SUMMARY
We propose a unified framework for the analysis of Chromatin (Ch) Immunoprecipitation (IP) microarray (ChIP-chip) data for detecting transcription factor binding sites (TFBSs) or motifs. ChIP-chip assays are used to focus the genome-wide search for TFBSs by isolating a sample of DNA fragments with TFBSs and applying this sample to a microarray with probes corresponding to tiled segments across the genome. Present analytical methods use a two-step approach: (i) analyze array data to estimate IP enrichment peaks then (ii) analyze the corresponding sequences independently of intensity information. The proposed model integrates peak finding and motif discovery through a unified Bayesian hidden Markov model (HMM) framework that accommodates the inherent uncertainty in both measurements. A Markov Chain Monte Carlo algorithm is formulated for parameter estimation, adapting recursive techniques used for HMMs. In simulations and applications to a yeast RAP1 dataset, the proposed method has favorable TFBS discovery performance compared to currently available two-stage procedures in terms of both sensitivity and specificity.
Keywords: Data augmentation, Gene regulation, Tiling array, Transcription factor binding site
1. Introduction
Chromatin (Ch) Immunoprecipitation (IP) microarray (ChIP-Chip) assays use microarrays of DNA sequences to measure specific DNA-protein interactions, with a goal of discovering the genomic locations of transcription factor binding sites (TFBSs). Transcription factors (TFs) are proteins that regulate the expression of nearby genes by binding to DNA. TFs bind to TFBSs that are usually on the order of 10–20 nucleotides in length, and even in relatively small genomes, the binding sites occur in hundreds of locations (Buck and Lieb, 2004). However, motif discovery methods cannot be directly applied to the genome in order to find TFBSs because of the number of false positive binding site matches that would result. ChIP-chip data allows one to narrow the search from the whole genome to only those regions where binding has likely occurred in vivo.
In chromatin immunoprecipitation experiments, the TF of interest binds to the DNA in vivo under controlled conditions, and the protein-DNA complexes are fixed or crosslinked and extracted. The DNA is sheared into approximately 1kb fragments by sonication. Next, an antibody specific to the TF of interest selectively binds to the protein-DNA complexes of interest, and this entire complex precipitates out of solution. The DNA precipitate is then extracted, the crosslinks are reversed, it is universally amplified, and fluorescently labeled. This IP sample is enriched for DNA fragments that contained a binding site. Reference samples of the input DNA fragments that do not go through the IP process are used as controls, and either two-color microarrays (Buck and Lieb, 2004) or high density oligonucleotide arrays (Kapranov et al., 2002; Cawley et al., 2004) compare the DNA present in the IP and the reference sample at each DNA segment that has a corresponding probe. If a probe or continuous region of many probes has higher intensity in the IP sample than the reference, it is said to be relatively enriched. The sequences of the enriched regions are often searched for the presence of TFBSs.
TFBSs generally do not match an exact sequence, and but are usually represented by a 4×w position specific weight matrix (PSWM) Θ where the four rows represent the nucleotides A, C, G and T and the w columns represent the w motif positions (Liu et al., 1995). The element Θij is the probability that the nucleotide at position j of the sequence is i, i ∈ {A,C,G,T}. Searching for patterns of several base pairs within segments of DNA that might be several thousand base pairs long can lead to many false positive matches because there are thousands of potentially similar sites within a single DNA segment. This multiplicity greatly increases the computational burden, especially if many DNA segments are considered simultaneously. Further, the “background” DNA sequence that does not contain binding sites generally has a highly non-random distribution of nucleotides. The computational and statistical challenges of motif discovery have led to the development of a number of statistical model-based methods for motif discovery (Liu et al., 1995; Gupta and Liu, 2003; Zhou and Liu, 2004) as well as computationally fast and partially heuristic methods (Liu et al., 2002; Buhler and Tompa, 2002).
The analysis of ChIP-chip data is typically done through a two step approach. The first step deals with the ChIP-chip array data, and it analyzes the probe intensities to find the regions of enrichment. The second step uses the genomic sequences of the regions found in the first step to estimate the motif. Next, we describe the main features and form of ChIP-chip data, and discuss currently available methods for its analysis.
1.1 Data structure
There are two main technologies used for ChIP-chip experiments: (i) a two-color system in which the IP sample is labeled with one fluorescent dye and the reference sample is labeled with a different dye and applied to the same array, and (ii) the oligonucleotide (e.g. Affymetrix) array, in which the IP sample is applied to one set of arrays, and the reference sample is applied to a different array set. In this article, we focus on two-color ChIP-chip data. The probes on the two-color arrays range from about 100 to 2,000 base pairs in length. For each probe p on each array, there are two measurements: one for the IP sample intensity IPp and one for the reference sample intensity Refp. The variation due to the random error of a specific probe’s measurement is reduced by taking the ratio of IPp/Refp which removes the multiplicative effect of probe p that is common to both IPp and Refp (Rocke and Durbin, 2001). Enrichment implies that log(IPp/Refp) > 0 for a given probe p.
