Abstract
Modification of the histone proteins that form the core around which chromosomal DNA is looped has profound epigenetic effects on the accessibility of the associated DNA for transcription, replication and repair. The SET domain is now recognized as generally having methyltransferase activity targeted to specific lysine residues of histone H3 or H4. There is considerable sequence conservation within the SET domain and within its flanking regions. Previous reviews have shown that SET proteins from Arabidopsis and maize fall into five classes according to their sequence and domain architectures. These classes generally reflect specificity for a particular substrate. SET proteins from rice were found to fall into similar groupings, strengthening the merit of the approach taken. Two additional classes, VI and VII, were established that include proteins with truncated/interrupted SET domains. Diverse mechanisms are involved in shaping the function and regulation of SET proteins. These include protein-protein interactions through both intra- and inter- molecular associations that are important in plant developmental processes, such as flowering time control and embryogenesis. Alternative splicing that can result in the generation of two to several different transcript isoforms is now known to be widespread. An exciting and tantalizing question is whether, or how, this alternative splicing affects gene function. For example, it is conceivable that one isoform may debilitate methyltransferase function whereas the other may enhance it, providing an opportunity for differential regulation. The review concludes with the speculation that modulation of SET protein function is mediated by antisense or sense-antisense RNA.
Keywords: Arabidopsis SET genes, alternative splicing, epigenetics, histone methylation, rice SET genes, SET domain classes
1. Introduction
In eukaryotes, chromosomal DNA is organized as chromatin, in which ~146-bp form almost 2 left-handed coils around an octamer of basic proteins comprised of a histone H3-H4 tetramer and two H2A-H2B dimers [1–3]. These fundamental units, nucleosomes, are linked together by ~50-bp of spacer DNA that is associated with histone H1 to yield a characteristic ‘beads-on-a-string’ structure that folds further to yield a very compact state in the nucleus [4, 5]. Recently, it has become evident that fast, long range, reversible conformational fluctuations in nucleosomal structure, together with specific chemical modifications of the histones, play vital roles in eukaryotic gene regulation [6–8]. Covalent modifications of the N-terminal histone tails include acetylation, phosphorylation, methylation, ubquitination and ADP-ribosylation [9–11]. These modifications form the basis of the histone code (or, possibly, codes) that regulate gene expression epigenetically through various mechanisms [9, 12–14]. For example, methylation of histone H3K9 provides an epigenetic mark for the binding of the chromodomain (protein domain structure that binds methylated lysine) containing protein, HP1 (heterochromatin protein 1), that leads to heterochromatin formation and gene repression [15]. In contrast, acetylation of histones tends to decrease interaction between histones and DNA, and facilitates binding of bromodomain (protein domain structure that binds acetylated lysine) containing proteins, thereby promoting an open chromatin conformation suitable for transcriptional activation [16–18].
Histone methylation is linked to multiple developmental processes including heterochromatin formation, cell cycle regulation, transcriptional silencing and transcriptional activation [19–25]. At least six lysine residues on histone H3 (K4, K9, K27, K36, K79) and H4 (K20) are targeted by histone lysine methyltransferases (HKMTs) [13, 24]. Except for H3K79 [26], SET domain-containing HKMTs have the ability to transfer one or multiple methyl groups to the ε-nitrogen of specific lysine residues on histones [10, 27]. Adding to the complexity of histone code, each lysine residue can be mono-, di- or tri-methylated [27, 28]. Moreover, unlike histone lysine acetylation which is generally associated with gene activation [29, 30], histone methylation at specific lysine residues can lead to either gene activation or repression [10].
Insight into the nature and regulation of enzymes responsible for modifications of specific amino acid residues in the nucleosomal core histones is essential towards deciphering the histone code. Whilst mammalian SUV39H1 was the first enzyme to be shown to possess HKMT activity towards H3K9 [19], its homolog, Su(var)3–9, in Drosophila was the first to be identified in a genetic screen for a suppressor of position effect variegation (PEV) [31]. The phenomenon of PEV was discovered by H. J. Muller in 1930 when describing the rearrangement of the white color eye gene from euchromatic to heterochromatic chromosomal regions. These conformational changes result in silencing and cell-to-cell variation of gene expression that lead to the mosaic eye color phenotype in Drosophila. Subsequent identification of HP1, that binds methylated H3K9, provided a bridge connecting H3K9 methylation to heterochromatin formation and hence gene repression [15, 32, 33]. In Drosophila, transcriptional repression or activation of homeotic gene expression is mediated by two distinct and antagonistic groups of protein complexes. Repression of homeotic gene expression is mediated by a Polycomb group (PcG) protein complex, containing Enhancer of zeste [E(Z)] and extra sex combs (ESC), that targets H3K27 and H3K9 methylation [34, 35]. In contrast, the trithorax group (trxG) proteins, ASH1 [36] and Trx [37], function in targeting H3K4 methylation [38–40] and hence in maintaining an active state of homeotic genes.
Identification of the SET domain followed from the observation [41] that the Drosophila E(Z) PcG protein includes a region with high sequence similarity to two previously identified trxG proteins: Drosophila Trx [37] and human acute lymphoblastic leukemia 1 (ALL-1) [42, 43]. The presence of this conserved region in these two groups of proteins with opposing functions (maintaining gene repression by PcG and activation by trxG proteins) led to the proposition [41] that it may comprise a domain that interacts with common nucleic acid or protein targets for gene regulation. Later, Su(var)3-9 was found to contain the C-terminal domain that is also shared by E(Z) and Trx [31]. The SET domain is named after these three founding Drosophila proteins: Suppressor of variegation 3-9 [Su(var)3-9], Enhancer of zeste [E(Z)], and Trithorax (Trx) [31]. The discovery of the evolutionarily conserved SET domain, present in the above-mentioned HKMTs, was an exciting step towards a comprehensive understanding of epigenetic regulation of gene expression through histone methylation. Here, we summarize current knowledge concerning plant SET proteins and compare the known functions of orthologous SET proteins of plants and other organisms. In addition to the existing classification of SET proteins in Arabidopsis and maize [22, 44], we have included 9 additional Arabidopsis SET proteins and 34 rice SET proteins from Plant Chromatin Database (ChromDB; http://www.chromdb.org). We also comment on recent evidence that alternative splicing may be relatively common for plant SET genes and consider the possibility that it has a functional role in regulating histone methylation. In addition, the possible role of antisense RNA in expression of Arabidopsis SET genes will be discussed.
2. Classification and functions of plant SET proteins
Proteins containing the conserved SET domain can be found in organisms ranging from virus to all three domains of life (Bacteria, Archaea, and Eukayota). At the time of writing, 1026 entries of SET proteins are cataloged in the Pfam sequence alignment database [45]. According to annotation from both Pfam [45] and ChromDB, at least 47, 37 and 35 SET proteins are present in Arabidopsis, rice and maize respectively. In plants, Arabidopsis SET (AtSET) proteins are the best annotated and characterized. Baumbusch et al. [44] were the first to classify 37 putative Arabidopsis SET genes into four distinct classes: 1) Enhancer of zeste [E(z)] homologs; 2) Ash1 homologs and related; 3) trithorax (trx) homologs and related; and 4) Suppressor of variegation [Su(var)] homologs and related. Subsequently, Springer et al. [22] classified 32 Arabidopsis and 22 maize SET proteins into 19 orthology groups distributed among five classes according to their phylogenetic relationships and domain organization.
We have added SET genes from rice to those of Arabidopsis and maize based on their sequence homology and phylogenetic relationships (Table 1). Except for the addition of two classes (VI and VII), this analysis is consistent with the classification by Springer et al. [22]. Although much remains to be experimentally verified, in general, it appears that the resulting groupings reflect the substrate specificities of the members.
Table 1.
SET domain-containing protein in plants.
