Abstract
The role of tRNA in translating the genetic code has received considerable attention over the last 50 years, and we now know in great detail how particular amino acids are specifically selected and brought to the ribosome in response to the corresponding mRNA codon. Over the same period, it has also become increasingly clear that the ribosome is not the only destination to which tRNAs deliver amino acids, with processes ranging from lipid modification to antibiotic biosynthesis all using aminoacyl-tRNAs as substrates. Here we review examples of alternative functions for tRNA beyond translation, which together suggest that the role of tRNA is to deliver amino acids for a variety of processes that includes, but is not limited to, protein synthesis.
Keywords: Amino acid, aminoacylation, protein synthesis, tRNA, translation
1. tRNA–dependent amino acid biosynthesis
The attachment of amino acids to the 3′-end of tRNAs is catalyzed by the aminoacyl-tRNA synthetase (aaRS) family of proteins [1]. aaRSs are ubiquitous and essential but only eukaryotes and a handful of bacteria have the full set of 20 enzymes, one for each canonical amino acid in the genetic code. Most bacteria and archaea lack asparaginyl-tRNA synthetase (AsnRS) and/or glutaminyl-tRNA synthetase (GlnRS) and some methanogenic archaea lack cysteinyl-tRNA synthetase (CysRS) [2]. Also, no aaRS for the rare amino acid selenocysteine has been found in any domain of life [3]. These organisms instead use indirect pathways to synthesize a number of amino acids (Asn, Cys, Gln and Sec) directly on their cognate tRNA: non discriminating aaRSs first form misacylated aminoacyl-tRNA (aa-tRNA), which is not used by the ribosome but instead converted to cognate aa-tRNA by various RNA-dependent modifying enzymes [4].
In organisms lacking GlnRS and AsnRS, Glu-tRNAGln and Asp-tRNAAsn are synthesized by non-discriminating aaRSs and converted to cognate Gln-tRNAGln and Asn-tRNAAsn by tRNA dependent amidotransferases (AdT). Two types of AdT exist, the heterotrimeric GatCAB present in both bacteria and archaea and the homodimeric GatDE present in archaea [5,6]. The tRNA moiety is recognized by the B and E kinase subunits of GatCAB and GatDE, respectively, which phosphorylate the mischarged tRNAs to form activated intermediates [7-9]. The glutaminase subunit (GatA/D) liberates ammonia from an amide donor and amidates Glu or Asp on the tRNA to form Gln or Asn, respectively. In both types of AdT, a 40 Å-long hydrophilic channel connects the glutaminase and kinase subunits [9,10]. It has been proposed, but remains to be proven, that ammonia liberated in the glutaminase active site is transported through the channel via a series of protonations and deprotonations to the kinase active site, and that binding of mischarged tRNA is required for opening the channel. Another open question concerns the precise in vivo mechanism by which misacylated aa-tRNA species are stabilized and escape detection, and subsequent delivery to the ribosome, by elongation factor Tu (EF-Tu). EF-Tu has a comparatively weak affinity for non-cognate aa-tRNA in vitro [11], but can bind and utilize such species under some circumstances in vivo when Adt is absent [12]. The presence of AdT seems to be critical; in Thermus thermophilus a ternary “transamidosome” complex is formed with aspartyl-tRNA synthetase and tRNAAsn, thereby channeling Asp- tRNAAsn directly to AdT [13]. Whether such a complex between AdT, aaRS and tRNA also forms during the indirect synthesis of Gln-tRNAGln remains an open question.
The formation of higher order complexes that facilitate substrate channeling, as seen with AdT, is also observed for the indirect synthesis of Cys-tRNACys in some archaea [14]. The first step of tRNA-dependent Cys biosynthesis is formation of O-phosphoseryl-tRNACys (Sep-tRNACys) by SepRS [15-18], which is then transformed to Cys-tRNACys by Sep-tRNA:Cys-tRNA synthase (SepCysS) in a pyridoxal phosphate (PLP)-dependent manner [15]. SepRS and SepCysS form a binary complex that not only promotes formation of the product but also protects the intermediate Sep-tRNACys from being captured by elongation factor and subsequently delivered to the ribosome [19]. The mechanisms by which the rare amino acid selenocysteine is incorporated into proteins share some similarities with the indirect Cys-tRNACys pathway, synthesis occurring via misaminoacylation of the unique tRNASec with serine [3]. The structure of human O-phosphoseryl-tRNA:selenocysteinyl-tRNA synthase (SepSecS) with tRNASec, phosphoserine and thiophosphate, a substrate analog of selenophostate, together with in vivo and in vitro studies, revealed the mechanism of selenocysteine formation in eukaryotes and archaea and how it differs from the well-characterized bacterial pathway [20-23]. In archaea and eukaryotes, SepSecS catalyzes the elimination of phosphate from O-phosphoseryl-tRNASec (Sep-tRNASec) forming an intermediate that is attacked by selenophosphate and subsequent hydrolysis yields selenocysteinyl-tRNASec [21,24]. Bacteria instead use water as the leaving group in the first step, a difference that could potentially be exploited to develop new transition state analog inhibitors that target Sec-tRNASec biosynthesis.
2. Proofreading and editing
The tRNA-dependent amino acid synthesis pathways described above all generate cognate aa-tRNAs as final products, which can then form ternary complexes with EF-Tu (EF-1a in eukaryotes) for use in ribosomal protein synthesis. In effect, the multiple steps of these indirect pathways provide a series of biosynthetic checkpoints that maintain the fidelity of aa-tRNA synthesis. This is not the case with direct aa-tRNA synthesis; many aaRSs have difficulty in discriminating against near-cognate amino acids, necessitating proofreading (“editing”) to clear non-cognate aa-tRNAs. Editing by aaRSs can occur before (pretransfer) and/or after (posttransfer) attachment of the non-cognate amino acid to tRNA, as explained below.
2.1. Pretransfer editing
Pretransfer editing occurs immediately following ATP-dependent activation of a non-cognate amino acid, before it is transferred to tRNA (Fig.1). Pretransfer editing can be either tRNA dependent or independent, and occurs via multiple pathways including selective release into solution, selective hydrolysis or cyclization of aminoacyl adenylate (aa-AMP) by the enzyme active site [25]. tRNA independent pre-transfer editing has been elucidated in aaRSs that either lack or require a separate posttransfer editing site for their canonical function [26-28]. Recently a dormant pretransfer editing pathway was uncovered in an E. coli leucyl-tRNA synthetase mutant deficient in posttransfer editing, underscoring the significance of redundancy of editing pathways in general [29,30]. Pre-transfer editing is enhanced in many aaRSs in the presence of a tRNA cofactor, which aids in translocation of misactivated aa-AMP to a distal editing site. For example, in isoleucyl-tRNA synthetase, hydrolysis of non-cognate Val-AMP is believed to occur through a conformational change induced upon misacylation of tRNA, which requires one round of the posttransfer editing reaction prior to subsequent pretransfer editing [31]. However, in many other examples, the exact nature of tRNA dependent aa-AMP hydrolysis remains unclear and the exact mechanisms are unknown.
Figure 1.
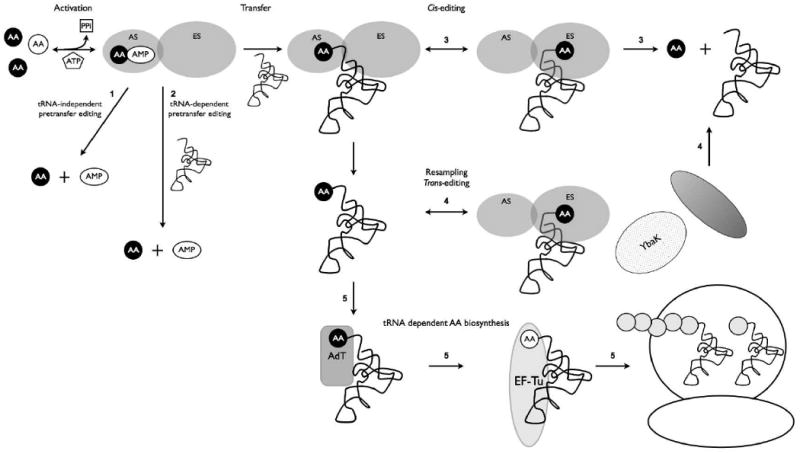
Fate of mischarged tRNAs in protein synthesis: Misactivated amino acids can be edited prior to their transfer to the tRNA either in a tRNA independent or dependent fashion (pathways 1 and 2 respectively). Following transfer mischarged aa-tRNA can be edited in cis (pathway 3) or trans (pathway 4) by the aaRS editing site. Free-standing editing factors such as YbaK and AlaXps can also hydrolyze mischarged aa-tRNAs (pathway 4). Mischarged Glu-tRNAGln or Asp-tRNAAsn are converted to Gln-tRNAGln or Asn-tRNAAsn via the transamidase reaction (tRNA-dependent amino acid biosynthesis, pathway 5).
