Abstract
The initiator tRNA must serve functions distinct from those of other tRNAs, evading binding to elongation factors and instead binding directly to the ribosomal P site with the aid of initiation factors. It also plays a key role in decoding the start codon, setting the frame for translation of the mRNA. Sequence elements and modifications of the initiator tRNA distinguish it from the elongator methionyl tRNA and help it to perform its varied tasks. These identity elements appear to finely tune the structure of the initiator tRNA, and growing evidence suggests that the body of the tRNA is involved in transmitting the signal that the start codon has been found to the rest of the pre-initiation complex.
Keywords: Initiator tRNA, translation initiation, ribosome
Introduction
The initiator tRNA plays a critical role in the cell. It reads the start codon, allowing the initiating ribosome to begin translation in the correct location. A properly located translational start is crucial, since by beginning translation in the wrong location the cell not only fails to produce the desired protein, but also creates an aberrant, possibly toxic one. In addition, the availability of the initiator tRNA may be limiting for translation; a roughly twofold decrease in initiator tRNA expression in yeast increased the doubling time approximately threefold [1], and a “modest” overexpression of the initiator tRNA resulted in greatly elevated rates of translation and allowed mouse embryonic fibroblasts to induce tumors in mice [2]. Thus, a detailed understanding of the mechanism of initiator tRNA function in translation may have important medical relevance.
In the course of its duties, the initiator tRNA must accomplish a number of tasks involving interactions with several binding partners: aminoacylation with methionine by methionyl-tRNA synthetase; in bacteria and eukaryotic organelles, formylation of the methionine moiety by methionyl-tRNA transformylase; interaction with one or more initiation factors; and binding to the ribosome. The initiator tRNA must perform functions different from those of any other tRNA. It is the only tRNA that binds directly to the P site of the ribosome during the translational cycle; it is also one of the only tRNAs that must avoid binding to elongation factor Tu (EF-Tu; eEF1A in eukaryotes). In addition, the initiator tRNA must be distinguished from the other methionine-bearing tRNA present in the cytoplasm, the elongator methionyl tRNA that contributes methionine residues during peptide chain elongation. (In animal mitochondria, a single methionine-bearing tRNA species appears to serve as both initiator and elongator [3].)
The initiation of protein synthesis is clearly a crucial process for the cell (reviewed in [4-6]). There are significant differences between how this process is accomplished in eukaryotes and in bacteria. Translation initiation in eukaryotes requires at least twelve initiation factors. Aminoacylated initiator tRNA is delivered to the P site of the ribosome as part of a ternary complex (TC) with GTP-bound eukaryotic initiation factor 2 (eIF2). TC binds to the small (40S) ribosomal subunit with the help of eIFs 1, 1A and 3, forming a 43S pre-initiation complex (PIC). The PIC can then bind the 5' end of an mRNA in a process facilitated by eIF3, eIF4F, and the poly(A) binding protein. The PIC is thought to scan along the message in search of the start codon. eIF2, with the aid of the GTPase activating protein eIF5, hydrolyzes GTP, and start codon recognition triggers release of inorganic phosphate (Pi) from the complex. Pi release is followed by dissociation of eIF2•GDP, leaving the initiator tRNA in the P site with its anticodon base-paired to the mRNA start codon. In addition to irreversible GTP hydrolysis, start codon recognition induces a conformational rearrangement in the complex from an open state, thought to be competent for scanning, to a closed one, thought to be locked down on the mRNA. At this stage, joining of the large (60S) subunit to the complex, with the help of eIF5B, forms a complete 80S complex that can continue on to the elongation phase of translation.
Two major differences, likely in part related to each other, exist between bacterial and eukaryotic initiation. First, bacteria have Shine-Dalgarno sequences upstream of most start codons. These sequences base pair with the 16S rRNA and localize the 30S subunit to the start codon. Eukaryotes lack the anti-Shine-Dalgarno sequences in their 18S rRNA and thus do not use Shine-Dalgarno sequences to locate initiation codons. Secondly, in contrast to the ≥12 initiation factors found in eukaryotes, bacteria possess only three. IF1 is the ortholog of eIF1A, promoting initiator tRNA binding to the P site and blocking the A site until the ribosome is ready to begin elongation. IF2 is not the ortholog of eIF2, but is instead the ortholog of eIF5B. Both IF2 and eIF5B facilitate the subunit joining step of initiation. However, IF2 also acts to promote initiator tRNA binding to the 30S subunit, whereas in eukaryotes, this role is filled by eIF2 rather than by eIF5B. Although IF2 binds initiator tRNA in vitro (with an affinity of approximately 1 μM) [7,8], it is thought to interact with the tRNA on the ribosome, rather than forming a complex with tRNA that then binds to the 30S subunit. This is in contrast to eukaryotic initiation, where initiator tRNA must bind to eIF2 before it can bind to the ribosome. The third bacterial factor, IF3, helps the ribosomal pre-initiation complex select the correct tRNA for initiation. It has a similar fold to that of eIF1, binds a similar site on the small ribosomal subunit and appears to be at least functionally orthologous to the eukaryotic factor [9,10].
In bacteria, after aminoacylation of the initiator tRNA, the methionine moiety is formylated by methionyl-tRNA transformylase [11,12]. This modification is important for the function of the tRNA in initiation, as it is used by bacterial initiation factor 2 (IF2) to check that the appropriate tRNA is in place for initiation [13]. IF2 recognizes the formylated amino acid on the initiator tRNA, and will bind other tRNAs bearing formylated or acetylated amino acids [13]. Formylation is not a determinant used in eukaryotes to identify the correct tRNA for initiation; while many eukaryotic initiator tRNAs can be formylated by this enzyme in vitro [13], the enzyme is not present in the cytoplasm of eukaryotes and the methionine on the initiator tRNA is not formylated.
In elongation, elongator aminoacyl-tRNAs are delivered to the ribosomal A site, not the P site. In eukaryotes, elongator tRNAs are delivered to the A site in complex with GTP-bound eukaryotic elongation factor 1A (eEF1A). In bacterial elongation, the homologous EF-Tu serves this role [14].
The sequence of the initiator tRNA gene is the most highly conserved among those of all tRNA species, across all three domains of life [15]. In fact, all known vertebrate initiator tRNAs have identical sequences [16]. A number of differences, however, are observed at the level of sequence conservation between eukaryotic and bacterial initiator tRNAs. As will be seen below, these differences in sequence elements occur in locations throughout the initiator tRNA body. The importance of a number of these elements has been investigated, and some differences have been observed between how bacterial and eukaryotic initiator tRNAs function. Although this review focuses primarily on eukaryotic initiator tRNAs, we will also compare the bacterial and eukaryotic tRNAs in order to help elucidate general principles behind their functions and to note instances of their divergence.
Exclusion from elongation
What special attributes allow the initiator tRNA to fulfill its role? The initiator tRNA is likely tuned for its particular function in the ribosomal P site, as opposed to elongator tRNAs, which have to fulfill different roles, in both the A and P sites. The cell acquires an additional degree of control by having a separate tRNA for initiation, and thus regulates the levels of initiator and elongator methionyl tRNAs separately [17]. It is important that each type of methionyl tRNA be restricted to its separate function, as competition for tRNA by the initiation and elongation machinery could lead to serious problems for the cell [18-20]. How is this accomplished? Conserved elements in the sequence, and in some cases modifications, of the initiator methionyl tRNA distinguish it from the elongator methionyl tRNA and allow each tRNA to perform its particular function (Figure 1).
Figure 1.