| Class1 | Gp2 | Arabidopsis thaliana3 | Zea Mays4 | Oryza sativa spp. Japonica5 |
|---|---|---|---|---|
| I | 1 | MEA (SDG5; At1g02580; 689aar) | - | - |
| 2 | CLF (SDG1; At2g23380; 902aar) | MEZ1 (AF443596; 931aar) | SDG711 (Os06g16390; 896aar) | |
| 3 | SWN (SDG10; At4g02020; 856aar) | MEZ2 (AF443597; 894aar), MEZ3 (AF443598; 895aar) | SDG718 (Os03g19480; 895aar) | |
| II | 1 | ASHH3 (SDG7; At2g44150; 363aar) | SDG110 (AF545814; 342aar) | SDG724 (Os09g13740; 340aar) |
| ASHH4 (SDG24; At3g59960; 352aar) | - | - | ||
| 2 | ASHR3 (SDG4; At4g30860; 497aar) | - | SDG736 (Os02g39800; 492aar) | |
| 3 | ASHH1 (SDG26; At1g 76710; 492aar) | SDG102 (AY122273; 513aar) | SDG708 (Os04g34980; 518aar) | |
| 4 | ASHH2 (SDG8; At1g77300; 1759aar) | - | SDG725 (Os02g34850; 2133aar) | |
| III | 1 | ATX1 (SDG27; At2g31650; 1062aar) | - | SDG723 (Os09g04890; 1022aar) |
| ATX2 (SDG30; At1g05830; 1193aar) | SDG106 (DR826986; 385aar)*, SDG128 (DV534215; 296aar)* | - | ||
| 2 | ATX3 (SDG14; At3g61740; 902aar) | - | - | |
| ATX4 (SDG16; At4g27910; 912aar) | SDG115 (DN233805; 147aar)* | SDG721 (Os01g11950; 991aar) | ||
| ATX5 (SDG29; At5g53430; 1043aar) | - | SDG705 (Os01g46700; 1057aar) | ||
| 3 | ATXR3 (SDG2; At4g15180; 2351aar) | SDG108 (DV025562; 614aar)* | SDG701 (Os08g08210; 2257aar) | |
| 4 | ATXR7 (SDG25; At5g42400; 1421aar) | SDG127 (DN204179; 141aar)* | SDG717 (Os12g41900; 1212aar) | |
| IV | ATXR5 (SDG15; At5g09790; 352aar) | SDG139 (DV172958; 231aar)* | SDG720 (Os01g73460; 385aar) | |
| ATXR6 (SDG34; At5g24330; 349aar) | SDG129 (BM736459; 109aar)* | SDG730 (Os02g03030; 361aar) | ||
| V | 1 | SUVH1 (SDG32; At5g04940; 670aar) | SDG101 (DV540845; 795aar) | SDG704 (Os11g38900; 813aar), SDG713 (Os03g20430; 627aar) |
| SUVH3 (SDG19; At1g73100; 669aar) | SDG105 (AY093419; 678aar), SDG111 (AY187718; 486aar) | SDG728 (Os05g41170; 672aar) | ||
| SUVH7 (SDG17; At1g17770; 693aar) | SDG113 (AF545813; 766aar) | SDG709 (Os01g59620; 736aar), | ||
| SUVH8 (SDG21; At2g24740; 755aar) | - | SDG733 (Os11g03700; 663aar), SDG734 (Os12g03460; 663aar) | ||
| 2 | SUVH4 (SDG33; At5g13960; 624aar) | SDG118 (AY122271; 696aar) | SDG714 (Os01g70220; 663aar) | |
| SUVH6 (SDG23; At2g22740; 790aar) | - | SDG727 (Os09g19830; 921aar) | ||
| 3 | SUVH2 (SDG3; At2g33290; 651aar) | SDG136 (DN217108; 662aar) | SDG726 (Os07g25450; 684aar) | |
| SUVH9 (SDG22; At4g13460; 650aar) | SDG135 (CF049431; 449aar),* SDG137 (DN232551; 685aar) | SDG715 (Os08g45130; 594aar) | ||
| 4 | SUVR3 (SDG20; At3g03750; 338aar) | SDG116 (CO520549; 128aar)* | SDG729 (Os01g56540; 338aar) | |
| 5 | SUVH5 (SDG9; At2g35160; 794aar) | SDG103 (DN214510, 654aar)*, SDG104 (AY122272; 886aar), SDG119 (DV031465, 508aar)* | SDG703 (Os04g34990; 842aar), SDG710 (Os08g30910; 1173aar) | |
| 6 | SUVR1 (SDG13; At1g04050; 630aar) | - | - | |
| SUVR2 (SDG18; At5g43990; 717aar) | - | SDG712 (Os02g40770; 741aar) | ||
| SUVR4 (SDG31; At3g04380; 492aar) | SDG107 (DV544186; 148aar)* | - | ||
| 7 | SUVR5 (SDG6; At2g23740; 203aar) | SDG117 (AY187719; 1198aar), SDG131 (DN228281; 131aar)* | SDG706 (Os02g47900; 1136aar) | |
| VI | ASHR1 (SDG37; At2g17900; 480aar) | SDG130 (AAL75997; 410aar) | SDG716 (Os03g49730; 403aar) | |
| ASHR2 (SDG39; At2g19640; 398aar) | - | SDG740 (Os08g10470; 392aar) | ||
| ATXR1 (SDG35; At1g26760; 969aar) | SDG122 (DY238779; 534aar) | SDG739 (Os03g07260; 536aar) | ||
| ATXR2 (SDG36; At3g21820; 473aar) | SDG140 (DV505249; 217aar)* | SDG722 (Os04g53700; 517aar) | ||
| ATXR4 (SDG38; At5g06620; 258aar) | SDG123 (AY172976; 303aar) | SDG741 (Os10g27060; 298aar) | ||
| VII | SDG40 (At5g17240; 491aar) | SDG731 (Os07g28840; 479aar) | ||
| Putative RuBisCo SSMT (At3g07670; 504aar) | ||||
| At2g18850 (543aar) | ||||
| At5g14260 (514aar) | ||||
| RuBisCo LSMT (At1g14030; 482aar) | ||||
| At1g01920 (572aar) | ||||
| At1g24610 (476aar) | ||||
| At3g55080 (463aar) | ||||
| At3g56570 (531aar) | ||||
| At4g20130 (483aar) | ||||
SET protein from Arabidopsis, maize and rice were classified into seven classes. Classification of Arabidopsis and maize Class I to V was reported in Springer et al. [22]. Class VI and VII include SET proteins with truncated or interrupted SET domains.
Different orthology groups of Class I to V SET proteins defined by Springer et al. [22].
Arabidopsis SET proteins listed in Pfam database, common name, ChromDB name, locus number and amino acid length are indicated.
Maize SET proteins listed in ChromDB. Asterisk denotes length of partial protein sequence and corresponding accession number of EST sequence.
Rice SET proteins from Chrom DB were queried against Arabidopsis protein database using NCBI BLAST and their domain architecture were determined using NCBI conserved domains database. Phylogenetic relationship between homologous proteins were determined by Vector NTI software (Invitrogen) and the proteins are grouped into different orthology groups corresponding to Arabidopsis and maize SET proteins based on both sequence homology and domain architecture. Locus number and amino acid length are indicated. Rice SDG701, SDG732 and SDG738 were not grouped.
2.1 Class I: Enhancer of zeste [E(Z)] homologs (H3K27)
In Drosophila, homeotic gene regulation (via H3K27 methylation) is mediated by the polycomb repressive complex 2 (PRC2) that contains the PcG proteins, E(Z), ESC, P55 and Su(z)12 [34, 35, 46–48]. As reviewed in Gutton and Berger [49], homologs corresponding to these PcG proteins are also present in Arabidopsis PcG complexes that function in mediating various aspects of plant development. In Arabidopsis, CURLY LEAF (CLF), SWINGER (SWN) and MEDEA (MEA) constitute the E(Z) homologs of SET domain-containing PcG proteins (Table 1). CLF was the first identified plant SET domain-containing protein within this class [50]. In Arabidopsis, CLF controls leaf and flower morphology as well as flowering time through repression of the floral homeotic genes AGAMOUS (AG) and SHOOTMERISTEMLESS (STM) [51, 52]. Its close relative, SWN, acts redundantly in mediating tri-methylation of histone H3K27 at both AG and STM loci [52, 53]. MEA plays a role in maintaining its own imprinting during endosperm development and methylation of H3K27 is associated with MEA silencing during vegetative development and male gametogenesis [54–56]. These recent findings provide further evidence that the E(Z) group of SET domain-containing proteins is primarily associated with H3K27 methylation [22] and that tri-methylation of H3K27 is present at specific repressed loci at different stages of plant development. In Drosophila, an additional polycomb repressive complex, PRC1, was found to impede chromatin remodeling mediated by hSWI/SNF [57, 58]. Although no complex corresponding to PRC1 has been identified in plants, the presence of a family of multiple PcG members in Arabidopsis suggested that they may constitute alternative functional PRC2 complexes [49, 53]. For example, Makarevich et al. [59] recently showed that at least two different MEA, CLF or SWN-containing PcG complexes are involved in histone H3K27 tri-methylation at PHERES1 (PHE1) or FUSCA3 (FUS3) loci during seed development. In addition, vegetative expression of PHE1 and FUS3 was upregulated only in the swn clf double mutant, suggesting their redundant role in constituting PRC2, and hence their participation in different PRC2 complexes, thereby repressing PHE1 [59].
A conserved role of SET proteins in plant development is reinforced by the finding that overexpression of the SET domain of the rice CLF homolog, OsSET (SDG711), affects shoot development in transgenic Arabidopsis [60]. In addition to the C-terminally located SET domain, alignment of Arabidopsis and maize E(Z) homologs (MEZ1, MEZ2 and MEZ3) revealed several domains that are unique to this class of proteins (Fig. 1). These include two domains of unknown function (EZD1 and EZD2), a SANT (SWI3, ADA2, N-CoR and TFIIIB″ DNA-binding domains) and a cysteine-rich region [61]. These domains can also be found in rice OsSET1 and OsiEZ1 (SDG718) [62]. Therefore, it is highly likely that these domains may facilitate the ability of the SET domain to modify histones, perhaps by contributing to the formation of the PcG complex that is functionally important for this class of proteins.
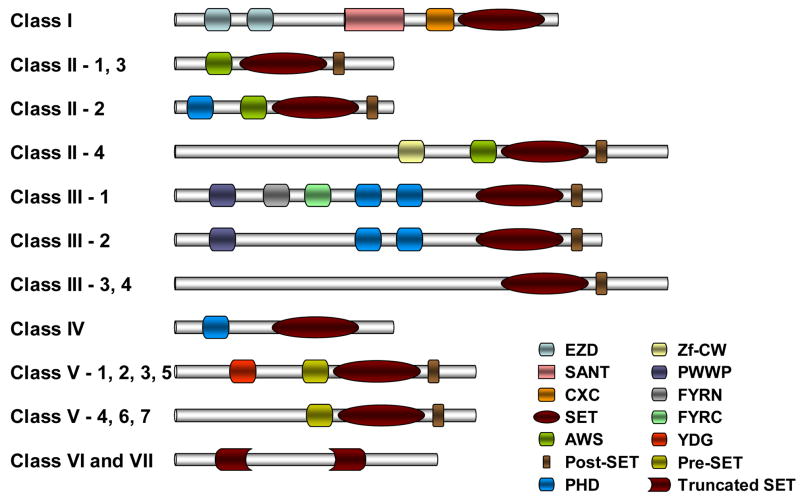
Fig. 1.