2.2. Posttransfer editing
Following aminoacylation of tRNA with a non-cognate amino acid, several aaRSs utilize a post-transfer editing mechanism to hydrolyze mischarged aa-tRNAs and prevent amino acid misincorporation during translation. The mechanisms of hydrolyzing mischarged aa-tRNA are known for several posttransfer editing aaRSs, and can occur either in cis or in trans (Fig. 1). An important prerequisite for the posttranfer editing reaction is movement of the 3′-CCA end of the mischarged tRNA from the aminoacylation site into the editing site [25]. Structural studies of class I aaRS have shown conformational changes in both the tRNA and enzyme between the aminoacylation and editing states, which suggests the 3′-CCA end of the tRNA directly translocates between the active and editing sites (Fig. 1 Pathway 3; [32-35]). In support of this model, aa-tRNA release has been shown to be the rate limiting step in aminoacylation for class I aaRSs, giving the 3′-CCA time to move between the sites before being released [36]. While direct translocation of the tRNA 3′-CCA end between the aminoacylation and editing sites is also thought to occur for class II aaRSs, dynamics of tRNA movement appear to be more complex. Initial evidence for phenylalanyl-tRNA synthetase (PheRS) supported the direct translocation model and further data has not refuted this. However, for both PheRS and alanyl-tRNA synthetase (AlaRS) low levels of mischarged species can be detected under certain conditions [37,38]. These mischarged species do not compromise translational fidelity; once Tyr-tRNAPhe is released from the active site, the PheRS editing site can compete effectively with EF-Tu for the mischarged tRNA and rebinds and edits the non-cognate species (Fig. 1 Pathway 4; [38]). While PheRS appears to utilize both tRNA translocation and rebinding after release, a translocation model is unlikely in AlaRS. Biochemical and structural data have shown that in AlaRS the activation site and editing site have independent tRNAAla binding capabilities and tRNA likely rebinds to the editing site after aminoacylation [39,40]. As product release is not the rate limiting step of aminoacylation for class II aaRSs the aa-tRNA is likely released following aminoacylation and then rebound at the editing site [36].
In addition to the editing domains present in some aaRSs, several freestanding editing factors homologous to class II editing domains can be found in all three kingdoms of life. These factors act in trans to clear mischarged aa-tRNAs (Fig. 1 Pathway 4; [25]). Examples include D-Tyr-tRNATyr deacylases [41], AlaXps, [42] and YbaK [43]. In the specific case of tRNAPro, trans-editing by YbaK constitutes an additional layer of quality control by prolyl-tRNA synthetase (ProRS). ProRS insertion domain (INS) cis-edits mischarged Ala-tRNAPro [44] whereas YbaK in complex with ProRS serves to trans-edit Cys-tRNAPro [45,46]. It is not known why editing functions have become associated with the aaRS in some instances while in other cases the editing domains have remained independent of the aaRS, although it may reflect the need to edit a wider range of non-cognate species for particular tRNA isoacceptors.
3. aa-tRNAs as markers of a cell's health
Aminoacylation, editing, and tRNA-dependent amino acid biosynthesis all serve a common goal, to generate a cellular pool of cognate aa-tRNA. Normally the majority of this aa-tRNA pool is delivered to the ribosome for protein synthesis, although some components of this pool also provide amino acids for other biosynthetic pathways (Fig. 2; see section 4 below). When the aa-tRNA pool becomes unbalanced the cell responds in a number of ways. For example, bacteria adjust their metabolism using mechanisms such as the stringent response to attempt to restore a complete and balanced pool of aa-tRNA. In eukaryotes the consequences of altering the aa-tRNA pool are more complex. For example overexpression of RNA pol III gene products, such as ribosomal 5S RNA and tRNA, and increased protein synthesis, are key characteristics of a large variety of tumor cells. In particular, the recent finding that increased expression of tRNAiMet is sufficient to drive cell proliferation and oncogenic transformation has implicated tRNAs in tumorigenesis [47].
Figure 2.
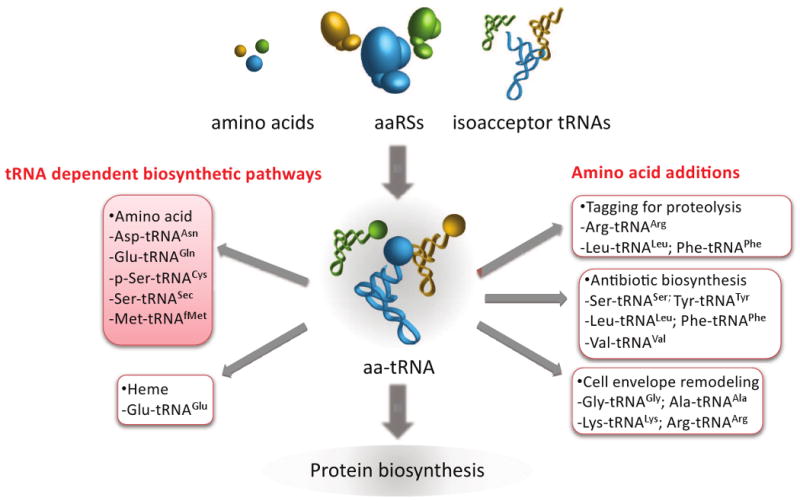
Cellular biosynthetic pathways that utilize aa-tRNAs. Examples are provided in the red shaded boxes of pathways other than protein synthesis, together with the corresponding substrates, that require aa-tRNAs as precursors.
Absence of extracellular amino acids causes a starvation response in cells leading to deactivation of one of the major factors driving cell growth and proliferation in eukaryotes, the TOR protein (target of rapamycin [48]). Nutrient deprivation, however, has also been shown to result in accumulation of tRNAs in the nucleus [49-51]. Macroautophagy, a major intracellular catabolic process of eukaryotes, is highly induced during starvation and functions mainly to provide sufficient nutrients (amino acids, lipids, sugars and nucleotides) and energy for the cells to survive a temporary depletion of nutrients [52]. As cellular levels of amino acids are re-established tRNAs are re-exported to the cytosol. The presence of tRNAs and amino acids in the cytosol and reactivation of mTOR restores global protein synthesis, cell growth and proliferation. Concurrently, autophagic activity returns to its basal cellular levels. Due to their almost insatiable capacity for growth and proliferation, cancer cells often deplete nutrients from their immediate surroundings, yet they still grow. It is believed that the autophagic process plays a major role in tumor progression, functioning as an essential survival mechanism that provides the deprived tumor cells with adequate nutrients and energy to sustain growth [53]. One important factor influencing a tumor cell's ability to grow and proliferate is the availability of sufficient aa-tRNAs in the cytosol. It is essential for tumor cells to have an adequate supply of aa-tRNAs in the near vicinity of the translational machinery in order to keep up with the high demand for newly synthesized proteins. This leads to the intriguing hypothesis that in proliferating tumor cells, despite nutrient limitation, fully aminoacylated tRNAs may be localized in the cytosol with the autophagic pathway supplying adequate amino acids to sustain robust protein synthesis.
If tRNAs are predominantly cytoplasmic in nutrient deprived tumor cells could this directly reflect a more global defect in nucleocytoplasmic trafficking? It is well known that nucleocytoplasmic shuttling of oncogenes and tumor suppressors is often disrupted in tumor cells resulting in mislocalization and alteration of their cellular activities. Defective nucleocytoplasmic trafficking can be highly advantageous for proliferating cells when transcription factors which function as tumor suppressors are mislocated to the cytosol [54]. Dysfunctional expression of human XPO-T or yeast Los1p, which specifically export newly synthesized tRNAs from the nucleus, is known to disrupt trafficking leading to nuclear accumulation of tRNAs [55,56]. Recent studies suggest that nuclear accumulation of tRNAs transduces a starvation signal within the cell most likely by signaling the mTOR pathway and activating autophagy (MPI, unpublished data). Whether these effects emerge directly from nuclear accumulation of tRNA or due to down-regulated protein synthesis remains to be resolved. Interestingly, DNA damage also affects nuclear export of tRNA, leading to accumulation of immature tRNAs and subsequent execution of G1 and cell cycle progression [57]. Taken together, recent data clearly suggest that subcellular localization of tRNA, and its aminoacylation state, are critical determinants of cell growth and proliferation providing potential new strategies for the development of anti-cancer therapeutics.