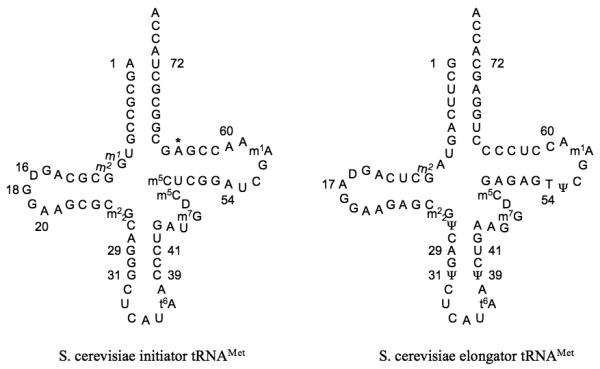
Cloverleaf diagram of S. cerevisiae initiator (left) and elongator (right) methionyl tRNAs. Identity elements in the sequence are indicated by numbers. The position 64 Oribosylphosphate modification is indicated by an asterisk. There is some confusion in the literature and databases about the identity of base 64 in the elongator tRNA. In most cases, including all elongator tRNA genes in the S. cerevisiae genome, this base is a C. However, in a few cases it is reported as a U. It is possible that this is the result of deamination of C in a fraction of the tRNAs; in this case, sequencing of the corresponding cDNA would give a U at this position.
The initiator tRNA is aminoacylated by methionyl tRNA synthetase, the same enzyme that charges the elongator methionyl tRNA. Initiator and elongator fates diverge after this step. The initiator binds eIF2•GTP and must not bind eEF1A. A key question, then, is how it is excluded from binding eEF1A.
The initiator tRNA binds eEF1A much less tightly than does the elongator methionyl tRNA (>100 nM versus 0.8 nM using wheat germ components) [14]. Several features of the initiator tRNA play a role in preventing it from binding to the elongation factor. For example, the base pair at position 1:72 in the acceptor stem is an important contributor to initiator/elongator discrimination. Bacterial initiator tRNAs contain a C:A mismatch at position 1:72, which has been shown to be a determinant for formylation of the methionine moiety by methionyl-tRNA transformylase [11,12], and the major determinant for exclusion of the bacterial initiator tRNA from elongation; changing this position to a Watson-Crick base pair causes the tRNA to bind to EF-Tu [21]. An A1:U72 base pair is invariant in eukaryotic and archaeal initiator tRNAs (the sole known exception being ψ1:A72 in S. pombe), while the elongator tRNAs of all three domains generally bear a G:C base pair in the 1:72 position. Drabkin and colleagues [22] showed that G1:C72 in the human initiator tRNA allowed the initiator tRNA to act in elongation in rabbit reticulocyte lysate. In yeast, an initiator tRNA containing G1:C72 could act as an elongator tRNA in vivo, and A1:U72 in the elongator tRNA decreased its elongation ability [23].
The eukaryotic initiator tRNA lacks the T loop's eponymous T (with one known exception, the parasite Encephalitozoon cuniculi), bearing the sequence AU at positions 54-55 instead of the Tψ found in other tRNAs, including the elongator methionyl tRNA. This is not the case in bacteria, where the Tψ sequence is present in the T loop of both elongator and initiator tRNAs [15]. As previously noted, this could therefore represent an important difference in initiation between bacteria and eukaryotes [24].
The T at position 54 in the elongator tRNA is an elongator determinant important for eEF1A recognition. In yeast, the function of an elongator tRNA containing A54 was dramatically compromised, and overexpression of eEF1A helped to restore the elongator function of this tRNA [23]. Substitution of A54 with U/T54 was shown to strongly affect the ability of the initiator tRNA to function in vivo: in yeast, U54 alone in the initiator tRNA was lethal, and the changes U54 C60 (which preserve a lack of base pairing between the bases at positions 54 and 60) resulted in lack of growth when provided on a low-copy vector to an initiator null strain, and poor growth even when present in high copy [24]. These data are consistent with the idea that A54 in the initiator tRNA may be an anti-elongator element that prevents the initiator tRNA from binding to the elongation factor [23-25]. In yeast, A54 did not appear to be a requirement for initiation, since G54 and C54 supported growth of an initiator null strain [24]. Evidence exists, however, that A54 in conjunction with other conserved bases may play a role in other aspects of initiator tRNA function, as discussed below.
Additional elements of the structure of the T stem and loop are involved in exclusion of the initiator tRNA from elongation. Vertebrate initiator tRNAs contain the base pairs A50:U64 and U51:A63, which are generally not contained in elongator tRNAs. These base pairs serve as an anti-elongator determinant that likely prevents the initiator tRNA from binding eEF1A [22]. The changes U50:A64 and G51:C63 allowed human initiator tRNA to function in elongation in rabbit reticulocyte lysate. These changes conferred greater elongation activity to the initiator tRNA than the G1:C72 change, and when coupled with G1:C72 resulted in a tRNA almost as active in elongation as the elongator methionyl tRNA. In bacteria, both the 50:64 and 51:63 base pairs are important for elongator tRNA binding to EF-Tu [26] and are anti-elongator determinants in the initiator tRNA [27]. It has been suggested that the bases at these positions affect the structure of the RNA backbone in this region in such a way that binding of the elongation factor is prevented [28]. This is consistent with the surprising observation [29] that E. coli initiator tRNA could act as elongator tRNA in rabbit reticulocyte lysate, and the authors' suggestion that the E. coli initiator tRNA therefore lacks structural features that exclude the eukaryotic initiator tRNA from elongation.
Thus, for eukaryotic initiator tRNAs, the major anti-elongator determinant resides in the T stem, and the A1:U72 base pair serves as a lesser anti-elongator determinant [22]. This relation is reversed in E. coli, where the main determinant preventing the initiator tRNA from acting in elongation is the C1 A72 mismatch, and an additional, secondary determinant is located in the T stem in the same location as in the eukaryotic initiator tRNA, the wobble base pair U50:G64 [27]. Thus, bacterial and eukaryotic initiator tRNAs use similar elements to prevent the initiator tRNA from acting as an elongator tRNA, but place the emphasis on these elements differently. It would be interesting to determine the evolutionary origins of this difference in emphasis.
Interestingly, in the same region of the T loop, at position 64, plant and fungal initiator tRNAs contain an O-ribosylphosphate modification that serves as an anti-elongator element. Yeast and wheat germ initiator tRNAs lacking the position 64 modification were able to support elongation in vitro [30], and the initiator tRNA was able to act as an elongator tRNA in a yeast strain lacking functional Rit1p, the phosphoribosyl transferase that makes the modification [31]. To ensure that it modifies the initiator and not elongator tRNA, Rit1p recognizes aspects of the T loop and stem, notably the bases A54 and A60 that are conserved in eukaryotic initiators [31]. The position 64 modification becomes important when the availability of TC components is compromised, as combining a lack of Rit1p with mutations in genes for the initiator tRNA or eIF2 resulted in synergistic growth defects, which were alleviated by high copy initiator tRNA but worsened by high copy eEF1A [20]. This modification achieves its role by preventing binding of the tRNA to eEF1A, presumably sterically [18,30,32,33]. Forster and colleagues showed that initiator tRNA lacking this modification could bind to eEF1A•GTP [18].
That determinants blocking binding to eEF1A are found in and at the base of the T stem and loop is consistent with work by Dreher and colleagues indicating that eEF1A interacts largely with the tRNA acceptor stem and T loop [14]. This is also the case for EF-Tu, as seen in the crystal structure of tRNAPhe bound to EF-Tu•GDPNP [34]. Hydrogen-bonding interactions were seen between nucleotides 63 and 64 and the protein backbone, at amino acid G391, of EF-Tu [35].
As mentioned above, bacteria and eukaryotes employ different yet related strategies to exclude the initiator tRNA from binding to the elongation factor. Elements in the T loop are important in many species, but in different ways. Curiously, within eukaryotes, additional methods have evolved to serve the same purpose; where the human initiator tRNA contains structural elements conferred by nucleotides in the T loop, the plant and fungal tRNAs contain a bulky modification instead. It would be interesting to know what distinct evolutionary pressures created these different solutions to the same problem.