Domain architecture of plant SET proteins. Representative schematic diagram (right) showing overall domain composition of various orthology groups of SET proteins from different classes (left). Diagrams for Arabidopsis and maize SET proteins are described in detail by Springer et al. [22]. Abbreviations: EZD, E(Z) domain; SANT, SWI3, ADA2, N-CoR and TFIIIB″ DNA-binding domain; CXC, cysteine-rich region; AWS, associated with SET domain; PHD, plant homeodomain; Zf-CW, a zinc finger with conserved Cys and Trp residues; PWWP, domain named after a conserved Pro-Trp-Trp-Pro motif; FYRN, F/Y-rich N-terminus; FYRC, F/Y-rich C-terminus; YDG, conserved Tyr-Asp-Gly motif.
2.2 Class II: The ASH1 homologs (H3K36)
All members within this class have a SET domain that is invariably preceded by an AWS (associated with SET) domain and followed by a cysteine-rich post-SET domain (Fig. 1). As reported by Springer et al. [22], this class of proteins can be grouped into several orthology groups, based on the position of the SET domain. As shown in Table 1, the rice homologs can be classified into corresponding subgroups.
Arabidopsis ASHH1, maize SDG102 and rice SDG708 contain an N-terminally located SET domain while ASHH3, ASHH4, SDG110 and SDG724 possess a more centrally located SET domain. Compared to other proteins within this class, ASHH2 (1759-aar) is significantly larger and it, like rice SDG725 (2133-aar), can be placed in a distinct orthology group. In addition to the presence of canonical domains (AWS, SET, post-SET), ASHH2 contains a CW (cysteine and tryptophan conserved) domain (as predicted by Pfam-A) that can be found in at least five other protein families in higher plants [63]. As for the orthology group that contains ASHR3 and SDG736, an additional PHD (plant homeodomain) element is located upstream of the AWS domain (Fig. 1). Therefore, it is possible that during the evolution of these SET domain-containing proteins, an additional domain was acquired to enable additional functions.
Although biochemical characterization of proteins within this class is not yet available, two lines of evidence suggest that this class of proteins is involved in H3K36 methylation. ASHH1 is the Arabidopsis homolog of yeast Set2 [44] which has been shown to exhibit H3K36 methyltransferase activity [64]. In addition, genetic studies revealed that ashh2 mutants show an early flowering phenotype that is accompanied by a decrease in H3K36 methylation at the FLOWERING LOCUS C (FLC) [65].
2.3 Class III: The trithorax homologs and related proteins (H3K4)
In contrast to PcG proteins, trxG proteins are positive regulators of homeotic gene expression [66, 67]. In Drosophila, Trx functions through the trithorax acetyltransferase complex (TAC1) in mediating homeotic gene expression [68]. H3K4 methyltransferase activity of Trx was only recently demonstrated experimentally [40]. In contrast, Set1, the Trx homolog in yeast, was the first and only H3K4 methyltransferase identified in yeast [69] and it is important for normal cell growth and silencing of rDNA [70]. Similar to Trx, Set1 functions in a large protein complex, COMPASS (Complex Proteins Associated with Set1), in mediating histone methylation [71, 72]. In Arabidopsis, it has been demonstrated that H3K4 methylation is associated with a permissive state of gene expression during plant development [73, 74]. ATX1 (subgroup III-1; Table 1) was the first protein confirmed to have H3K4 methyltransferase activity in plants and it is involved in floral development through activating flower homeotic genes [75, 76].
Chromatin immunoprecipitation experiments showed that tri-methylation of H3K4 is associated with active transcription from the seed-specific promoter, phaseolin (phas) through the activity of a family of B3 domain-containing transcription factors, including FUS3 (D.W-K Ng and T.C. Hall, unpublished data) and PvALF (Phaseolus vulgaris ABI3-like factor) [77–79]. In addition, an increase in H3K4me3 at the phas chromatin was detected within one hour upon phas activation, illustrating the dynamic aspects of chromatin remodeling in mediating the activation of this seed-specific promoter [79]. Moreover, LEC1 (LEAFY COTYLEDON1; [80]) and FUS3 (transcription factors involved in embryo development) are up-regulated in the clf swn mutant [59]. Therefore, further studies on the roles of the various classes SET proteins in modulating epigenetic regulation of gene expression during seed development are likely to provide new insight to developmental processes.
As shown in Fig. 1, in addition to the SET domain, Class III proteins bear several highly conserved protein domains (PWWP, FYRN, FYRC) and the PHD domain (often found in chromatin-associated proteins [81, 82]). While all these conserved domains can be located in proteins within subgroup III-1 (ATX1 and ATX2), subgroup III-2 proteins (ATX3, ATX4 and ATX5) lack the FYRN and FYRC domains. In contrast to other subgroups of proteins within the same class, ATXR3, ATXR7 (subgroups III-3 and -4) and their corresponding rice homologs (SDG701 and SDG717) lack all of the highly conserved domains, and they possess only a C-terminally located SET domain and a post-SET domain. Although biochemical characterization is lacking for the other members of the plant Trx class, the presence of various highly conserved domains within this class of proteins may suggest diverse functions for these SET domain-containing proteins. In fact, Alvarez-Venegas et al. [83] showed that ATX1 interacts with phosphatidylinositol 5-phosphate (PI5P) through its PHD finger. PI5P is a component of the cell lipid pool and acts as secondary messenger mediating several plant processes, such as hyperosmotic stress response and membrane trafficking, via lipid signal transduction [84]. Such interaction between PHD finger and PI5P has also been demonstrated in animals [85]. These findings support the idea that SET domain proteins may act as transducers that bind signaling molecules such as PI5P, modifying enzymatic activity either by complete deactivation or by changing substrate specificity.
2.4 Class IV: Proteins with a SET and a PHD domain
This class of proteins is characterized based on the presence of a PHD finger and a C-terminally located SET domain (Fig. 1). Corresponding rice homologs, SDG720 and SDG730, can be located in the ChromDB in addition to the Arabidopsis (ATXR5 and ATXR6) and maize (SDG139 and SDG129) members of this class. In contrast to Class III proteins, Springer et al. [22] suggested that Arabidopsis ATXR5 and ATXR6 arose from an independent duplication of Arabidopsis SET genes and that certain conserved amino acid residues within the SET domain important for HKMT activity are different. Nevertheless, the presence of the PHD finger and recent findings that this domain can bind methylated H3K4 [86, 87], suggested that this class of proteins, like those in Class III, is closely related to the trithorax class of SET-domain containing proteins. Although little is known about the function of this class of proteins, recent data suggested that ATXR5 and ATXR6 may contribute to cell cycle regulation or DNA replication through interactions with proliferating cell nuclear antigen (PCNA) [88].
2.5 Class V: Suppressor of variegation [Su(var)] homologs and relatives (H3K9)
This large class of proteins is characterized by the presence of pre-SET, SET and post-SET domains (Fig. 1). In addition to the already classified Arabidopsis and maize Su(var) proteins, corresponding rice homologs are sorted in Table 1 into the various orthologous groups. In all the Su(var) homologs (subgroups V-1, -2 and -5), an additional conserved YDG (conserved Tyr-Asp-Gly motif) domain is present upstream of the pre-SET domain.
Members of this class are likely to be involved in heterochromatin formation mediated by H3K9 methylation [33, 89, 90]. In Arabidopsis, SUVH4 (KYP), the best studied member of this class, represses expression of the floral homeotic gene, SUPERMAN (SUP), through its H3K9 methyltransferase activity [90]. Naumann et al. [91] reported that the N-terminus and YDG domain in SUVH2 are involved in directing DNA methylation and subsequent histone methylation at its target locus so that heterochromatic methylation marks are affected in suvh2 mutant plants. Recently, Ebbs et al. [92, 93] showed that there is a hierarchical and locus-specific control of H3K9 di-methylation as well as non-CG DNA methylation by SUVH4, SUVH5 and SUVH6 HMT in Arabidopsis. The involvement of this class of proteins in heterochromatin formation was demonstrated for NtSET1 in tobacco [94], and reinforced by the subsequent finding that NtSET1 interacts with LHP1 (Arabidopsis homolog of HP1) [95]. In addition, Yu et al. [95] showed that NtSET1 contains both H3K9 and H3K27 methylation activities in vitro and that overexpression of NtSET1 leads to an increase in H3K9 dimethylation in tobacco BY2 cells. Overall, the highly conserved, plant-specific distribution of domains (YDG, Pre-SET and SET) found in subgroups V-1, -2 and -5 (Fig. 1) of proteins suggests that they share a recent, plant-specific common ancestor [61].
Unlike the Su(var) homologs, all the Su(var)-related homologs (subgroups V-4, -6 and -7) lack the YDG domain (Fig. 1). However, Thorstensen et al. [96] have identified a novel conserved plant specific N-terminal domain, WIYLD, that is present in Arabidopsis SUVR1, SUVR2, SUVR4 as well as in other SUVR-homologs of other plant species. Interestingly, the authors also showed that recombinant SUVR4 acts as a di-methyltransferase with preference for mono-methylated H3K9 as substrate. Therefore, this finding may suggest that the SUVH and SUVR proteins can act in concert in achieving various functional H3K9 methylation states that will eventually lead to DNA methylation in a locus-specific manner [97].
2.6 Class VI: Proteins with an interrupted SET domain
Proteins from both Classes VI and VII lack the diverse domains that can be found in other classes of SET proteins and the SET domain is either truncated or interrupted in the SET-I region (Fig. 1). Class VI consists of two ASH-related proteins (ASHR1 and ASHR2) and three Trx-related (ATXR1, ATXR2 and ATXR4) proteins. Homologous SET genes can be found in maize (SDG122, SDG123, SDG130 and SDG140) and rice (SDG716, SDG740, SDG739, SDG722 and SDG741). This suggests that this class of proteins existed before the divergence of monocot and dicot plants. The SET domain of ASHR1 is interrupted by a Zf-MYND [zinc finger MYND (Myeloid, Nervy, and DEAF-1)] domain. Brown et al. [98] recently reported that Smyd2, the human homolog of Arabidopsis ASHR1, restricts cell-cycle progression through di-methylation of H3K36. Although not yet established in plants, it is likely that, like its human homolog, ASHR1 will be shown to have H3K36 methyltransferase activity.