4. tRNA dependent addition of amino acids
Beyond their essential roles in protein synthesis, several aa-tRNAs are also used in other amino acid addition pathways (Fig. 2). These reactions involve aa-tRNAs of different specificities and several types of acceptor molecules such as membrane lipids, peptidoglycan precursors, proteins and intermediates for the biosynthesis of antimicrobial molecules.
4.1. tRNA dependent addition of amino acids to membrane lipids
Initially discovered in Staphylococcus aureus, lysylphophatidylglycerol synthase (LysPGS) transfers lysine from Lys-tRNALys to phosphatidylglycerol (PG) within the cytoplasmic membrane to form lysyl-phosphatidylglycerol (Lys-PG) [58]. LysPGS, encoded by the gene mprF (multipeptide resistance factor), is composed of a membrane domain consisting of several transmembrane alpha-helices and a cytoplasmic domain containing the active site of the enzyme [59]. aaPGS homologs are widespread among bacteria and are also found in certain archaea of the Methanosarcina genus [59,60]. tRNA mediated addition of amino acids to the membrane is one of the multiple mechanisms that has evolved in bacteria to remodel their cell wall as they adapt to changing environments [61]. Addition of positively charged amino acids to the highly negatively charged phospholipids of the membrane diminishes the affinity of the bacterial envelope for cationic molecules within the growth environment. Thus, Lys-PG in the membrane is associated with a wide variety of resistance phenotypes against various cationic antibacterial peptides (CAMs) and several other antimicrobial molecules (for review see ref. [60]). In addition, LysPGS has been associated with the virulence of several pathogenic microorganisms in cell line models [58,60], and in animal [62,63] or plant models [64].
Additional aa-tRNA specificities have been recently discovered in the aaPGS protein family. AlaPGS in C. perfringens [65] and in P. aeruginosa [66], and more recently an ArgPGS in E. faecium [59] have all been characterized. These new modifications provide bacteria with an enhanced repertoire of amino acids with which to modify phospholipids and to further adapt their membrane to environmental cues. In spite of the lack of addition of a net positive charge to the membrane, the addition of Ala not only provides P. aeruginosa with resistance to CAMs and beta-lactams, but also to acidic and osmotic stress growth conditions [66,67]. The nature of the different phenotypes linked to the addition of amino acids to lipid polar head groups illustrates that in addition to changing the net charge of the membrane and the affinity of the membrane for CAMs, these modifications may also change more general biochemical and permeability properties of the cellular barrier.
AaPGSs and EF-Tu have comparable affinities for aa-tRNA, suggesting that canonical species can effectively supply amino acids for both protein synthesis and lipid modification (Fig. 3; [68]). The specificity of aaPGSs for each moiety of aa-tRNAs (i.e., amino acid and tRNA) is less stringent than observed for aaRSs. AlaPGS and LysPGS from C. perfringens can efficiently catalyze PG modification with a broad selection of RNA (heterologous tRNAs, minihelices) acylated with the cognate amino acid (i.e., Ala or Lys respectively) [65]. In addition, certain aaPGSs can process more than one aa-tRNA species in vitro. For example, the aaPGS from E. faecium is triple specific and modifies PG with Arg-tRNAArg, Lys-tRNALys and Ala-tRNAAla, and the LysPGS from B. subtilis is able to use Lys-tRNALys and Ala-tRNAAla as amino acid donors [59]. The physiological significance of these multiple-specificity aaPGS activities remains an open question.
Figure 3.
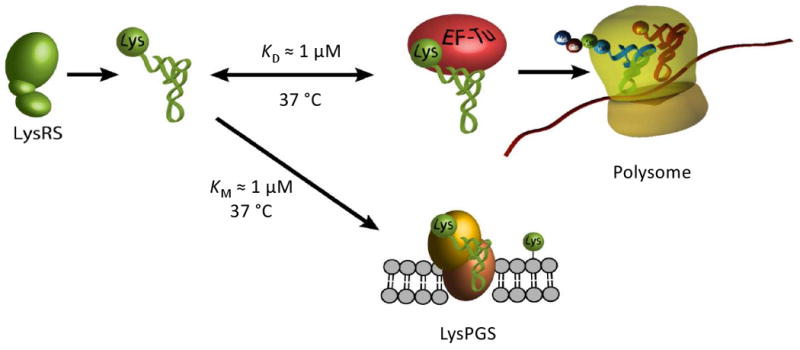
Partitioning of Lys-tRNALys between protein synthesis and lipid remodeling. Lys-tRNALys shows comparable affinities for LysPGS and EF-Tu, allowing aa-tRNAs to potentially enter different biosynthetic pathways simultaneously.
4.2. tRNA dependent addition of amino acids to the amino-terminus of proteins
Another protein family able to utilize a range of aa-tRNAs is the aminoacyl-transferase (aa-transferase), which initiate recycling of damaged or excess peptides via the N-end rule. Found in bacteria, fungi and mammals, the N-end rule relates the identity of the N-terminal residue of a protein to its in vivo half-life [69]. This pathway relies on an aa-transferase to recognize a secondary destabilizing residue at the N-terminus of a target peptide and utilize an aminoacyl-tRNA to transfer a primary destabilizing amino acid to the N-terminal residue, making the protein a target for the cellular degradation machinery [69]. In prokaryotes, the aat-encoded leucylphenylalanyl-transferase (L/FK,R-transferase) demonstrates affinity for the secondary destabilizing residues Lys and Arg and attaches a primary destabilizing residue of either Leu [70] or Phe [71]. The presence of the primary destabilizing residue marks the peptide as a target for ClpS, which transfers the protein to the ClpAP machinery where it is degraded [69]. In eukaryotes, this hierarchy differs with oxidized cystiene (Cys*), Asp, or Glu constituting secondary destabilizing residues that are attached to the primary destabilizing residue Arg by the ATE1-encoded arginyltransferase (RC*,D,E-transferase) [72,73]. The primary destabilizing residue marks the peptide as a target for ubiquitin conjugation and subsequent degradation by the eukaryotic proteasome [69]. Recently, two aa-transferases that vary in specificity have been discovered. ATEL1 arginyl-transferase (RD,E-transferase) in the eukaryotic pathogen Plasmodium falciparum has sequence similarity to the prokaryotic L/FK,R-transferase but the specificity of the eukaryotic RD,E-transferase [74]. Bpt leucyl-transferase (L-D,E-transferase), found in the prokaryotic human pathogen Vibrio vulnificus, exhibits hybrid specificity conjugating leucine to the eukaryotic secondary destabilizing residues of Asp and Glu [72].
Recently, the structure of E. coli L/FK,R-transferase has been determined in complex with Leu-tRNALeu and an amino-terminal Arg protein substrate [75-77]. This structure not only allowed further verification of the substrate binding sites but also the elucidation of the catalytic mechanism, which previously remained unknown. L/FK,R-transferase is a monomer consisting of two domains [77]. The N-terminal domain adopts a novel fold while the C-terminal domain resembles the GCN5-related N-acetyltransferase (GNAT) family [77]. Conserved residues lie along the central cleft that exists at the interface of the two domains. At the bottom of the cleft a deep hydrophobic pocket recognizes the side chain of the bound tRNA while the backbone of the 3′-region of a tRNA-acceptor helix is recognized by a cluster of highly conserved positively charged resides [76]. The hydrophobic nature of the pocket selects for hydrophobic side chains while the size allows for the selection of amino acid residues lacking a branched β-carbon [75,76]. Recognition of primary substrate Leu-tRNALeu or the alternate substrate Phe-tRNAPhe occurs in this manner. The main mode of L/FK,R-transferase protein acceptor recognition is the presence of an N-terminal basic amino acid residue [75]. This recognition is achieved by the highly negatively charged surface area adjacent to the binding site of the Arg or Lys side chain within the hydrophobic pocket [75]. Peptide bond formation by L/FK,R-transferase involves a protein-based reaction via an electron relay from a donor Asp 186 residue to acceptor Gln 188 [75]. This mechanism most closely resembles the reverse-acylation step of proteolysis by serine proteases and is distinct from ribosomal peptide-bond formation.