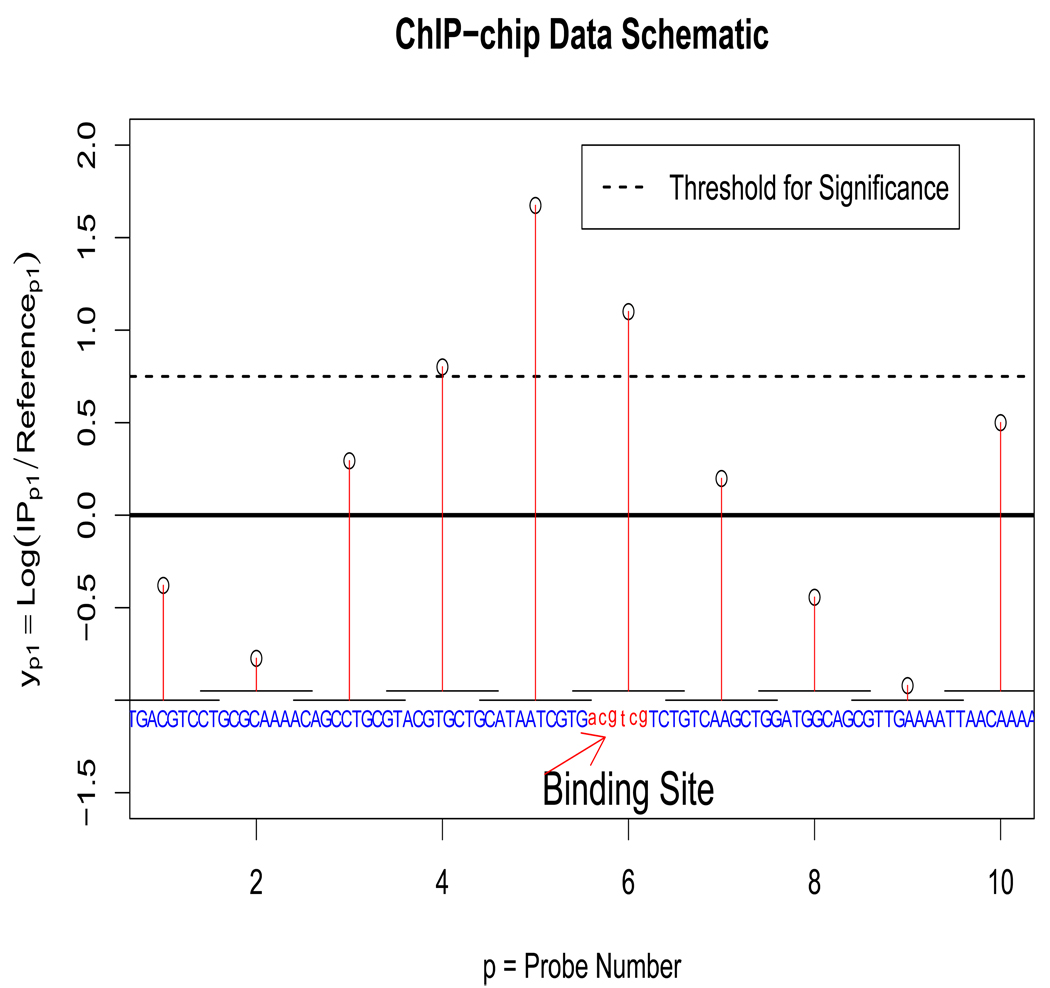
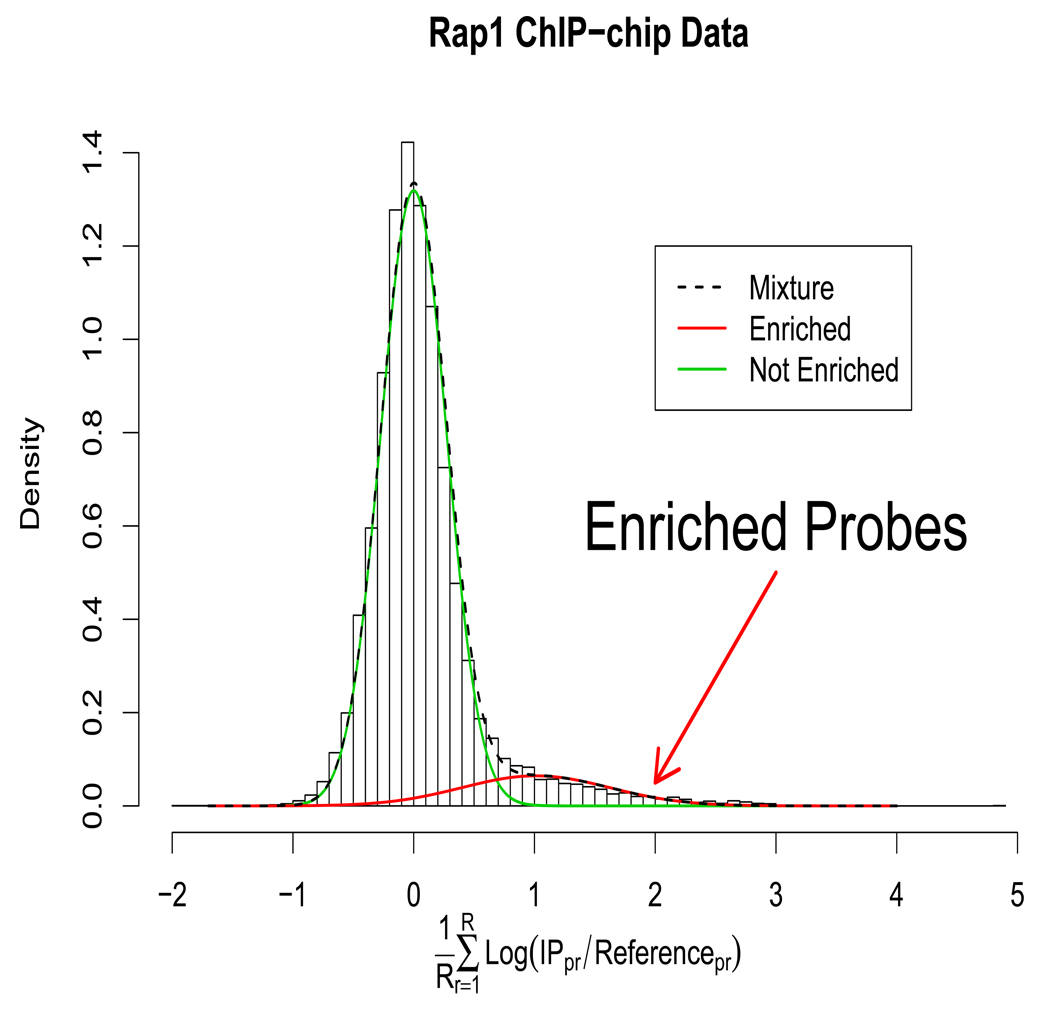
The full ChIP-chip experiment can be represented as an P × R matrix Y where microarray replicates are indexed r ∈ [1 … R], and the probes are indexed by p ∈ [1 … P]. A row of this matrix which contains all measurements from a probe is denoted as yp. The number of probes P ranges from 10,000 to 1,000,000 in different experiments, and the number of replicates R is small, usually between 1 and 10. The rth element of yp is denoted as ypr and is the log-ratio of the IP sample intensity and the reference sample intensity, that is, ypr = log(IPpr/Refpr). A schematic of the data is shown in Figure 1. The values of ypr that are higher are more likely to be IP enriched. The histogram of average values of yp (Figure 2) from a yeast RAP1 experiment (Lieb et al., 2001) shows that the averages can be thought of as a mixture of the enriched and the not enriched probes. The sequence that corresponds to probe p will be denoted as xp. The consecutive probes are adjacent on the genome, and the fragments which hybridize to the probes correspond to the complementary sequence. xp is a sequence of A’s, C’s, G’s, and T’s with length Kp. A subsequence of xp from position j to position k will be denoted as xp[j : k]. The probe sequences can range from a few hundred to several thousand base pairs in length, but the resolution of each probe is limited by the size of the applied DNA fragments. ChIP-chip analysis should also consider the spatial correlation between probes that represent adjacent loci. Probes are correlated if the genomic distance between the probes is less than the length of the DNA fragments in the sample. Correlation between adjacent probes is a prominent feature of the data because the DNA fragments applied to the arrays may span two or more probes (Buck and Lieb, 2004).