Interaction with eIF2
In eukaryotes, the initiator tRNA must bind eIF2•GTP before it can be delivered to the ribosome. This marks a significant difference between eukaryotes and bacteria: in the current model of bacterial initiation, the tRNA can bind directly to the P site and subsequently interact with IF2 on the ribosome. Binding of the initiator tRNA to eIF2•GTP is fifteen-fold tighter (10 nM) than binding to eIF2•GDP (150 nM), as measured using yeast components [36]. This is important for release of the tRNA by eIF2•GDP, leaving the tRNA in the P site following GTP hydrolysis after start site selection.
In addition to its function in preventing binding of the tRNA to eEF1A, the A1:U72 base pair plays roles in the initiation pathway. A number of studies have shown that A1:U72 is a key element for initiator tRNA function in vivo [23,24] and in vitro [25,36,37]. Farrugio and colleagues showed that the A1:U72 base pair is important specifically for binding to eIF2 [38]. Using human tRNA in vitro, they observed that changing this base pair to G1:C72 reduced affinity for eIF2 16-fold (from 50 nM to 790 nM) but did not affect later steps in the initiation pathway. The affinity observed for the G1:C72 initiator tRNA was not as low as that of yeast elongator methionyl tRNA for eIF2 (which did not bind detectably), so the authors noted that eIF2 must also interact with other elements of the initiator tRNA.
Indeed, the methionine moiety is important for eIF2•GTP binding, providing a means to keep unacylated tRNA from entering the initiation pathway. Using an in vitro reconstituted system of yeast components, Kapp and colleagues saw that the methionine residue made a positive contact with eIF2•GTP but not with eIF2•GDP, while interactions between the tRNA body and the factor were the same regardless of whether the tRNA was aminoacylated [36]. This work also showed that A1:U72 is necessary for recognition of the methionine moiety, likely by helping to position it in its recognition pocket on eIF2. The authors suggested that the identity or geometry of the base pair at 1:72, rather than the presence of an easily disrupted base pair, is needed to position the methionine moiety properly in its binding pocket on eIF2.
Several studies have examined the importance of the identity of the initiating amino acid. Yeast initiator tRNA charged with isoleucine binds poorly to eIF2 [39]. Human initiator tRNA with the altered anticodon sequence CUA and charged with glutamine cannot initiate in vivo or in vitro, whereas the bacterial version can initiate from a UAG codon (using formylglutamine) [40,41]. Yet, an initiator tRNA variant with a GAC anticodon is aminoacylated with valine and can initiate translation from a GUC codon in mammalian cells [41]. Thus, whereas in bacteria a formylated amino acid is recognized, in eukaryotes an aspect of the methionine itself is recognized. Kapp [36] suggested that this could be the ability of the side chain to fit in the binding pocket on eIF2, or perhaps a contact with other groups of the amino acid.
Identity elements in the T loop and the anticodon stem were seen to jointly affect binding of the tRNA to eIF2. In vitro work using yeast components showed that a change in the T loop to U54,C60 did not affect binding by itself, but in combination with U31:U39 in the anticodon stem a threefold reduction in affinity of initiator tRNA for eIF2 was observed [37]. This is consistent with the existence of a functional interaction between the anticodon stem and T loop of the initiator tRNA, as discussed below. In addition, several combinations of initiator identity elements conferred a greater affinity for eIF2•GTP to the elongator methionyl tRNA. Substituting the initiator identity elements A1:U72 along with G31:C39 into the elongator tRNA increased the binding affinity for eIF2•GTP to nearly that of the initiator tRNA, and A1:U72 with A54 and A60 in the T loop brought the affinity to within two-fold of that of the initiator tRNA [37]. These observations suggest that the structure of the initiator tRNA is coordinated by elements in distant spatial locations in the molecule.
Binding to the ribosomal P site and start codon recognition
The initiator tRNA is thought to bind directly to the P site of the small ribosomal subunit and to play a critical role in recognizing the start codon in the mRNA. Although the initiation factors clearly help mediate these events, the structure of the tRNA itself also plays a key role.
Structural insights
The first crystal structure of an initiator tRNA was that of the yeast tRNA, solved to 6 Å by Schevitz and coworkers in 1975, and followed by a 4.5 Å structure in 1979 [42]. In these structures the anticodon arm was the most poorly defined part of the molecule, suggesting the existence of a “flexible hinge” between the D stem and the anticodon stem. In the 3 Å crystal structure of the yeast initiator tRNA determined by Basavappa and Sigler [33] (Figure 2), the anticodon stem and loop regions remained poorly defined, despite the improved resolution of the structure overall. The higher resolution, however, did enable visualization of the O-ribosylphosphate modification at position 64, identified only a year or two earlier [30,32].
Figure 2.
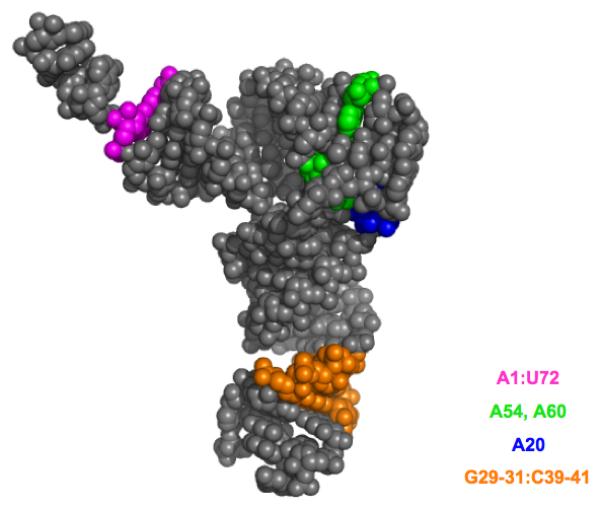
Space-filling model of the yeast initiator tRNA crystal structure [33], with identity elements shown in color.
In addition, a special substructure was observed in the elbow of the tRNA (Figure 3), formed by conserved residues located in the D and T loops that are found in combination exclusively in eukaryotic initiator tRNAs. The D and T loops contain a number of characteristic eukaryotic initiator tRNA features. The D loop is marked by the lack of nucleotide 17, which is present in all other tRNAs, including the bacterial initiator; the D loop also contains A20, also found in the yeast elongator, whereas E. coli initiator and most elongator tRNAs contain dihydrouridine at this position. The T loop contains A54, which is found exclusively in eukaryotic initiator tRNAs; m1A58; and A60, where a pyrimidine is typically found in bacterial and archaeal initiators and all elongator tRNAs. These nucleotides participate in hydrogen bonding interactions that create a stronger connection between the D and T loops than that seen in elongator tRNA structures, with three hydrogen bonds between A20 in the D loop and bases G57, A59, and A60 in the T loop. Hydrogen bonding was also observed within the T loop between m1A58 and A60, and between m1A58 and A54. It has been suggested that this substructure might keep the tRNA from entering elongation, perhaps by preventing structural changes in the D-loop-T-loop region necessary for interactions of the tRNA with the ribosome and factors in elongation. Alternatively, or in addition, this unique structure may play a role in the initiation process itself (see below).
Figure 3.
