Abstract
The availability and utility of genome-scale metabolic reconstructions have exploded since the first genome-scale reconstruction was published a decade ago. Reconstructions have now been built for a wide variety of organisms, and have been used toward five major ends: (1) contextualization of high-throughput data, (2) guidance of metabolic engineering, (3) directing hypothesis-driven discovery, (4) interrogation of multi-species relationships, and (5) network property discovery. In this review, we examine the many uses and future directions of genome-scale metabolic reconstructions, and we highlight trends and opportunities in the field that will make the greatest impact on many fields of biology.
Keywords: computational biology, metabolic model, modeling, network, systems biology
Introduction
Biochemistry has long been occupied with the reconstruction of metabolic pathways. With modern genome-sequencing capabilities, these pathway reconstructions have been increasingly integrated into genome-scale metabolic models. Ten years ago, a metabolic model of Haemophilus influenza became the first genome-scale metabolic reconstruction to be published (Edwards and Palsson, 1999). In the decade since, the field of genome-scale metabolic network analysis has expanded rapidly, and today >50 genome-scale metabolic reconstructions have been published (see Figure 1A). With the growing influence of these reconstructions on biomedical and biological research, and with the field now shifting from an inward focus on method development to an outward focus on application development, it is timely to review the various uses of genome-scale metabolic reconstructions and the future potential of systems-based analyses of metabolism.
Figure 1.
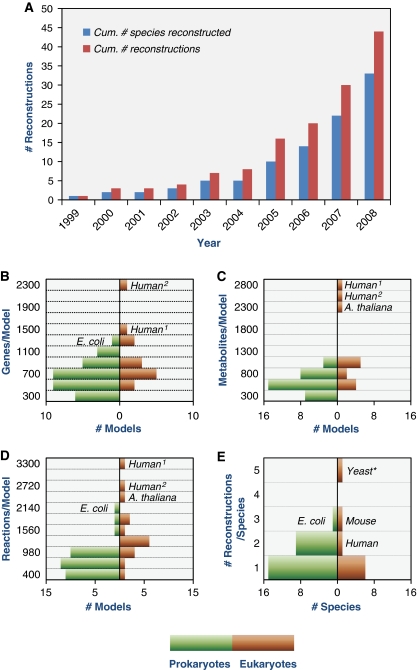
Reconstruction statistics. The cumulative number of metabolic GENREs published over the past decade is shown in (A). (B–D) Histograms of the number of metabolic GENREs containing varying numbers of genes (B), metabolites (C), and reactions (D). (E) Histogram of the number of reconstructions published per species. All histograms display prokaryotic (green) and eukaryotic (brown) statistics. *Yeast, S. cerevisiae; Human1,2, human reconstructions (Duarte et al, 2007; Ma et al, 2007).
Of all organisms that have been analyzed through a constraint-based metabolic reconstruction, Escherichia coli has gained the most attention as a model organism. As the applications of E. coli genome-scale models have been reviewed earlier in detail (Feist and Palsson, 2008), we specifically exclude E. coli from this review, and instead focus on the many other target organisms that have been studied.
A survey of papers citing metabolic reconstructions over the last 10 years revealed several themes in uses of the reconstructions. Following these themes, this review assigns uses of genome-scale metabolic reconstructions (metabolic GENREs; Becker and Palsson, 2005) to five major categories: (1) contextualization of high-throughput data, (2) guidance of metabolic engineering, (3) directing hypothesis-driven discovery, (4) interrogation of multi-species relationships, and (5) network property discovery (see Figure 2). These categories were chosen because they cover the majority of topics that have been addressed using metabolic GENREs, and they each represent a significantly different aim of these models. Areas of research not covered in these categories represent directions which in general have not been as well developed, and which may be possible avenues for future study. Each section describes the importance and historical context of that use, ways that metabolic GENREs have accelerated the knowledge gained in the given field, and finally the drawbacks or current hurdles in applying metabolic GENREs to the given problem. These applications include both applied and theoretical approaches, and represent a broad range of problems that have been engaged using genome-scale metabolic reconstructions.
Figure 2.
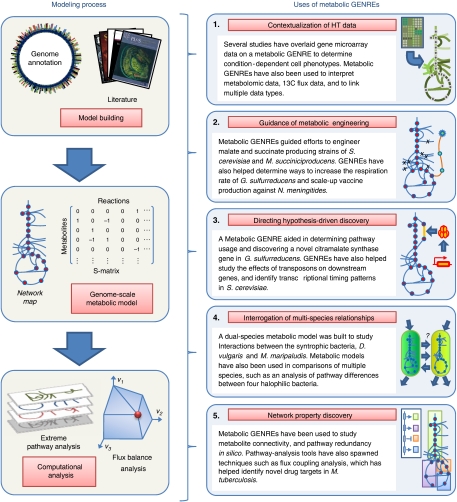
Uses of metabolic GENREs. The building and analysis of metabolic GENREs are shown in the left panels, and the five categories of uses of metabolic GENREs as highlighted in this paper are described in the right set of panels. Each panel on the right includes a representative example from literature.
Model building and analysis
Methods for developing metabolic reconstructions have been reviewed in the past (Reed et al, 2006; Durot et al, 2009; Feist et al, 2009) and several resources exist for model building and analysis (Becker et al, 2007), so those methods are not presented in detail here. Although the majority of this review focuses on uses of completed metabolic GENREs, we will nevertheless quickly overview the reconstruction process itself to highlight ways in which it can be intrinsically useful.
To date, all high-confidence genome-scale metabolic reconstructions have been built manually through a four-step process (Oberhardt et al, 2008). First, an initial reconstruction is built from gene-annotation data coupled with information from online databases such as KEGG (Kanehisa et al, 2006) and EXPASY (Gasteiger et al, 2003), which link known genes to functional categories and help bridge the genotype–phenotype gap. Second, the initial reconstruction is curated through an examination of the primary literature. Then, the reconstruction as a knowledge base is converted into a mathematical model that can be analyzed through constraint-based approaches. Third, the reconstruction is validated through comparison of model predictions to phenotypic data. In a final fourth step, a metabolic reconstruction is subjected to continued wet- and dry-lab cycles, which improve accuracy and allow investigation of key hypotheses.
Generally, a reconstruction includes semi-automated gene-annotation data based on BLAST-homology scores from a sequenced genome, augmented by detailed, manually collected data from organism-specific literature. One of the most immediate contributions of metabolic GENREs to biological knowledge comes in the process of gap analysis during model building, whereby formerly un-annotated gene functions are incorporated into gene-annotation knowledge by analysis of incomplete but essential metabolic pathways (Gonzalez et al, 2008; Oberhardt et al, 2008; Chavali et al, 2008b). The gap-analysis process can be beneficial both by stimulating literature searches that reveal previously overlooked phenotypic data and by posing hypotheses for enzymes that likely exist in the organism but for which no corresponding gene is currently annotated. Aside from offering hypotheses for future experiments, this process serves to crystallize the work done on a particular organism and highlight major areas still left for investigation. The gap-analysis step is also crucial for conversion of a genome-scale reconstruction as a knowledge base into the metabolic GENRE as a functional model, toward whose analysis the full suite of network tools can be applied (Manichaikul et al, 2009).
Through gap analysis and subsequent pathway analysis, studies have elucidated both the stoichiometry of certain reactions (Feist et al, 2006) and the most efficient pathways for production of certain metabolites (Kjeldsen and Nielsen, 2009), and in some cases have even proposed methods for engineering more efficient strains (Puchalka et al, 2008). Also, it is common for reconstruction efforts to provide high-quality estimates of cellular parameters such as growth yield, specific fluxes (Senger and Papoutsakis, 2008b), P/O ratio, and ATP maintenance costs (Famili et al, 2003), and these theoretical values are often used for hypothesis building or validation in biological studies (Lee et al, 2007; Hiratsuka et al, 2008). Several published metabolic reconstruction studies also include in silico predictions for minimal medium design (Oliveira et al, 2005; Baart et al, 2007b; Chavali et al, 2008b).
A completed metabolic GENRE can be used for a variety of applications. Many of the constraint-based analyses possible on metabolic GENREs rely on the theory that evolution selects for fitness-optimizing organisms, a concept crystallized with the development of flux balance analysis (FBA) (Maynard Smith, 1978; Lee et al, 2006). FBA involves optimization of a network for a given objective function, often a ‘biomass' reaction, to predict in silico flux values and/or growth. This optimization process outputs an optimal set of metabolic flux values that are consistent with maximization (or minimization) of the chosen objective. Another class of constraint-based approaches, termed pathway analysis methods, enables an accounting of all possible flux pathways in a given network, and have been used to examine network properties such as flexibility and variability of flux distributions (Papin et al, 2004). These techniques underlie most model validation performed on metabolic GENREs, and have opened up a wide variety of applications and new directions for model use.
Currently available reconstructions
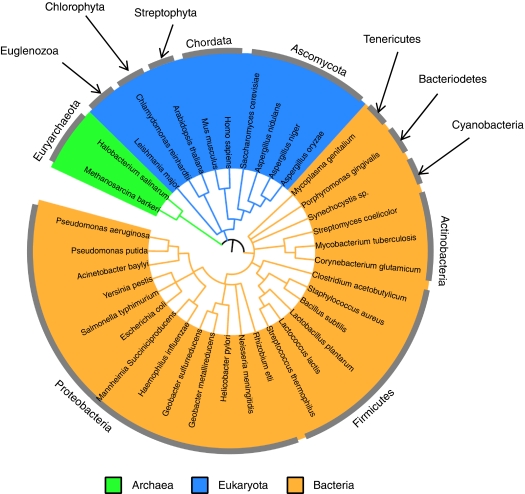
Metabolic GENREs of prokaryotes encompass an average of 600 metabolites, 650 genes, and 800 reactions, whereas metabolic GENREs of eukaryotes include on average 1200 metabolites, 1000 genes, and 1500 reactions. Excluding the two existing reconstructions of Homo sapiens metabolism lowers the average eukaryotic network size to 800, 800, and 1300, metabolites, genes, and reactions, respectively, a closer but still higher distribution to that of prokaryotes (see Figure 1B–D). Depending on whether the mouse, human, and Arabidopsis thaliana metabolic reconstructions are included in the statistic, between 6 and 13% of all ORFs in a eukaryotic genome are generally included in a metabolic GENRE, whereas metabolic GENREs of prokaryotes include on average 18% of all ORF's. Existing reconstructions span the domains Eukaryota, Bacteria, and Archaea. The most represented domain is bacteria, with 25 species reconstructed. The phylogenetic tree in Figure 3 reveals a conspicuous lack of plant metabolic reconstructions, with a preliminary reconstruction of A. thaliana (Radrich et al, http://hdl.handle.net/10101/npre.2009.3309.1) as the only plant metabolic GENRE released so far. This gap indicates an important direction for future efforts.