Figure 1.
ChIP-chip data schematic is shown for one ChIP-chip replicate. The genomic sequence is shown in blue, and the segments corresponding to the probes is indicated by bars over the sequence. The number of base pairs has been greatly reduced for clarity. Note that log(IPp1/Refp1) is increased for the probes close to a binding site, and the region corresponding to the significant probes contains a binding site. Also, note the correlation between adjacent probes. This figure appears in color in the electronic version of the article.
Figure 2.
Histogram of average probe intensities from Rap1 yeast experiment. The density estimates from the proposed model fit are overlayed, and the two component mixture of both Enriched and not Enriched probes is evident. This figure appears in color in the electronic version of the article.
1.2 Current methods for analyzing ChIP-chip data
Sliding window approaches were suggested by Cawley et al. (2004); Keles et al. (2004); Ji and Wong (2005) and Buck et al. (2005). Cawley et al. (2004) proposed using a Wilcoxon rank sum statistic for each probe, while Keles et al. (2004) used a Welch t-statistic, and Ji and Wong (2005) used a t-like-statistic which has a shrunken variance estimate. These methods identify regions or peaks of intensity as IP enriched when the moving average of the statistic exceeds a threshold, and give an False Discovery Rate (FDR) for each peak.
Another approach to finding regions of enrichment is to use Hidden Markov Models (HMMs). Ji and Wong (2005) developed a nonparametric method called Unbalanced Mixture Subtraction (UMS) to estimate the emission densities and the FDR within an HMM. Li et al. (2005) proposed an HMM with the same state space, but used normal distribution models for the emission densities. Li et al. (2005) and Ji and Wong (2005) both demonstrate the superior performance of the HMM method over moving average models in terms of power for detecting IP enrichment for small sample sizes. One limitation with both of these methods is that the transition probabilities and the emission densities are not estimated simultaneously by the HMM so that the user must select values which may lead to suboptimal performance. Keles (2007) proposed a hierarchical mixture model for detecting regions of IP enrichment, that considers regions of adjacent probes collectively in order to take advantage of the correlation between probes. A similar robust hierarchical method was proposed by Gottardo et al. (2006), allowing for probe specific differences in error variance, and using a normalization procedure called Model-based Analysis of Tiling-array (MAT) for oligonucleotide ChIP-chip (Johnson et al., 2006). Normalization methods for two color ChIP-chip data are inherently difficult, due to the skewed nature of ChIP-chip log ratios Buck and Lieb (2004). Recently, Zheng et al. (2007) advanced a model for the estimating for the shape of the probe intensity enrichment peaks. This approach has the advantage adaptively pooling information from adjacent probes based upon data-driven peak shape estimates.
In most two step procedures, the first step estimates the probe’s enrichment probability based on the intensity information. Probes that exceed some cutoff based upon the intensity model are submitted to a second step that estimates the probes’ binding site probability based on the probe sequence. However, the probe intensity and sequence are not independent–that is, probes with with or near TFBSs have a higher likelihood of being enriched. Using a intensity based cutoff in the first step that does not consider the probe’s surrounding sequence is likely to miss some probes with TFBSs. Ignoring the probe level information when considering the sequence could also lead to biases, as the probe measurement may be informative towards the extent of actual binding. It thus seems reasonable to consider a joint model of the ChIP-chip measurements and sequence for more accurate estimation.
Shim and Keles (2007) propose a procedure that analyzes the genome sequence conditionally upon the probe intensity information. Their method uses the measurements from the probes within a conditional two-component mixture model. The model estimates the probability of a TFBS occurrence at a particular base pair in the genomic sequence conditionally upon a smoothed average of the probe level intensity statistics. The relationship between the ChIP-chip value of probe i, Ti, and the indicator variable, Zi, denoting whether a motif starts at position i, is modeled as logit[Pr(Zi = 1|Ti)] = β0 + β1Ti. Averaging of Ti’s between probes only approximately accounts for spatial correlations, unlike an HMM. A more debatable assumption is that the ChIP-chip value, Ti is taken to be without error despite the uncertainty inherent in estimation. The estimation procedure appears likely to suffer from limitations of the EM-based algorithm, for example multimodality traps, as well as the inability to capture multiple binding sites close together. Shim and Keles (2007) use their technique as a refinement procedure after the bound regions have already been selected, rather than allowing the sequence model to inform the probe measurement model.
In this article, we propose a joint model for ChIP-chip and sequence data for TFBS discovery, accounting for the uncertainty inherent in both steps. The probe level information, followed by a motif discovery step, is used to generate initial motif candidates under the assumption that the “best” binding sites are more likely to be present close to the highly enriched probes. However, once the initial estimates are obtained, the entire set of data is fit using a novel joint model (Section 2). As we see later in data analyses (Section 4), our framework succeeds in finding many TFBSs that are missed by two-step procedures. The motivation for the joint model is to minimize the number of probes close to binding sites, that fail the intensity cutoff. We use the probe level information to allow (i) probe level experimental/measurement errors not to bias the observations in case a functional binding site is truly present, but observed binding is not significant enough, and (ii) the intensity of binding to have an influence on our assessment of the strength of a binding site. There could be minor errors due to some regions containing motif matches that show no high intensities, but the joint model should not allow too many of these regions to bias the analysis. In addition, our model may be easily extended to more than one motif of interest.
2. The general model
In this section, we first describe the models used for the probe intensity, the probe sequences, and the full joint HMM framework for the probe intensity and sequence data.
2.1 Probe intensity model
The probe level data is modeled through a hidden Markov model (HMM). HMMs are Markov random processes with latent states that emit random variables whose distributions depend on the state. The hidden states of the HMM at the probe level are the binding states of probe p denoted as sp where sp = 1 if the pth probe is IP enriched and sp = 0 otherwise. The log-ratio of the intensities for the pth probe, yp, are assumed to have the density fsp(yp) (sp = 0, 1), with ypr, the observation for replicate r of probe p distributed as , where μp is a probe-specific mean, and the replicates are independently distributed. The Gaussian assumption is justifiable if one considers the raw intensity values to have approximately a gamma or lognormal distribution so that the log transformation yields an approximately Gaussian random variable. Next, we assume a hierarchical model for μp, with . In other words, enrichment implies that the probes’ average intensity is relatively high (> 0) whereas the non-enriched probes will have mean intensities close to 0. Figure 2 demonstrates that the observed probe averages ¯yp may be accurately fit by a mixture of normal densities for μp. The density for yp can be written as . Integration with respect to the parameter μp yields a compound symmetric multivariate Gaussian density, , where 1R and IR are the R-dimensional vector of 1’s and identity matrix, respectively.