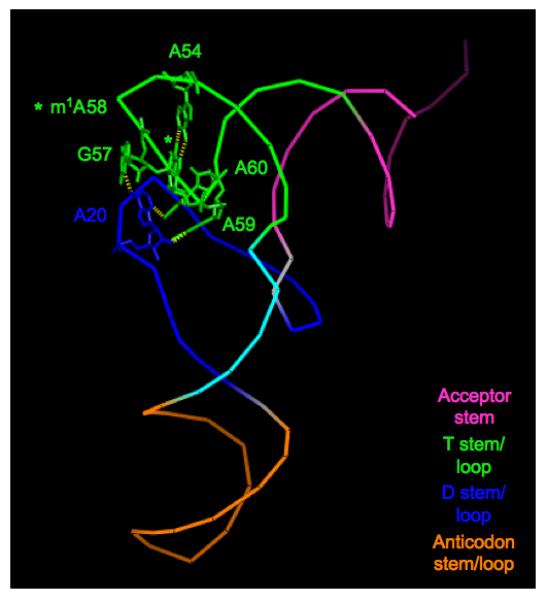
Ribbon diagram of the yeast initiator tRNA crystal structure [33], showing the special hydrogen bonding substructure observed between the D and T loops. Residues participating in hydrogen bonding are shown as sticks; hydrogen bonds are shown as yellow dashes. The substructure is created by hydrogen bonding within the T loop (involving residues A54, m1A58, and A60), as well as between the D and T loops (involving A20, G57, A59, and A60).
The crystal structure of a bacterial initiator tRNA was solved to 3.5 Å by Woo et al. in 1980 [43]. Comparing this structure to that of a yeast elongator tRNA, tRNAPhe, the authors observed differences between the two tRNAs in the anticodon loop, CCA end, and D loop. The low resolution, however, prevented a conclusive determination of tertiary interactions. A more compact D loop structure was observed in the initiator tRNA, even though in the E. coli initiator tRNA this loop contains an additional base relative to yeast tRNAPhe. In the anticodon stem, the base 5′ to the anticodon, U33, faced out of the anticodon loop, instead of inward as in the tRNAPhe structure. The anticodon loop appeared “skewed,” as opposed to the more symmetrical anticodon loop of tRNAPhe. The authors proposed that the conformation of the anticodon might be affected by hydrogen bonding involving U33; in the initiator tRNA the ribose moiety appeared to participate in a hydrogen bond with phosphate 36, while in the elongator the hydrogen bond was between the uracil base and phosphate 36. The authors noted that this could give insight into the presence of C33 in the initiator tRNAs of higher eukaryotes (the only tRNAs that contain C instead of U at this position), since C33 is unable to hydrogen bond with phosphate 36 but a hydrogen bond remains possible with the ribose.
The idea that the initiator tRNA may have a unique anticodon loop conformation is long-standing. In 1979 Wrede et al. [44] saw strikingly different S1 nuclease cleavage patterns between initiator and elongator tRNAs. They observed that the three initiator tRNAs examined – mammalian, E. coli, and yeast – cleaved at the same two positions in the anticodon loop (C34, A35), whereas for six elongator tRNAs from E. coli, yeast, and S. pombe, cleavage was typically seen at four positions in the anticodon loop (U33, C34, A35, and U36). This suggested that the anticodon loop of the initiator tRNA took on a conformation different from that of elongator tRNAs. Since the initiator and elongator tRNAs have the same anticodon loop sequence (save the difference at base 33 in higher eukaryotic initiators) and modifications, the authors suggested that this special conformation is imparted to the anticodon loop by three conserved G:C base pairs in the anticodon stem of the initiator tRNA, which are not all present in elongator tRNAs (these base pairs are discussed below). In support of this idea, RajBhandary and coworkers observed that changing the 29:41 base pair, both 29:41 and 30:40, and then all three to their respective elongator identities in E. coli initiator tRNA resulted in a progressive increase in the susceptibility of anticodon loop bases to cleavage by S1 nuclease, yielding a cleavage pattern similar to that of elongator tRNA for the tRNAs with two and three G:C base pairs changed [45]. In contrast, in work using an initiator tRNA bearing a CUA anticodon and capable of initiating translation in vivo in E. coli from a stop codon [40], changing the G:C base pairs affected initiation activity, but not the S1 nuclease pattern observed [46]. Thus, the G:C base pairs may influence, but not exclusively determine, the special anticodon conformation of the initiator tRNA.
As mentioned above, in the initiator tRNA crystal structures the anticodon stem/loop region was not well-defined enough to confirm that its conformation differed from that of the elongator tRNA. Several pieces of evidence did not appear to square with this idea. NMR solution structures showed synthetic yeast initiator and E. coli elongator anticodon stem-loops adopted similar conformations [47], and the authors proposed that the more extensive S1 cleavage of the elongator anticodon loop resulted from a greater conformational flexibility relative to the initiator anticodon loop. In this study, however, possible evidence of another conformational state was observed but could not be examined in detail. In addition, in structures of the bacterial ribosome with tRNA and mRNA bound [48-50], the conformation of the initiator tRNA anticodon loop was not notably different from that of the elongator tRNA.
A recent 3.1 Å crystal structure of the E. coli initiator tRNA [51] showed a special, more compact, anticodon conformation. In this structure, Cm32 and A38 formed a wobble base pair, extending the anticodon stem, in an interaction stabilized by stacking with G31:C39, although this base pair might rely on the low pH at which the structure was determined. In addition, A37 was seen to be involved in a base triple with G29 and C41. In this structure, however, the observed conformation may have been stabilized by a significant interaction of the anticodon loops in the crystal lattice. The authors suggest that a switch between two conformations of the initiator tRNA (this compact conformation, and that seen in ribosome structures with mRNA) could be important for first allowing initiator tRNA selection on the ribosome – the unique, compact conformation allowing only the initiator tRNA to bind in the constrained P site – and, subsequently, signaling through a conformational change that the start codon has been found.
Tuning of tRNA structure
While a number of identity elements contribute to initiator tRNA function, there is evidence that many of these elements work in combination. It appears that no one particular interaction causes the tRNA to bind specifically to the P site of the small ribosomal subunit. Instead, the tRNA structure is finely tuned by multiple elements in distinct regions of the tRNA that together impart the appropriate structure for specific P site binding.
As mentioned above, A54 and A60 influenced the ability of the yeast initiator tRNA to bind to eIF2 only in combination with identity elements in other regions of the molecule, indicating that elements distant in space have long-range effects on the structure of the initiator tRNA [37]. Consistent with this idea, this type of structural tuning may be important for bacterial initiator tRNA binding in the P site. Shoji and colleagues showed that the rate constant (koff) for dissociation of unacylated initiator tRNA from the 70S ribosome was unaffected by 16S rRNA mutations in the P site, including G1338U, A790G, and m2G966U, while the koff for all other unacylated tRNAs increased [52]. The authors point out that elements in the body of the tRNA must contribute to this effect, since the koff value for the elongator methionyl tRNA, with the same anticodon, increased with these rRNA changes. The authors suggest that the initiator tRNA is uniquely tuned to bind specifically in the P site of the ribosome, and that a combination of features is likely responsible for this tuning; little or no effect was seen on koff with a single change at position 1:72 or even with the anticodon stem of the initiator tRNA switched to that of glutamate elongator tRNA.
Additional identity element roles
The A1:U72 base pair may play a role in the context of the ribosomal complex. Kapp and coworkers expanded the known role of the A1:U72 base pair by showing that changing it affected steps after eIF2 binding in a reconstituted system of yeast components. Changing A1:U72 to the elongator identity element G1:C72 had an approximately ten-fold effect on binding of TC to the 40S subunit, and a small effect on peptide bond formation [37]. Given that this base pair affects the binding of the tRNA to eIF2, it is possible that its alteration could result in improper presentation of the tRNA and thus its improper positioning in the ribosomal complex.
Work by Astrom and colleagues suggested that bases in the acceptor stem besides the 1:72 base pair might be important for initiator tRNA identity. Elongator methionyl tRNA with the initiator acceptor stem was able to rescue a yeast strain lacking any wild-type initiator tRNA genes, while elongator tRNA with A1:U72 was not [23]. This acceptor stem transplant also rescued more efficiently than the elongator tRNA containing the initiator elements A1:U72, A54, G29:C41, and G31:C39 together, although this combination of elements did rescue partially. One candidate element within the acceptor stem could be the C3:G70 base pair, which is the only acceptor-stem base pair that is conserved in initiator tRNAs in all three domains of life [15]. In elongator tRNAs, although there may be a preference for a C:G base pair, the base pair found in this position varies. C3:G70 was shown to be important for formylation in bacteria [11,12]. It will be interesting to investigate whether this base pair might be involved in other ways in initiator function. In an in vivo study in yeast, changing C3:G70 failed to show an effect [24], but this was also the case for the anticodon stem G:C base pairs, which are known to be important for initiation.