2.7 Class VII: RBCMT and other SET-related proteins
While the majority of SET domain-containing proteins possess histone-methylating activity, Class VII SET proteins methylate non-histone proteins (Table 1). Rubisco large (RBLSMT) and small (RBSSMT) subunit methyltransferases (RBCMTs) exemplify SET-related proteins that methylate non-histone targets [99–101]. While the N- and C-termini of these proteins bear high similarity to those of other SET domain proteins (Fig. 1), the SET-I region has only weak similarity to, and is longer than that of canonical SET proteins. Nevertheless, the structural features within the SET-I region essential for substrate-specific methylation are retained [100, 102]. We include Arabidopsis SDG40, two unnamed proteins (corresponding to At2g18850 and At5g14260) and rice SDG731 in this class as they show some similarity to RBSSMT. The remaining five uncharacterized Arabidopsis proteins were classified into this class as they contain domain architecture resembling that of RBLSMT.
3. Regulation of plant SET proteins
3.1. Regulation of SET protein functions through protein-protein interactions
At least some SET proteins are capable of self-association [103–105]. This property suggests that the lysine-methyltransferase activity, including varying degrees of methylation (mono-, di- or tri-methylation), substrate specificity and localization can be subjected to regulation by intra- and inter-molecular interactions with other proteins. Indeed, modulation of the SET domain-mediated lysine-methyltransferase activity by interacting proteins has been demonstrated for yeast SET1-COMPASS [106, 107] and for human MLL1 trithorax complex [108].
Regulation of histone methylation by SET domain-containing methyltransferases via their interaction with other proteins has been demonstrated in yeast and mammalian systems [48, 72, 109]. In plants, the regulation of SET protein-mediated histone methylation through protein-protein interactions is poorly understood. However, the involvement of E(Z) class proteins in PcG complexes that contain various non-SET proteins provides an avenue to investigate the role, if any, of interacting proteins in regulating the methyltransferase activity. In Arabidopsis, the involvement of MEA, CLF and SWN in developmental processes may be linked to their interactions with other non-SET proteins, as mutations in the interacting partners display phenotypes indicative of affecting similar developmental pathways. During seed development, MEA is part of a PcG complex that includes several other regulators of early seed development such as FERTILIZATION-INDEPENDENT ENDOSPERM (FIE) and FERTILIZATION-INDEPENDENT SEED2 (FIS2) [110–112]. FIE is a WD40 domain-containing protein that shows similarity to the Drosophila EXTRA SEX COMB (ESC). FIS2 is a Zn-finger protein similar to Su(Z)12 [35, 47]. Physical interactions between MEA and FIE [113, 114]), and between MEA (or SWN) and FIS2 [53, 115] have been demonstrated using yeast-two-hybrid assays. Therefore, based on the similarities in the mutant phenotypes and their requirement for the repression of PHE1 during endosperm development [59], complex formation is a prerequisite for MEA or SWN to function as HMTs in planta.
Recently, Kohler et al. [116] showed that Arabidopsis MULTICOPY SUPPRESSOR OF IRA 1 (MSI1) interacts with FIE and is required for seed development. Like FIE, MSI1 is a WD40 domain-containing protein that shares similarity with Drosophila NURF55 [35]. This group further demonstrated that MSI1 is a component of a 600 kDa protein complex that includes MEA and FIE. The high degree of sequence similarity and the functional association of various relevant subunits strongly suggest that the MEA-FIE-FIS2-MSI1 complex may be the plant homolog of the fly and mammalian ESC-E(Z) complexes. Interestingly, FIE also physically interacts with CLF [51]. Based on the similarity of mutant phenotypes, EMF2 (a zinc-finger protein) has been proposed to be an additional component of a putative complex containing FIE and CLF [53]. Recently, Wood et al. [117] showed that VERNALIZATION2 (VRN2), a Zn-finger domain-containing protein, and VERNALIZATION INSENSITIVE 3 (VIN3), a PHD domain-containing protein, also interact with the FIE-CLF-SWN complex, providing a mechanism for the repression of FLC expression during vernalization. This finding further supports the idea that the functionality of SET proteins is diversified through protein-protein interactions.
In addition to its methyltransferase activity, the substrate specificity of a particular SET protein may be mediated by protein interactions. For example, the WDR5 (a WD40 domain-containing protein) component of MLL1 trithorax complex has been shown to confer substrate specificity by interacting with di-methylated H3K4 [118, 119]. By analogy, the importance of the WD40 domain-containing protein, FIE, to overall plant development can be explained by its association with MEA and CLF in the regulation of sporophytic and gametophytic stages, respectively [51, 120]. It can be speculated that FIE may regulate HMT activity by maintaining complex integrity and (or) by providing substrate specificity similar to that observed for WDR5 in the human MLL1 complex. The presence of an additional WD40 domain protein, MSI1, in the MEA-FIE-FIS2-MSI1 complex may confer the ability to recognize two different methylated lysine residues on histones.
3.2. Alternative splicing of Arabidopsis SET genes
The presence of introns in eukaryotic RNAs confers multiple degrees of flexibility in gene expression via various molecular mechanisms, such as developmental and cell-specific regulation of transcription [121], generation of non-coding RNAs [122], and alternative splicing [123]. Among these mechanisms, alternative splicing is of special interest in that it can produce two or more forms of mature mRNA from the same precursor transcript and hence generate transcripts bearing diverse information.
In general, alternative splicing in non-translated regions can have an impact on protein expression levels [124]. Conversely, in coding regions, it may alter protein structure and function. In extreme cases, alternative splicing can lead to the production of two proteins with antagonistic functions [125]. It is estimated that 60–80% of human genes undergo alternative splicing [126], contributing to human genome complexity. A smaller percentage of plant genes, ~22% in Arabidopsis and 10% in rice [127], also experience alternative splicing. It is possible that these numbers will increase in the future, as more plant cDNAs or ESTs are characterized. Any statement about alternative splicing events should take into consideration the fact that they may be the result of experimental artifacts such as sequencing errors, splicing errors, and RNA degradation events [127, 128]. However, despite these possibilities, some alternative splicing events, especially those that are evolutionarily conserved, may be biologically important and contribute to organismal fitness [129]. Expression of alternatively spliced transcripts can be tissue specific [130] and can be differentially regulated by physical [131] or chemical [132] stimuli, arguing for the involvement of multiple regulatory systems in the expression of individual isoforms.
Alternative splicing in the coding region of Arabidopsis SET genes may generate proteins with distinct functions. Such effects have been reported for Drosophila Su(var)3-9, which can express two distinct transcripts (2.4 kb and 2.0 kb), encoding two proteins that share similarity only for the first 80 amino acid residues (aar). Whereas the 2.4 kb transcript is translated to yield a SET domain-containing HMT, the 2.0 kb transcript encodes the γ subunit of eukaryotic translation initiation factor 2 [133]. Conversely, protein isoforms from alternative splicing in coding regions may have redundant functions if the conserved domains are not affected. An example is zebrafish SET gene, SmyD1. Inactivation of either isoform of this gene results in no morphological phenotype whereas debilitation of both isoforms have severe effects on myofibril organization [134]. The possibility also exists that SET protein isoforms resulting from alternative splicing assemble into higher order protein complexes. Homodimerization or heterodimerization has been reported for the virus SET protein vSET [135], human G9a, and GLP [136], and Drosophila ASH1 and Trx [105]. If plant SET proteins form dimers, alternative splicing of SET genes could generate proteins forming various homo- and hetero-dimers having distinct biological activities.
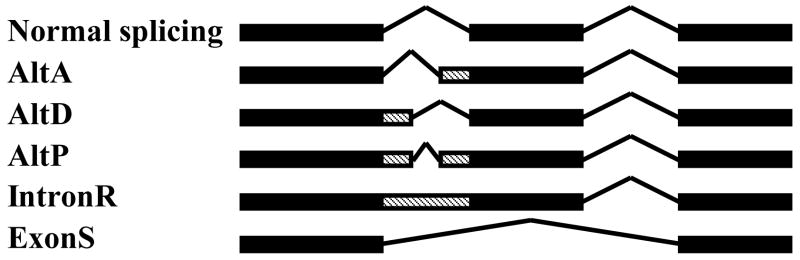
Scanning all 47 Arabidopsis SET genes against the Alternative-Splicing-in-Arabidopsis database (ASIP) [127], the UniProt consortium [137], and published literature [96], led to the identification of 18 (38%) Arabidopsis SET genes with alternative splicing (Table 2). This is a substantially higher percentage than the overall percentage (22%) of splicing for Arabidopsis genes [127], and suggests that alternative splicing is important in generating SET protein complexity and functionality. Alternative splicing in SET genes occurs through various mechanisms (Fig. 2) including alternative donor (AltD), alternative acceptor (AltA), alternative position (AltP), intron retention (IntronR), and exon skipping (ExonS) [127]. A search in the ASIP database revealed two pairs of SET genes from Arabidopsis and rice (At2g19640 and OS08g10470; At5g17240 and Os07g28840) for which the same alternative splicing mechanism (intron retention) is employed to produce mature transcripts encoding the orthologous proteins. This provides a specific example of evolutionary conservation, presumably as a result of some functional advantages.
Table 2.
