Abstract
Bacteria communicate using secreted chemical signaling molecules called autoinducers in a process known as quorum sensing. The quorum-sensing network of the marine bacterium Vibrio harveyi uses three autoinducers, each known to encode distinct ecological information. Yet how cells integrate and interpret the information contained within these three autoinducer signals remains a mystery. Here, we develop a new framework for analyzing signal integration on the basis of information theory and use it to analyze quorum sensing in V. harveyi. We quantify how much the cells can learn about individual autoinducers and explain the experimentally observed input–output relation of the V. harveyi quorum-sensing circuit. Our results suggest that the need to limit interference between input signals places strong constraints on the architecture of bacterial signal-integration networks, and that bacteria probably have evolved active strategies for minimizing this interference. Here, we analyze two such strategies: manipulation of autoinducer production and feedback on receptor number ratios.
Keywords: biophysics, information theory, quorum sensing, signal integration, signal transduction
Introduction
Unicellular organisms live in complex and dynamic environments. They sense and respond to both external environmental cues and to each other through quorum sensing, that is, cell-to-cell communication. Adapting to changing environments often requires cells to simultaneously integrate information from multiple environmental inputs, and cells have developed elaborate signaling networks to accomplish this feat. However, the design principles underlying the architectures of these networks remain largely mysterious. For example, in the model quorum-sensing bacterium Vibrio harveyi, three chemical communication signals are integrated to regulate gene expression, but the logic and mechanism underlying this integration are poorly understood. Such open questions highlight the need for new conceptual and theoretical tools to supplement ongoing experimental work. Here, we present a new theoretical framework for understanding signal integration based on information theory (Shannon, 1948) and we use it to study information processing in the V. harveyi quorum-sensing circuit.
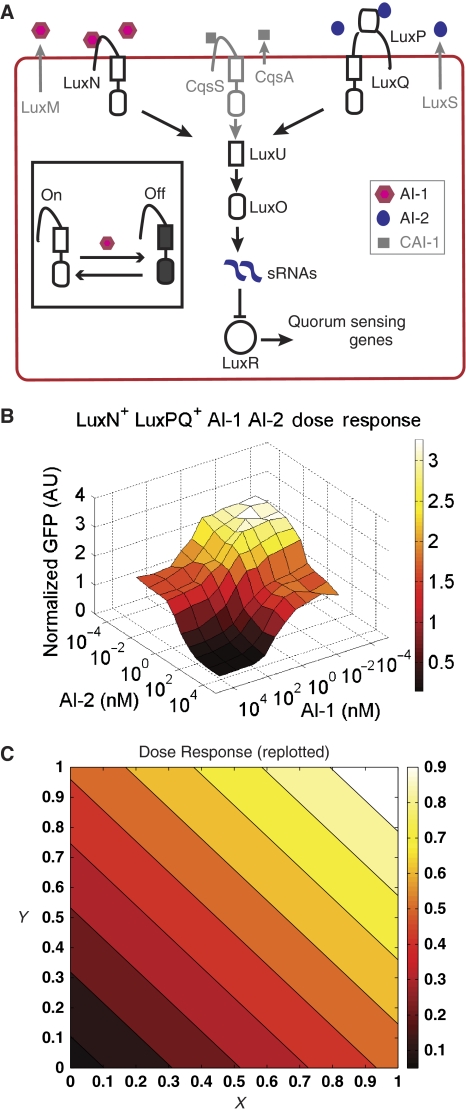
Quorum sensing is widespread in the bacterial world and can occur both within and between bacterial species, and even between bacteria and their eukaryotic hosts (Waters and Bassler, 2005). Quorum sensing enables bacteria to alter their behavior depending on the number and/or species of bacteria present and is important for a variety of collective behaviors, such as biofilm formation, bioluminescence, virulence, as well as stress response (Waters and Bassler, 2005, 2006; Bassler and Losick, 2006). The V. harveyi quorum-sensing circuit is among the best characterized of all quorum-sensing networks (Figure 1A). V. harveyi produces and detects three chemical signaling molecules called autoinducers (AIs), AI-1, CAI-1, and AI-2. Although AI-1 is produced only by V. harveyi, CAI-1 is produced by other Vibrio species, and AI-2 is produced by a large variety of both Gram-negative and Gram-positive bacteria and probably functions as a universal signaling molecule. Thus, the use of multiple AIs potentially provides bacteria with information about the local density of V. harveyi, all Vibrio species, and total bacteria (Waters and Bassler, 2005). Sensory information from the three AIs is channeled through a common phosphorelay (see Figure 1). The three autoinducers, AI-1, AI-2, and CAI-1, are detected by cognate transmembrane receptors, LuxN, LuxPQ, and CqsS, respectively (Henke and Bassler, 2004), and they collectively control production of the master quorum-sensing transcriptional regulator LuxR (see Figure 1A; Tu and Bassler, 2007).