2.2 Sequence Model
Next, we formulate the model for the sequence data in detail. The vast majority of the DNA that does not contain the binding sites of interest is referred to as the background sequence. Subsequent letters of this background sequence may depend on the previous letters, this dependence is often modeled as having a Markov structure (Liu et al., 2002). We propose using PSWMs representing repeats to allow for the modeling of the low complexity background patterns, resulting in fewer parameters than high-dimensional Markov models. Specifically, the PSWMs of the proposed background model include one-letter words (A, C, G, and T) as well as repeats of A’s and T’s. PSWMs in the model will be denoted as Θυ (υ ∈ [1 … V]), with ΘV representing the PSWM corresponding to the motif of interest. Let Θυ have length wυ, and let π be the vector of the prevalences πυ of PSWM υ. The emission densities of the sequence are denoted as psp(xp) (sp = 0, 1) for the non-enriched and enriched states, respectively. Let Θ(sp) denote the set of PSWMs that can be involved in generating the sequence under state sp, with Θ(0) = (Θ1, ‥, ΘV−1), and Θ(1) = (Θ1, ‥, ΘV) = (Θ(0), ΘV). The probability of observing sequence xp is denoted by psp(xp|Θ(sp)) (sp = 0, 1). The sets of PSWMs in Θ(0) and Θ(1) can be considered as words that are part of a stochastic dictionary (Gupta and Liu, 2003). Let the motif site locations be denoted by the indicator variables A = (Aij), where Aij = 1(0) if position j of probe i is (is not) the start of a motif site. The full likelihood of the joint model can be written as
In order to estimate parameters of the model under the presence of a huge amount of missing data: A, the unknown site locations, and s = (s1, …, sP), the latent emission states, we formulate a Data Augmentation (DA) sampling scheme for fitting the full HMM, which is given in the Appendices.
2.3 Priors
The final part of the model specification involves prior elicitation. The prior for the intensity parameter μ1 was taken to be noninformative (∝ 1). The priors for were also noninformative, with . The priors for each row of the HMM transition matrix (τij) (i, j = 0, 1) are taken to be Dirichlet distributions with hyperparameters denoted as δij. More precisely, [τi0, τi1] ∼ Dirichlet(δi0, δi1). The δij are equal for all transitions so that δij = δi′j′ and are small (0.1) relative to the total number of transitions ∼ P = 11,575, and therefore, minimally informative.
One difficulty in estimating the motif is that the motif and prevalence of the motif may be jointly nonidentifiable in practice. The less conserved a motif is, the more prevalent it may be. If there is no prior placed on the motif prevalence, then the model often tends to converge to a highly prevalent and non-specific motif which contradicts the biological understanding of the specificity of transcription factor binding. A relatively strong prior may be implemented for πV to avoid this problem and hasten convergence. We assume a prior for the motif prevalence as πV ∼ Beta(δV (1 − γ), δV γ) where δ0 is a large pseudocount and γ (with 0 < γ < 1) indicates the prior expected value. The conditional prior for the other components of π, (i.e. π1, …, πV −1) can then be drawn from the prior Dirichlet distribution Dir(δ1, ‥, δV−1) and scaled by 1−πV (δ1, ‥, δV−1 represent small non-zero pseudocount values). Let δ = (δ0, ‥, δV). The prior for the motif matrix of interest ΘV is taken to be the product Dirichlet distribution Pir(B) where B = (bij) is a 4 × wV matrix of pseudocounts, bij denoting the count of the symbol i at motif position j which is also set to a small non-zero value, uniform across letters.
3. Application to Yeast RAP1 data
We applied our method to a yeast dataset from Lieb et al. (2001) which involved a ChIP-chip experiment for the Rap1 transcription factor. The data consist of four arrays and 11,575 non-telomeric probes of various lengths spanning the yeast genome of 17 chromosomes with a total of 12 million base pairs. The array data and the sequence data were preprocessed, and details are in the supplementary file.
3.1 Model initialization
We used an initialization phase similar to the first step of the two stage procedure in which the segments of highest enrichment are selected using the IO model, and used to provide the initial estimate of the motif matrix. Once the motif estimate is initialized, the full original set of probes was analyzed by the joint model. For initialization, the probes that were selected by the IO model were ranked according to the log-ratio of the intensity probabilities in favor of enrichment log(f1(yp)/f0(yp)). The sequences of the probes in the highest 1% of likelihood ratios were then selected for the search for the initial motif estimate.
The initialization of the sequence model requires a reasonable estimate of the TFBS motif to facilitate convergence. The sequences selected by the above procedure are likely to have the highest concentration of the motif binding sites, but it is evident that there are many non-random patterns in the DNA that correspond to different modes in the likelihood and can lead to the failure of the stochastic dictionary model to find the motif which gives the highest likelihood for these sequences. To get the initial motif estimate, an accumulating stochastic dictionary model was fit to the sequences in which successive motifs are estimated and added to the dictionary. First, the dictionary was initialized with PSWMs of length one representing A’s, C’s, G’s, and T’s as well as repeat words of A’s and T’s of both of length 4 and length 8, which appeared sufficient to capture the dependence in the background, i.e. did not lead to further “repeat” motifs being predicted. These 8 motifs were considered part of the fixed background model with motif matrices Θ1, … Θ8. The search for the “interesting” motif (ΘV) was restricted to the assumed motif width of 13 (Lieb et al., 2001), and a motif of length 13 with uniform probability across all letters at all positions was added to the dictionary and updated using the data augmentation method described in Section 2 (V = 9). This motif is considered the foreground motif Θ* and is the only motif updated in each cycle of the DA sampler. After approximate convergence, the updated motif is added to the fixed background dictionary, and another motif of length 13 with uniform probability across all letters at all positions is added to the dictionary so that V = 10, and this new word becomes the new foreground motif. The procedure of iteratively adding words to the background allows the model to consider different modes in the space of potential motifs.
Two likelihoods of the sequences are plotted across the iterations in order to find a reasonable motif for initialization. The first is the likelihood of the sequences given the full dictionary up to that point which may be denoted as ΠXi∈Top Sequence p(Xi|Θ1, …, Θ8+m, Θ*) where m ≥ 0 is the number of accumulated words and Θ* is the updated motif. The likelihood increases as motifs are added to the dictionary, and after a few iterations a plateau is reached signifying entrapment in a likelihood mode. The second likelihood computed is based on the original eight-PSWM background with only the current foreground motif and may be denoted as ΠXi∈Top Sequence p(Xi|Θ1, …Θ8, Θ*). This likelihood is an indication of the improvement in model fit given the addition of only the current foreground motif (Figure 1 in the Supplementary material). The motif that gives the largest increase in sequence likelihood is taken as a reasonable choice for the initial estimate of ΘV in the joint sequence and intensity model, while the PSWMs Θ1, …, Θ8 are used in the background model.