Although there is currently no structure or mechanism for the eukaryotic RC*,D,E-transferase, the roles it plays in cellular development are well studied. Previously implicated in cardiovascular development, the fidelity of chromosome segregation, and the control of signaling by nitric oxide, RC*,D,E-transferase has also recently been linked to leaf senescence and leaf and shoot development in plants [73], [70,78-81]. These effects seem to be the result of varying expression levels of different isoforms with different protein targets in tissues [73,82]. Representative crystal structures of the different isoforms are needed to determine the specificity determinants of protein acceptor recognition, as substrate recognition by prokaryotic L/FK,R-transferase seems to be sequence-independent. Additional specific in vivo targets of prokaryotic L/FK,R-transferase have yet to be determined. The only characterized target, PATase, implicates L/FK,R-transferase in putrescine homeostasis as well as in increased specificity for the modification of an N-terminal methionine residue [83]. The recently discovered dual aa-transferases, L/FK,R-transferase and LD,E-transferase, in the prokaryotic pathogen Vibrio vulnificus provide intriguing targets for the identification of additional potential in vivo targets.
4.3. tRNA dependent addition of aa of to peptidoglycan precursors
Similar in structure, but not specificity or function to the aminoacyltransferases are a group of peptidytransferases required for peptidoglycan biosynthesis. First discovered in Staphylococcus aureus [84], factors essential for methicillin resistance (Fem) are a type of non-ribosomal peptidyltransferase that aid in cell wall biosynthesis by extending the interpeptide bridge of the peptidoglycan molecule [85,86]. The cross linking of short intrastrand peptides provides the cell with additional rigidity allowing for resistance to β-lactam antibiotics, including methicillin. This process requires not only FemABX enzymes, but also a selection of amino acids donated by their corresponding tRNA isoacceptors [87]. To build the interpeptide chain, the amino acid is activated and transferred to its tRNA by an aaRS and then moved to a hexapeptide lipid intermediate. FemABX catalyzes the addition of the amino acid to build the interpeptide bridge. The type and number of residues constituting this chain varies by species, as do enzyme and substrate specificities [88]. For example, S. aureus exhibits a pentaglycine interpeptide chain and the three Fem proteins act individually to link amino acids: FemX attaches the first glycine residue to the ε amino group of the lysine side chain, FemA adds two glycine residues, and FemB links the final two residues [85]. Following the formation of the interpeptide chain, D,D-transpeptidases finalize the cross linking of strands. This step is required, as incomplete or incorrect formation of the interpeptide bridge leads to increased antibiotic susceptibility or cell death [88].
In order for particular aa-tRNAs to be available for peptidoglycan biosynthesis, they must escape the protein biosynthesis machinery. Some aa-tRNA isoacceptors sequestered for peptidoglycan biosynthesis cannot complex with EF-Tu:GTP due to the absence of certain GTψC and GG sequences, and therefore are unable to bind to the ribosome [87]. Recent studies involving chemical acylation of RNA helicies [89] and site-directed mutagenesis [90] in Weissella viridescens FemXwv have indicated some of the modes of recognition of aa-tRNA by FemX. The FemXwv active site excludes all amino acids, with the exception of Gly and Ala, based on steric hindrance. The enzyme then safeguards against misincorporation of Gly by identifying the discriminator base of the tRNAGly and tRNAAla acceptor stems and binds tRNAAla, but not tRNAGly [89].
Along with the sequences of the aa-tRNAs, the structures of the Fem proteins also dictate how they catalyze the transfer of amino acid residues. The X-ray structure of S. aureus FemA was the first of the Fem family to be solved and it was found to include several known protein folds that facilitate both peptide and aa-tRNA binding [86]. Fem A is composed of two domains: a globular domain, which is divided into subdomains, 1A and 1B, and a domain consisting of two helical arms. An L-shaped cavity in domain 1B serves as FemA's lone binding pocket and accommodates hexapeptide lipid intermediate binding. Similar to bacterial seryl-tRNA synthetases, the antiparallel coiled-coil structure of domain 2 docks aa-tRNA as glycine is added to the growing pentaglycine chain [86]. As more is discovered about the specificities of the FemABX family, as well as the isoacceptor tRNAs they utilize, they are also becoming attractive targets for the development of antibiotics that specifically target β-lactam resistant bacteria [90,91].
4.4. tRNA and antibiotic biosynthesis
The involvement of aa-tRNA in antibiotic biosynthesis provides another role for this versatile molecule, particularly in diverse environmental microorganisms such as Streptomyces, the soil-dwelling multicellular bacteria that produce a myriad of bioactive metabolites. Biosynthesis of complex antibiotics from simple metabolic precursors involves tens to hundreds of cellular factors that are mostly encoded by horizontally acquired genes in biosynthetic clusters. These species-specific activities of secondary metabolism display substantial cross talk with the more conserved network of primary metabolism that determines the onset and supports the process of antibiotic production. tRNAs are required both for the canonical biosynthesis of ribosomal peptide antibiotics [92] and as regulators or alternative carrier molecules for nonribosomal antibiotic synthesis.
Genetic analysis of a mutant (bldA) of Streptomyces coelicolor A3(2) implicated a tRNA specific for the leucine codon UUA - the rarest codon in all Streptomyces genes - in antibiotic biosynthesis and morphological development [92-94]. Deletion of the bldA gene resulted in loss of two bioactive pigments, actinorhodin and undecylprodigiosin, produced by S. coelicolor [95]. The bldA tRNA was accumulated only when the antibiotics started to be produced, which implied that the levels of charged tRNA might determine the level of the products of TTA-containing genes in an adaptive manner [95]. It was found that TTA codon-containing pathway-specific regulatory genes are required for the expression of the biosynthetic genes in the two gene clusters [96]. Recently, bioinformatic analyses with four Streptomyces genomes showed that the majority of TTA-containing genes are species specific and relatively recently acquired [97]. Nearly half of the pathway-specific regulatory genes in the genomes analyzed contain TTA codons, suggesting a way of limiting the biosynthesis of antibiotic in the host to physiologically appropriate circumstances. Using rare codon tRNAs to control antibiotic biosynthesis in Streptomyces could be a general mechanism due to the uniqueness of the genomic contexts. The developmentally regulated use of rare codon tRNAs is not limited to antibiotic biosynthesis. Rare tRNAs are also important for biofilm formation and dispersal in E. coli, and it seems likely that still other roles remain to be discovered [98].
Recently the foundation has been laid for exploring additional roles for tRNA in Streptomyces by the discovery of two biosynthetic enzymes that can use aminoacyl-tRNAs. The antibiotic valanimycin produced by Streptomcyes viridifaciens is derived from L-valine and L-serine. L-valine is first transformed to isobutylhydroxylamine that must be reacted with L-serine during valanimycin biosynthesis [99]. A seryl-tRNA synthetase gene unexpectedly identified in the valanimycin biosynthetic gene cluster (vlm) gave rise to the idea that the seryl residue could be transferred from seryl-tRNA to the hydroxyl group of isobutylhydroxylamine [100]. When the VlmA protein, which is distantly related to enzymes involved in the peptidyl modification of bacterial cell wall, was tested in an in vitro reaction with the two substrates, O-seryl-isobutylhydroxylamine was indeed formed and the formation was shown to be seryl-tRNA dependent. Another tRNA-dependent example is the biosynthesis of albonoursin (alb) in Streptomyces noursei [101]. Albonoursin is a cyclodipeptide antibiotic made from L-phenylalanine and L-leucine. A novel enzyme AlbC identified from the analysis of the alb gene cluster was shown to catalyze the formation of two peptide bonds at the same time to render the cyclic product. This reaction only occurred when soluble extracts of E. coli cells, but not free amino acids, were present. It was subsequently found that charged E. coli tRNAPhe and tRNALeu were the required substrates. AlbC homologs were identified in several other organisms including firmicutes, actinobacteria and γ-proteobacteria, all of which produced cyclo(Tyr-Tyr) or cyclo(Leu-Leu) in the presence of ATP, the corresponding amino acids and E. coli cell extracts. Treatment of the extracts with RNases abolished the formation of cyclopeptide, strongly indicating a dependency on aminoacyl-tRNAs for synthesis. Furthermore, numerous cyclodipeptides with the incorporation of L-alanine, L-valine and L-methionine were detected, indicating that AlbC and its homologs could use other tRNAs as substrates besides tRNAPhe, tRNALeu and tRNATyr. These examples clearly demonstrate that tRNA likely has many more roles that remain to be uncovered by studying the dazzling variety of biochemical reactions found in antibiotic biosynthetic pathways. In particular, biochemical characterization of Streptomyces tRNAs may hold the promise to discovering new antibiotic biosyntheses [102].