Arabidopsis SET genes that undergo alternative splicing and/or with naturally occurring antisense transcripts.
| Class1 | SET gene (locus)2 | Alternative splicing patterns3 | Possible effects on proteins4 | Antisense5 |
|---|---|---|---|---|
| II-2 | ASHR3 (At4g30860) | IntronR | use a different stop codon | - |
| II-3 | ASHH1 (At1g76710) | AltA | no effect on domain structure | - |
| II-4 | ASHH2 (At1g77300) | AltA; ExonS | no effect on domain structure | Y |
| III-1 | ATX1 (At2g31650) | AltD | no effect on domain structure | - |
| III-1 | ATX2 (At1g05830) | AltA | affect 3′UTR | - |
| III-2 | ATX3 (At3g61740) | IntronR | use a different stop codon | - |
| III-2 | ATX5 (At5g53430) | - | - | Y |
| IV | ATXR5 (At5g09790) | AltA; ExonS | affect SET domain | Y |
| V-1 | SUVH1 (At5g04940) | IntronR | affect 3′UTR | Y |
| V-1 | SUVH3 (At1g73110) | - | - | Y |
| V-1 | SUVH8 (At2g24740) | - | - | Y |
| V-2 | SUVH6 (At2g22740) | - | - | Y |
| V-4 | SUVR3 (At3g03750) | AltP | affect SET domain | - |
| V-5 | SUVH5 (At2g35160) | AltA | use a different start codon | - |
| V-6 | SUVR1 (At1g04050) | IntronR | use a different start codon | - |
| V-6 | SUVR2 (At5g43990) | AltA; AltD | use a different start codon | - |
| V-6 | SUVR4 (At3g04380) | IntronR | no effect on domain structure | - |
| VI | ASHR2 (At2g19640) | IntronR | no effect on domain structure | - |
| VI | ATXR1 (At1g26760) | - | - | Y |
| VII | SDG40 (At5g17240) | IntronR | affect SET domain | - |
| VII | At5g14260 | AltD; IntronR | affect 3′UTR | - |
| VII | At1g01920 | AltA; IntronR | affect SET domain | - |
| VII | At1g24610 | - | - | Y |
| VII | At3g55080 | AltA; ExonS | affect SET domain | - |
Orthology groups of Arabidopsis SET proteins listed in Table 1.
Arabidopsis SET genes, common name and/or locus number are indicated
Abbreviations for alternative splicing mechanisms described in Fig. 2.
The possible effects of alternative splicing on proteins according to the conceptual translation of alternatively spliced transcripts.
Fig 2.
Alternative splicing patterns. Schematic diagram showing normal and five major alternative splicing patterns for a representative gene containing three exons (black boxes). Shaded boxes denote alternative exonic regions: AltA, alternative acceptor (3′ side of introns); AltD, alternative donor (5′ side of introns); AltP, alternative positions (both 5′ and 3′ side of introns); IntronR, intron retention; ExonS, exon skipping. Angled lines denote the intronic regions. Modified from: Wang and Brendel [127].
Alternative splicing of Arabidopsis SET genes could have important consequences in regard to protein functions. For 3 (ATX2, SUVH1, At5g14260) of the 18 alternatively spliced SET genes, examination of ESTs in the ASIP database revealed that splicing differences occurred in the 3′ UTRs (Table 2). The 3′ (or 5′) UTRs are known to contain elements that can control protein stability [138], protein translation efficiency [139], and spatial [140] or subcellular distribution of proteins [141]. Casas-Mollano et al. [142] showed that an intron located in the 5′UTR of the Arabidopsis SUVH3 regulates both constitutive and high levels of expression. Therefore, alternative splicing events observed in ATX2, SUVH1 and At5g14260 may affect the localization or abundance of transcripts or encoded proteins, rather than causing a change in the structure of encoded proteins.
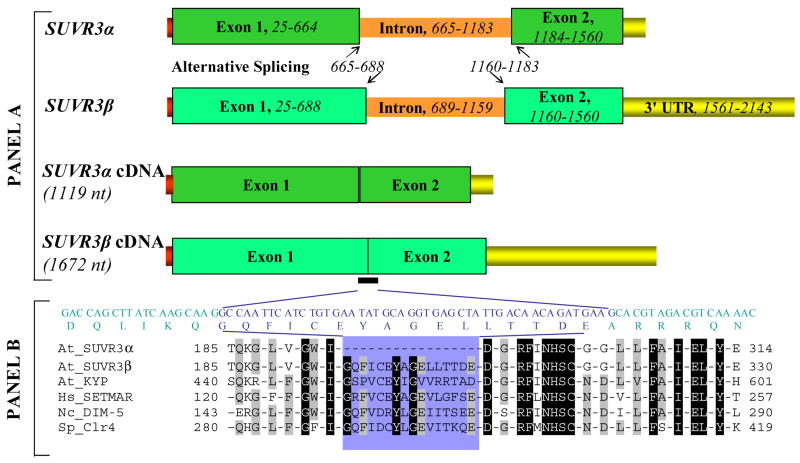
Alternative splicing for the other 15 Arabidopsis SET genes is predicted by conceptual translation to occur at the protein-encoding region of transcripts (Table 2). For 10 SET genes, alternative splicing events appear to affect the start/stop codon or a region of the protein that lies outside of a conserved domain of sequence similarity. However, the other 5 SET genes (At1g01920, At3g55080, At5g17240, ATXR5 and SUVR3) have alternative splicing affecting the SET domain. In the case of At1g01920, ~ 40-aar are added/removed from the SET-N motif of the encoded protein. The splicing event of At3g55080 seems to directly regenerate or eliminate the SET domain coding region. In At5g17240, a 79-nt intron is retained in one isoform, resulting in a frameshift near the beginning of the SET domain coding region. Similarly, a 59-nt exon deletion from one isoform of ATXR5 is likely to cause a frameshift at the SET domain coding region. In the case of SUVR3, the consequences resulting from alternative splicing may be dramatic. We have found that RNAi-mediated debilitation of SUVR3 in transgenic Arabidopsis results in aberrant flower formation and other abnormalities (Wang, T. et al., unpublished data). During analysis of these plants, we found that two different SUVR3 transcripts were present. Sequences identical to those of the transcripts are present in cDNA and EST libraries [143, 144], each comprising two exons and one intron (Fig. 3). It became evident that these RNAs encoded SUVR3 isoforms, with the total coding sequence length of isoform SUVR3α being 48-nt shorter than that in SUVR3β. This results in a 16-aar difference between the two isoforms, with a loss of 3 invariant and 3 conserved residues from SUVR3α, suggesting that the SUVR3 isoforms may differ in their ability to methylate histone H3. If so, alternative splicing could be seen to have a significant regulatory role in regard to chromatin remodeling.
Fig. 3.
Structures of SUVR3α and SUVR3β isoforms. Panel A: The red, green, orange and boxes denote 5′ UTR, exon, intron and 3′ UTR regions, respectively. Their positions in SUVR3α and SUVR3β are indicated. The 48-nt fragment represented by the black bar, which belongs to the exonic regions in SUVR3β but belongs to the intronic region in SUVR3α, and its corresponding 16-aar are shown in blue letters. Panel B: Sequence alignment of the SET domain of three Arabidopsis SET proteins: SUVR3α, SUVR3β, Arabidopsis KRYPTONITE (At_KYP, AAK28969); human SET domain mariner transposase (Hs_SETMAR, AAH11635); N. crassa (Nc) DIM-5 (CAF06044) and S. pombe (Sp) CLR4 (NP_595186). The aar highlighted are invariant (white on black) or conserved (white on gray) among all six SET proteins.
3.3. Anti-sense RNA expression of Arabidopsis SET genes
Our understanding of anti-sense transcription in all organisms is very limited. In mouse, genome-wide analysis of transcripts suggests that antisense transcription is widespread and not restricted to imprinted genes, as previously postulated [145]. In Arabidopsis, the ratio of expression of the sense (S) to the antisense (AS) transcripts fluctuates markedly among different tissues during development [146]. In addition, sense-antisense transcripts (SAT) tend to be poly(A)-negative and nuclear localized in Arabidopsis, as in mouse [147].
Naturally occurring AS transcripts (NATs) corresponding to nine Arabidopsis SET genes were found in the ASIP [127] and Arabidopsis Cis-NAT Pairs Databases [148] (Table 2). This opens the possibility that the expression of the sense transcript for some of these genes might be under the control of the anti-sense message. Sense-antisense association would result in double-stranded RNA (dsRNA), a natural substrate for a Dicer ribonuclease [149]. The small RNAs produced by this reaction could direct the formation of a silent chromatin state, thus silencing the expression of the homologous genomic region.
NATs can be found in both prokaryotes and eukaryotes. In eukaryotic cells, such RNAs are known to be mostly involved in genomic imprinting [150] and X chromosome inactivation [151]. The discovery that AS transcription is more widespread than previously thought challenges this view and suggests that dsRNA may play a more general role (or perhaps a wider role) in regulation of gene expression. Therefore, it would be quite informative to determine if NATs affect the expression and chromatin state of one or more AtSET genes and then to elucidate the mechanism by which NATs act at these loci. NATs could facilitate a sensitive, rapid and dynamic control over AtSET expression under different environmental cues. Determination of the spatial and temporal expression pattern of these NATs and their corresponding genes will provide some clues to the role of AtSET NATs in regulating plant development. Alternatively, NATs could assist in the permanent imprinting of AtSET genes. Except for MEA, a self-regulated imprinting SET gene [54–56] devoid of antisense transcript, no AtSET has been reported to be imprinted. Therefore, the nine AtSET genes for which AS transcript information is listed in Table 2 could be additional targets for genomic imprinting.