3.2 Data analysis
The next phase of the analysis is the application of the joint model that is applied to the full original dataset. An assessment of the sensitivity to the selection of hyperparameters was performed as well as a comparison of the results with other ChIP-chip analysis methods.
3.2.1 Sensitivity analysis
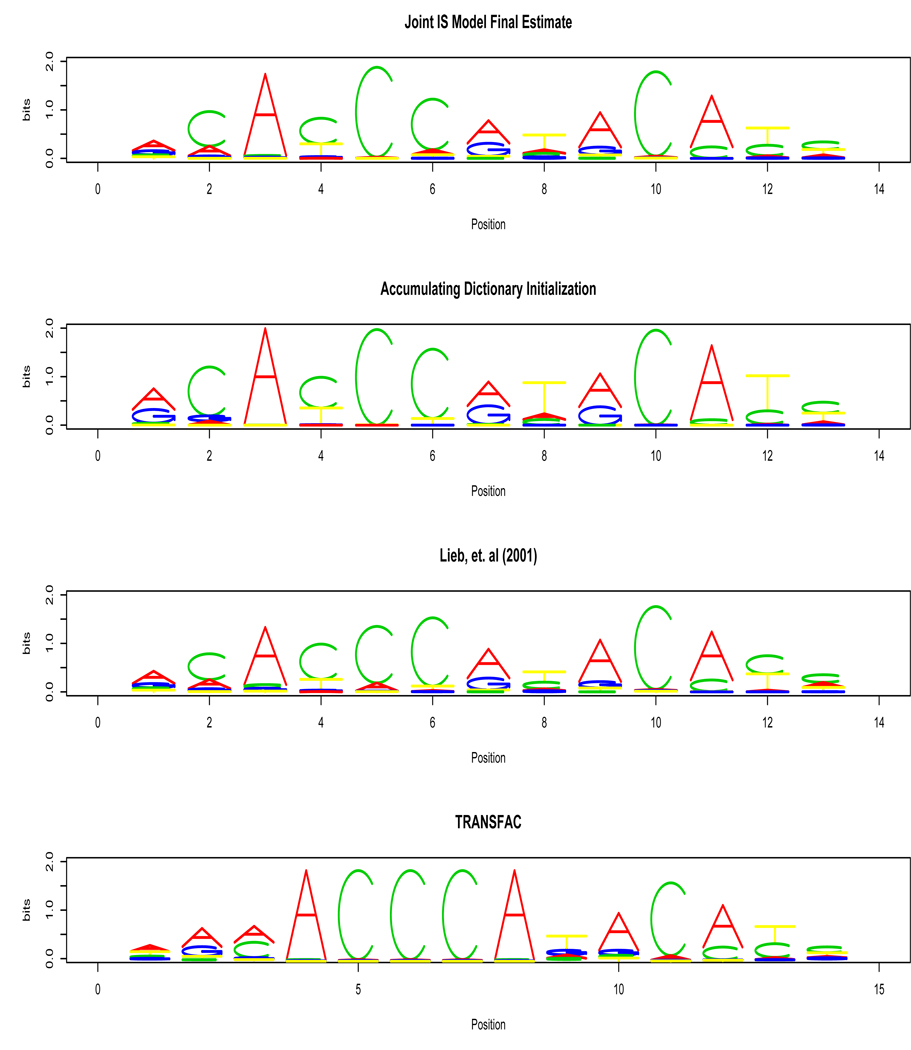
We first did a sensitivity analysis to examine the dependence of the final estimates on the choice of the prior hyperparameters. The hyperparameters for the pseudocounts δij, δυ, and the elements of the pseudocount matrix B were set to 0.1. These pseudocounts are quite small compared to the number of observed counts, and do not greatly affect the inference. We first fixed δ0 = 106 and varied the prior parameter for the expected motif prevalence γ ∈ [5, 6, 7, 8, 9, 10, 20]×10−5 to assess the sensitivity to this prior. MCMC convergence of the DA sampler was diagnosed with parallel chains by using criterion that the Gelman and Rubin statistic was less than 1.18. The parameters monitored included all of the intensity model parameters, the HMM transition parameters, and the parameters for the most probable letter at each position in the estimated motif. The IS model DA sampler ran for 2000 iterations, and the last 50% were sampled for posterior inference. The corresponding numbers of binding sites found by the IS model for each value of γ were [283, 284, 295, 299, 305, 309, > 1000] respectively. The last value indicated that the model did not converge to the correct mode of the posterior distribution. The number of TFBSs was about 300 in the range γ ∈ [7.0, 10.0]×10−5. The positions of the binding sites discovered were also very consistent, the intersection of the binding site lists for the first six consecutive values of γ being [281, 283, 293, 296, 304]. In other words, 281 of the 283 TFBSs found when γ = 5.0 × 10−5 were also found when γ = 6.0 × 10−5. A similar sensitivity analysis was performed by varying the δ0 hyperparameter (δ0 ∈ [10, 9, 8, 7, 6, 5, 4] × 105) while fixing γ = 0.0001. The number of sites found were [314, 312, 312,, 318, 321, > 1000] respectively and the intersections of the binding site lists for consecutive values of δ0 were [316, 312, 306, 308]. This indicates that the sites were consistent for δ0 ∈ [5, 10] × 105. Increasing γ and decreasing δ0 both had the effect of slightly increasing the number of sites found at the expense of model convergence. However, the model gave consistent results for a broad range of hyperparameter values, and the final motif estimate (Figure 3) has a strong resemblance to the motifs reported previously by (Lieb et al., 2001) and in the TRANSFAC database (Matys et al., 2003). The sensitivity of the two-stage models to the parameters was also considered, and comparative tables of the motif sites discovered is given in the supplementary material. See Supplementary Tables 1–5. The two-stage methods may have the benefit of additional stability as γ is increased above 0.0001, but the posterior probability of the sites found decreases with increasing γ.
Figure 3.
Comparison of motif logos of the model estimates and literature. The final motif estimate for Rap1 by the joint IS model is at the top. The bottom two plots show the motif discovered by Lieb et al. (2001) and the motif listed in the TRANSFAC database. This figure appears in color in the electronic version of the article.
3.2.2 Comparisons with other approaches
We chose the largest value of γ = 10−4 for which convergence was observed to compare the IS method with three two step methods. The first method is the intensity only (IO) model which is the proposed method without the sequence component, the second method is the ChIPOTle method (Buck et al., 2005), and the third method is TileMap (Ji and Wong, 2005). The ChIPOTle method requires one to choose a normal approximation or a nonparametric model to estimate the p-value for rejection of the “No Enrichment” null hypothesis, and one must decide on a p-value cutoff for selecting regions for the motif finding stage. We chose the normal approximation method and a p-value cutoff of 0.001. There does not seem to be an objective rule for choosing this cutoff, but this conservative value is consistent with the other models.