5. tRNAs that don't function as aa-tRNA
While many roles of tRNA outside translation take advantage of its capacity to carry activated amino acids, several important functions do not require the aminoacyl form. Uncharged tRNAs function as sensors of amino acid concentration and as regulators of global gene expression in response to changes in amino acid concentration. In Gram-positive bacteria, this regulation often occurs via the T box transcription termination system in which an uncharged tRNA binds to the 5′ untranslated leader mRNA to induce formation of an antiterminator over the competing intrinsic transcriptional terminator [103]. Formation of the antiterminator allows for continued transcription of the downstream genes, such as those encoding the aminoacyl-tRNA synthetases and proteins involved in amino acid biosynthesis and transport [103]. In response to amino acid starvation in Gram-negative bacteria, such as E. coli, uncharged tRNAs stimulate the stringent response by inducing the production of the global transcriptional regulator ppGpp, which regulates tRNA and rRNA concentrations and genes involved in amino acid biosynthesis [104]. Similarly, in certain eukaryotic cells under amino acid starvation, uncharged tRNAs activate the protein Gcn2p, which reduces overall protein translation by phosphorylating eIF2 and increases amino acid production by activating the transcriptional regulator Gcn4p [105].
Uncharged tRNAs are often cleaved in half in response to amino acid starvation to rapidly reduce tRNA levels, thereby reducing protein translation [106,107]. However, tRNA cleavage is developmentally regulated and not induced by amino acid starvation in the Gram-positive bacterium S. coelicolor [109]. tRNA cleavage is also induced by oxidative stress in many eukaryotic cells, including mammals and plants [109]. A recent study on Giardia lamblia found that aa-tRNAs are cleaved in response to stress and development to a dormant cyst [110]. Unlike cleaved uncharged tRNAs, aa-tRNA cleavage products are retained at a constant level throughout the stress response, and therefore may have functions in addition to maintaining a low metabolic rate. It has been suggested that uncharged tRNAs may act as regulators of gene expression, possibly as antisense RNAs. Uncharged tRNAs have also been shown to act as primers for DNA synthesis of certain retrovirus genomes and serve a regulatory role in replication of the ColE1 plasmid in E. coli by inducing cleavage of RNA I [111,112]. Further exploration is now needed to more fully understand how tRNAs and their cleavage products function as regulatory RNAs.
The function of a tRNA within translation can be predicted with software programs that base tRNA prediction on the anticodon sequence, predicted secondary structure, and additional identity elements [113-115]. However, many of these annotated tRNA genes lack canonical features, such as a predicted conserved secondary structure, and are classified as pseudo-tRNAs [114,115]. These pseudo-tRNAs may be relics of tRNAs that now maintain a different function, whether in biosynthesis of cell walls or antibiotics, regulation of gene expression, or genome replication. One example is tRNAOther, a tRNA-like small RNA found in Bacillus cereus [116,117]. Deletion of tRNAOther is not deleterious to growth, but causes the cell to lose resistance to cationic antibiotics, ionophores, and detergents, and alters transcript levels of one of two trpS genes during stationary phase. These unusual roles of tRNAOther suggest that other pseudo-tRNAs may also have unexpected functions outside translation.
6. Future prospects
The observation that binding to EF-Tu does not constitute an irreversible commitment to protein synthesis [38] suggests that aa-tRNAs might also provide a source of activated amino acids for processes beyond those described above. In particular, the existence of aaRS paralogs in antibiotic synthesis gene clusters implicates aa-tRNA as common precursors in antibiotic biosynthesis [118]. Possible roles for tRNAs and pseudo-tRNAs outside translation are also starting to emerge, for example as members of the broader class of regulatory RNAs [119,120]. Overall, recent advances have expanded the functional repertoire of aa-tRNAs, tRNAs, and pseudo-tRNAs, and it seems likely that many more functions await discovery in the post-genomic era.
Acknowledgments
Work in these areas in the authors' laboratories is supported by grants from NIGMS (65183, MI), National Science Foundation (744791, MI), American Heart Association (GA0016068, SC; 09BGIA2230347, MPI), American Cancer Society (ODSR 2009-2, MPI), and a pre-doctoral fellowship from the OSU Center for RNA Biology (NMR).
List of abbreviations
- AaRS
aminoacyl tRNA-synthetase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Woese CR, Olsen GJ, Ibba M, Söll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol Mol Biol Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ibba M, Söll D. Aminoacyl-tRNAs: setting the limits of the genetic code. Genes Dev. 2004;18:731–738. doi: 10.1101/gad.1187404. [DOI] [PubMed] [Google Scholar]
- 3.Böck A, Thanbichler M, Rother M, Resch A. Selenocysteine. In: Ibba M, Francklyn C, Cusack S, editors. The Aminoacyl-tRNA Synthetases. Landes Bioscience; Georgetown, Texas, USA: 2005. pp. 320–327. [Google Scholar]
- 4.Sheppard K, Yuan J, Hohn MJ, Jester B, Devine KM, Söll D. From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res. 2008;36:1813–1825. doi: 10.1093/nar/gkn015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curnow AW, Hong K, Yuan R, Kim S, Martins O, Winkler W, Henkin TM, Söll D. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tumbula DL, Becker HD, Chang WZ, Söll D. Domain-specific recruitment of amide amino acids for protein synthesis. Nature. 2000;407:106–110. doi: 10.1038/35024120. [DOI] [PubMed] [Google Scholar]
- 7.Wilcox M. Gamma-phosphoryl ester of glu-tRNA-GLN as an intermediate in Bacillus subtilis glutaminyl-tRNA synthesis. Cold Spring Harb Symp Quant Biol. 1969;34:521–528. doi: 10.1101/sqb.1969.034.01.059. [DOI] [PubMed] [Google Scholar]
- 8.Feng L, Sheppard K, Tumbula-Hansen D, Söll D. Gln-tRNAGln formation from Glu-tRNAGln requires cooperation of an asparaginase and a Glu-tRNAGln kinase. J Biol Chem. 2005;280:8150–8155. doi: 10.1074/jbc.M411098200. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura A, Yao M, Chimnaronk S, Sakai N, Tanaka I. Ammonia channel couples glutaminase with transamidase reactions in GatCAB. Science. 2006;312:1954–1958. doi: 10.1126/science.1127156. [DOI] [PubMed] [Google Scholar]
- 10.Oshikane H, et al. Structural basis of RNA-dependent recruitment of glutamine to the genetic code. Science. 2006;312:1950–1954. doi: 10.1126/science.1128470. [DOI] [PubMed] [Google Scholar]
- 11.Roy H, Becker HD, Mazauric MH, Kern D. Structural elements defining elongation factor Tu mediated suppression of codon ambiguity. Nucleic Acids Res. 2007;35:3420–3430. doi: 10.1093/nar/gkm211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruan B, Palioura S, Sabina J, Marvin-Guy L, Kochhar S, Larossa RA, Söll D. Quality control despite mistranslation caused by an ambiguous genetic code. Proc Natl Acad Sci USA. 2008;105:16502–16507. doi: 10.1073/pnas.0809179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailly M, Blaise M, Lorber B, Becker HD, Kern D. The transamidosome: a dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis. Mol Cell. 2007;28:228–239. doi: 10.1016/j.molcel.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 14.Hauenstein SI, Perona JJ. Redundant synthesis of cysteinyl-tRNACys in Methanosarcina mazei. J Biol Chem. 2008;283:22007–22017. doi: 10.1074/jbc.M801839200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sauerwald A, et al. RNA-dependent cysteine biosynthesis in archaea. Science. 2005;307:1969–1972. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- 16.Fukunaga R, Yokoyama S. Structural insights into the first step of RNA-dependent cysteine biosynthesis in archaea. Nat Struct Mol Biol. 2007;14:272–279. doi: 10.1038/nsmb1219. [DOI] [PubMed] [Google Scholar]