4. Future directions
The extensive genome duplication observed in plants appears to provide them with the plasticity necessary to successfully adapt to the various environmental conditions they encounter as sessile organisms. This is best exemplified by the diversification of genes involve in plant secondary metabolism through gene duplication [152, 153]. Such gene duplication events offer a great opportunity for studying gene evolution [154, 155]. In particular, studying the diverse functions of families of paralogous plant SET genes in plant development is likely to provide new insight to the molecular and functional evolution of proteins containing the SET and other auxiliary domains. In addition, although considerable attention has been paid to the SET domain proteins as histone methyltransferases, their activity as protein methyltransferases capable of modifying non-histone substrates and modulating their functions is still poorly understood. The paucity of knowledge is not restricted to plant biology, as only a few instances have been reported even in yeast and mammalian systems. For example, methylation of p53 by mammalian SET9 has been shown to cause the retention of p53 in the nucleus and increase the stability of an otherwise highly unstable protein and, consequently, regulate p53-dependent gene expression [156]. SET9 has also been demonstrated to methylate TAF10, a general transcription factor, thereby increasing its affinity to RNA polymerase II [157]. The methylation of p53 and TAF10 shows how SET domain proteins can play a role in transcription by methylating non-histone proteins. Furthermore, the SET domain-containing methyltransferases can contribute to cellular processes other than transcription. For instance, Set1 a histone H3 Lys4 methyltransferase has recently been shown to mediate methylation of Dam1, a kinetochore protein [158]. Therefore, it is possible Set1 may play an unknown function during mitosis. Determining the histone and non-histone protein substrates for the plant SET domain-containing methyltransferases is an exciting area of research that will continue to provide novel insight into the contribution of SET proteins to plant development.
Acknowledgments
This work was supported in part by NSF grants MCB 0110477 and MCB 0346681to TCH and U.S. Public Health Service Grant GM58770 to R. A. We thank Xiangyu Shi for help and discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Wolffe AP, Guschin D. Review: chromatin structural features and targets that regulate transcription. J Struct Biol. 2000;129:102–122. doi: 10.1006/jsbi.2000.4217. [DOI] [PubMed] [Google Scholar]
- 4.Finch JT, Noll M, Kornberg RD. Electron microscopy of defined lengths of chromatin. Proc Natl Acad Sci U S A. 1975;72:3320–3322. doi: 10.1073/pnas.72.9.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oudet P, Gross-Bellard M, Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975;4:281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- 6.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 7.Nemeth A, Langst G. Chromatin higher order structure: opening up chromatin for transcription. Brief Funct Genomic Proteomic. 2004;2:334–343. doi: 10.1093/bfgp/2.4.334. [DOI] [PubMed] [Google Scholar]
- 8.Tomschik M, Zheng H, van Holde K, Zlatanova J, Leuba SH. Fast, long-range, reversible conformational fluctuations in nucleosomes revealed by single-pair fluorescence resonance energy transfer. Proc Natl Acad Sci U S A. 2005;102:3278–3283. doi: 10.1073/pnas.0500189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 11.Iizuka M, Smith MM. Functional consequences of histone modifications. Curr Opin Genet Dev. 2003;13:154–160. doi: 10.1016/s0959-437x(03)00020-0. [DOI] [PubMed] [Google Scholar]
- 12.Luger K, Richmond TJ. The histone tails of the nucleosome. Curr Opin Genet Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 13.Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 14.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 15.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 16.Haynes SR, Dollard C, Winston F, Beck S, Trowsdale J, Dawid IB. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992;20:2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 18.Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 19.Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 20.Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 21.Sims RJ, 3rd, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 22.Springer NM, Napoli CA, Selinger DA, Pandey R, Cone KC, Chandler VL, Kaeppler HF, Kaeppler SM. Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol. 2003;132:907–925. doi: 10.1104/pp.102.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 25.Volkel P, Angrand PO. The control of histone lysine methylation in epigenetic regulation. Biochimie. 2007;89:1–20. doi: 10.1016/j.biochi.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 26.Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 27.Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- 28.Bannister AJ, Kouzarides T. Histone methylation: recognizing the methyl mark. Methods Enzymol. 2004;376:269–288. doi: 10.1016/S0076-6879(03)76018-2. [DOI] [PubMed] [Google Scholar]
- 29.Wade PA, Pruss D, Wolffe AP. Histone acetylation: chromatin in action. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 30.Lusser A, Kolle D, Loidl P. Histone acetylation: lessons from the plant kingdom. Trends Plant Sci. 2001;6:59–65. doi: 10.1016/s1360-1385(00)01839-2. [DOI] [PubMed] [Google Scholar]
- 31.Tschiersch B, Hofmann A, Krauss V, Dorn R, Korge G, Reuter G. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994;13:3822–3831. doi: 10.1002/j.1460-2075.1994.tb06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eissenberg JC, James TC, Foster-Hartnett DM, Hartnett T, Ngan V, Elgin SC. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1990;87:9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 35.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 36.Tripoulas NA, Hersperger E, La Jeunesse D, Shearn A. Molecular genetic analysis of the Drosophila melanogaster gene absent, small or homeotic discs1 (ash1) Genetics. 1994;137:1027–1038. doi: 10.1093/genetics/137.4.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazo AM, Huang DH, Mozer BA, Dawid IB. The trithorax gene, a trans-acting regulator of the bithorax complex in Drosophila, encodes a protein with zinc-binding domains. Proc Natl Acad Sci USA. 1990;87:2112–2116. doi: 10.1073/pnas.87.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beisel C, Imhof A, Greene J, Kremmer E, Sauer F. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature. 2002;419:857–862. doi: 10.1038/nature01126. [DOI] [PubMed] [Google Scholar]
- 39.Byrd KN, Shearn A. ASH1, a Drosophila trithorax group protein, is required for methylation of lysine 4 residues on histone H3. Proc Natl Acad Sci U S A. 2003;100:11535–11540. doi: 10.1073/pnas.1933593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith ST, Petruk S, Sedkov Y, Cho E, Tillib S, Canaani E, Mazo A. Modulation of heat shock gene expression by the TAC1 chromatin-modifying complex. Nat Cell Biol. 2004;6:162–167. doi: 10.1038/ncb1088. [DOI] [PubMed] [Google Scholar]
- 41.Jones RS, Gelbart WM. The Drosophila Polycomb-group gene Enhancer of zeste contains a region with sequence similarity to trithorax. Mol Cell Biol. 1993;13:6357–6366. doi: 10.1128/mcb.13.10.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce CM, Canaani E. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 43.Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 44.Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–4333. doi: 10.1093/nar/29.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ng J, Hart CM, Morgan K, Simon JA. A Drosophila ESC-E(Z) protein complex is distinct from other polycomb group complexes and contains covalently modified ESC. Mol Cell Biol. 2000;20:3069–3078. doi: 10.1128/mcb.20.9.3069-3078.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tie F, Furuyama T, Prasad-Sinha J, Jane E, Harte PJ. The Drosophila Polycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development. 2001;128:275–286. doi: 10.1242/dev.128.2.275. [DOI] [PubMed] [Google Scholar]
- 48.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 49.Guitton AE, Berger F. Control of reproduction by Polycomb Group complexes in animals and plants. Int J Dev Biol. 2005;49:707–716. doi: 10.1387/ijdb.051990ag. [DOI] [PubMed] [Google Scholar]
- 50.Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature. 1997;386:44–51. doi: 10.1038/386044a0. [DOI] [PubMed] [Google Scholar]
- 51.Katz A, Oliva M, Mosquna A, Hakim O, Ohad N. FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J. 2004;37:707–719. doi: 10.1111/j.1365-313x.2003.01996.x. [DOI] [PubMed] [Google Scholar]
- 52.Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, Jenuwein T, Goodrich J. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 2006;25:4638–4649. doi: 10.1038/sj.emboj.7601311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development. 2004;131:5263–5276. doi: 10.1242/dev.01400. [DOI] [PubMed] [Google Scholar]
- 54.Baroux C, Gagliardini V, Page DR, Grossniklaus U. Dynamic regulatory interactions of Polycomb group genes: MEDEA autoregulation is required for imprinted gene expression in Arabidopsis. Genes Dev. 2006;20:1081–1086. doi: 10.1101/gad.378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jullien PE, Katz A, Oliva M, Ohad N, Berger F. Polycomb group complexes self-regulate imprinting of the Polycomb group gene MEDEA in Arabidopsis. Curr Biol. 2006;16:486–492. doi: 10.1016/j.cub.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 57.Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 58.Francis NJ, Saurin AJ, Shao Z, Kingston RE. Reconstitution of a functional core polycomb repressive complex. Mol Cell. 2001;8:545–556. doi: 10.1016/s1097-2765(01)00316-1. [DOI] [PubMed] [Google Scholar]