Figure 3.
Phylogenetic tree of reconstructed species. This figure shows a phylogenetic tree of all species for which metabolic GENREs have been built. Sections are colored by superkingdom, and phyla are noted on the outer ring of the tree. The phylogenetic tree was generated using semi-automated software at http://itol.embl.de/ (Ciccarelli et al, 2006), and phyla were determined using the NCBI taxonomy browser.
Category 1: Contextualization of high-throughput data
With biology increasingly becoming a data-rich field, an emerging challenge has been determining how to organize, sort, interrelate, and contextualize all of the high-throughput datasets now available. This challenge has motivated the field of top–down systems biology, wherein statistical analyses of high-throughput data are used to infer biochemical network structures and functions. In top–down modeling, determination of network structure poses a major technological and computational hurdle (Stark et al, 2003). However, many of the weaknesses of top–down modeling, such as lower accuracy and confidence in the resulting models, can be alleviated by comparison or merging with carefully built bottom–up models, such as metabolic GENREs. By serving as a framework on which other data types can be overlaid, the metabolic reconstruction has served as a powerful tool for contextualizing high-throughput data and aiding top–down approaches, as described below.
High-throughput data can be overlaid onto a metabolic GENRE in several ways. One highly functional way to use a metabolic GENRE for contextualization of gene expression data, protein expression data, C13 flux data, or high performance liquid chromatography derived byproduct secretion profiles is by directly imposing constraints on the GENRE based on the values in the experimental dataset. For instance, if an experimental dataset indicates that glycolytic enzymes are highly active under a given condition, flux can be funneled through glycolysis by constraining the relevant fluxes in silico, thereby forcing flux through the activated reactions and allowing evaluation of changes in global flux distributions. Gene microarray data can be similarly used to constrain metabolic fluxes and can give tremendous insight into conditional changes in metabolic activity (Shlomi et al, 2008), despite a nonperfect correlation between gene expression and protein expression (Ideker et al, 2001; Chechik et al, 2008). In addition to altering the constraints in a functioning metabolic model, high-throughput data can also simply be overlaid on a metabolic network to foster insight into metabolic hotspots or pathways that are significantly altered under certain conditions (Usaite et al, 2006). This process can elucidate otherwise inscrutable relationships between datapoints. Other types of high-throughput data can be analyzed in context of a metabolic GENRE as well, including rapid phenotyping data (e.g. BIOLOG phenotype microarrays, which test metabolic phenotypes of cells under thousands of growth conditions simultaneously), and whole or partial-genome gene essentiality data (Oberhardt et al, 2008). These data describe physiological states, which can directly be compared with in silico phenotypes, often in a qualitative or binary way: an organism grows on substrate X or it does not. In this sense, contextualization of these data represents a refinement of traditional taxonomic methods, which often use growth profiles of an organism for identification but generally lack mechanistic details or explanations for the majority of assayed phenotypes (Boone et al, 2001).
Metabolic reconstructions have been particularly useful as context for gene expression data. Many examples exist of gene microarrays being used in conjunction with genome-scale metabolic reconstructions to give a deeper understanding of why certain changes in expression occur in different environments. In particular, Saccharomyces cerevisiae has been used as a model organism for this type of analysis. A comparison of in silico metabolic fluxes versus microarray gene expression data in E. coli and S. cerevisiae revealed that metabolic genes whose fluxes are directionally coupled generally show similar expression patterns, share transcriptional regulators, and reside in the same operon (Notebaart et al, 2008). Expression data has also been coupled with various generations of S. cerevisiae metabolic reconstructions to determine which portions of metabolism are most sensitive to nitrogen limitation (Usaite et al, 2006) and to compare metabolic states during growth on glucose, maltose, ethanol, and acetate (Daran-Lapujade et al, 2004). In these studies, expression states of metabolic genes were overlaid on the reactions their protein products catalyze, and expression patterns of metabolic enzymes were then compared against the fluxes predicted in silico under the given growth condition. Without a model to lay these expression data on, it would be difficult to characterize the global expression states. In another striking example, a metabolic model of S. cerevisiae was augmented with 55 regulatory transcription factors regulating 348 metabolic genes to form a regulatory-metabolic network (Herrgard et al, 2006). From an initial regulatory network, ChIP-chip and binding-site motif data were used to expand the regulatory rule-set, and this expanded network was shown to have higher predictive power of gene expression when evaluated with 12 microarray datasets. The use of a regulatory-metabolic model to predict gene expression changes is a powerful direction for further research with metabolic reconstructions, one which pushes closer toward modeling the function of an entire cell (Lee et al, 2008b).
High-throughput technologies to determine the intracellular metabolic state of cells have also been aided by the development of metabolic GENREs. Intracellular metabolic fluxes can be determined through the use of 13C-labeled glucose experiments, in which labeled carbon is tracked during growth of cells in a chemostat culture and computational methods are used to reconstruct the paths that carbon took inside the cells during growth. Although 13C isotopomer tracking has been performed without the aid of a metabolic GENRE, the comprehensive coverage of metabolic pathways enabled by the genome-scale reconstructions has made these attractive frameworks for 13C tracking experiments (Vo et al, 2007; Panagiotou et al, 2008). Metabolic GENREs have also been used as frameworks for interpreting metabolite concentration data. In one study (Cakir et al, 2006), a high-throughput GC-MS method was used to determine concentrations of 52 metabolites in S. cerevisiae. Differences in metabolite concentrations under known environmental conditions were mapped onto a modified S. cerevisiae metabolic GENRE, and this mapping was then combined with transcriptome data to investigate the effectors of metabolic regulation in the cell.
In many cases such as the one highlighted above, multiple high-throughput data types are analyzed in concert through the framework of the metabolic GENRE, which allows for a highly integrated picture of cell function to emerge. Transcriptomic data in particular is often linked with other data types, such as protein expression data (Shlomi et al, 2008), protein–protein interaction data, protein–metabolite interaction data, and physical interaction data (Prinz et al, 2004). Particularly in light of multiple data types, the metabolic GENRE can be a valuable tool for data interpretation. For instance, multiple data types were used in concert with a metabolic GENRE to determine tissue-specific metabolic activities in H. sapiens (Shlomi et al, 2008) and to compare the filamentous-form versus the yeast form of S. cerevisiae (Prinz et al, 2004).
Metabolic GENREs are natural frameworks for contextualizing genome-scale data, and as a result there have been many studies that use metabolic GENREs in this manner. High-throughput data have even been used to aid in building metabolic GENREs; for instance, the fact that some mitochondrial genes lie outside the mitochondrion made proteomic data key to building a model of human mitochondrial metabolism, in combination with biochemical data from literature (Vo et al, 2004). However, a major challenge still lies in determining an optimal strategy for interpreting high-throughput data. For instance, while a link has been established between expression of the gene for a metabolic enzyme and the bounds on metabolite flux through that enzyme (Chechik et al, 2008), establishing a scalable, reliable heuristic for bounding reaction fluxes using transcriptomic or proteomic data remains an unanswered challenge in the field. Noise also has a function in obscuring the relationship between high-throughput data and flux-related phenotypes. Particularly in situations where multiple noisy high-throughput datasets are considered at once, the question arises of how to integrate all of the data into one cohesive mathematical framework. These difficulties will have to be addressed through development of rigorous quality control measures for standardizing data analysis in the future.
Category 2: Guidance of metabolic engineering
Metabolic engineering involves the use of recombinant DNA technology to selectively alter cell metabolism and improve a targeted cellular function (Bailey et al, 1990). Traditionally, metabolic engineering has been performed on a small scale through manipulation of a few genes to affect yield of a target metabolite. Enzymatic targets are chosen through analysis of literature-derived central metabolic pathway maps, or intuitive engineering based on local metabolic knowledge. These local approaches have yielded success in the past, enabling engineering of new metabolic pathways and improvement of existing processes in E. coli (Bailey et al, 1990), S. cerevisiae (Nevoigt, 2008), and other microorganisms (Park and Lee, 2008). However, the complexity of metabolic networks, compounded by multiple layers of transcriptional, protein, and substrate-level regulation of metabolic enzymes, renders predictable metabolic engineering extremely difficult, and often causes unwanted consequences or sub-optimal outcomes when local network maps or intuitive knowledge are the basis of engineering decisions (Kim et al, 2008; Nevoigt, 2008). The inherent drawbacks of using local analysis tools to guide cell-scale metabolic engineering efforts have motivated the use of metabolic GENREs and other genome-scale technologies, in what has been termed ‘systems metabolic engineering' (Park and Lee, 2008). The use of metabolic GENREs represents a major evolution for the field, wherein whole-cell networks and systems-level analyses are for the first time being leveled to determine optimal engineering strategies on a whole-cell basis (Park and Lee, 2008).
Because of the industrial importance and metabolic centrality of TCA intermediates such as malic acid and succinic acid, many recent metabolic engineering efforts have focused on increasing production of these metabolites. In a recent study, S. cerevisiae was engineered to produce 59 g/l of malate, an amount five times higher than earlier efforts (Zelle et al, 2008). This remarkable improvement was validated by 13C-NMR flux determination, using a metabolic GENRE as the basis for the 13C flux model. Several other examples of metabolic GENRE-guided metabolic engineering involve genome-scale reconstructions of the succinic acid producing bacterium Mannheimia succiniciproducens. In the initial publication of the M. succiniciproducens genome sequence, a constraint-based metabolic model was presented including 373 reactions and 352 metabolites (Hong et al, 2004). This model was used to compare metabolic flux distributions between M. succiniciproducens and E. coli, and to identify a combination of three pyruvate-forming enzymes whose removal from E. coli would likely increase succinic acid production. Genetic engineering efforts targeting those three genes were successful, and a succinic acid producing strain of E. coli was generated (Lee et al, 2008c). Later, an expanded metabolic GENRE was published for M. succiniciproducens, including 686 reactions and 519 metabolites (Kim et al, 2007). This expanded reconstruction, which was used to predict succinic acid production in a variety of experimental circumstances, is currently being used to further investigate the metabolic capabilities and guide engineering strategies in M. succiniciproducens (Lee et al, 2008c).