Modifications
Modifications may also play a role in allowing the initiator tRNA to serve its function. tRNAs in general bear many nucleotide modifications. The S. cerevisiae initiator tRNA contains eleven modifications, but most are nonessential and of unknown function. Exceptions to this include m1A58, which in yeast is known to be essential for the stability of the initiator tRNA [53]. This modification is made by the enzyme 1-methyladenosine 58 methyltransferase, which is comprised of the Gcd10p/Gcd14p complex [53,54]. In yeast, initiator tRNA lacking m1A58 is degraded via the nuclear Trf4/Rrp6 surveillance pathway [55]. This modification is not found in most bacteria, with mycobacterial tRNAs being an exception [56].
The modified nucleotide N6-threonylcarbamoyladenosine (t6A) is found in position 37, immediately 3′ to the anticodon, in eukaryotic initiator tRNA. This modification may be involved in decoding of the start codon. t6A37 is also found in tRNALysUUU, where it helps to decode the UUU codon, stabilizing the codon-anticodon pairing through stacking interactions (as well as preventing an internal tRNA base pair) [57]. A37 is not modified in the E. coli initiator tRNA. It has been suggested that this is important for allowing the E. coli initiator to read the codons UUG and GUG, which are known to serve as start codons in E. coli, in addition to AUG. It was seen that E. coli initiator tRNA with this modification could no longer decode UUG or GUG [58]. If this modification helps to exclude those codons, the presence of t6A37 in the eukaryotic initiator tRNA would be consistent with the fact that the start codon of eukaryotes is overwhelmingly (although not exclusively) AUG.
G:C base pairs in the anticodon stem
The major identity element implicated in specific P site binding of the initiator tRNA is found in the anticodon stem. Initiator tRNAs in all three domains of life have three G:C base pairs at positions 29-31:39-41 in the anticodon stem, with few exceptions.
A number of studies have investigated the role of these three G:C base pairs in bacteria. The anticodon stem G:C base pairs were shown to be important for binding of initiator tRNA to the P site in vitro using E. coli components [45]. Changing the 29:41 base pair, both 29:41 and 30:40, and then all three to their respective elongator identities progressively diminished the activity of the tRNA in initiation, without affecting binding to IF2. The G:C base pairs were demonstrated to be important for initiator tRNA function in vivo in E. coli, using a tRNA with a CUA anticodon mutation that was able to initiate from a stop codon [40]. Using a CAT reporter, a change to the middle G:C base pair (30:40) had a more severe effect than a change to either of the other two base pairs (20-fold versus about 5-fold), while changing two base pairs reduced translation about 30-fold and no detectable initiation was observed (>200 fold reduction) with all three base pair changes [46]. The effect of the 30:40 base pair change was most likely due to a problem at or following the step of binding in the P site, as aminoacylation and formylation of the tRNA were unaffected by these changes.
In bacteria, IF3 is known to be important for selecting the initiator tRNA over other tRNAs, and the G:C base pairs have been proposed to be necessary for IF3-dependent initiator tRNA discrimination [59]. Genetic work suggested that the effect of the G:C base pairs in initiation had to do with discrimination by IF3, as compromising the function of IF3 led to increased activity of a three-G:C mutant initiator tRNA in initiation [60]. The binding site for IF3 on the 30S subunit, as determined by footprinting and cryoEM, is too far from these anticodon stem base pairs for the factor to contact them directly [61,62]. In the crystal structure of the 70S ribosome [48,49], two bases in the 16S rRNA, G1338 and A1339, were seen to contact the top two G:C base pairs, making type II and type I interactions (respectively) with the minor groove of G29:C41 and G30:C40. Dallas and Noller [62] proposed that IF3 binding induces a conformational change in the 30S subunit that allows G1338 and A1339 to check tRNA identity through their interaction with the anticodon stem G:C base pairs. In vivo and in vitro studies using site-directed mutagenesis of G1338 and/or A1339 were consistent with the interactions seen in the crystal structures, and with a role for these rRNA bases in tRNA binding to the P site [52,63,64]. Whether these bases play a role in discrimination between initiator and elongator tRNAs during initiation remains unclear, however.
The anticodon loop of the initiator tRNA also appears to be involved in discrimination by IF3, as changes in the loop of a synthetic initiator anticodon stem-loop (swapping the two bases 3′ of the anticodon with the two 5′ of the anticodon) were shown to eliminate selection by IF3 [59]. Noller and Lancaster [63] proposed that IF3 helps to recognize some feature of the anticodon loop – excluding many elongator tRNAs – and the interaction of the top two G:C pairs with ribosomal RNA bases G1338 and A1339 upon IF3 binding helps to discriminate in particular between the initiator and elongator methionine tRNAs; thus, by these two criteria, the initiator methionyl tRNA would be uniquely identified out of all tRNAs. However, IF3 may play an indirect role in initiator discrimination by promoting an increased rate of dissociation of all tRNAs from the 30S subunit [65], and, as mentioned above, the role of G1338 and A1339 in initiator/elongator discrimination remains hypothetical.
Modification of the ribosomal RNA may be involved in the response of the complex to IF3. An E. coli strain deficient in rRNA methylations allowed efficient initiation with an initiator tRNA with changes in the G:C base pairs (even all three) [66]. The loss of one particular methylation (1207) increased initiation with an initiator tRNA with all three G:C base pairs changed, while the loss of others (966, 967) decreased initiation with this tRNA. This suggests that rRNA methylations may play a role in selection of the initiator tRNA in the P site. As some of the rRNA methylations are in the IF3 binding region of the 30S subunit, the authors suggested that these methylations could contribute by helping to mediate the conformational changes proposed to be induced by IF3 that allow inspection of the G:C base pairs.
Thus, a growing body of evidence shows the importance of the initiator tRNA anticodon stem G:C base pairs in bacteria. Do these base pairs play the same role in eukaryotes?
Changing the G:C base pairs in human initiator tRNA to their elongator identities lowered translation efficiency in rabbit reticulocyte and wheat germ lysates, although a smaller effect was observed in the wheat germ system [25]. Astrom and coworkers saw that the G:C base pairs were involved in conferring initiator tRNA identity in yeast. Elongator methionyl tRNA with a combination of the three G:C base pairs and A1:U72 was able to complement an initiator null yeast strain, albeit with low efficiency, if the and subunits of eIF2 were overexpressed; adding A54 to this combination of elements increased the ability of this elongator tRNA to complement the initiator null strain [23].
However, an initiator tRNA with an altered 31:39 base pair (U31:U39), or with both the 29:41 and 31:39 base pairs changed together (A29:U41, U31:U39), allowed a yeast strain with an otherwise initiator null background to grow normally [24]. Are the G:C base pairs less important in eukaryotes? Perhaps they serve a more subtle, but still important, function.
Using purified yeast components, Kapp et al. [37] found that G29:C41 and G31:C39 were required for thermodynamic coupling between binding of TC and a model mRNA containing an AUG codon to the 40S subunit. Since this coupling likely reflects interactions in the complex relevant to start codon recognition, this result suggested that these G:C base pairs may influence this important step. It was proposed that the G:C base pairs influence a conformational change in the tRNA that occurs upon pairing between the anticodon and the start codon, and that this conformational change is partly responsible for triggering downstream events in initiation.