- 17.Kamtekar S, Hohn MJ, Park HS, Schnitzbauer M, Sauerwald A, Söll D, Steitz TA. Toward understanding phosphoseryl-tRNACys formation: the crystal structure of Methanococcus maripaludis phosphoseryl-tRNA synthetase. Proc Natl Acad Sci USA. 2007;104:2620–2625. doi: 10.1073/pnas.0611504104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauenstein SI, Hou YM, Perona JJ. The homotetrameric phosphoseryl-tRNA synthetase from Methanosarcina mazei exhibits half-of-the-sites activity. J Biol Chem. 2008;283:21997–22006. doi: 10.1074/jbc.M801838200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang CM, Liu C, Slater S, Hou YM. Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNACys. Nat Struct Mol Biol. 2008;15:507–514. doi: 10.1038/nsmb.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forchhammer K, Böck A. Selenocysteine synthase from Escherichia coli. Analysis of the reaction sequence. J Biol Chem. 1991;266:6324–6328. [PubMed] [Google Scholar]
- 21.Palioura S, Sherrer RL, Steitz TA, Söll D, Simonovic M. The human SepSecS-tRNASec complex reveals the mechanism of selenocysteine formation. Science. 2009;325:321–325. doi: 10.1126/science.1173755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan J, et al. RNA-dependent conversion of phosphoserine forms selenocysteine in eukaryotes and archaea. Proc Natl Acad Sci USA. 2006;103:18923–18927. doi: 10.1073/pnas.0609703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu XM, et al. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007;5:e4. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mueller EG. Se-ing into selenocysteine biosynthesis. Nat Chem Biol. 2009;5:611–612. doi: 10.1038/nchembio0909-611. [DOI] [PubMed] [Google Scholar]
- 25.Ling J, Reynolds N, Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Annu Rev Microbiol. 2009;63:61–78. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- 26.Hati S, et al. Pre-transfer editing by class II prolyl-tRNA synthetase: role of aminoacylation active site in “selective release” of noncognate amino acids. J Biol Chem. 2006;281:27862–27872. doi: 10.1074/jbc.M605856200. [DOI] [PubMed] [Google Scholar]
- 27.Gruic-Sovulj I, Rokov-Plavec J, Weygand-Durasevic I. Hydrolysis of non-cognate aminoacyl-adenylates by a class II aminoacyl-tRNA synthetase lacking an editing domain. FEBS letters. 2007;581:5110–5114. doi: 10.1016/j.febslet.2007.09.058. [DOI] [PubMed] [Google Scholar]
- 28.Splan KE, Ignatov ME, Musier-Forsyth K. Transfer RNA modulates the editing mechanism used by class II prolyl-tRNA synthetase. J Biol Chem. 2008;283:7128–7134. doi: 10.1074/jbc.M709902200. [DOI] [PubMed] [Google Scholar]
- 29.Boniecki MT, Vu MT, Betha AK, Martinis SA. CP1-dependent partitioning of pretransfer and posttransfer editing in leucyl-tRNA synthetase. Proc Natl Acad Sci U S A. 2008;105:19223–19228. doi: 10.1073/pnas.0809336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yadavalli SS, Musier-Forsyth K, Ibba M. The return of pretransfer editing in protein synthesis. 2008;105:19031–19032. doi: 10.1073/pnas.0810781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nordin BE, Schimmel P. Transiently misacylated tRNA is a primer for editing of misactivated adenylates by class I aminoacyl-tRNA synthetases. Biochemistry. 2003;42:12989–12997. doi: 10.1021/bi035052q. [DOI] [PubMed] [Google Scholar]
- 32.Tukalo M, Yaremchuk A, Fukunaga R, Yokoyama S, Cusack S. The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation. Nat Struct Mol Biol. 2005;12:923–930. doi: 10.1038/nsmb986. [DOI] [PubMed] [Google Scholar]
- 33.Fukunaga R, Yokoyama S. Crystal structure of leucyl-tRNA synthetase from the archaeon Pyrococcus horikoshii reveals a novel editing domain orientation. J Mol Biol. 2005;346:57–71. doi: 10.1016/j.jmb.2004.11.060. [DOI] [PubMed] [Google Scholar]
- 34.Silvian LF, Wang J, Steitz TA. Insights into editing from an Ile-tRNA synthetase structure with tRNAIle and mupirocin. Science. 1999;285:1074–1077. [PubMed] [Google Scholar]
- 35.Fukai S, Nureki O, Sekine S, Shimada A, Tao J, Vassylyev DG, Yokoyama S. Structural basis for double-sieve discrimination of L-valine from L- isoleucine and L-threonine by the complex of tRNAVal and valyl-tRNA synthetase. Cell. 2000;103:793–803. doi: 10.1016/s0092-8674(00)00182-3. [DOI] [PubMed] [Google Scholar]
- 36.Zhang CM, Perona JJ, Ryu K, Francklyn C, Hou YM. Distinct kinetic mechanisms of the two classes of aminoacyl-tRNA synthetases. J Mol Biol. 2006;361:300–311. doi: 10.1016/j.jmb.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 37.Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 38.Ling J, So BR, Yadavalli SS, Roy H, Shoji S, Fredrick K, Musier-Forsyth K, Ibba M. Resampling and editing of mischarged tRNA prior to translation elongation. Mol Cell. 2009;33:654–660. doi: 10.1016/j.molcel.2009.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beebe K, Mock M, Merriman E, Schimmel P. Distinct domains of tRNA synthetase recognize the same base pair. Nature. 2008;451:90–93. doi: 10.1038/nature06454. [DOI] [PubMed] [Google Scholar]
- 40.Sokabe M, Ose T, Nakamura A, Tokunaga K, Nureki O, Yao M, Tanaka I. The structure of alanyl-tRNA synthetase with editing domain. Proc Natl Acad Sci USA. 2009;106:11028–11033. doi: 10.1073/pnas.0904645106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calendar R, Berg P. D-Tyrosyl RNA: formation, hydrolysis and utilization for protein synthesis. J Mol Biol. 1967;26:39–54. doi: 10.1016/0022-2836(67)90259-8. [DOI] [PubMed] [Google Scholar]
- 42.Ahel I, Korencic D, Ibba M, Söll D. Trans-editing of mischarged tRNAs. Proc Natl Acad Sci USA. 2003;100:15422–15427. doi: 10.1073/pnas.2136934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.An S, Musier-Forsyth K. Trans-editing of Cys-tRNA Pro by Haemophilus influenzae YbaK protein. J Biol Chem. 2004;279:42359–42362. doi: 10.1074/jbc.C400304200. [DOI] [PubMed] [Google Scholar]
- 44.Beuning PJ, Musier-Forsyth K. Hydrolytic editing by a class II aminoacyl-tRNA synthetase. Proc Natl Acad Sci USA. 2000;97:8916–8920. doi: 10.1073/pnas.97.16.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.An S, Musier-Forsyth K. Cys-tRNAPro editing by Haemophilus influenzae YbaK via a novel synthetase/YbaK/tRNA ternary complex. J Biol Chem. 2005;280:34465–34472. doi: 10.1074/jbc.M507550200. [DOI] [PubMed] [Google Scholar]
- 46.Ruan B, Söll D. The bacterial YbaK protein is a Cys-tRNAPro and Cys-tRNACys deacylase. J Biol Chem. 2005;280:25887–25891. doi: 10.1074/jbc.M502174200. [DOI] [PubMed] [Google Scholar]
- 47.Marshall L, Kenneth NS, White RJ. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- 48.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 49.Takano A, Endo T, Yoshihisa T. tRNA actively shuttles between the nucleus and cytosol in yeast. Science. 2005;309:140–142. doi: 10.1126/science.1113346. [DOI] [PubMed] [Google Scholar]
- 50.Shaheen HH, Hopper AK. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2005;102:11290–11295. doi: 10.1073/pnas.0503836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shaheen HH, Horetsky RL, Kimball SR, Murthi A, Jefferson LS, Hopper AK. Retrograde nuclear accumulation of cytoplasmic tRNA in rat hepatoma cells in response to amino acid deprivation. Proc Natl Acad Sci USA. 2007;104:8845–8850. doi: 10.1073/pnas.0700765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He C, Klionsky DJ. Regulation Mechanisms and Signaling Pathways of Autophagy Annu Rev Genet. 2009 doi: 10.1146/annurev-genet-102808-114910. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levine B. Cell biology: autophagy and cancer. Nature. 2007;446:745–747. doi: 10.1038/446745a. [DOI] [PubMed] [Google Scholar]