Metabolic GENREs have been used to guide other types of genetic engineering efforts aside from increasing production of value-added chemicals. In one study, an algorithm called Optknock (Burgard et al, 2003) was applied to the metabolic GENRE of Geobacter sulfurreducens to determine optimal gene knockouts to maximally increase respiration rates (Izallalen et al, 2008). G. sulfurreducens is a bacterium whose ability to oxidize organic compounds using metals as terminal electron acceptors has made it highly attractive for bioremediation efforts. The Optknock analysis predicted that increasing ATP demand would increase NADPH oxidation rates, and subsequent alteration of membrane-bound F0F1 ATP synthase achieved the predicted increase in G. sulfurreducens respiration rate (Izallalen et al, 2008). Metabolic GENRE-guided genetic engineering has also been used to aid the scale-up for bulk production of a vaccine against the pathogen, Neisseria meningitides (Baart et al, 2007a). These examples serve to highlight the various ways in which metabolic GENREs have aided genetic engineering by casting the engineering efforts in context of the whole-cell metabolism of the organism being studied.
Although metabolic GENREs are powerful tools of great utility for metabolic engineering, issues such as pleiotropy (a single gene that affects multiple phenotypic traits), unaccounted for or inactive isozymes, and mis-annotation of critical genes can weaken the efficacy of computational predictions in determining engineering targets. In addition, it is sometimes not clear whether the considerable effort it takes to build a metabolic reconstruction is a good investment when developing a metabolic engineering strategy, as issues such as allosteric enzyme regulation can necessitate detailed dynamic modeling of a specific pathway to accurately predict phenotypes (Stephanopoulos and Vallino, 1991; Contador et al, 2009). Still, even when such approaches are necessary for the engineering process, metabolic GENREs are uniquely capable of predicting secondary effects of a given metabolic perturbation on other, often nonobvious portions of metabolism (Nevoigt, 2008), and therefore nearly always have the potential to be useful in developing such strategies. Further, kinetic constants for metabolic interactions can be extremely difficult to ascertain, so constraint-based modeling remains an attractive alternative to these methods (Contador et al, 2009). Another hurdle in metabolic engineering is the importance of transcriptional regulation in determining metabolic phenotypes. Regulatory genes are often important targets in metabolic engineering efforts due to their primary function in determining the distribution of metabolic flux (Bailey et al, 1990). With the inclusion of some regulatory rules, the usefulness of a metabolic GENRE can be significantly increased for guiding these efforts. However, regulatory networks are generally less well characterized than metabolic networks, and tend to be far more species and strain specific (Herrgard et al, 2004). This makes it difficult to reliably predict good metabolic engineering targets, as some crucial regulatory information is generally unknown for a given organism. Therefore, an increased effort into the reconstruction and analysis of regulatory networks will be of major utility for GENRE-guided metabolic engineering efforts in the future.
Category 3: Directing hypothesis-driven discovery
Much of what is known in biology today is the result of meticulous, hypothesis-driven discovery. This research has been guided by heuristic, informal models of biology developed in the minds of experts during years of work in a particular field or on a particular problem. However, with the tremendous expansion of biological data in recent years, the need has arisen for new method development to integrate high-throughput data with the biological discovery process. Gene microarrays serve as a prime example; a traditional hypothesis-driven study might include examination of 1 or 2 genes in a microarray that are of particular interest. This approach would ignore the thousands of other genes on the chip, however, and could miss important information or trends embedded in those data. Therefore, a systematic framework for incorporating genome-scale data available from multiple high-throughput methods would allow hypothesis-driven biology to benefit from the full range of tools available today.
Metabolic GENREs represent concise collections of existing hypotheses, and taken together as a broad context they enable systematic identification of new hypotheses that can be tested and resolved. Therefore, they represent a crucial framework for incorporating the flood of biological data now available into the biological discovery process.
Beyond computational predictions, metabolic GENREs have been used to frame investigations into specific biological questions, using a mix of traditional biological approaches and computational systems-level thinking. This type of mixed analysis has been used extensively in G. sulfurreducens to determine pathway usage in redundant cellular systems. In one study, metabolic GENRE-derived flux predictions were compared with growth phenotypes of G. sulfurreducens to elucidate which of eight sets of redundant pathways are used in vivo (Segura et al, 2008), and several functions were shown to be carried out by only one gene despite the existence of seemingly redundant pathways. This finding bolstered an earlier study, in which it was shown through computational analysis and in vivo deletion studies that isozymes are often not perfectly redundant (Harrison et al, 2007). Another study used 13C-labeling to track fluxes through a known threonine-associated isoleucine biosynthetic pathway in G. sulfurreducens (Risso et al, 2008). With the discovery that only a fraction of isoleucine is generated through this pathway, a putative citramalate synthase gene was tested and shown through knockout experiments to comprise part of a previously uncharacterized citramalate-associated isoleucine synthesis pathway, which produces the majority of isoleucine in the cell. The initial search for the citramalate synthase gene was motivated by model simulations in which addition of a citramalate pathway significantly improved agreement of experimental and predicted isotopomer distributions in the 13C experiment (Risso et al, 2008).
Some biological questions investigated using metabolic GENREs involve cellular-level phenomena difficult to approach without a whole-cell model of metabolism. In one analysis, the proximity of in vivo transposon-disrupted genes to downstream essential genes was compared with the in silico essentiality of the transposon-disrupted genes using a metabolic GENRE of Pseudomonas aeruginosa (Oberhardt et al, 2008). It was shown that transposon inserts in one published genome-wide transposon study affected downstream genes, whereas transposon inserts in another published study did not, consistent with claims in the two studies. In another analysis, the transcriptional timing of metabolic genes in defined sub-networks of a metabolic GENRE of S. cerevisiae was studied using time courses of transcriptomic and proteomic data, as well as protein binding affinity data from ChIP-chip assays (Chechik et al, 2008). This analysis suggested that under relatively static environmental conditions, metabolism is primarily controlled through protein-level regulation, whereas during times of environmental change, transcriptional control guides metabolic function. Transcriptional control has also been studied by identification of ‘reporter metabolites,' which represent the most highly transcriptionally regulated metabolites in a system (Patil and Nielsen, 2005; Raghevendran et al, 2006; David et al, 2008). Like the analysis of transcriptional timing in S. cerevisiae, reporter metabolites represent a marriage of metabolic GENRE-network analysis and traditional biological investigation, and they enable study of functional phenomena in cells that would be difficult to assess otherwise.
Metabolic GENREs intrinsically represent a simplification of cellular function. The distinct biochemical networks categorized by scientists (e.g. metabolism, regulation, and signaling) blend together in a living cell, creating a far more complicated web of interactions than is convenient or possible to model (Featherstone and Broadie, 2002). This web is fundamentally stochastic, and co-habits the cell with many other simultaneous phenomena including transcription and translation, protein modification, cell division, adhesion, motility, and mechanical transduction of external forces. The very simplifications that make metabolic GENREs powerful tools also make them challenging to use for the study of totally unknown or novel phenomena.
Ostensibly, these challenges would limit the usefulness of metabolic GENREs to analyzing purely metabolic processes and refining our knowledge of already characterized cellular systems. However, the examples highlighted above in this section suggest otherwise. Metabolic GENREs enable integration of large datasets for analysis of whole-cell phenotypes, and when wielded effectively, these analyses can be targeted to answer profound questions in biology. The key to unlocking the potential of metabolic GENREs is to ask tractable questions, and to understand well the limitations of the technology used to determine the answers.
Category 4: Interrogation of multi-species relationships
Few cells grow in pure cultures outside of the laboratory, and in many cases it is through the interactions of species that the most interesting phenotypes emerge (Riedel et al, 2001; Filoche et al, 2004; Fernandez et al, 2008). Metagenomics studies particularly have shown most ecosystems to be extremely diverse, including up to thousands of distinct taxa (Vieites et al, 2009). Further, higher-eukaryotic biology necessitates the study of multi-cellular systems, as it inherently focus on interactions between different cell types. Metabolic GENREs are increasingly being applied to these multi-cell problems, as well as to the study of functional differences between species. These avenues of research offer exciting prospects for deepening our understanding of the workings of multi-cellular communities and bridging the phenotype–genotype gap in the future.
A promising direction for computational systems biology is the incorporation of network-level analysis into the field of comparative genomics, which is currently driven by bioinformatics. Comparative network-level analysis is not completely new, as some of the first metabolic reconstructions generated were compared with discern phenotypic differences between species (Schilling et al, 2002; Forster et al, 2003). However, several studies have emerged in recent years that either compare metabolic reconstructions of highly related species or use models of interacting species to predict communal phenotypes. As more metabolic GENREs become available, these comparisons become more feasible and relevant for a wide variety of organisms. However, most multi-species analyses reported to date have involved either sub-genome-scale metabolic models or models that have not been carefully annotated. For instance, an analysis of the syntrophic bacteria Desulfovibrio vulgaris and Methanococcus maripaludis included creation of a dual-species stoichiometric model, including 170 reactions and 147 metabolites that comprised the central metabolism of both species (Stolyar et al, 2007). This work represents an important step in applying scalable computational methods to mutualistic bacterial communities, but it is focused on central metabolism rather than the genome scale.
Several multi-species analyses have focused on highly related cell types, attempting to discern differences in metabolic phenotypes based on network analysis. One group developed genome-level reconstructions of four halophilic bacteria, and compared their metabolic phenotypes by highlighting differences in various pathways between the four organisms (Falb et al, 2008). Although the group used computational flux methods in a separate publication to analyze a highly developed metabolic GENRE of a halophilic bacterium (Gonzalez et al, 2008), the comparison study itself included no in silico flux analysis. Another recent study did use in silico flux methods to compare two whole-cell metabolic networks, those of the human fibroblast and the diseased fibroblast suffering from Leigh's syndrome (Vo et al, 2007). The networks were derived from the global human metabolic network reconstruction (Duarte et al, 2007), and 13C-flux, literature-derived phenotypic data, and in silico computed flux states were used to discern differences between the normal and diseased states. Other efforts have included highly automated comparisons between species, sometimes taking many species into account (Verkhedkar et al, 2007; Borenstein et al, 2008; Lee et al, 2009).