The 18S rRNA bases equivalent to G1338 and A1339 also appear to be important for initiator tRNA binding in eukaryotes. Mutations in these bases in S. cerevisiae affected TC binding to the ribosome and recognition of the AUG codon [67]. As in the bacterial case, it is not clear whether these bases play a role in specifically interacting with the initiator tRNA or if they have a more general effect on binding of tRNAs to the P site.
Conformational coupling between T loop and anticodon stem
As mentioned above, evidence suggests that the initiator tRNA could be involved in start codon recognition. In eukaryotes, start codon recognition has several steps. One of these steps is a conformational change of the 40S subunit from an open state competent to scan the mRNA to a closed state that is arrested on the mRNA after start codon selection [68-70]. The initial signal that causes the complex to undergo this conformational change to the closed state upon start codon recognition appears to be the matching base pairing of the initiator tRNA anticodon with the start codon [10,41,71,72]. How is this signal transferred from the base-pairing event to bring about these large-scale changes? The codon/anticodon duplex could be sensed directly by the ribosome and/or some combination of initiation factors. Alternatively, the tRNA itself might transmit the signal. It has been shown [73] that elongator tRNAs are capable of this function in the A site during the decoding phase of protein synthesis. For this to be possible, communication must occur between the anticodon region and distant parts of the tRNA making contacts with factors and/or the ribosome.
In fact, Kapp and colleagues [37] showed that the effect of changes in the anticodon stem G:C base pairs on thermodynamic coupling between TC and mRNA binding to the 40S subunit was influenced by identity elements in the T loop. Changing initiator tRNA identity elements A54 and A60, which participate in the hydrogen bonding network observed to form a tight connection between the D and T loops [33], to their elongator counterparts (U54, C60) suppressed the effect of changes to the top or bottom anticodon stem G:C base pairs (although not both together), restoring the coupling between binding of TC and mRNA to the 40S subunit. Changing A54 and A60 on their own had no effect. This suggests a conformational coupling between the anticodon stem and the D-loop-T-loop substructure of the initiator tRNA during start codon selection.
In elongator tRNAs in the A site, there is evidence that codon/anticodon pairing results in a conformational change of the tRNA that loosens the D-loop-T-loop association [73-79], and a “kinking” or bending of the anticodon stem about the D-stem-anticodon stem hinge region was observed in A site and P site tRNAs base-paired with mRNA in several bacterial and yeast ribosome structures [50,80,81]. Base pairing with the start codon could result in a similar conformational change in the initiator tRNA involving these regions. If changes to the anticodon stem G:C base pairs affect anticodon stem mobility, a loosening of the D-loop-T-loop substructure by the substitutions U54 and C60 could compensate by lowering the energy barrier for changing the D-loop-T-loop interaction, maintaining the ability of the tRNA to assume the post-codon-recognition conformation. This conformational change in the initiator tRNA may be involved in transmitting the signal that the start codon has been located from the anticodon-start codon duplex to the rest of the complex, causing the conformational change to the “closed” state of the pre-initiation complex. An alternative, but not mutually exclusive, possibility is that these elements coordinate a movement of the initiator tRNA within the complex from a pre-start codon recognition state (e.g., P/I [82,83]) to a post-recognition state fully engaged in the P site, and that this movement is a key element of pre-initiation complex closure.
Conclusions
Evidence points to the idea that the initiator tRNA is structurally tuned to perform its function, not just preventing it from serving as an elongator tRNA and allowing it to bind specifically in the P site, but also allowing it to respond within the complex, serving as an active player in start codon recognition. It appears likely that part of the function of the initiator tRNA is to transmit the signal that the start codon has been recognized. The special conformation and/or dynamics of the anticodon loop, the special substructure observed between the D and T loops, and the evidence for a conformational coupling within the tRNA that affects start codon selection, add up to suggest that the tRNA body is an active player in sending the signal that anticodon/start codon base pairing has taken place. This is in keeping with the active roles observed for tRNAs in decoding in the A site during elongation [73], and adds to the growing evidence that is changing the traditional view of tRNAs as merely static adaptors [84].
Given the similarities between the bacterial and eukaryotic initiator tRNAs, it seems likely that the proposed active role played by the eukaryotic tRNA in start codon recognition is also played by its bacterial counterpart. In eukaryotes, many complicated extra steps culminating in irreversible GTP hydrolysis and pre-initiation complex closure govern selection of the start codon. Clearly, the details of how this signal is sensed and responded to are different in bacteria and eukaryotes. To what extent does the mechanism of initiator tRNA involvement in start codon selection differ between eukaryotes and bacteria? What is the route of transmission from the initiator tRNA body to the rest of the complex that precipitates further events necessary for start site selection? In eukaryotes, the signal might go to eIF2 directly, as it is in contact with the tRNA; this would imply a significantly different method of transmission from the case in bacteria, which lack eIF2. Or, it could be that the signal is sensed directly by the ribosome, which is highly conserved in both systems, in which case the conformational change in the initiator tRNA might be sensed similarly by bacterial and eukaryotic complexes. If so, then despite all the additional components required for eukaryotic start codon recognition, this would mark a fundamental similarity in the process throughout the domains of life.
List of abbreviations
- TC
Ternary complex
- IF
initiation factor
- eIF
eukaryotic initiation factor
- PIC
pre-initiation complex
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Francis MA, RajBhandary UL. Expression and function of a human initiator tRNA gene in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:4486–94. doi: 10.1128/mcb.10.9.4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marshall L, Kenneth NS, White RJ. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- 3.Takeuchi N, Vial L, Panvert M, Schmitt E, Watanabe K, Mechulam Y, Blanquet S. Recognition of tRNAs by Methionyl-tRNA transformylase from mammalian mitochondria. J Biol Chem. 2001;276:20064–8. doi: 10.1074/jbc.M101007200. [DOI] [PubMed] [Google Scholar]
- 4.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 5.Pestova TV, Lorsch JR, Hellen CUT. The Mechanism of Translation Initiation in Eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 87–128. [Google Scholar]
- 6.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–45. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petersen HU, Roll T, Grunberg-Manago M, Clark BF. Specific interaction of initiation factor IF2 of E. coli with formylmethionyltRNA f Met. Biochem Biophys Res Commun. 1979;91:1068–74. doi: 10.1016/0006-291x(79)91989-2. [DOI] [PubMed] [Google Scholar]
- 8.Wu XQ, RajBhandary UL. Effect of the amino acid attached to Escherichia coli initiator tRNA on its affinity for the initiation factor IF2 and on the IF2 dependence of its binding to the ribosome. J Biol Chem. 1997;272:1891–5. doi: 10.1074/jbc.272.3.1891. [DOI] [PubMed] [Google Scholar]