- 54.Kau TR, Way JC, Silver PA. Nuclear transport and cancer: from mechanism to intervention. Nat Rev Cancer. 2004;4:106–117. doi: 10.1038/nrc1274. [DOI] [PubMed] [Google Scholar]
- 55.Hellmuth K, Lau DM, Bischoff FR, Kunzler M, Hurt E, Simos G. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol. 1998;18:6374–6386. doi: 10.1128/mcb.18.11.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arts GJ, Kuersten S, Romby P, Ehresmann B, Mattaj IW. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J. 1998;17:7430–7441. doi: 10.1093/emboj/17.24.7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghavidel A, Kislinger T, Pogoutse O, Sopko R, Jurisica I, Emili A. Impaired tRNA nuclear export links DNA damage and cell-cycle checkpoint. Cell. 2007;131:915–926. doi: 10.1016/j.cell.2007.09.042. [DOI] [PubMed] [Google Scholar]
- 58.Peschel A, et al. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J Exp Med. 2001;193:1067–1076. doi: 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roy H, Ibba M. Broad range amino acid specificity of RNA-dependent lipid remodeling by multiple peptide resistance factors. J Biol Chem. 2009 doi: 10.1074/jbc.M109.046367. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roy H. Tuning the properties of the bacterial membrane with aminoacylated phosphatidylglycerol. IUBMB Life. 2009;61:940–953. doi: 10.1002/iub.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peschel A, Sahl HG. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nature Microbiol Rev. 2006;4:529–536. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- 62.Weidenmaier C, Peschel A, Kempf VA, Lucindo N, Yeaman MR, Bayer AS. DltABCD- and MprF-mediated cell envelope modifications of Staphylococcus aureus confer resistance to platelet microbicidal proteins and contribute to virulence in a rabbit endocarditis model. Infect Immun. 2005;73:8033–8038. doi: 10.1128/IAI.73.12.8033-8038.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thedieck K, Hain T, Mohamed W, Tindall BJ, Nimtz M, Chakraborty T, Wehland J, Jansch L. The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol Microbiol. 2006;62:1325–1339. doi: 10.1111/j.1365-2958.2006.05452.x. [DOI] [PubMed] [Google Scholar]
- 64.Vinuesa P, Neumann-Silkow F, Pacios-Bras C, Spaink HP, Martinez-Romero E, Werner D. Genetic analysis of a pH-regulated operon from Rhizobium tropici CIAT899 involved in acid tolerance and nodulation competitiveness. Mol Plant Microbe Interact. 2003;16:159–168. doi: 10.1094/MPMI.2003.16.2.159. [DOI] [PubMed] [Google Scholar]
- 65.Roy H, Ibba M. RNA-dependent lipid remodeling by bacterial multiple peptide resistance factors. Proc Natl Acad Sci USA. 2008;105:4667–4672. doi: 10.1073/pnas.0800006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klein S, et al. Adaptation of Pseudomonas aeruginosa to various conditions includes tRNA-dependent formation of alanyl-phosphatidylglycerol. Mol Microbiol. 2009;71:551–565. doi: 10.1111/j.1365-2958.2008.06562.x. [DOI] [PubMed] [Google Scholar]
- 67.Roy H, Dare K, Ibba M. Adaptation of the bacterial membrane to changing environments using aminoacylated phospholipids. Mol Microbiol. 2009;71:547–550. doi: 10.1111/j.1365-2958.2008.06563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roy H, Ibba M. RNA-dependent lipid remodeling by bacterial multiple peptide resistance factors. Proc Natl Acad Sci USA. 2008;105:4667–4672. doi: 10.1073/pnas.0800006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mogk A, Schmidt R, Bukau B. The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol. 2007;17:165–172. doi: 10.1016/j.tcb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 70.Rai R, et al. Arginyltransferase regulates alpha cardiac actin function, myofibril formation and contractility during heart development. Development. 2008;135:3881–3889. doi: 10.1242/dev.022723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shrader TE, Tobias JW, Varshavsky A. The N-end rule in Escherichia coli: cloning and analysis of the leucyl, phenylalanyl-tRNA-protein transferase gene aat. J Bacteriol. 1993;175:4364–4374. doi: 10.1128/jb.175.14.4364-4374.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Balzi E, Choder M, Chen WN, Varshavsky A, Goffeau A. Cloning and functional analysis of the arginyl-tRNA-protein transferase gene ATE1 of Saccharomyces cerevisiae. J Biol Chem. 1990;265:7464–7471. [PubMed] [Google Scholar]
- 73.Rai R, Kashina A. Identification of mammalian arginyltransferases that modify a specific subset of protein substrates. Proc Natl Acad Sci USA. 2005;102:10123–10128. doi: 10.1073/pnas.0504500102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Graciet E, Hu RG, Piatkov K, Rhee JH, Schwarz EM, Varshavsky A. Aminoacyl-transferases and the N-end rule pathway of prokaryotic/eukaryotic specificity in a human pathogen. Proc Natl Acad Sci USA. 2006;103:3078–3083. doi: 10.1073/pnas.0511224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watanabe K, Toh Y, Suto K, Shimizu Y, Oka N, Wada T, Tomita K. Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase. Nature. 2007;449:867–871. doi: 10.1038/nature06167. [DOI] [PubMed] [Google Scholar]
- 76.Suto K, Shimizu Y, Watanabe K, Ueda T, Fukai S, Nureki O, Tomita K. Crystal structures of leucyl/phenylalanyl-tRNA-protein transferase and its complex with an aminoacyl-tRNA analog. EMBO J. 2006;25:5942–5950. doi: 10.1038/sj.emboj.7601433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dong X, Kato-Murayama M, Muramatsu T, Mori H, Shirouzu M, Bessho Y, Yokoyama S. The crystal structure of leucyl/phenylalanyl-tRNA-protein transferase from Escherichia coli. Protein Sci. 2007;16:528–534. doi: 10.1110/ps.062616107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Graciet E, Walter F, Maoileidigh DO, Pollmann S, Meyerowitz EM, Varshavsky A, Wellmer F. The N-end rule pathway controls multiple functions during Arabidopsis shoot and leaf development. Proc Natl Acad Sci USA. 2009;106:13618–13623. doi: 10.1073/pnas.0906404106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yoshida S, Ito M, Callis J, Nishida I, Watanabe A. A delayed leaf senescence mutant is defective in arginyl-tRNA:protein arginyltransferase, a component of the N-end rule pathway in Arabidopsis. Plant J. 2002;32:129–137. doi: 10.1046/j.1365-313x.2002.01407.x. [DOI] [PubMed] [Google Scholar]
- 80.Kwon YT, Kashina AS, Davydov IV, Hu RG, An JY, Seo JW, Du F, Varshavsky A. An essential role of N-terminal arginylation in cardiovascular development. Science. 2002;297:96–99. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- 81.Karakozova M, Kozak M, Wong CC, Bailey AO, Yates JR, 3rd, Mogilner A, Zebroski H, Kashina A. Arginylation of beta-actin regulates actin cytoskeleton and cell motility. Science. 2006;313:192–196. doi: 10.1126/science.1129344. [DOI] [PubMed] [Google Scholar]
- 82.Kwon YT, Kashina AS, Varshavsky A. Alternative splicing results in differential expression, activity, and localization of the two forms of arginyl-tRNA-protein transferase, a component of the N-end rule pathway. Mol Cell Biol. 1999;19:182–193. doi: 10.1128/mcb.19.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ninnis RL, Spall SK, Talbo GH, Truscott KN, Dougan DA. Modification of PATase by L/F-transferase generates a ClpS-dependent N-end rule substrate in Escherichia coli. EMBO J. 2009;28:1732–1744. doi: 10.1038/emboj.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Biarrotte-Sorin S, Maillard AP, Delettre J, Sougakoff W, Arthur M, Mayer C. Crystal structures of Weissella viridescens FemX and its complex with UDP-MurNAc-pentapeptide: insights into FemABX family substrates recognition. Structure. 2004;12:257–267. doi: 10.1016/j.str.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 85.Bumsted RM, Dahl JL, Söll D, Strominger JL. Biosynthesis of the peptidoglycan of bacterial cell walls. X. Further study of the glycyl transfer ribonucleic acids active in peptidoglycan synthesis in Staphylococcus aureus. J Biol Chem. 1968;243:779–782. [PubMed] [Google Scholar]