Of the five categories of uses of metabolic GENREs described in this paper, multi-species studies have been represented the least in literature so far. With more genome-scale metabolic models being built and an increased focus on studying multi-cellular systems, however, we anticipate that this field will see a major increase in activity in the coming years. The difficulty and time required to build a well-curated metabolic GENRE is a major bottleneck toward these efforts, as a comparison of multiple species or the interactions between a host and a pathogen requires the building of two or more genome-scale models. Also, meaningful modeling of multi-species interactions often necessitates augmentation of a metabolic GENRE with some regulatory and signaling information, which further increases the complexity and difficulty of this type of modeling (de Kievit and Iglewski, 2000).
Along with intracellular signaling and regulatory pathways, spatial and temporal constraints can also become important for interactions between different cell types in a biofilm or tissue, leading to other computational and experimental challenges. Various modeling methods have been proposed to address these issues (Robertson et al, 2007; Chavali et al, 2008a), but coupling metabolic GENREs with models at varying spatiotemporal scales will require novel computational approaches (Jiang et al, 2005; Lee et al, 2008b; Zhang et al, 2009). A merging of metabolic GENREs, regulatory and signaling networks into multi-cell or multi-species models will be a major achievement of systems biology, and will move us closer to the goal of accurately modeling cells in therapeutically and environmentally important contexts.
Category 5: Network property discovery
With the development of powerful molecular biology tools over the last half century, a reductionist mindset has dominated the practice of biology (Singh, 2003). However, scientists have long understood the importance of holistic thinking when approaching biological systems, as complex cellular networks can spawn emergent phenomena that would be undetectable by reductionist approaches (Waliszewski et al, 1998; Westerhoff and Palsson, 2004). Conditionally essential genes (i.e. ‘synthetic lethals') would be overlooked, for instance, if genes were studied purely in isolation (Scherens and Goffeau, 2004). In recent years, some of the same tools that underpinned reductionist biology have been expanded to high-throughput methods, enabling the development of gene-based holistic network analysis techniques (Westerhoff and Palsson, 2004). Metabolic GENREs have enabled analysis of emergent phenomena through a focus on whole networks rather than individual pathways or genes, and many computational techniques have been developed to probe network properties. These types of network-level analyses will be critical to fully unravel the complex genotype–phenotype relationships in cells.
One of the most direct contributions of metabolic GENREs to our understanding of metabolism has been its enabling of the study of otherwise inaccessible network properties. Metabolic properties such as the existence of loops (Kun et al, 2008; Wright and Wagner, 2008), optimal pathway usage (Nishikawa et al, 2008), metabolite connectivity (Becker et al, 2006; Samal et al, 2006; Guimera et al, 2007), and pathway redundancy (Papin et al, 2002b; Mahadevan and Lovley, 2008) have all been studied in metabolic GENREs using computational methods. Many of these network analyses are performed through variants of FBA (see ‘Model building and analysis' section). A primary end toward which network analyses have been used is the improvement of existing genome annotations, such as in the FBA-driven gap-analysis process of model building. However, other more systematic methods have been developed for improving genome annotations based on the analyses afforded by a metabolic GENRE. One such method used a metabolic GENRE of S. cerevisiae to derive condition-dependent annotations of metabolic genes, achieving higher accuracy than gene ontology annotations in determining gene function (Rokhlenko et al, 2007; Shlomi et al, 2007). Another major application of network analysis tools has been the discovery of co-regulated genes, through a computational process called flux coupling analysis (Burgard et al, 2004). This computational method has been validated recently through NMR-derived metabolite profiles of single-gene knockouts in yeast (Bundy et al, 2007). Coupled reaction sets have been used for a variety of purposes, including prediction of novel drug targets in Mycobacterium tuberculosis (Jamshidi and Palsson, 2007).
The field of computational systems biology has produced a rich array of methods for network-based analysis, offering tremendous insight into the functioning of metabolic networks. However, many of these methods produce results that can be difficult to link to observable phenotypes. Forging this link poses the greatest challenge toward development of useful network-based tools. For instance, several methods exist to analyze redundancy in metabolic networks (Price et al, 2002, 2003; Papin et al, 2002a, 2002b; Mahadevan and Lovley, 2008). Although these techniques define ‘redundancy' intuitively in terms of the number of available paths between a given set of inputs and outputs, relating ‘redundancy' to an observable phenotype poses a difficult challenge.
Although some network analysis techniques focus on phenotypes that are currently difficult to measure, there still exists great value in this research. With improvements in experimental technology, some currently unobservable phenotypes will become measurable, and the gap between in silico and observable phenotypes will shrink. Also, some network analyses have already deeply influenced biological thinking, even in some cases where the methods yield no easily measured phenotypes (Jeong et al, 2000). Network analysis tools will continue to be critical to the success of systems biology, by both expanding the scope of what is thought possible, and by anticipating the emergence of cutting-edge wet-lab technologies.
Exploring evolutionary relationships
Significant interest exists in using metabolic GENREs to investigate functional evolution of metabolic and regulatory networks, especially considering the strongly evolutionary-based assumptions underlying analysis techniques such as FBA. Some studies have examined the phenomenon of short-term adaptive evolution, and have shown that as a particular strain adapts to media it is grown in, its growth characteristics will converge toward the FBA-predicted optimal solution (Fong and Palsson, 2004; Fong et al, 2005). Although these studies support the theory that evolution has honed organisms to optimize for fitness-related phenotypes (Maynard Smith, 1978), the historical evolutionary process itself remains fairly unstudied in context of metabolic GENREs. This fascinating research direction represents a promising area of study. There have been a few recent attempts to use metabolic models toward this end. In particular, it has been suggested that certain topological properties of metabolic networks (such as degree distribution) might be formed as a byproduct of selection for some other phenotypes (such as growth rate), rather than because the topological properties themselves elicit a selective advantage (Pfeiffer et al, 2001; Papp et al, 2009). Genome-scale metabolic models have yet to be significantly used toward answering these important questions, but the breadth of organisms for which GENREs are available today (see Figure 3) represent a tremendous opportunity to merge evolutionary genomics with network-based metabolic analysis, and gain unique insight into the evolutionary forces that have on metabolism.
Current status of genome-scale metabolic reconstructions
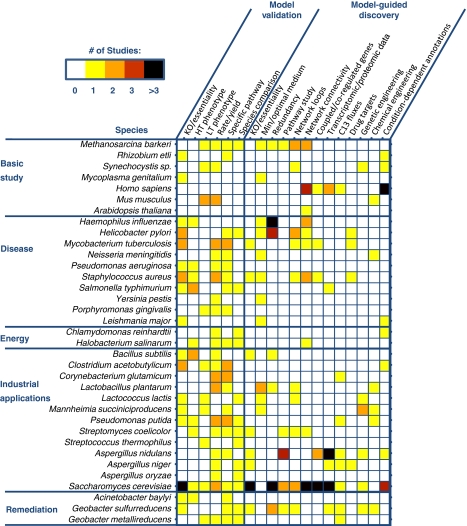
Metabolic GENREs are contributing to the development of predictive, mechanistic models of an entire cell, an unrealistic goal before the genomic revolution. These models provide a broad framework whose predictions can be continually refined as more data and computational methods become available. Figure 4 shows the analyses that have already been performed on metabolic GENRE of various species, and also highlights many gaps where some possibly informative analyses have yet to be done. These gaps represent a roadmap for future efforts. Metabolic GENREs are best viewed as low-resolution blueprints on top of which other systems, constraints, and perturbations can be overlaid. As these features are overlaid on metabolic GENREs, they continually improve the resolution of model predictions. With incorporation of regulatory and signaling data as well as other high-order systems into the constraint sets, metabolic GENREs are becoming increasingly agile and expressive of realistic cell phenotypes.
Figure 4.
Analyses of metabolic GENREs. A heat map of studies that have been published for all reconstructed species (y-axis) using a variety of analysis techniques (x-axis). Analyses are broken into two categories: ‘model validation,' indicating the use of a technique in a publication of a new metabolic GENRE to establish the validity of a model, and ‘model-guided discovery,' indicating the use of a technique either in an original metabolic GENRE publication or in a follow-up study to perform one of the five types of studies outlined in this paper. Species are grouped according to the most relevant broad category to which the metabolic GENRE has been applied. Colors in the heat map indicate the number of publications performing the given analysis on a metabolic GENRE of the organism. References for each species in the figure are as follows: M. barkeri (Becker et al, 2006; Feist et al, 2006; Kun et al, 2008; Mahadevan and Lovley, 2008; Wright and Wagner, 2008), R. etli (Resendis-Antonio et al, 2007), Synechocystis sp. (Kun et al, 2008), M. genitalium (Suthers et al, 2009), H. sapiens (Duarte et al, 2007; Ma et al, 2007; Vo et al, 2007; Shlomi et al, 2008, 2009; Veeramani and Bader, 2009), M. musculus (Sheikh et al, 2005; Quek and Nielsen, 2008; Selvarasu et al, 2009), A. thaliana (Radrich et al, http://hdl.handle.net/10101/npre.2009.3309.1), H. influenza (Edwards and Palsson, 1999; Papin et al, 2002a; Papin et al, 2002b; Price et al, 2002; Price et al, 2003; Schilling and Palsson, 2000), H. pylori (Price et al, 2002, 2003; Schilling et al, 2002; Papin et al, 2002b; Thiele et al, 2005; Becker et al, 2006; Guimera et al, 2007; Kun et al, 2008; Wright and Wagner, 2008), M. tuberculosis (Beste et al, 2009; Beste et al, 2007; Jamshidi and Palsson, 2007; Kun et al, 2008), N. meningitides (Baart et al, 2007a, 2007b), P. aeruginosa (Oberhardt et al, 2008), S. aureus (Becker and Palsson, 2005; Becker et al, 2006; Heinemann et al, 2005; Kun et al, 2008; Samal et al, 2006), S. typhimurium (Abuoun et al, 2009; Raghunathan et al, 2009), Y. pestis (Navid and Almaas, 2009), P. gingivalis (Mazumdar et al, 2009), L. major (Chavali et al, 2008b), C. reinhardtii (Boyle and Morgan, 2009), H. salinarum (Gonzalez et al, 2008), B. subtilis (Oh et al, 2007; Henry et al, 2009a, 2009b), C. acetobutylicum (Lee et al, 2008a; Senger and Papoutsakis, 2008a), C. glutamicum (Kjeldsen and Nielsen, 2009; Shinfuku et al, 2009), L. plantarum (Teusink et al, 2006, 2009; Stevens et al, 2008), L. lactis (Oliveira et al, 2005; Kun et al, 2008), M. succiniciproducens (Kim et al, 2007; Song et al, 2008; Lee et al, 2008c), P. putida (Nogales et al, 2008; Puchalka et al, 2008), S. coelicolor (Borodina et al, 2005; Hiratsuka et al, 2008; Kun et al, 2008), S. thermophilus (Pastink et al, 2009), A. nidulans (David et al, 2006, 2008; Panagiotou et al, 2008, 2009), A. niger (Andersen et al, 2008; Thykaer et al, 2009), A. oryzae (Vongsangnak et al, 2008), S. cerevisiae (Famili et al, 2003; Forster et al, 2003; Daran-Lapujade et al, 2004; Duarte et al, 2004; Prinz et al, 2004; Kuepfer et al, 2005; Patil and Nielsen, 2005; Becker et al, 2006; Cakir et al, 2006; Herrgard et al, 2006, 2008; Raghevendran et al, 2006; Samal et al, 2006; Usaite et al, 2006; Bundy et al, 2007; Harrison et al, 2007; Rokhlenko et al, 2007; Shlomi et al, 2007; Chechik et al, 2008; Deutscher et al, 2008; Kun et al, 2008; Mahadevan and Lovley, 2008; Nookaew et al, 2008; Notebaart et al, 2008; Wright and Wagner, 2008; Zelle et al, 2008; Cimini et al, 2009; Mintz-Oron et al, 2009; Mo et al, 2009), A. baylyi (Durot et al, 2008), G. sulfurreducens (Izallalen et al, 2008; Kun et al, 2008; Leang et al, 2009; Mahadevan et al, 2006; Mahadevan and Lovley, 2008; Risso et al, 2008; Scheibe et al, 2009; Segura et al, 2008), G. metallireducens (Sun et al, 2009).