The three two-step methods produced estimates of the regions of IP enrichment to which the stochastic dictionary model was applied with γ = 10−4 and δ0 = 106 to obtain lists of estimated TFBSs as in Section 4.1. The estimates for the parameters common to the IO and the IS models (Table 1) appear similar in both. The comparisons of estimated TFBSs (Table 2) show a marked agreement between the four methods, with the IS model finding the most TFBSs and the IO model the next highest. However, the TileMap method found roughly half of the TFBSs of the other methods. The TFBS found by the joint IS model included 98.2%, 94.5%, and 96.9% of the TFBS found by the IO model, ChIPOTle, and TileMap respectively. Also, the IS model was highly consistent in that it found a much larger number of sites compared to TileMap, for example, that found only 52.9% of the ChIPOTle sites. This might indicate a higher sensitivity of the IS model, but the higher specificity cannot be directly assessed because the locations of all “true” binding sites are not known.
Table 1.
Parameter estimates from IO and IS methods
| Intensity Only | Intensity with Sequence | ||||
|---|---|---|---|---|---|
| Parameter | Estimate | (SD) | Estimate | (SD) | |
| μ1 | 0.982 | (0.03) | 1.02 | (0.03) | |
| σa | 0.1172 | (0.001) | 0.1173 | (0.001) | |
| 0.045 | (0.001) | 0.045 | (0.001) | ||
| 0.364 | (0.021) | 0.344 | (0.021) | ||
| τ00 | 0.97 | (0.002) | 0.97 | (0.002) | |
| τ11 | 0.71 | (0.02) | 0.70 | (0.02) | |
Table 2.
Estimated binding site overlaps between the four methods
| - | TileMap | ChIPOTle | Intensity Only | Intensity with Sequence |
|---|---|---|---|---|
| TileMap | 159 | |||
| ChIPOTle | 152 | 293 | ||
| Intensity Only | 152 | 267 | 284 | |
| Intensity with Sequence | 152 | 277 | 279 | 309 |
An analysis of the differences between the probe enrichment probabilities estimated by the IO model and the joint IS model was performed to examine the effect of adding the sequence component to the model. The enrichment probabilities for the probes that were identified as enriched by the IS model and not the IO model have IO model enrichment probabilities in the range [0.049, 0.746]. These probes are neither definitely enriched nor definitely not enriched according to the IO model, and the intensity information dominates the probe calls. This suggests that the IS model has only a moderate effect in altering enrichment probabilities due to sequence information. Despite fewer probes being identified as enriched by the joint IS model, this model found more binding sites which is consistent with the idea that the probe measurement information contributes significantly to prediction of binding sites. Most of the probes have smaller enrichment probabilities under the joint IS model. The IO model selected 934 probes as enriched while the IS model selected 922. These results are also consistent with the idea that including sequence in the model can help to classify some of the probes with ambiguous posterior enrichment probabilities so that more probes corresponding to binding sites are identified as enriched.
4. Simulation studies
In order to explore the performance of our approach more critically, we next conducted simulation studies designed to assess (i) how the joint model performs compared to standard two-step procedures and (ii) how the priors for motif prevalence affect the performance of the joint model, in a variety of data settings.
Simulated datasets were generated to assess the operating characteristics of the proposed method in three situations: when the probe enrichment values correspond to (i) the intensity only HMM, (ii) a misspecified model, and (iii) the TileMap nonparametric ChIP-chip model (Ji and Wong, 2005). The real intensity data was used instead of simulated intensity data in order to mimic the structure and the informativeness of the true experiment, and binding sites simulated and inserted into the corresponding sequence as described below. A HMM was fit to the 11,575 probe intensities on the 17 yeast chromosomes for a total of 12 million base pairs. To simulate the sequence data, we used the probe intensity data from the Rap1 dataset (Lieb et al., 2001) with four independent arrays described in Section 3, and applied the intensity-only model and the TileMap model which gave the probe enrichment probability estimate ŝp. The enrichment state for each of the probes were then simulated by Bernoulli random variables with probability ŝp, that is, sp,Simulated ∼ Bernoulli(ŝp). For the probes that were selected as enriched (sp,Simulated = 1), motif realizations were randomly inserted into the corresponding genomic sequences. We also considered a case where the intensity model is misspecified so that sp,Simulated ∼ Bernoulli(ŝp*) where ŝp* = Bernoulli(ŝp) with probability 0.9 and otherwise Logit(ŝp*) = Logit(ŝp) + ϵ where ϵ ∼ N(0, 1).
There were four simulation scenarios with two types of motif (a highly conserved artificial motif and the Rap1 binding motif taken from the literature), and two levels of motif site prevalence (0.0005: High, and 0.0002: Low). The highly conserved motif consisted of a 13 length sequence with each position having a 99% probability of the consensus letter and the rest of the letters with equal probability.
4.1 Analysis of simulated data
The accuracy of the binding site estimates is used to assess the models. The proposed joint intensity-sequence (IS) model gives the binding site probabilities directly, but the two step ChIP-chip methods like TileMap (TM) give only the enrichment probabilities of the probe sequences, not specific binding sites within those sequences. In order to get binding site estimates for TileMap, we used the following procedure. If a probe sequence had a posterior probability > 0.5 for enrichment, then it was included in the set of selected sequences. These selected sequences were searched for binding sites by fitting the stochastic dictionary model (Gupta and Liu, 2003). The primary aim of the analysis is to locate the binding sites of the TF, and these sites may be estimated by the posterior probability that each position on the genome corresponds to a sampled motif binding site (that is Ai = 1). This probability is estimated by averaging the indicators Ai at each position on the genome at each iteration of the DA sampler. A position on the genome was included in a list of binding sites if the posterior probability of being sampled as a TFBS was > 0.5.
When fitting motif discovery models with real DNA used as background, there are multiple motifs that represent multiple modes in the likelihood surface which may result in poor convergence. Multimodality issues when one does not know the true motif are discussed in Section 3. In the low prevalence scenarios, a strong prior was placed on the prevalence of the motif to prevent divergence as described in the Section 2.3, with δ0 = 106 and γ = 0.0001. Sensitivity analyses demonstrated that model estimation was robust to prior specifications within a moderately large range of the set values (more details in Section 3).