- 86.Benson TE, et al. X-ray crystal structure of Staphylococcus aureus FemA. Structure. 2002;10:1107–1115. doi: 10.1016/s0969-2126(02)00807-9. [DOI] [PubMed] [Google Scholar]
- 87.Kawakami M, Tanada S, Takemura S. Properties of alanyl-oligonucleotide, puromycin, and Staphylococcus epidermidis glycyl-tRNA in interaction with elongation factor Tu:GTP complex. FEBS Lett. 1975;51:321–324. doi: 10.1016/0014-5793(75)80917-3. [DOI] [PubMed] [Google Scholar]
- 88.Giannouli S, Kyritsis A, Malissovas N, Becker HD, Stathopoulos C. On the role of an unusual tRNAGly isoacceptor in Staphylococcus aureus. Biochimie. 2009;91:344–351. doi: 10.1016/j.biochi.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 89.Fonvielle M, Chemama M, Villet R, Lecerf M, Bouhss A, Valery JM, Etheve-Quelquejeu M, Arthur M. Aminoacyl-tRNA recognition by the FemXWv transferase for bacterial cell wall synthesis. Nucleic Acids Res. 2009;37:1589–1601. doi: 10.1093/nar/gkn1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maillard AP, Biarrotte-Sorin S, Villet R, Mesnage S, Bouhss A, Sougakoff W, Mayer C, Arthur M. Structure-based site-directed mutagenesis of the UDP-MurNAc-pentapeptide-binding cavity of the FemX alanyl transferase from Weissella viridescens. J Bacteriol. 2005;187:3833–3838. doi: 10.1128/JB.187.11.3833-3838.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hegde SS, Shrader TE. FemABX family members are novel nonribosomal peptidyltransferases and important pathogen-specific drug targets. J Biol Chem. 2001;276:6998–7003. doi: 10.1074/jbc.M008591200. [DOI] [PubMed] [Google Scholar]
- 92.Nolan EM, Walsh CT. How nature morphs peptide scaffolds into antibiotics. ChemBioChem. 2009;10:34–53. doi: 10.1002/cbic.200800438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lawlor EJ, Baylis HA, Chater KF. Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2) Genes Dev. 1987;1:1305–1310. doi: 10.1101/gad.1.10.1305. [DOI] [PubMed] [Google Scholar]
- 94.Leskiw BK, Lawlor EJ, Fernandez-Abalos JM, Chater KF. TTA codons in some genes prevent their expression in a class of developmental, antibiotic-negative, Streptomyces mutants. Proc Nat Acad Sci USA. 1991;88:2461–2465. doi: 10.1073/pnas.88.6.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Leskiw BK, Mah R, Lawlor EJ, Chater KF. Accumulation of bldA-specified tRNA is temporally regulated in Streptomyces coelicolor A3(2) J Bacteriol. 1993;175:1995–2005. doi: 10.1128/jb.175.7.1995-2005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fernández-Moreno MA, Caballero JL, Hopwood DA, Malpartida F. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell. 1991;66:769–780. doi: 10.1016/0092-8674(91)90120-n. [DOI] [PubMed] [Google Scholar]
- 97.Chandra G, Chater K. Evolutionary flux of potentially bldA-dependent Streptomyces genes containing the rare leucine codon TTA. Antonie van Leeuwenhoek. 2008;94:111–126. doi: 10.1007/s10482-008-9231-5. [DOI] [PubMed] [Google Scholar]
- 98.Garcia-Contreras R, Zhang XS, Kim Y, Wood TK. Protein Translation and Cell Death: The Role of Rare tRNAs in Biofilm Formation and in Activating Dormant Phage Killer Genes. PLoS ONE. 2008;3:e2394. doi: 10.1371/journal.pone.0002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Garg RP, Qian XL, Alemany LB, Moran S, Parry RJ. Investigations of valanimycin biosynthesis: Elucidation of the role of seryl-tRNA. Proc Natl Acad Sci USA. 2008;105:6543–6547. doi: 10.1073/pnas.0708957105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Garg RP, Gonzalez JM, Parry RJ. Biochemical Characterization of VlmL, a Seryl-tRNA Synthetase Encoded by the Valanimycin Biosynthetic Gene Cluster. J Biol Chem. 2006;281:26785–26791. doi: 10.1074/jbc.M603675200. [DOI] [PubMed] [Google Scholar]
- 101.Gondry M, et al. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes. Nat Chem Biol. 2009;5:414–420. doi: 10.1038/nchembio.175. [DOI] [PubMed] [Google Scholar]
- 102.Kowtoniuk WE, Shen Y, Heemstra JM, Agarwal I, Liu DR. A chemical screen for biological small molecule—RNA conjugates reveals CoA-linked RNA. Proc Nat Acad Sci USA. 2009;106:7768–7773. doi: 10.1073/pnas.0900528106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gutierrez-Preciado A, Henkin TM, Grundy FJ, Yanofsky C, Merino E. Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol Mol Biol Rev. 2009;73:36–61. doi: 10.1128/MMBR.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH. Dissection of the mechanism for the stringent factor RelA. Mol Cell. 2002;10:779–788. doi: 10.1016/s1097-2765(02)00656-1. [DOI] [PubMed] [Google Scholar]
- 105.Zaborske JM, Narasimhan J, Jiang L, Wek SA, Dittmar KA, Freimoser F, Pan T, Wek RC. Genome-wide analysis of tRNA charging and activation of the eIF2 kinase Gcn2p. J Biol Chem. 2009;284:25254–25267. doi: 10.1074/jbc.M109.000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jochl C, Rederstorff M, Hertel J, Stadler PF, Hofacker IL, Schrettl M, Haas H, Huttenhofer A. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008;36:2677–2689. doi: 10.1093/nar/gkn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hao S, et al. Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex. Science. 2005;307:1776–1778. doi: 10.1126/science.1104882. [DOI] [PubMed] [Google Scholar]
- 108.Haiser HJ, Karginov FV, Hannon GJ, Elliot MA. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res. 2008;36:732–741. doi: 10.1093/nar/gkm1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li Y, Luo J, Zhou H, Liao JY, Ma LM, Chen YQ, Qu LH. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008;36:6048–6055. doi: 10.1093/nar/gkn596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hargittai MR, Gorelick RJ, Rouzina I, Musier-Forsyth K. Mechanistic insights into the kinetics of HIV-1 nucleocapsid protein-facilitated tRNA annealing to the primer binding site. J Mol Biol. 2004;337:951–968. doi: 10.1016/j.jmb.2004.01.054. [DOI] [PubMed] [Google Scholar]
- 112.Wang Z, Yuan Z, Xiang L, Shao J, Wegrzyn G. tRNA-dependent cleavage of the ColE1 plasmid-encoded RNA I. Microbiology. 2006;152:3467–3476. doi: 10.1099/mic.0.29134-0. [DOI] [PubMed] [Google Scholar]
- 113.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Taquist H, Cui Y, Ardell DH. TFAM 1.0: an online tRNA function classifier. Nucleic Acids Res. 2007;35:W350–353. doi: 10.1093/nar/gkm393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ataide SF, Jester BC, Devine KM, Ibba M. Stationary-phase expression and aminoacylation of a transfer-RNA-like small RNA. EMBO Rep. 2005;6:742–747. doi: 10.1038/sj.embor.7400474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ataide SF, Rogers TE, Ibba M. The CCA anticodon specifies separate functions inside and outside translation in Bacillus cereus. RNA Biol. 2009;6:479–487. doi: 10.4161/rna.6.4.9332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zeng Y, Roy H, Patil PB, Ibba M, Chen S. Characterization of two seryl-tRNA synthetases in albomycin-producing Streptomyces sp ATCC 700974 Antimicrob. Agents Chemother. 2009 doi: 10.1128/AAC.00782-09. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Geslain R, Ribas de Pouplana L. Regulation of RNA function by aminoacylation and editing? Trends Genet. 2004;20:604–610. doi: 10.1016/j.tig.2004.09.012. [DOI] [PubMed] [Google Scholar]