As one of the simplest and most informative methods in constraint-based modeling, FBA has become a standard in the field, with a biomass reaction usually serving as the objective. Although the biomass objective yields accurate predictions under simple growth conditions for prokaryotic cells (Fong and Palsson, 2004), it is unclear to what degree optimization of biomass is descriptive of growth conditions in nature. Some studies have explored this by examining whether cells follow nonbiomass objective functions under certain conditions (Schuetz et al, 2007; Gianchandani et al, 2008), but even these studies stop short of questioning the optimization premise itself. Furthermore, although FBA predicts metabolic flux values through a network, FBA notably produces only one optimal solution, whereas it is quite common for multiple equally valid optima exist. This concept has been examined through an extension of FBA called flux variability analysis, which explores the entire optimal solution space as opposed to picking just one optimal solution (Mahadevan and Schilling, 2003), but it is an important caveat that should curb over-interpretation of FBA results.
Given a fully defined metabolic, regulatory, and signaling network, we would hypothesize that no extrinsic objective would be necessary, but rather that what we call an ‘objective' would be intrinsically built into the rules of the network itself. Therefore, as metabolic GENREs are increasingly augmented with regulatory and signaling rules, at what point does the ‘objective function' hypothesis break down entirely? Where exactly in a cell is the information content we call an ‘objective' held? These questions will become increasingly crucial and addressable as systems biology models become more sophisticated, and will stand to provide enormous insight into evolutionary biology.
An assumption underlying most approaches with metabolic GENREs is that networks modeled at steady state can still yield valuable information, regardless of the lack of detailed kinetic data. However, the steady state flux approximation unravels at the edges of metabolic activity, where stochasticity, enzyme kinetics, spatial distributions, and varying levels of metabolic regulation become dominant forces in cell activity. These edge effects become increasingly relevant as more accurate results are demanded from metabolic GENREs, and they represent the major challenges to building models of an entire cell. We have already described some efforts to incorporate dynamics and transcriptional control into metabolic GENREs (Herrgard et al, 2006; Lee et al, 2008b). A framework was also recently proposed for incorporating genome-scale kinetic data into metabolic GENREs for whole-cell dynamic modeling (Jamshidi and Palsson, 2008). However, a standing challenge is the determination of enough kinetic parameters to enable use of such a framework. In some cases, it has been shown that knowledge of a few key parameters can be sufficient for predicting metabolic and regulatory dynamics (Lee et al, 2008b), but it is unclear how effective these reduced-parameter models can be. Also, we are only now beginning to tackle different temporal and spatial scales at work in cells, and efforts to incorporate stochasticity into genome-scale regulatory and metabolic models are in their infancy (Zhu et al, 2004; Ghosh et al, 2007).
With transcriptional regulatory interactions, stochastic effects, nonlinear enzyme regulation, and other unaccounted-for cellular events intrinsically limiting the accuracy of predictions, validation of metabolic GENREs becomes a difficult and open-ended goal. Metabolic GENREs are often validated with comparisons between in silico phenotypes and various sets of in vivo data. However, no standard exists for how a model should be validated, which is apparent from the scattered representation of methods in validation of existing models (see Figure 4). Recent efforts have been made to quantify the level of discrepancy expected between in silico and in vivo metabolic phenotypes. In one notable study, 465 single-gene mutants of S. cerevisiae were grown and quantified under 16 different growth conditions each (Snitkin et al, 2008). An analysis of the performance of two published S. cerevisiae metabolic GENREs revealed sensitivity (correctly predicted nonessential genes versus the total number of nonessential genes) to be on the order of 95%, and specificity (correctly predicted essentials versus the total number of essential genes) to range between 50 and 60%. These numbers were significantly improved to approximately 95–98% and 69–86% (respectively) through disqualification of some in vivo experiments, which were discovered on further analysis to be in error. These final numbers are a gauge of the accuracy that might be expected from metabolic GENREs of well-studied organisms, given a preponderance of well-verified experimental data for comparison of in vivo and in silico results.
Many of the most pressing questions in basic and applied biology involve studying relationships between multiple cell types. Comparisons of diseased versus normal networks in the human will continue to yield insight into disease and drug activity (Vo et al, 2007), whereas development of more sophisticated modeling methods for interacting species (Stolyar et al, 2007) will enable increasingly realistic prediction of communal phenotypes. Evolutionary genomics will be enriched by the incorporation of network data into the analysis of species relationships. Also, methods developed to examine dynamic model activity (Klipp et al, 2005; Jamshidi and Palsson, 2008), to integrate metabolic models with regulatory and signaling information (Lee et al, 2008b), and to model interactions on multiple spatial or temporal scales (Chavali et al, 2008a; Zhang et al, 2009) have yet to be significantly applied to metabolic GENREs. These extensions of metabolic GENREs will likely yield great insight in the future. Metabolic GENREs represent a bold attempt to characterize the ‘black box' of genotype–phenotype relationships within a fully mechanistic model. These models have already led to many advances, ranging from theoretical to highly practical applications. The five categories outlined in this paper show many of these uses, and explain both the limitations and the promise of metabolic GENREs. As systems biology matures and continues to deepen the marriage between cutting-edge wet-lab technology and sophisticated computational modeling, metabolic GENRE will serve a crucial function in the years to come.