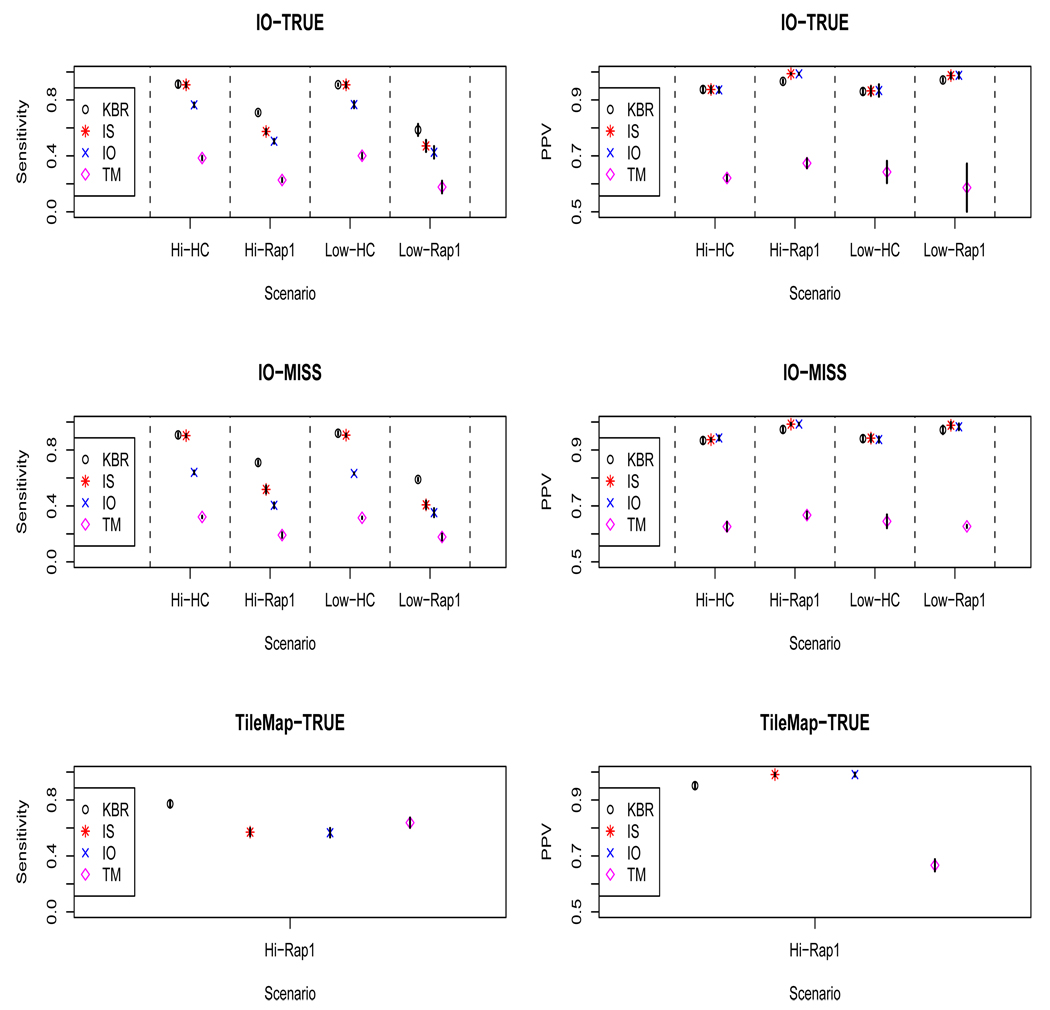
Four models were applied to each dataset (Figure 4). In the first model, the motif sites were sampled with the stochastic dictionary model conditioning upon the true enrichment region, and we call this the Known Binding Region (KBR) model. Second, the two step procedure was applied by fitting the Intensity Only (IO) model, and third, the two step procedure was applied using the TileMap method (TM). The TileMap method was not originally designed for two-color arrays, but it is flexible enough to use a test statistic for probe enrichment computed by another method. The test statistics for each probe were computed separately as the p-value under the null hypothesis that where is given by the IO model, and is a shrinkage estimate for the variance suggested by Ji and Wong (2005). Lastly, the proposed joint intensity and sequence (IS) model was applied. The model performance measures were sensitivity and Positive Predictive Value (PPV) for detection of simulated binding sites. PPV is the probability that an identified site was a true site.
Figure 4.
The plots show the results of simulations when the underlying true model is either the Intensity Only model (IO-TRUE), a misspecified Intensity Only model (IO-MISS), or the TileMap model (TileMap-TRUE). The four scenarios have High or Low motif prevalence (Hi/Low) with either a Highly Conserved (HC) or the Rap1 motif. The error bars are +/− 1 standard deviation. This figure appears in color in the electronic version of the article.
Figure 4 shows that the highly conserved motif was detected more accurately than the Rap1 motif for all models. Also, decreasing the prevalence of the Rap1 motif negatively impacted the sensitivities of all models. However, the effects of motif conservation were the strongest. The IS model was almost equivalent to the KBR model with the artificial motif. The IS model gives superior performance compared with the IO model in terms sensitivity for a given level of specificity for all four scenarios. Most notably, the sensitivity is enhanced by the joint IS model for all scenarios from for the IO model compared with for the IS model given a similar level of specificity. This implies that the motif matrix estimation is more accurate for the IS because this estimation is directly related to the accuracy of binding site estimation. In the low prevalence Rap1 motif scenario, the TileMap procedure failed to find any binding sites in 2 of the 5 simulations. The fits to simulated data that failed to converge were removed from analysis.
4.2 Simulated data based on the misspecified models
Next, in order to do a fair comparison, we simulated sequences based upon a misspecified IO model and the nonparametric TileMap intensity model enrichment estimates. First we discuss the misspecified model. For the highly conserved motif, the IS model performs almost as well as the KBR model, but the IO model loses some sensitivity under misspecification. For the high frequency Rap1 motif, the IS model loses less sensitivity (56% to 52%) than the IO model (50% to 40%). Further, the sensitivity (without loss of PPV) of the IS model is better preserved under misspecification than the IO model for all simulation scenarios.
The TileMap intensity model selected fewer regions to be enriched than the IO model, and a higher prevalence of binding sites within these regions was needed in order to estimate the motif accurately. The Rap1 motif was randomly inserted into the selected regions with a prevalence of 0.001. This scenario was repeated 5 times (Lower 2 panels of Figure 4). The TileMap (TM) model has the highest sensitivity, but the joint IS model is still demonstrated to comparable to the IO model The nonparametric intensity model of TileMap assumes that the probe intensity component of the proposed model may be misspecified, but the proposed joint model still shows an excellent performance.