- 59.Makarevich G, Leroy O, Akinci U, Schubert D, Clarenz O, Goodrich J, Grossniklaus U, Kohler C. Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 2006;7:947–952. doi: 10.1038/sj.embor.7400760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang YK, Wang Y, Zhang Y, Li SG, Lu XC, Li H, Zou C, Xu ZH, Bai SN. OsSET1, a novel SET-domain-containing gene from rice. J Exp Bot. 2003;54:1995–1996. doi: 10.1093/jxb/erg201. [DOI] [PubMed] [Google Scholar]
- 61.Springer NM, Danilevskaya ON, Hermon P, Helentjaris TG, Phillips RL, Kaeppler HF, Kaeppler SM. Sequence relationships, conserved domains, and expression patterns for maize homologs of the polycomb group genes E(z), esc, and E(Pc) Plant Physiol. 2002;128:1332–1345. doi: 10.1104/pp.010742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thakur JK, Malik MR, Bhatt V, Reddy MK, Sopory SK, Tyagi AK, Khurana JP. A POLYCOMB group gene of rice (Oryza sativa L. subspecies indica), OsiEZ1, codes for a nuclear-localized protein expressed preferentially in young seedlings and during reproductive development. Gene. 2003;314:1–13. doi: 10.1016/s0378-1119(03)00723-6. [DOI] [PubMed] [Google Scholar]
- 63.Perry J, Zhao Y. The CW domain, a structural module shared amongst vertebrates, vertebrate-infecting parasites and higher plants. Trends Biochem Sci. 2003;28:576–580. doi: 10.1016/j.tibs.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 64.Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, Allis CD. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao Z, Yu Y, Meyer D, Wu C, Shen WH. Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat Cell Biol. 2005;7:1256–1260. doi: 10.1038/ncb1329. [DOI] [PubMed] [Google Scholar]
- 66.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 67.Grimaud C, Negre N, Cavalli G. From genetics to epigenetics: the tale of Polycomb group and trithorax group genes. Chromosome Res. 2006;14:363–375. doi: 10.1007/s10577-006-1069-y. [DOI] [PubMed] [Google Scholar]
- 68.Petruk S, Sedkov Y, Smith S, Tillib S, Kraevski V, Nakamura T, Canaani E, Croce CM, Mazo A. Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science. 2001;294:1331–1334. doi: 10.1126/science.1065683. [DOI] [PubMed] [Google Scholar]
- 69.Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci U S A. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci U S A. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 74.Bouveret R, Schonrock N, Gruissem W, Hennig L. Regulation of flowering time by Arabidopsis MSI1. Development. 2006;133:1693–1702. doi: 10.1242/dev.02340. [DOI] [PubMed] [Google Scholar]
- 75.Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, Avramova Z. ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr Biol. 2003;13:627–637. doi: 10.1016/s0960-9822(03)00243-4. [DOI] [PubMed] [Google Scholar]
- 76.Alvarez-Venegas R, Avramova Z. Methylation patterns of histone H3 Lys 4, Lys 9 and Lys 27 in transcriptionally active and inactive Arabidopsis genes and in atx1 mutants. Nucleic Acids Res. 2005;33:5199–5207. doi: 10.1093/nar/gki830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bobb AJ, Eiben HG, Bustos MM. PvAlf, an embryo-specific acidic transcriptional activator enhances gene expression from phaseolin and phytohemagglutinin promoters. Plant J. 1995;8:331–343. doi: 10.1046/j.1365-313x.1995.08030331.x. [DOI] [PubMed] [Google Scholar]
- 78.Li G, Bishop KJ, Chandrasekharan MB, Hall TC. β-phaseolin gene activation is a two-step process: PvALF- facilitated chromatin modification followed by abscisic acid-mediated gene activation. Proc Natl Acad Sci U S A. 1999;96:7104–7109. doi: 10.1073/pnas.96.12.7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ng DW, Chandrasekharan MB, Hall TC. Ordered histone modifications are associated with transcriptional poising and activation of the phaseolin promoter. Plant Cell. 2006;18:119–132. doi: 10.1105/tpc.105.037010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.West M, Yee KM, Danao J, Zimmerman JL, Fischer RL, Goldberg RB, Harada JJ. LEAFY COTYLEDON1 Is an Essential Regulator of Late Embryogenesis and Cotyledon Identity in Arabidopsis. Plant Cell. 1994;6:1731–1745. doi: 10.1105/tpc.6.12.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aasland R, Gibson TJ, Stewart AF. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 82.Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 83.Alvarez-Venegas R, Sadder M, Hlavacka A, Baluska F, Xia Y, Lu G, Firsov A, Sarath G, Moriyama H, Dubrovsky JG, Avramova Z. The Arabidopsis homolog of trithorax, ATX1, binds phosphatidylinositol 5-phosphate, and the two regulate a common set of target genes. Proc Natl Acad Sci U S A. 2006;103:6049–6054. doi: 10.1073/pnas.0600944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang X. Lipid signaling. Curr Opin Plant Biol. 2004;7:329–336. doi: 10.1016/j.pbi.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 85.Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, Villasenor J, Mehrotra B, Chen J, Rao VR, Brugge JS, Ferguson CG, Payrastre B, Myszka DG, Cantley LC, Wagner G, Divecha N, Prestwich GD, Yuan J. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- 86.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 88.Raynaud C, Sozzani R, Glab N, Domenichini S, Perennes C, Cella R, Kondorosi E, Bergounioux C. Two cell-cycle regulated SET-domain proteins interact with proliferating cell nuclear antigen (PCNA) in Arabidopsis. Plant J. 2006;47:395–407. doi: 10.1111/j.1365-313X.2006.02799.x. [DOI] [PubMed] [Google Scholar]
- 89.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 90.Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 91.Naumann K, Fischer A, Hofmann I, Krauss V, Phalke S, Irmler K, Hause G, Aurich AC, Dorn R, Jenuwein T, Reuter G. Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J. 2005;24:1418–1429. doi: 10.1038/sj.emboj.7600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ebbs ML, Bartee L, Bender J. H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol Cell Biol. 2005;25:10507–10515. doi: 10.1128/MCB.25.23.10507-10515.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ebbs ML, Bender J. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell. 2006;18:1166–1176. doi: 10.1105/tpc.106.041400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shen WH. NtSET1, a member of a newly identified subgroup of plant SET-domain-containing proteins, is chromatin-associated and its ectopic overexpression inhibits tobacco plant growth. Plant J. 2001;28:371–383. doi: 10.1046/j.1365-313x.2001.01135.x. [DOI] [PubMed] [Google Scholar]
- 95.Yu Y, Dong A, Shen WH. Molecular characterization of the tobacco SET domain protein NtSET1 unravels its role in histone methylation, chromatin binding, and segregation. Plant J. 2004;40:699–711. doi: 10.1111/j.1365-313X.2004.02240.x. [DOI] [PubMed] [Google Scholar]
- 96.Thorstensen T, Fischer A, Sandvik SV, Johnsen SS, Grini PE, Reuter G, Aalen RB. The Arabidopsis SUVR4 protein is a nucleolar histone methyltransferase with preference for monomethylated H3K9. Nucleic Acids Res. 2006;34:5461–5470. doi: 10.1093/nar/gkl687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mutskov V, Felsenfeld G. Silencing of transgene transcription precedes methylation of promoter DNA and histone H3 lysine 9. EMBO J. 2004;23:138–149. doi: 10.1038/sj.emboj.7600013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brown MA, Sims RJ, 3rd, Gottlieb PD, Tucker PW. Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol Cancer. 2006;5:26–36. doi: 10.1186/1476-4598-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ying Z, Mulligan RM, Janney N, Houtz RL. Rubisco small and large subunit N-methyltransferases. Bi- and mono-functional methyltransferases that methylate the small and large subunits of Rubisco. J Biol Chem. 1999;274:36750–36756. doi: 10.1074/jbc.274.51.36750. [DOI] [PubMed] [Google Scholar]
- 100.Trievel RC, Beach BM, Dirk LM, Houtz RL, Hurley JH. Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell. 2002;111:91–103. doi: 10.1016/s0092-8674(02)01000-0. [DOI] [PubMed] [Google Scholar]
- 101.Trievel RC, Flynn EM, Houtz RL, Hurley JH. Mechanism of multiple lysine methylation by the SET domain enzyme Rubisco LSMT. Nat Struct Biol. 2003;10:545–552. doi: 10.1038/nsb946. [DOI] [PubMed] [Google Scholar]
- 102.Marmorstein R. Structure of SET domain proteins: a new twist on histone methylation. Trends Biochem Sci. 2003;28:59–62. doi: 10.1016/S0968-0004(03)00007-0. [DOI] [PubMed] [Google Scholar]
- 103.Cui X, De Vivo I, Slany R, Miyamoto A, Firestein R, Cleary ML. Association of SET domain and myotubularin-related proteins modulates growth control. Nat Genet. 1998;18:331–337. doi: 10.1038/ng0498-331. [DOI] [PubMed] [Google Scholar]
- 104.Rozenblatt-Rosen O, Rozovskaia T, Burakov D, Sedkov Y, Tillib S, Blechman J, Nakamura T, Croce CM, Mazo A, Canaani E. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc Natl Acad Sci U S A. 1998;95:4152–4157. doi: 10.1073/pnas.95.8.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rozovskaia T, Rozenblatt-Rosen O, Sedkov Y, Burakov D, Yano T, Nakamura T, Petruck S, Ben-Simchon L, Croce CM, Mazo A, Canaani E. Self-association of the SET domains of human ALL-1 and of Drosophila TRITHORAX and ASH1 proteins. Oncogene. 2000;19:351–357. doi: 10.1038/sj.onc.1203307. [DOI] [PubMed] [Google Scholar]
- 106.Morillon A, Karabetsou N, Nair A, Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol Cell. 2005;18:723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 107.Mueller JE, Canze M, Bryk M. The requirements for COMPASS and Paf1 in transcriptional silencing and methylation of histone H3 in Saccharomyces cerevisiae. Genetics. 2006;173:557–567. doi: 10.1534/genetics.106.055400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 109.Simon J, Bornemann D, Lunde K, Schwartz C. The extra sex combs product contains WD40 repeats and its time of action implies a role distinct from other Polycomb group products. Mech Dev. 1995;53:197–208. doi: 10.1016/0925-4773(95)00434-3. [DOI] [PubMed] [Google Scholar]