Acknowledgments
We thank Daniel Segre for suggesting the phylogenetic tree analysis, and Andres Pinzon for helping construct the figure. We also acknowledge Erwin Gianchandani, Arvind Chavali, and Ani Manichaikul for helpful comments on the figures. We also thank our funding sources National Science Foundation (NSF) (CAREER Grant 0643548 to JP) and National Institutes of Health (NIH) (GM08715/NIH Biotechnology Training Grant). BOP is funded by the NIGMS and the NIAID.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abuoun M, Suthers PF, Jones GI, Carter BR, Saunders MP, Maranas CD, Woodward MJ, Anjun MF (2009) Genome scale reconstruction of a Salmonella metabolic model: comparison of similarity and differences with a commensal Escherichia coli strain. J Biol Chem (e-pub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen MR, Nielsen ML, Nielsen J (2008) Metabolic model integration of the bibliome, genome, metabolome and reactome of Aspergillus niger. Mol Syst Biol 4: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baart GJ, de Jong G, Philippi M, van't Riet K, van der Pol LA, Beuvery EC, Tramper J, Martens DE (2007a) Scale-up for bulk production of vaccine against meningococcal disease. Vaccine 25: 6399–6408 [DOI] [PubMed] [Google Scholar]
- Baart GJ, Zomer B, de Haan A, van der Pol LA, Beuvery EC, Tramper J, Martens DE (2007b) Modeling Neisseria meningitidis metabolism: from genome to metabolic fluxes. Genome Biol 8: R136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JE, Birnbaum S, Galazzo JL, Khosla C, Shanks JV (1990) Strategies and challenges in metabolic engineering. Ann NY Acad Sci 589: 1–15 [DOI] [PubMed] [Google Scholar]
- Becker SA, Feist AM, Mo ML, Hannum G, Palsson BO, Herrgard MJ (2007) Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat Protoc 2: 727–738 [DOI] [PubMed] [Google Scholar]
- Becker SA, Palsson BO (2005) Genome-scale reconstruction of the metabolic network in Staphylococcus aureus N315: an initial draft to the two-dimensional annotation. BMC Microbiol 5: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker SA, Price ND, Palsson BO (2006) Metabolite coupling in genome-scale metabolic networks. BMC Bioinformatics 7: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beste DJ, Espasa M, Bonde B, Kierzek AM, Stewart GR, McFadden J (2009) The genetic requirements for fast and slow growth in mycobacteria. PLoS One 4: e5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beste DJ, Hooper T, Stewart G, Bonde B, Avignone-Rossa C, Bushell ME, Wheeler P, Klamt S, Kierzek AM, McFadden J (2007) GSMN-TB: a web-based genome-scale network model of Mycobacterium tuberculosis metabolism. Genome Biol 8: R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone DR, Castenholz RW, Garrity GM (2001) In Bergey's Manual of Systematic Bacteriology, Garrity GM (ed), 2 edn New York: Springer [Google Scholar]
- Borenstein E, Kupiec M, Feldman MW, Ruppin E (2008) Large-scale reconstruction and phylogenetic analysis of metabolic environments. Proc Natl Acad Sci USA 105: 14482–14487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodina I, Krabben P, Nielsen J (2005) Genome-scale analysis of Streptomyces coelicolor A3(2) metabolism. Genome Res 15: 820–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle NR, Morgan JA (2009) Flux balance analysis of primary metabolism in Chlamydomonas reinhardtii. BMC Syst Biol 3: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundy JG, Papp B, Harmston R, Browne RA, Clayson EM, Burton N, Reece RJ, Oliver SG, Brindle KM (2007) Evaluation of predicted network modules in yeast metabolism using NMR-based metabolite profiling. Genome Res 17: 510–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgard AP, Nikolaev EV, Schilling CH, Maranas CD (2004) Flux coupling analysis of genome-scale metabolic network reconstructions. Genome Res 14: 301–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgard AP, Pharkya P, Maranas CD (2003) Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng 84: 647–657 [DOI] [PubMed] [Google Scholar]
- Cakir T, Patil KR, Onsan Z, Ulgen KO, Kirdar B, Nielsen J (2006) Integration of metabolome data with metabolic networks reveals reporter reactions. Mol Syst Biol 2: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavali AK, Gianchandani EP, Tung KS, Lawrence MB, Peirce SM, Papin JA (2008a) Characterizing emergent properties of immunological systems with multi-cellular rule-based computational modeling. Trends Immunol 29: 589–599 [DOI] [PubMed] [Google Scholar]
- Chavali AK, Whittemore JD, Eddy JA, Williams KT, Papin JA (2008b) Systems analysis of metabolism in the pathogenic trypanosomatid Leishmania major. Mol Syst Biol 4: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chechik G, Oh E, Rando O, Weissman J, Regev A, Koller D (2008) Activity motifs reveal principles of timing in transcriptional control of the yeast metabolic network. Nat Biotechnol 26: 1251–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P (2006) Toward automatic reconstruction of a highly resolved tree of life. Science 311: 1283–1287 [DOI] [PubMed] [Google Scholar]
- Cimini D, Patil KR, Schiraldi C, Nielsen J (2009) Global transcriptional response of Saccharomyces cerevisiae to the deletion of SDH3. BMC Syst Biol 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contador CA, Rizk ML, Asenjo JA, Liao JC (2009) Ensemble modeling for strain development of L-lysine-producing Escherichia coli. Metab Eng 11: 221–233 [DOI] [PubMed] [Google Scholar]
- Daran-Lapujade P, Jansen ML, Daran JM, van Gulik W, de Winde JH, Pronk JT (2004) Role of transcriptional regulation in controlling fluxes in central carbon metabolism of Saccharomyces cerevisiae. A chemostat culture study. J Biol Chem 279: 9125–9138 [DOI] [PubMed] [Google Scholar]
- David H, Hofmann G, Oliveira AP, Jarmer H, Nielsen J (2006) Metabolic network driven analysis of genome-wide transcription data from Aspergillus nidulans. Genome Biol 7: R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David H, Ozcelik IS, Hofmann G, Nielsen J (2008) Analysis of Aspergillus nidulans metabolism at the genome-scale. BMC Genomics 9: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kievit TR, Iglewski BH (2000) Bacterial quorum sensing in pathogenic relationships. Infect Immun 68: 4839–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher D, Meilijson I, Schuster S, Ruppin E (2008) Can single knockouts accurately single out gene functions? BMC Syst Biol 2: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BO (2007) Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci USA 104: 1777–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte NC, Herrgard MJ, Palsson BO (2004) Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res 14: 1298–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durot M, Bourguignon PY, Schachter V (2009) Genome-scale models of bacterial metabolism: reconstruction and applications. FEMS Microbiol Rev 33: 164–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durot M, Le Fevre F, de Berardinis V, Kreimeyer A, Vallenet D, Combe C, Smidtas S, Salanoubat M, Weissenbach J, Schachter V (2008) Iterative reconstruction of a global metabolic model of Acinetobacter baylyi ADP1 using high-throughput growth phenotype and gene essentiality data. BMC Syst Biol 2: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JS, Palsson BO (1999) Systems properties of the Haemophilus influenzae Rd metabolic genotype. J Biol Chem 274: 17410–17416 [DOI] [PubMed] [Google Scholar]
- Falb M, Muller K, Konigsmaier L, Oberwinkler T, Horn P, von Gronau S, Gonzalez O, Pfeiffer F, Bornberg-Bauer E, Oesterhelt D (2008) Metabolism of halophilic archaea. Extremophiles 12: 177–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famili I, Forster J, Nielsen J, Palsson BO (2003) Saccharomyces cerevisiae phenotypes can be predicted by using constraint-based analysis of a genome-scale reconstructed metabolic network. Proc Natl Acad Sci USA 100: 13134–13139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone DE, Broadie K (2002) Wrestling with pleiotropy: genomic and topological analysis of the yeast gene expression network. Bioessays 24: 267–274 [DOI] [PubMed] [Google Scholar]
- Feist AM, Herrgard MJ, Thiele I, Reed JL, Palsson BO (2009) Reconstruction of biochemical networks in microorganisms. Nat Rev Microbiol 7: 129–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist AM, Palsson BO (2008) The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat Biotechnol 26: 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist AM, Scholten JC, Palsson BO, Brockman FJ, Ideker T (2006) Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri. Mol Syst Biol 2: 2006.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez N, Diaz EE, Amils R, Sanz JL (2008) Analysis of microbial community during biofilm development in an anaerobic wastewater treatment reactor. Microb Ecol 56: 121–132 [DOI] [PubMed] [Google Scholar]
- Filoche SK, Anderson SA, Sissons CH (2004) Biofilm growth of Lactobacillus species is promoted by Actinomyces species and Streptococcus mutans. Oral Microbiol Immunol 19: 322–326 [DOI] [PubMed] [Google Scholar]
- Fong SS, Burgard AP, Herring CD, Knight EM, Blattner FR, Maranas CD, Palsson BO (2005) In silico design and adaptive evolution of Escherichia coli for production of lactic acid. Biotechnol Bioeng 91: 643–648 [DOI] [PubMed] [Google Scholar]
- Fong SS, Palsson BO (2004) Metabolic gene-deletion strains of Escherichia coli evolve to computationally predicted growth phenotypes. Nat Genet 36: 1056–1058 [DOI] [PubMed] [Google Scholar]
- Forster J, Famili I, Fu P, Palsson BO, Nielsen J (2003) Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res 13: 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31: 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P, Ghosh S, Basu K, Das SK (2007) A markov model based analysis of stochastic biochemical systems. Comput Syst Bioinformatics Conf 6: 121–132 [PubMed] [Google Scholar]
- Gianchandani EP, Oberhardt MA, Burgard AP, Maranas CD, Papin JA (2008) Predicting biological system objectives de novo from internal state measurements. BMC Bioinformatics 9: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez O, Gronau S, Falb M, Pfeiffer F, Mendoza E, Zimmer R, Oesterhelt D (2008) Reconstruction, modeling & analysis of Halobacterium salinarum R-1 metabolism. Mol Biosyst 4: 148–159 [DOI] [PubMed] [Google Scholar]
- Guimera R, Sales-Pardo M, Amaral LA (2007) A network-based method for target selection in metabolic networks. Bioinformatics 23: 1616–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R, Papp B, Pal C, Oliver SG, Delneri D (2007) Plasticity of genetic interactions in metabolic networks of yeast. Proc Natl Acad Sci USA 104: 2307–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann M, Kummel A, Ruinatscha R, Panke S (2005) In silico genome-scale reconstruction and validation of the Staphylococcus aureus metabolic network. Biotechnol Bioeng 92: 850–864 [DOI] [PubMed] [Google Scholar]
- Henry CS, Xia F, Stevens R (2009a) Application of high-performance computing to the reconstruction, analysis, and optimization of genome-scale metabolic models. In J Phys Conference Series 180 [Google Scholar]
