supports the paradigm of the relationship between Twist expression and the acquisition of stem-like properties. In the future, we hope to establish the utility of Twist expression as an additional marker for breast cancer stem cells along with the more established CD44+/CD24-/low/ALDH+ phenotype.
Acknowledgments
The authors thank Michele Doucet and John Domek for technical help, Paul Winnard Jr. for critically reading the manuscript, and Richard Blosser and Ada Tam for help with flow cytometry.
Footnotes
This work was supported by the Maryland Stem Cell Research Foundation (MDTSCRC0072 to F.V. and MDTSCRC0064 to V.R.) and by the National Institutes of Health grant RO1CA097226 and P50 CA103175 (to V.R.).
References
- 1.Knudson AG, Jr, Strong LC, Anderson DE. Heredity and cancer in man. Prog Med Genet. 1973;9:113–158. [PubMed] [Google Scholar]
- 2.Passegue E, Jamieson CH, Ailles LE, Weissman IL. Normal and leukemic hematopoiesis: are leukemias a stem cell disorder or a reacquisition of stem cell characteristics? Proc Natl Acad Sci USA. 2003;100(Suppl 1):11842–11849. doi: 10.1073/pnas.2034201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smalley M, Ashworth A. Stem cells and breast cancer: a field in transit. Nat Rev Cancer. 2003;3:832–844. doi: 10.1038/nrc1212. [DOI] [PubMed] [Google Scholar]
- 4.Campbell LL, Polyak K. Breast tumor heterogeneity: cancer stem cells or clonal evolution? Cell Cycle. 2007;6:2332–2338. doi: 10.4161/cc.6.19.4914. [DOI] [PubMed] [Google Scholar]
- 5.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 6.Greenwood MJ, Lansdorp PM. Telomeres, telomerase, and hematopoietic stem cell biology. Arch Med Res. 2003;34:489–495. doi: 10.1016/j.arcmed.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Croker AK, Allan AL. Cancer stem cells: implications for the progression and treatment of metastatic disease. J Cell Mol Med. 2008;12:374–390. doi: 10.1111/j.1582-4934.2007.00211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 9.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 10.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 11.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 12.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheridan C, Kishimoto H, Fuchs RK, Mehrotra S, Bhat-Nakshatri P, Turner CH, Goulet R, Jr, Badve S, Nakshatri H. CD44+/CD24- breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 2006;8:R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kastan MB, Schlaffer E, Russo JE, Colvin OM, Civin CI, Hilton J. Direct demonstration of elevated aldehyde dehydrogenase in human hematopoietic progenitor cells. Blood. 1990;75:1947–1950. [PubMed] [Google Scholar]
- 15.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stingl J, Eaves CJ, Kuusk U, Emerman JT. Phenotypic and functional characterization in vitro of a multipotent epithelial cell present in the normal adult human breast. Differentiation. 1998;63:201–213. doi: 10.1111/j.1432-0436.1998.00201.x. [DOI] [PubMed] [Google Scholar]
- 17.Mironchik Y, Winnard PT, Jr, Vesuna F, Kato Y, Wildes F, Pathak AP, Kominsky S, Artemov D, Bhujwalla Z, Van Diest P, et al. Twist overexpression induces in vivo angiogenesis and correlates with chromosomal instability in breast cancer. Cancer Res. 2005;65:10801–10809. doi: 10.1158/0008-5472.CAN-05-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vesuna F, van Diest P, Chen JH, Raman V. Twist is a transcriptional repressor of E-cadherin gene expression in breast cancer. Biochem Biophys Res Commun. 2008;367:235–241. doi: 10.1016/j.bbrc.2007.11.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vesuna F, Winnard P, Jr, Glackin C, Raman V. Twist overexpression promotes chromosomal instability in the breast cancer cell line MCF-7. Cancer Genet Cytogenet. 2006;167:189–191. doi: 10.1016/j.cancergencyto.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rose CS, Malcolm S. A TWIST in development. Trends Genet. 1997;13:384–387. doi: 10.1016/s0168-9525(97)01296-1. [DOI] [PubMed] [Google Scholar]
- 21.Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, Lin A, Kluger HM, Berger AJ, Cheng E, Trombetta ES, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64:5270–5282. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- 22.van Doorn R, Dijkman R, Vermeer MH, Out-Luiting JJ, van der Raaij-Helmer EM, Willemze R, Tensen CP. Aberrant expression of the tyrosine kinase receptor EphA4 and the transcription factor twist in Sezary syndrome identified by gene expression analysis. Cancer Res. 2004;64:5578–5586. doi: 10.1158/0008-5472.CAN-04-1253. [DOI] [PubMed] [Google Scholar]