- 110.Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- 111.Luo M, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chaudhury AM. Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 1999;96:296–301. doi: 10.1073/pnas.96.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ohad N, Yadegari R, Margossian L, Hannon M, Michaeli D, Harada JJ, Goldberg RB, Fischer RL. Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell. 1999;11:407–416. doi: 10.1105/tpc.11.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A. Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci U S A. 2000;97:10637–10642. doi: 10.1073/pnas.170292997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Spillane C, MacDougall C, Stock C, Kohler C, Vielle-Calzada JP, Nunes SM, Grossniklaus U, Goodrich J. Interaction of the Arabidopsis polycomb group proteins FIE and MEA mediates their common phenotypes. Curr Biol. 2000;10:1535–1538. doi: 10.1016/s0960-9822(00)00839-3. [DOI] [PubMed] [Google Scholar]
- 115.Wang D, Tyson MD, Jackson SS, Yadegari R. Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc Natl Acad Sci U S A. 2006;103:13244–14249. doi: 10.1073/pnas.0605551103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kohler C, Hennig L, Bouveret R, Gheyselinck J, Grossniklaus U, Gruissem W. Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 2003;22:4804–4814. doi: 10.1093/emboj/cdg444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA. The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci U S A. 2006;103:14631–14636. doi: 10.1073/pnas.0606385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 119.Schuetz A, Allali-Hassani A, Martin F, Loppnau P, Vedadi M, Bochkarev A, Plotnikov AN, Arrowsmith CH, Min J. Structural basis for molecular recognition and presentation of histone H3 By WDR5. EMBO J. 2006;25:4245–4252. doi: 10.1038/sj.emboj.7601316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Preuss D. Chromatin silencing and Arabidopsis development: A role for polycomb proteins. Plant Cell. 1999;11:765–768. doi: 10.1105/tpc.11.5.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Reddy VS, Reddy AS. Developmental and cell-specific expression of ZWICHEL is regulated by the intron and exon sequences of its gene. Plant Mol Biol. 2004;54:273–293. doi: 10.1023/B:PLAN.0000028793.88757.8b. [DOI] [PubMed] [Google Scholar]
- 122.Hube F, Guo J, Chooniedass-Kothari S, Cooper C, Hamedani MK, Dibrov AA, Blanchard AA, Wang X, Deng G, Myal Y, Leygue E. Alternative splicing of the first intron of the steroid receptor RNA activator (SRA) participates in the generation of coding and noncoding RNA isoforms in breast cancer cell lines. DNA Cell Biol. 2006;25:418–428. doi: 10.1089/dna.2006.25.418. [DOI] [PubMed] [Google Scholar]
- 123.Castells E, Puigdomenech P, Casacuberta JM. Regulation of the kinase activity of the MIK GCK-like MAP4K by alternative splicing. Plant Mol Biol. 2006;61:747–756. doi: 10.1007/s11103-006-0046-3. [DOI] [PubMed] [Google Scholar]
- 124.Hughes TA. Regulation of gene expression by alternative untranslated regions. Trends Genet. 2006;22:119–122. doi: 10.1016/j.tig.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 125.Mumberg D, Lucibello FC, Schuermann M, Muller R. Alternative splicing of fosB transcripts results in differentially expressed mRNAs encoding functionally antagonistic proteins. Genes Dev. 1991;5:1212–1223. doi: 10.1101/gad.5.7.1212. [DOI] [PubMed] [Google Scholar]
- 126.Lee C, Wang Q. Bioinformatics analysis of alternative splicing. Brief Bioinform. 2005;6:23–33. doi: 10.1093/bib/6.1.23. [DOI] [PubMed] [Google Scholar]
- 127.Wang BB, Brendel V. Genomewide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci USA. 2006;103:7175–7180. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Neverov AD, Artamonova, Nurtdinov RN, Frishman D, Gelfand MS, Mironov AA. Alternative splicing and protein function. BMC Bioinformatics. 2005;6:266. doi: 10.1186/1471-2105-6-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yeo GW, Van Nostrand E, Holste D, Poggio T, Burge CB. Identification and analysis of alternative splicing events conserved in human and mouse. Proc Natl Acad Sci USA. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xu Q, Modrek B, Lee C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 2002;30:3754–3766. doi: 10.1093/nar/gkf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Colot HV, Loros JJ, Dunlap JC. Temperature-modulated alternative splicing and promoter use in the Circadian clock gene frequency. Mol Biol Cell. 2005;16:5563–5571. doi: 10.1091/mbc.E05-08-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bagnasco SM, Peng T, Nakayama Y, Sands JM. Differential expression of individual UT-A urea transporter isoforms in rat kidney. J Am Soc Nephrol. 2000;11:1980–1986. doi: 10.1681/ASN.V11111980. [DOI] [PubMed] [Google Scholar]
- 133.Krauss V, Reuter G. Two genes become one: the genes encoding heterochromatin protein Su(var)3-9 and translation initiation factor subunit eIF-2gamma are joined to a dicistronic unit in holometabolic insects. Genetics. 2000;156:1157–1167. doi: 10.1093/genetics/156.3.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tan X, Rotllant J, Li H, De Deyne P, Du SJ. SmyD1, a histone methyltransferase, is required for myofibril organization and muscle contraction in zebrafish embryos. Proc Natl Acad Sci USA. 2006;103:2713–2718. doi: 10.1073/pnas.0509503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Manzur KL, Farooq A, Zeng L, Plotnikova O, Koch AW, Sachchidanand, Zhou MM. A dimeric viral SET domain methyltransferase specific to Lys27 of histone H3. Nat Struct Biol. 2003;10:187–196. doi: 10.1038/nsb898. [DOI] [PubMed] [Google Scholar]
- 136.Ueda J, Tachibana M, Ikura T, Shinkai Y. Zinc finger protein Wiz links G9a/GLP histone methyltransferases to the co-repressor molecule CtBP. J Biol Chem. 2006;281:20120–20128. doi: 10.1074/jbc.M603087200. [DOI] [PubMed] [Google Scholar]
- 137.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Mazumder R, O’Donovan C, Redaschi N, Suzek B. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Touriol C, Morillon A, Gensac MC, Prats H, Prats AC. Expression of human fibroblast growth factor 2 mRNA is post-transcriptionally controlled by a unique destabilizing element present in the 3′-untranslated region between alternative polyadenylation sites. J Biol Chem. 1999;274:21402–21408. doi: 10.1074/jbc.274.30.21402. [DOI] [PubMed] [Google Scholar]
- 139.Knirsch L, Clerch LB. A region in the 3′ UTR of MnSOD RNA enhances translation of a heterologous RNA. Biochem Biophys Res Commun. 2000;272:164–168. doi: 10.1006/bbrc.2000.2754. [DOI] [PubMed] [Google Scholar]
- 140.Ng DW, Chandrasekharan MB, Hall TC. The 5′ UTR negatively regulates quantitative and spatial expression from the ABI3 promoter. Plant Mol Biol. 2004;54:25–38. doi: 10.1023/B:PLAN.0000028767.06820.34. [DOI] [PubMed] [Google Scholar]
- 141.Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Casas-Mollano JA, Lao NT, Kavanagh TA. Intron-regulated expression of SUVH3, an Arabidopsis Su(var)3-9 homologue. J Exp Bot. 2006;57:3301–3311. doi: 10.1093/jxb/erl093. [DOI] [PubMed] [Google Scholar]
- 143.Asamizu E, Nakamura Y, Sato S, Tabata S. A large scale analysis of cDNA in Arabidopsis thaliana: generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res. 2000;7:175–180. doi: 10.1093/dnares/7.3.175. [DOI] [PubMed] [Google Scholar]
- 144.Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, Pham P, Cheuk R, Karlin-Newmann G, Liu SX, Lam B, Sakano H, Wu T, Yu G, Miranda M, Quach HL, Tripp M, Chang CH, Lee JM, Toriumi M, Chan MM, Tang CC, Onodera CS, Deng JM, Akiyama K, Ansari Y, Arakawa T, Banh J, Banno F, Bowser L, Brooks S, Carninci P, Chao Q, Choy N, Enju A, Goldsmith AD, Gurjal M, Hansen NF, Hayashizaki Y, Johnson-Hopson C, Hsuan VW, Iida K, Karnes M, Khan S, Koesema E, Ishida J, Jiang PX, Jones T, Kawai J, Kamiya A, Meyers C, Nakajima M, Narusaka M, Seki M, Sakurai T, Satou M, Tamse R, Vaysberg M, Wallender EK, Wong C, Yamamura Y, Yuan S, Shinozaki K, Davis RW, Theologis A, Ecker JR. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 145.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 146.Llave C, Kasschau KD, Rector MA, Carrington JC. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kiyosawa H, Mise N, Iwase S, Hayashizaki Y, Abe K. Disclosing hidden transcripts: mouse natural sense-antisense transcripts tend to be poly(A) negative and nuclear localized. Genome Res. 2005;15:463–474. doi: 10.1101/gr.3155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang XJ, Gaasterland T, Chua NH. Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol. 2005;6:R30. doi: 10.1186/gb-2005-6-4-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci U S A. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Moore T, Constancia M, Zubair M, Bailleul B, Feil R, Sasaki H, Reik W. Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc Natl Acad Sci U S A. 1997;94:12509–12514. doi: 10.1073/pnas.94.23.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 152.Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchell-Olds T. Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell. 2001;13:681–693. doi: 10.1105/tpc.13.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ober D. Seeing double: gene duplication and diversification in plant secondary metabolism. Trends Plant Sci. 2005;10:444–449. doi: 10.1016/j.tplants.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 154.Pichersky E, Gang DR. Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci. 2000;5:439–445. doi: 10.1016/s1360-1385(00)01741-6. [DOI] [PubMed] [Google Scholar]
- 155.Wink M. Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry. 2003;64:3–19. doi: 10.1016/s0031-9422(03)00300-5. [DOI] [PubMed] [Google Scholar]
- 156.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, Barlev NA, Reinberg D. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 157.Kouskouti A, Scheer E, Staub A, Tora L, Talianidis I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol Cell. 2004;14:175–182. doi: 10.1016/s1097-2765(04)00182-0. [DOI] [PubMed] [Google Scholar]
- 158.Zhang K, Lin W, Latham JA, Riefler GM, Schumacher JM, Chan C, Tatchell K, Hawke DH, Kobayashi R, Dent SY. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell. 2005;122:723–734. doi: 10.1016/j.cell.2005.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]