- Henry CS, Zinner JF, Cohoon MP, Stevens RL (2009b) iBsu1103: a new genome-scale metabolic model of Bacillus subtilis based on SEED annotations. Genome Biol 10: R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrgard MJ, Covert MW, Palsson BO (2004) Reconstruction of microbial transcriptional regulatory networks. Curr Opin Biotechnol 15: 70–77 [DOI] [PubMed] [Google Scholar]
- Herrgard MJ, Lee BS, Portnoy V, Palsson BO (2006) Integrated analysis of regulatory and metabolic networks reveals novel regulatory mechanisms in Saccharomyces cerevisiae. Genome Res 16: 627–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrgard MJ, Swainston N, Dobson P, Dunn WB, Arga KY, Arvas M, Bluthgen N, Borger S, Costenoble R, Heinemann M, Hucka M, Le Novere N, Li P, Liebermeister W, Mo ML, Oliveira AP, Petranovic D, Pettifer S, Simeonidis E, Smallbone K et al. (2008) A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat Biotechnol 26: 1155–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka T, Furihata K, Ishikawa J, Yamashita H, Itoh N, Seto H, Dairi T (2008) An alternative menaquinone biosynthetic pathway operating in microorganisms. Science 321: 1670–1673 [DOI] [PubMed] [Google Scholar]
- Hong SH, Kim JS, Lee SY, In YH, Choi SS, Rih JK, Kim CH, Jeong H, Hur CG, Kim JJ (2004) The genome sequence of the capnophilic rumen bacterium Mannheimia succiniciproducens. Nat Biotechnol 22: 1275–1281 [DOI] [PubMed] [Google Scholar]
- Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L (2001) Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292: 929–934 [DOI] [PubMed] [Google Scholar]
- Izallalen M, Mahadevan R, Burgard A, Postier B, Didonato R Jr, Sun J, Schilling CH, Lovley DR (2008) Geobacter sulfurreducens strain engineered for increased rates of respiration. Metab Eng 10: 267–275 [DOI] [PubMed] [Google Scholar]
- Jamshidi N, Palsson BO (2007) Investigating the metabolic capabilities of Mycobacterium tuberculosis H37Rv using the in silico strain iNJ661 and proposing alternative drug targets. BMC Syst Biol 1: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamshidi N, Palsson BO (2008) Formulating genome-scale kinetic models in the post-genome era. Mol Syst Biol 4: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL (2000) The large-scale organization of metabolic networks. Nature 407: 651–654 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Pjesivac-Grbovic J, Cantrell C, Freyer JP (2005) A multiscale model for avascular tumor growth. Biophys J 89: 3884–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M (2006) From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34: D354–D357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TY, Kim HU, Park JM, Song H, Kim JS, Lee SY (2007) Genome-scale analysis of Mannheimia succiniciproducens metabolism. Biotechnol Bioeng 97: 657–671 [DOI] [PubMed] [Google Scholar]
- Kim TY, Sohn SB, Kim HU, Lee SY (2008) Strategies for systems-level metabolic engineering. Biotechnol J 3: 612–623 [DOI] [PubMed] [Google Scholar]
- Kjeldsen KR, Nielsen J (2009) In silico genome-scale reconstruction and validation of the Corynebacterium glutamicum metabolic network. Biotechnol Bioeng 102: 583–597 [DOI] [PubMed] [Google Scholar]
- Klipp E, Nordlander B, Kruger R, Gennemark P, Hohmann S (2005) Integrative model of the response of yeast to osmotic shock. Nat Biotechnol 23: 975–982 [DOI] [PubMed] [Google Scholar]
- Kuepfer L, Sauer U, Blank LM (2005) Metabolic functions of duplicate genes in Saccharomyces cerevisiae. Genome Res 15: 1421–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kun A, Papp B, Szathmary E (2008) Computational identification of obligatorily autocatalytic replicators embedded in metabolic networks. Genome Biol 9: R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leang C, Krushkal J, Ueki T, Puljic M, Sun J, Juarez K, Nunez C, Reguera G, DiDonato R, Postier B, Adkins RM, Lovley DR (2009) Genome-wide analysis of the RpoN regulon in Geobacter sulfurreducens. BMC Genomics 10: 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DS, Burd H, Liu J, Almaas E, Wiest O, Barabasi AL, Oltvai ZN, Kapatral V (2009) Comparative genome-scale metabolic reconstruction and flux balance analysis of multiple Staphylococcus aureus genomes identify novel anti-microbial drug targets. J Bacteriol 191: 4015–4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Yun H, Feist AM, Palsson BO, Lee SY (2008a) Genome-scale reconstruction and in silico analysis of the Clostridium acetobutylicum ATCC 824 metabolic network. Appl Microbiol Biotechnol 80: 849–862 [DOI] [PubMed] [Google Scholar]
- Lee JM, Gianchandani EP, Eddy JA, Papin JA (2008b) Dynamic analysis of integrated signaling, metabolic, and regulatory networks. PLoS Comput Biol 4: e1000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Gianchandani EP, Papin JA (2006) Flux balance analysis in the era of metabolomics. Brief Bioinform 7: 140–150 [DOI] [PubMed] [Google Scholar]
- Lee SM, Ender M, Adhikari R, Smith JM, Berger-Bachi B, Cook GM (2007) Fitness cost of staphylococcal cassette chromosome mec in methicillin-resistant Staphylococcus aureus by way of continuous culture. Antimicrob Agents Chemother 51: 1497–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Kim JM, Song H, Lee JW, Kim TY, Jang YS (2008c) From genome sequence to integrated bioprocess for succinic acid production by Mannheimia succiniciproducens. Appl Microbiol Biotechnol 79: 11–22 [DOI] [PubMed] [Google Scholar]
- Ma H, Sorokin A, Mazein A, Selkov A, Selkov E, Demin O, Goryanin I (2007) The Edinburgh human metabolic network reconstruction and its functional analysis. Mol Syst Biol 3: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan R, Bond DR, Butler JE, Esteve-Nunez A, Coppi MV, Palsson BO, Schilling CH, Lovley DR (2006) Characterization of metabolism in the Fe(III)-reducing organism Geobacter sulfurreducens by constraint-based modeling. Appl Environ Microbiol 72: 1558–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan R, Lovley DR (2008) The degree of redundancy in metabolic genes is linked to mode of metabolism. Biophys J 94: 1216–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan R, Schilling CH (2003) The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng 5: 264–276 [DOI] [PubMed] [Google Scholar]
- Manichaikul A, Ghamsari L, Hom EF, Lin C, Murray RR, Chang RL, Balaji S, Hao T, Shen Y, Chavali AK, Thiele I, Yang X, Fan C, Mello E, Hill DE, Vidal M, Salehi-Ashtiani K, Papin JA (2009) Metabolic network analysis integrated with transcript verification for sequenced genomes. Nat Methods 6: 589–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J (1978) Optimization theory in evolution. Ann Rev Ecol Syst 9: 31–56 [Google Scholar]
- Mazumdar V, Snitkin ES, Amar S, Segre D (2009) Metabolic network model of a human oral pathogen. J Bacteriol 191: 74–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz-Oron S, Aharoni A, Ruppin E, Shlomi T (2009) Network-based prediction of metabolic enzymes' subcellular localization. Bioinformatics 25: i247–i252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo ML, Palsson BO, Herrgard MJ (2009) Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst Biol 3: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navid A, Almaas E (2009) Genome-scale reconstruction of the metabolic network in Yersinia pestis, strain 91001. Mol Biosyst 5: 368–375 [DOI] [PubMed] [Google Scholar]
- Nevoigt E (2008) Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiol Mol Biol Rev 72: 379–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa T, Gulbahce N, Motter AE (2008) Spontaneous reaction silencing in metabolic optimization. PLoS Comput Biol 4: e1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales J, Palsson BO, Thiele I (2008) A genome-scale metabolic reconstruction of Pseudomonas putida KT2440: iJN746 as a cell factory. BMC Syst Biol 2: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nookaew I, Jewett MC, Meechai A, Thammarongtham C, Laoteng K, Cheevadhanarak S, Nielsen J, Bhumiratana S (2008) The genome-scale metabolic model iIN800 of Saccharomyces cerevisiae and its validation: a scaffold to query lipid metabolism. BMC Syst Biol 2: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notebaart RA, Teusink B, Siezen RJ, Papp B (2008) Co-regulation of metabolic genes is better explained by flux coupling than by network distance. PLoS Comput Biol 4: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhardt MA, Puchalka J, Fryer KE, Martins dos Santos VA, Papin JA (2008) Genome-scale metabolic network analysis of the opportunistic pathogen Pseudomonas aeruginosa PAO1. J Bacteriol 190: 2790–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh YK, Palsson BO, Park SM, Schilling CH, Mahadevan R (2007) Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data. J Biol Chem 282: 28791–28799 [DOI] [PubMed] [Google Scholar]
- Oliveira AP, Nielsen J, Forster J (2005) Modeling Lactococcus lactis using a genome-scale flux model. BMC Microbiol 5: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagiotou G, Andersen MR, Grotkjaer T, Regueira TB, Hofmann G, Nielsen J, Olsson L (2008) Systems analysis unfolds the relationship between the phosphoketolase pathway and growth in Aspergillus nidulans. PLoS ONE 3: e3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagiotou G, Andersen MR, Grotkjaer T, Regueira TB, Nielsen J, Olsson L (2009) Studies of the production of fungal polyketides in Aspergillus nidulans by using systems biology tools. Appl Environ Microbiol 75: 2212–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papin JA, Price ND, Edwards JS, Palsson BB (2002a) The genome-scale metabolic extreme pathway structure in Haemophilus influenzae shows significant network redundancy. J Theor Biol 215: 67–82 [DOI] [PubMed] [Google Scholar]
- Papin JA, Price ND, Palsson BO (2002b) Extreme pathway lengths and reaction participation in genome-scale metabolic networks. Genome Res 12: 1889–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papin JA, Stelling J, Price ND, Klamt S, Schuster S, Palsson BO (2004) Comparison of network-based pathway analysis methods. Trends Biotechnol 22: 400–405 [DOI] [PubMed] [Google Scholar]
- Papp B, Teusink B, Notebaart RA (2009) A critical view of metabolic network adaptations. HFSP J 3: 24–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Lee SY (2008) Towards systems metabolic engineering of microorganisms for amino acid production. Curr Opin Biotechnol 19: 454–460 [DOI] [PubMed] [Google Scholar]
- Pastink MI, Teusink B, Hols P, Visser S, de Vos WM, Hugenholtz J (2009) Genome-scale model of Streptococcus thermophilus LMG18311 for metabolic comparison of lactic acid bacteria. Appl Environ Microbiol 75: 3627–3633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil KR, Nielsen J (2005) Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc Natl Acad Sci USA 102: 2685–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer T, Schuster S, Bonhoeffer S (2001) Cooperation and competition in the evolution of ATP-producing pathways. Science 292: 504–507 [DOI] [PubMed] [Google Scholar]