- 23.Kwok WK, Ling MT, Lee TW, Lau TC, Zhou C, Zhang X, Chua CW, Chan KW, Chan FL, Glackin C, et al. Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res. 2005;65:5153–5162. doi: 10.1158/0008-5472.CAN-04-3785. [DOI] [PubMed] [Google Scholar]
- 24.Rosivatz E, Becker I, Specht K, Fricke E, Luber B, Busch R, Hofler H, Becker KF. Differential expression of the epithelial-mesenchymal transition regulators snail, SIP1, and twist in gastric cancer. Am J Pathol. 2002;161:1881–1891. doi: 10.1016/S0002-9440(10)64464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maestro R, Dei Tos AP, Hamamori Y, Krasnokutsky S, Sartorelli V, Kedes L, Doglioni C, Beach DH, Hannon GJ. Twist is a potential oncogene that inhibits apoptosis. Genes Dev. 1999;13:2207–2217. doi: 10.1101/gad.13.17.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stasinopoulos IA, Mironchik Y, Raman A, Wildes F, Winnard P, Jr, Raman V. HOXA5-twist interaction alters p53 homeostasis in breast cancer cells. J Biol Chem. 2005;280:2294–2299. doi: 10.1074/jbc.M411018200. [DOI] [PubMed] [Google Scholar]
- 27.Gerecht-Nir S, Dazard JE, Golan-Mashiach M, Osenberg S, Botvinnik A, Amariglio N, Domany E, Rechavi G, Givol D, Itskovitz-Eldor J. Vascular gene expression and phenotypic correlation during differentiation of human embryonic stem cells. Dev Dyn. 2005;232:487–497. doi: 10.1002/dvdy.20247. [DOI] [PubMed] [Google Scholar]
- 28.Rohwedel J, Horak V, Hebrok M, Fuchtbauer EM, Wobus AM. M-twist expression inhibits mouse embryonic stem cell-derived myogenic differentiation in vitro. Exp Cell Res. 1995;220:92–100. doi: 10.1006/excr.1995.1295. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Morales JR, Henrich T, Ramialison M, Wittbrodt J. New genes in the evolution of the neural crest differentiation program. Genome Biol. 2007;8:R36. doi: 10.1186/gb-2007-8-3-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villavicencio EH, Walterhouse DO, Iannaccone PM. The sonic hedgehog-patched-gli pathway in human development and disease. Am J Hum Genet. 2000;67:1047–1054. doi: 10.1016/s0002-9297(07)62934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim M, Turnquist H, Jackson J, Sgagias M, Yan Y, Gong M, Dean M, Sharp JG, Cowan K. The multidrug resistance transporter ABCG2 (breast cancer resistance protein 1) effluxes Hoechst 33342 and is overexpressed in hematopoietic stem cells. Clin Cancer Res. 2002;8:22–28. [PubMed] [Google Scholar]
- 32.Lizard G, Roignot P, Maynadie M, Lizard-Nacol S, Poupon MF, Bordes M. Flow cytometry evaluation of the multidrug-resistant phenotype with functional tests involving uptake of daunorubicin, Hoechst 33342, or Rhodamine 123: a comparative study. Cancer Detect Prev. 1995;19:527–534. [PubMed] [Google Scholar]
- 33.Wang X, Ling MT, Guan XY, Tsao SW, Cheung HW, Lee DT, Wong YC. Identification of a novel function of TWIST, a bHLH protein, in the development of acquired taxol resistance in human cancer cells. Oncogene. 2004;23:474–482. doi: 10.1038/sj.onc.1207128. [DOI] [PubMed] [Google Scholar]
- 34.Loe DW, Deeley RG, Cole SP. Characterization of vincristine transport by the M(r) 190,000 multidrug resistance protein (MRP): evidence for cotransport with reduced glutathione. Cancer Res. 1998;58:5130–5136. [PubMed] [Google Scholar]
- 35.Grimshaw MJ, Cooper L, Papazisis K, Coleman JA, Bohnenkamp HR, Chiapero-Stanke L, Taylor-Papadimitriou J, Burchell JM. Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Res. 2008;10:R52. doi: 10.1186/bcr2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loeb LA. Cancer cells exhibit a mutator phenotype. Adv Cancer Res. 1998;72:25–56. doi: 10.1016/s0065-230x(08)60699-5. [DOI] [PubMed] [Google Scholar]
- 37.Wright MH, Calcagno AM, Salcido CD, Carlson MD, Ambudkar SV, Varticovski L. Brca1 breast tumors contain distinct CD44+/CD24- and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008;10:R10. doi: 10.1186/bcr1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trimboli AJ, Fukino K, de Bruin A, Wei G, Shen L, Tanner SM, Creasap N, Rosol TJ, Robinson ML, Eng C, et al. Direct evidence for epithelial-mesenchymal transitions in breast cancer. Cancer Res. 2008;68:937–945. doi: 10.1158/0008-5472.CAN-07-2148. [DOI] [PubMed] [Google Scholar]
- 40.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]