- 9.Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes & Development. 2003;17:2786–2797. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, Pestova TV. The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. Embo J. 2006;25:196–210. doi: 10.1038/sj.emboj.7600904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee CP, Seong BL, RajBhandary UL. Structural and sequence elements important for recognition of Escherichia coli formylmethionine tRNA by methionyl-tRNA transformylase are clustered in the acceptor stem. J Biol Chem. 1991;266:18012–7. [PubMed] [Google Scholar]
- 12.Guillon JM, Meinnel T, Mechulam Y, Lazennec C, Blanquet S, Fayat G. Nucleotides of tRNA governing the specificity of Escherichia coli methionyl-tRNA(fMet) formyltransferase. J Mol Biol. 1992;224:359–67. doi: 10.1016/0022-2836(92)91000-f. [DOI] [PubMed] [Google Scholar]
- 13.RajBhandary UL, Chow CM. Initiator tRNAs and Initiation of Protein Synthesis. In: Soll D, RajBhandary UL, editors. tRNA: Structure, Biosynthesis and Function. American Society for Microbiology; Washington, D.C.: 1995. pp. 511–528. [Google Scholar]
- 14.Dreher TW, Uhlenbeck OC, Browning KS. Quantitative assessment of EF-1alpha.GTP binding to aminoacyl-tRNAs, aminoacylviral RNA, and tRNA shows close correspondence to the RNA binding properties of EF-Tu. J Biol Chem. 1999;274:666–72. doi: 10.1074/jbc.274.2.666. [DOI] [PubMed] [Google Scholar]
- 15.Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–53. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanduc D. Changes of tRNA population during compensatory cell proliferation: differential expression of methionine-tRNA species. Arch Biochem Biophys. 1997;342:1–5. doi: 10.1006/abbi.1996.9869. [DOI] [PubMed] [Google Scholar]
- 18.Forster C, Chakraburtty K, Sprinzl M. Discrimination between initiation and elongation of protein biosynthesis in yeast: identity assured by a nucleotide modification in the initiator tRNA. Nucleic Acids Res. 1993;21:5679–83. doi: 10.1093/nar/21.24.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guillon JM, Heiss S, Soutourina J, Mechulam Y, Laalami S, Grunberg-Manago M, Blanquet S. Interplay of methionine tRNAs with translation elongation factor Tu and translation initiation factor 2 in Escherichia coli. J Biol Chem. 1996;271:22321–5. doi: 10.1074/jbc.271.37.22321. [DOI] [PubMed] [Google Scholar]
- 20.Astrom SU, Nordlund ME, Erickson FL, Hannig EM, Bystrom AS. Genetic interactions between a null allele of the RIT1 gene encoding an initiator tRNA-specific modification enzyme and genes encoding translation factors in Saccharomyces cerevisiae. Mol Gen Genet. 1999;261:967–76. doi: 10.1007/s004380051045. [DOI] [PubMed] [Google Scholar]
- 21.Seong BL, RajBhandary UL. Mutants of Escherichia coli formylmethionine tRNA: a single base change enables initiator tRNA to act as an elongator in vitro. Proc Natl Acad Sci U S A. 1987;84:8859–63. doi: 10.1073/pnas.84.24.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drabkin HJ, Estrella M, RajBhandary UL. Initiator-elongator discrimination in vertebrate tRNAs for protein synthesis. Mol. Cell. Biol. 1998;18:1459–1466. doi: 10.1128/mcb.18.3.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Astrom SU, von Pawel-Rammingen U, Bystrom AS. The yeast initiator tRNAMet can act as an elongator tRNA(Met) in vivo. J Mol Biol. 1993;233:43–58. doi: 10.1006/jmbi.1993.1483. [DOI] [PubMed] [Google Scholar]
- 24.von Pawel-Rammingen U, Astrom S, Bystrom AS. Mutational analysis of conserved positions potentially important for initiator tRNA function in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1432–42. doi: 10.1128/mcb.12.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drabkin HJ, Helk B, RajBhandary UL. The role of nucleotides conserved in eukaryotic initiator methionine tRNAs in initiation of protein synthesis. J Biol Chem. 1993;268:25221–8. [PubMed] [Google Scholar]
- 26.Schrader JM, Chapman SJ, Uhlenbeck OC. Understanding the sequence specificity of tRNA binding to elongation factor Tu using tRNA mutagenesis. J Mol Biol. 2009;386:1255–64. doi: 10.1016/j.jmb.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stortchevoi A, Varshney U, RajBhandary UL. Common location of determinants in initiator transfer RNAs for initiator-elongator discrimination in bacteria and in eukaryotes. J Biol Chem. 2003;278:17672–9. doi: 10.1074/jbc.M212890200. [DOI] [PubMed] [Google Scholar]
- 28.Mayer C, Stortchevoi A, Kohrer C, Varshney U, RajBhandary UL. Initiator tRNA and its role in initiation of protein synthesis. Cold Spring Harb Symp Quant Biol. 2001;66:195–206. doi: 10.1101/sqb.2001.66.195. [DOI] [PubMed] [Google Scholar]
- 29.Wagner T, Rundquist C, Gross M, Sigler PB. Structural features that underlie the use of bacterial Met-tRNAfMet primarily as an elongator in eukaryotic protein synthesis. J Biol Chem. 1989;264:18506–11. [PubMed] [Google Scholar]
- 30.Kiesewetter S, Ott G, Sprinzl M. The role of modified purine 64 in initiator/elongator discrimination of tRNA(iMet) from yeast and wheat germ. Nucleic Acids Res. 1990;18:4677–82. doi: 10.1093/nar/18.16.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Astrom SU, Bystrom AS. Rit1, a tRNA backbone-modifying enzyme that mediates initiator and elongator tRNA discrimination. Cell. 1994;79:535–46. doi: 10.1016/0092-8674(94)90262-3. [DOI] [PubMed] [Google Scholar]
- 32.Desgres J, Keith G, Kuo KC, Gehrke CW. Presence of phosphorylated O-ribosyl-adenosine in T-psi-stem of yeast methionine initiator tRNA. Nucleic Acids Res. 1989;17:865–82. doi: 10.1093/nar/17.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basavappa R, Sigler PB. The 3 A crystal structure of yeast initiator tRNA: functional implications in initiator/elongator discrimination. Embo J. 1991;10:3105–11. doi: 10.1002/j.1460-2075.1991.tb07864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BF, Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–72. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 35.Nissen P, Thirup S, Kjeldgaard M, Nyborg J. The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure Fold Des. 1999;7:143–56. doi: 10.1016/s0969-2126(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 36.Kapp LD, Lorsch JR. GTP-dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J Mol Biol. 2004;335:923–36. doi: 10.1016/j.jmb.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 37.Kapp LD, Kolitz SE, Lorsch JR. Yeast initiator tRNA identity elements cooperate to influence multiple steps of translation initiation. RNA. 2006;12:751–64. doi: 10.1261/rna.2263906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farruggio D, Chaudhuri J, Maitra U, RajBhandary UL. The A1 x U72 base pair conserved in eukaryotic initiator tRNAs is important specifically for binding to the eukaryotic translation initiation factor eIF2. Mol Cell Biol. 1996;16:4248–56. doi: 10.1128/mcb.16.8.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner T, Gross M, Sigler PB. Isoleucyl initiator tRNA does not initiate eucaryotic protein synthesis. J Biol Chem. 1984;259:4706–9. [PubMed] [Google Scholar]
- 40.Varshney U, RajBhandary UL. Initiation of protein synthesis from a termination codon. Proc Natl Acad Sci U S A. 1990;87:1586–90. doi: 10.1073/pnas.87.4.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drabkin HJ, RajBhandary UL. Initiation of protein synthesis in mammalian cells with codons other than AUG and amino acids other than methionine. Mol Cell Biol. 1998;18:5140–7. doi: 10.1128/mcb.18.9.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schevitz RW, Podjarny AD, Krishnamachari N, Hughes JJ, Sigler PB, Sussman JL. Crystal structure of a eukaryotic initiator tRNA. Nature. 1979;278:188–90. doi: 10.1038/278188a0. [DOI] [PubMed] [Google Scholar]
- 43.Woo NH, Roe BA, Rich A. Three-dimensional structure of Escherichia coli initiator tRNAfMet. Nature. 1980;286:346–51. doi: 10.1038/286346a0. [DOI] [PubMed] [Google Scholar]
- 44.Wrede P, Woo NH, Rich A. Initiator tRNAs have a unique anticodon loop conformation. Proc Natl Acad Sci U S A. 1979;76:3289–93. doi: 10.1073/pnas.76.7.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seong BL, RajBhandary UL. Escherichia coli formylmethionine tRNA: mutations in GGGCCC sequence conserved in anticodon stem of initiator tRNAs affect initiation of protein synthesis and conformation of anticodon loop. Proc Natl Acad Sci U S A. 1987;84:334–8. doi: 10.1073/pnas.84.2.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mandal N, Mangroo D, Dalluge JJ, McCloskey JA, RajBhandary UL. Role of the three consecutive G:C base pairs conserved in the anticodon stem of initiator tRNAs in initiation of protein synthesis in Escherichia coli. RNA. 1996;2:473–82. [PMC free article] [PubMed] [Google Scholar]