- Price ND, Papin JA, Palsson BO (2002) Determination of redundancy and systems properties of the metabolic network of Helicobacter pylori using genome-scale extreme pathway analysis. Genome Res 12: 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price ND, Reed JL, Papin JA, Famili I, Palsson BO (2003) Analysis of metabolic capabilities using singular value decomposition of extreme pathway matrices. Biophys J 84: 794–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S, Avila-Campillo I, Aldridge C, Srinivasan A, Dimitrov K, Siegel AF, Galitski T (2004) Control of yeast filamentous-form growth by modules in an integrated molecular network. Genome Res 14: 380–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchalka J, Oberhardt MA, Godinho M, Bielecka A, Regenhardt D, Timmis KN, Papin JA, Martins dos Santos VA (2008) Genome-scale reconstruction and analysis of the Pseudomonas putida KT2440 metabolic network facilitates applications in biotechnology. PLoS Comput Biol 4: e1000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quek LE, Nielsen LK (2008) On the reconstruction of the Mus musculus genome-scale metabolic network model. Genome Inform 21: 89–100 [PubMed] [Google Scholar]
- Raghevendran V, Patil KR, Olsson L, Nielsen J (2006) Hap4 is not essential for activation of respiration at low specific growth rates in Saccharomyces cerevisiae. J Biol Chem 281: 12308–12314 [DOI] [PubMed] [Google Scholar]
- Raghunathan A, Reed J, Shin S, Palsson B, Daefler S (2009) Constraint-based analysis of metabolic capacity of Salmonella typhimurium during host-pathogen interaction. BMC Syst Biol 3: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JL, Famili I, Thiele I, Palsson BO (2006) Towards multidimensional genome annotation. Nat Rev Genet 7: 130–141 [DOI] [PubMed] [Google Scholar]
- Resendis-Antonio O, Reed JL, Encarnacion S, Collado-Vides J, Palsson BO (2007) Metabolic reconstruction and modeling of nitrogen fixation in Rhizobium etli. PLoS Comput Biol 3: 1887–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel K, Hentzer M, Geisenberger O, Huber B, Steidle A, Wu H, Hoiby N, Givskov M, Molin S, Eberl L (2001) N-acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 147: 3249–3262 [DOI] [PubMed] [Google Scholar]
- Risso C, Van Dien SJ, Orloff A, Lovley DR, Coppi MV (2008) Elucidation of an alternate isoleucine biosynthesis pathway in Geobacter sulfurreducens. J Bacteriol 190: 2266–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson SH, Smith CK, Langhans AL, McLinden SE, Oberhardt MA, Jakab KR, Dzamba B, DeSimone DW, Papin JA, Peirce SM (2007) Multiscale computational analysis of Xenopus laevis morphogenesis reveals key insights of systems-level behavior. BMC Syst Biol 1: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokhlenko O, Shlomi T, Sharan R, Ruppin E, Pinter RY (2007) Constraint-based functional similarity of metabolic genes: going beyond network topology. Bioinformatics 23: 2139–2146 [DOI] [PubMed] [Google Scholar]
- Samal A, Singh S, Giri V, Krishna S, Raghuram N, Jain S (2006) Low degree metabolites explain essential reactions and enhance modularity in biological networks. BMC Bioinformatics 7: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibe TD, Mahadevan R, Fang Y, Garg S, Long PE, Lovley DR (2009) Coupling a genome-scale metabolic model with a reactive transport model to describe in situ uranium bioremediation. Microb Biotechnol 2: 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherens B, Goffeau A (2004) The uses of genome-wide yeast mutant collections. Genome Biol 5: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling CH, Covert MW, Famili I, Church GM, Edwards JS, Palsson BO (2002) Genome-scale metabolic model of Helicobacter pylori 26695. J Bacteriol 184: 4582–4593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling CH, Palsson BO (2000) Assessment of the metabolic capabilities of Haemophilus influenzae Rd through a genome-scale pathway analysis. J Theor Biol 203: 249–283 [DOI] [PubMed] [Google Scholar]
- Schuetz R, Kuepfer L, Sauer U (2007) Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol Syst Biol 3: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura D, Mahadevan R, Juarez K, Lovley DR (2008) Computational and experimental analysis of redundancy in the central metabolism of Geobacter sulfurreducens. PLoS Comput Biol 4: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvarasu S, Wong VV, Karimi IA, Lee DY (2009) Elucidation of metabolism in hybridoma cells grown in fed-batch culture by genome-scale modeling. Biotechnol Bioeng 102: 1494–1504 [DOI] [PubMed] [Google Scholar]
- Senger RS, Papoutsakis ET (2008a) Genome-scale model for Clostridium acetobutylicum: Part I. Metabolic network resolution and analysis. Biotechnol Bioeng 101: 1036–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger RS, Papoutsakis ET (2008b) Genome-scale model for Clostridium acetobutylicum: Part II. Development of specific proton flux states and numerically determined sub-systems. Biotechnol Bioeng 101: 1053–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh K, Forster J, Nielsen LK (2005) Modeling hybridoma cell metabolism using a generic genome-scale metabolic model of Mus musculus. Biotechnol Prog 21: 112–121 [DOI] [PubMed] [Google Scholar]
- Shinfuku Y, Sorpitiporn N, Sono M, Furusawa C, Hirasawa T, Shimizu H (2009) Development and experimental verification of a genome-scale metabolic model for Corynebacterium glutamicum. Microb Cell Fact 8: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomi T, Cabili MN, Herrgard MJ, Palsson BO, Ruppin E (2008) Network-based prediction of human tissue-specific metabolism. Nat Biotechnol 26: 1003–1010 [DOI] [PubMed] [Google Scholar]
- Shlomi T, Cabili MN, Ruppin E (2009) Predicting metabolic biomarkers of human inborn errors of metabolism. Mol Syst Biol 5: 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomi T, Herrgard M, Portnoy V, Naim E, Palsson BO, Sharan R, Ruppin E (2007) Systematic condition-dependent annotation of metabolic genes. Genome Res 17: 1626–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RS (2003) Darwin to DNA, molecules to morphology: the end of classical population genetics and the road ahead. Genome 46: 938–942 [DOI] [PubMed] [Google Scholar]
- Snitkin ES, Dudley AM, Janse DM, Wong K, Church GM, Segre D (2008) Model-driven analysis of experimentally determined growth phenotypes for 465 yeast gene deletion mutants under 16 different conditions. Genome Biol 9: R140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Kim TY, Choi BK, Choi SJ, Nielsen LK, Chang HN, Lee SY (2008) Development of chemically defined medium for Mannheimia succiniciproducens based on its genome sequence. Appl Microbiol Biotechnol 79: 263–272 [DOI] [PubMed] [Google Scholar]
- Stark J, Callard R, Hubank M (2003) From the top down: towards a predictive biology of signalling networks. Trends Biotechnol 21: 290–293 [DOI] [PubMed] [Google Scholar]
- Stephanopoulos G, Vallino JJ (1991) Network rigidity and metabolic engineering in metabolite overproduction. Science 252: 1675–1681 [DOI] [PubMed] [Google Scholar]
- Stevens MJ, Wiersma A, de Vos WM, Kuipers OP, Smid EJ, Molenaar D, Kleerebezem M (2008) Improvement of Lactobacillus plantarum aerobic growth as directed by comprehensive transcriptome analysis. Appl Environ Microbiol 74: 4776–4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolyar S, Van Dien S, Hillesland KL, Pinel N, Lie TJ, Leigh JA, Stahl DA (2007) Metabolic modeling of a mutualistic microbial community. Mol Syst Biol 3: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Sayyar B, Butler JE, Pharkya P, Fahland TR, Famili I, Schilling CH, Lovley DR, Mahadevan R (2009) Genome-scale constraint-based modeling of Geobacter metallireducens. BMC Syst Biol 3: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthers PF, Dasika MS, Kumar VS, Denisov G, Glass JI, Maranas CD (2009) A genome-scale metabolic reconstruction of Mycoplasma genitalium, iPS189. PLoS Comput Biol 5: e1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teusink B, Wiersma A, Jacobs L, Notebaart RA, Smid EJ (2009) Understanding the adaptive growth strategy of Lactobacillus plantarum by in silico optimisation. PLoS Comput Biol 5: e1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teusink B, Wiersma A, Molenaar D, Francke C, de Vos WM, Siezen RJ, Smid EJ (2006) Analysis of growth of Lactobacillus plantarum WCFS1 on a complex medium using a genome-scale metabolic model. J Biol Chem 281: 40041–40048 [DOI] [PubMed] [Google Scholar]
- Thiele I, Vo TD, Price ND, Palsson BO (2005) Expanded metabolic reconstruction of Helicobacter pylori (iIT341 GSM/GPR): an in silico genome-scale characterization of single- and double-deletion mutants. J Bacteriol 187: 5818–5830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thykaer J, Andersen MR, Baker SE (2009) Essential pathway identification: from in silico analysis to potential antifungal targets in Aspergillus fumigatus. Med Mycol 47(Suppl 1): S80–S87 [DOI] [PubMed] [Google Scholar]
- Usaite R, Patil KR, Grotkjaer T, Nielsen J, Regenberg B (2006) Global transcriptional and physiological responses of Saccharomyces cerevisiae to ammonium, L-alanine, or L-glutamine limitation. Appl Environ Microbiol 72: 6194–6203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeramani B, Bader JS (2009) Metabolic flux correlations, genetic interactions, and disease. J Comput Biol 16: 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhedkar KD, Raman K, Chandra NR, Vishveshwara S (2007) Metabolome based reaction graphs of M. tuberculosis and M. leprae: a comparative network analysis. PLoS ONE 2: e881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieites JM, Guazzaroni ME, Beloqui A, Golyshin PN, Ferrer M (2009) Metagenomics approaches in systems microbiology. FEMS Microbiol Rev 33: 236–255 [DOI] [PubMed] [Google Scholar]
- Vo TD, Greenberg HJ, Palsson BO (2004) Reconstruction and functional characterization of the human mitochondrial metabolic network based on proteomic and biochemical data. J Biol Chem 279: 39532–39540 [DOI] [PubMed] [Google Scholar]
- Vo TD, Paul Lee WN, Palsson BO (2007) Systems analysis of energy metabolism elucidates the affected respiratory chain complex in Leigh′s syndrome. Mol Genet Metab 91: 15–22 [DOI] [PubMed] [Google Scholar]
- Vongsangnak W, Olsen P, Hansen K, Krogsgaard S, Nielsen J (2008) Improved annotation through genome-scale metabolic modeling of Aspergillus oryzae. BMC Genomics 9: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waliszewski P, Molski M, Konarski J (1998) On the holistic approach in cellular and cancer biology: nonlinearity, complexity, and quasi-determinism of the dynamic cellular network. J Surg Oncol 68: 70–78 [DOI] [PubMed] [Google Scholar]
- Westerhoff HV, Palsson BO (2004) The evolution of molecular biology into systems biology. Nat Biotechnol 22: 1249–1252 [DOI] [PubMed] [Google Scholar]
- Wright J, Wagner A (2008) Exhaustive identification of steady state cycles in large stoichiometric networks. BMC Syst Biol 2: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelle RM, de Hulster E, van Winden WA, de Waard P, Dijkema C, Winkler AA, Geertman JM, van Dijken JP, Pronk JT, van Maris AJ (2008) Malic acid production by Saccharomyces cerevisiae: engineering of pyruvate carboxylation, oxaloacetate reduction, and malate export. Appl Environ Microbiol 74: 2766–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang Z, Sagotsky JA, Deisboeck TS (2009) Multiscale agent-based cancer modeling. J Math Biol 58: 545–559 [DOI] [PubMed] [Google Scholar]
- Zhu H, Huang S, Dhar P (2004) The next step in systems biology: simulating the temporospatial dynamics of molecular network. Bioessays 26: 68–72 [DOI] [PubMed] [Google Scholar]