5. Discussion
The proposed HMM for transcription binding site detection is a preliminary approach towards jointly analyzing the sequence data and ChIP-chip experimental measurements rather than the implementation of a two stage procedure. A sequence likelihood based on a stochastic dictionary model is included within the emission densities of the HMM. The joint Intensity Sequence (IS) model was shown to significantly out-perform the two-stage procedures for binding site discovery in terms of the sensitivity with a comparable specificity in the simulated data, indicating that using ChIP-chip binding information in the sequence motif discovery procedure improves estimation of TFBSs.
Several additional issues should be considered in ChIP-chip analysis. First, the low number of replicates is likely to be a continued feature of this type of data with some models even designed for experiments without replication (Johnson et al., 2006). Another issue is the effect of the variation in probe lengths– it was observed that longer probe lengths in general had a greater probability for enrichment. Future models could consider probe length explicitly. Also, the probes are only approximately equally spaced. A continuous-time HMM may be more appropriate to model probes that are unevenly distributed or contain large gaps.
This preliminary work presents a scenario for several possible variations and extensions of the IS model. The proposed model currently estimates probe specific enrichment probabilities. With the availability of higher resolution oligonucleotide arrays, future methods could consider genome-wide base-pair specific enrichment probabilities so that the genomic sequence is the fundamental unit of analysis rather than the probes that are an imperfect sampling of the genome. However, with arrays of higher resolution for larger genomes that contain possibly millions of probes (for example, human or mouse data), the dynamic programming techniques employed in the current MCMC procedure may not longer be feasible and alternative techniques would need to be explored. This could range from employing approximations to the model likelihood calculated through the recursive summations, to initial filtering steps that could reduce the total number of probes being considered using the joint model. Also, the bulk of the computational time is spent computing probe-specific sequence likelihoods and in sampling non-overlapping sequence segments, and this part of the algorithm may be done in parallel.
The simple binding model proposed is a reductionist perspective of the TF binding process. One possible limitation of using the current joint model in certain situations is that it may miss a number of real binding events that lack the motif sequence, as the binding is a result of interactions between the transcription factor with collaborating factors. For example, the Tup1 protein interacts with DNA indirectly in association with several different motifs (Buck and Lieb, 2006). The model could be extended to include the possibility of alternative binding motifs for the TF of interest, by introducing additional hidden states, each characterized by an alternative sequence motif. Biological insight into TFs working in conjunction are likely to motivate successful model extensions.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Jason Lieb, Michael Buck, and Paul Giresi for helpful discussions of the science and analysis of ChIP-chip experiments, and for the availability of the Rap1 data; and also the Associate editor and two anonymous referees whose insightful comments have greatly improved the content and presentation of the article.
Footnotes
Supplementary Materials
These materials may be accessed at the Biometrics website http://www.biometrics.tibs.org.
Contributor Information
Jonathan A. L. Gelfond, Department of Epidemiology and Biostatistics, University of Texas Health Science Center at San Antonio, USA.
Mayetri Gupta, Department of Biostatistics, University of North Carolina at Chapel Hill, NC, USA.
Joseph G. Ibrahim, Department of Biostatistics, University of North Carolina at Chapel Hill, NC, USA.
References
- Buck MJ, Lieb JD. Chip-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics. 2004;83:349–360. doi: 10.1016/j.ygeno.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Buck MJ, Lieb JD. A chromatin-mediated mechanism for specification of conditional transcription factor targets. Nature Genetics. 2006;38:1446–1451. doi: 10.1038/ng1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MJ, Nobel AB, Lieb JD. ChIPOTle: a user-friendly tool for the analysis of ChIP-chip data. Genome Biol. 2005;6:R97. doi: 10.1186/gb-2005-6-11-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler J, Tompa M. Finding motifs using random projections. J. Comput. Biol. 2002;9:225–242. doi: 10.1089/10665270252935430. [DOI] [PubMed] [Google Scholar]
- Cawley S, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Gottardo R, Li W, Liu S, Johnson E. University of British Columbia; A flexible and powerful Bayesian hierarchical model for ChIP-chip experiments. Technical report. 2006 doi: 10.1111/j.1541-0420.2007.00899.x. [DOI] [PubMed]
- Gupta M, Liu JS. Discovery of conserved sequence patterns using a stochastic dictionary model. J. Am. Statistical Association. 2003;98:55–66. [Google Scholar]
- Ji HK, Wong WH. Tilemap: create chromosomal map of tiling array hybridizations. Bioinformatics. 2005;21:3629–3636. doi: 10.1093/bioinformatics/bti593. [DOI] [PubMed] [Google Scholar]
- Johnson WE, et al. Model-based analysis of tiling-arrays for ChIP-chip. Proc. Nat. Acad. Sc. USA. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- Keles S. Mixture modeling for genome-wide localization of transcription factors. Biometrics. 2007;63:10–21. doi: 10.1111/j.1541-0420.2005.00659.x. [DOI] [PubMed] [Google Scholar]
- Keles S, van der Laan MJ, Dudoit S, Cawley SE. Multiple testing methods for ChIP-Chip high density oligonucleotide array data. J. Comput. Biol. 2004;13:579–613. doi: 10.1089/cmb.2006.13.579. [DOI] [PubMed] [Google Scholar]
- Li W, Meyer CA, Liu XS. A hidden Markov model for analyzing ChIP-chip experiments on genome tiling arrays and its application to p53 binding sequences. Bioinformatics. 2005;21:I274–I282. doi: 10.1093/bioinformatics/bti1046. [DOI] [PubMed] [Google Scholar]
- Lieb JD, Liu XL, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nature Genetics. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- Liu JS, Neuwald AF, Lawrence CE. Bayesian models for multiple local sequence alignment and Gibbs sampling strategies. J Amer Statist Assoc. 1995;90:1156–1170. [Google Scholar]
- Liu XS, Brutlag DL, Liu JS. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nature Biotechnology. 2002;20:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- Matys V, et al. Transfac (R): transcriptional regulation, from patterns to profiles. Nucleic Acids Research. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocke DM, Durbin B. A model for measurement error for gene expression arrays. J. Comput. Biol. 2001;8:557–569. doi: 10.1089/106652701753307485. [DOI] [PubMed] [Google Scholar]
- Shim H, Keles S. Integrating quantitative information from ChIP-chip experiments into motif finding. Biostatistics. 2007 doi: 10.1093/biostatistics/kxm014. (in press) [DOI] [PubMed] [Google Scholar]
- Zheng M, Barrera LO, Ren B, Wu YN. Chip-chip: Data, model, and analysis. Biometrics. 2007 doi: 10.1111/j.1541-0420.2007.00768.x. (in press) [DOI] [PubMed] [Google Scholar]
- Zhou Q, Liu JS. Modeling within-motif dependence for transcription factor binding site predictions. Bioinformatics. 2004;20:909–916. doi: 10.1093/bioinformatics/bth006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.