- 47.Schweisguth DC, Moore PB. On the conformation of the anticodon loops of initiator and elongator methionine tRNAs. J Mol Biol. 1997;267:505–19. doi: 10.1006/jmbi.1996.0903. [DOI] [PubMed] [Google Scholar]
- 48.Selmer M, Dunham CM, Murphy FV, 4th, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–42. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 49.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–77. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 50.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–4. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 51.Barraud P, Schmitt E, Mechulam Y, Dardel F, Tisné C. A unique conformation of the anticodon stem-loop is associated with the capacity of tRNAfMet to initiate protein synthesis. Nucleic Acids Res. 2008;36:4894–901. doi: 10.1093/nar/gkn462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shoji S, Abdi NM, Bundschuh R, Fredrick K. Contribution of ribosomal residues to P-site tRNA binding. Nucleic Acids Res. 2009;37:4033–42. doi: 10.1093/nar/gkp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson J, et al. The essential Gcd10p-Gcd14p nuclear complex is required for 1- methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998;12:3650–62. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anderson J, Phan L, Hinnebusch AG. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1- methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2000;97:5173–8. doi: 10.1073/pnas.090102597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–40. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Varshney U, Ramesh V, Madabushi A, Gaur R, Subramanya HS, RajBhandary UL. Mycobacterium tuberculosis Rv2118c codes for a single-component homotetrameric m1A58 tRNA methyltransferase. Nucleic Acids Res. 2004;32:1018–27. doi: 10.1093/nar/gkh207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murphy FV, 4th, Ramakrishnan V, Malkiewicz A, Agris PF. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat Struct Mol Biol. 2004;11:1186–91. doi: 10.1038/nsmb861. [DOI] [PubMed] [Google Scholar]
- 58.Mangroo D, Limbach PA, McCloskey JA, RajBhandary UL. An anticodon sequence mutant of Escherichia coli initiator tRNA: possible importance of a newly acquired base modification next to the anticodon on its activity in initiation. J Bacteriol. 1995;177:2858–62. doi: 10.1128/jb.177.10.2858-2862.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hartz D, Binkley J, Hollingsworth T, Gold L. Domains of initiator tRNA and initiation codon crucial for initiator tRNA selection by Escherichia coli IF3. Genes Dev. 1990;4:1790–800. doi: 10.1101/gad.4.10.1790. [DOI] [PubMed] [Google Scholar]
- 60.O'Connor M, Gregory ST, RajBhandary UL, Dahlberg AE. Altered discrimination of start codons and initiator tRNAs by mutant initiation factor 3. RNA. 2001;7:969–78. doi: 10.1017/s1355838201010184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCutcheon JP, Agrawal RK, Philips SM, Grassucci RA, Gerchman SE, Clemons WM, Jr., Ramakrishnan V, Frank J. Location of translational initiation factor IF3 on the small ribosomal subunit. Proc Natl Acad Sci U S A. 1999;96:4301–6. doi: 10.1073/pnas.96.8.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dallas A, Noller HF. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol Cell. 2001;8:855–64. doi: 10.1016/s1097-2765(01)00356-2. [DOI] [PubMed] [Google Scholar]
- 63.Lancaster L, Noller HF. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol Cell. 2005;20:623–32. doi: 10.1016/j.molcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 64.Qin D, Abdi NM, Fredrick K. Characterization of 16S rRNA mutations that decrease the fidelity of translation initiation. RNA. 2007;13:2348–55. doi: 10.1261/rna.715307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Antoun A, Pavlov MY, Lovmar M, Ehrenberg M. How initiation factors maximize the accuracy of tRNA selection in initiation of bacterial protein synthesis. Mol Cell. 2006;23:183–93. doi: 10.1016/j.molcel.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 66.Das G, Thotala DK, Kapoor S, Karunanithi S, Thakur SS, Singh NS, Varshney U. Role of 16S ribosomal RNA methylations in translation initiation in Escherichia coli. Embo J. 2008;27:840–51. doi: 10.1038/emboj.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dong J, Nanda JS, Rahman H, Pruitt MR, Shin BS, Wong CM, Lorsch JR, Hinnebusch AG. Genetic identification of yeast 18S rRNA residues required for efficient recruitment of initiator tRNA(Met) and AUG selection. Genes Dev. 2008;22:2242–55. doi: 10.1101/gad.1696608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–22. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maag D, Algire MA, Lorsch JR. Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal pre-initiation complex plays a role in start site selection. J Mol Biol. 2006;356:724–37. doi: 10.1016/j.jmb.2005.11.083. [DOI] [PubMed] [Google Scholar]
- 70.Fekete CA, et al. N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. Embo J. 2007;26:1602–14. doi: 10.1038/sj.emboj.7601613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cigan AM, Feng L, Donahue TF. tRNAi(met) functions in directing the scanning ribosome to the start site of translation. Science. 1988;242:93–7. doi: 10.1126/science.3051379. [DOI] [PubMed] [Google Scholar]
- 72.Kolitz SE, Takacs JE, Lorsch JR. Kinetic and thermodynamic analysis of the role of start codon/anticodon base pairing during eukaryotic translation initiation. RNA. 2009;15:138–52. doi: 10.1261/rna.1318509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cochella L, Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–80. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schwarz U, Menzel HM, Gassen HG. Codon-dependent rearrangement of the three-dimensional structure of phenylalanine tRNA, exposing the T-psi-C-G sequence for binding to the 50S ribosomal subunit. Biochemistry. 1976;15:2484–90. doi: 10.1021/bi00656a035. [DOI] [PubMed] [Google Scholar]
- 75.Farber N, Cantor CR. Comparison of the structures of free and ribosome-bound tRNAPhe by using slow tritium exchange. Proc Natl Acad Sci U S A. 1980;77:5135–9. doi: 10.1073/pnas.77.9.5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bertram S, Goringer U, Wagner R. Structural investigation of Phe-tRNAPhe from E.coli bound to the ribosomal A-site. Nucleic Acids Res. 1983;11:575–89. doi: 10.1093/nar/11.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moras D, Dock AC, Dumas P, Westhof E, Romby P, Ebel JP, Giege R. Anticodon-anticodon interaction induces conformational changes in tRNA: yeast tRNAAsp, a model for tRNA-mRNA recognition. Proc Natl Acad Sci U S A. 1986;83:932–6. doi: 10.1073/pnas.83.4.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rodnina MV, Fricke R, Wintermeyer W. Transient conformational states of aminoacyl-tRNA during ribosome binding catalyzed by elongation factor Tu. Biochemistry. 1994;33:12267–75. doi: 10.1021/bi00206a033. [DOI] [PubMed] [Google Scholar]
- 79.Dabrowski M, Spahn CM, Nierhaus KH. Interaction of tRNAs with the ribosome at the A and P sites. Embo J. 1995;14:4872–82. doi: 10.1002/j.1460-2075.1995.tb00168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Valle M, Sengupta J, Swami NK, Grassucci RA, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. Embo J. 2002;21:3557–67. doi: 10.1093/emboj/cdf326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FV, 4th, Weir JR, Ramakrishnan V. The Crystal Structure of the Ribosome Bound to EF-Tu and Aminoacyl-tRNA. Science. 2009 doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005;121:703–12. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 83.Simonetti A, Marzi S, Myasnikov AG, Fabbretti A, Yusupov M, Gualerzi CO, Klaholz BP. Structure of the 30S translation initiation complex. Nature. 2008;455:416–20. doi: 10.1038/nature07192. [DOI] [PubMed] [Google Scholar]
- 84.Woese CR. Translation: in retrospect and prospect. RNA. 2001;7:1055–67. doi: 10.1017/s1355838201010615. [DOI] [PMC free article] [PubMed] [Google Scholar]