Figure 1.
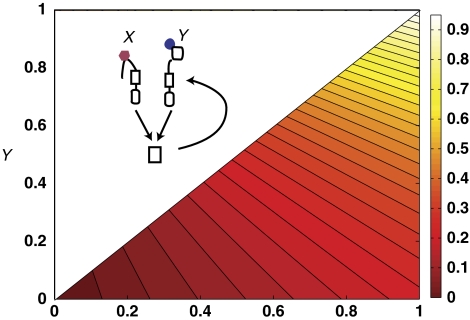
Information theoretic approach to signal integration in the Vibrio harveyi quorum-sensing circuit. (A) V. harveyi produces three distinct quorum-sensing signaling molecules, called (AIs), which are all detected by a single phosphorelay circuit that controls the expression of downstream target genes. Each signaling molecule, AI-1 (red hexagons), AI-2 (blue ovals), and CAI-1 (gray squares), is detected by a cognate receptor. The receptors phosphorylate a shared phosphorelay protein, LuxU, which in turn phosphorylates LuxO. In the absence of AIs, LuxO is phosphorylated and activates expression of genes encoding five small regulatory RNAs (sRNAs) that work in conjuction with Hfq to destabilize the mRNA of LuxR, the master regulator of quorum-sensing genes. In the presence of the AIs, LuxO is not phosphorylated, the sRNAs are not produced, and LuxR is expressed. (Inset) The receptors can exist in two states: a kinase ‘on' state and kinase ‘off' state with ligand binding favoring the ‘off' state. (B) Dose–response surface of V. harveyi to various combinations of AI-1 and AI-2 reproduced from Figure 3C in Long et al (2009). Each vertex of the grid is the averaged normalized GFP fluorescence intensity obtained from a population of 100 cells exposed to the specified AI-1 and AI-2 concentrations using a qrr4-gfp transcriptional reporter fusion that is activated by phosphorylated LuxO. (C) Dose–response surface re-plotted as a contour plot. The straight lines are constant output Z contours as function of the receptor on-state probabilities X ≈ 1 /( 1 + [AI- 1] / KIAI-1) and Y ≈ 1 /( 1 + [AI- 2] / KIAI-2). Output is normalized by the maximum of Z.
As all information about the AIs is channeled through a common phosphorelay, it is unclear how much bacteria can learn about each individual input. Even less clear is how the architecture and kinetic parameters (e.g. kinase and phosphatase rates) of the quorum-sensing network affect its signal-transduction properties. To address these questions, we have developed a new mathematical framework for analyzing signal integration in cells on the basis of information theory (Shannon, 1948; MacKay, 2003). Information theory provides a natural language for formulating questions about information processing and signal integration. It is used extensively in engineering to model signaling in man-made communication devices and has also proven to be a powerful tool in neuroscience (Rieke et al, 1997; Borst and Theunissen, 1999). Very recently, information theory has been applied to genetic networks to study development in fruit fly embryos and to investigate properties of small stochastic biochemical networks (Ziv et al, 2007; Tkacik et al, 2008a, 2008b; Tostevin and ten Wolde, 2009; Walczak et al, 2009). Here, we adapt information theory to study a biological circuit with multiple inputs and a single output.
One of the advantages of using information theory to describe cellular signaling is that, in principle, no detailed knowledge of the components and kinetic parameters that constitute the signaling circuit is required. Rather, the signaling circuit is modeled by its input–output relationship, often called the transfer function, which describes how the output varies as a function of the input signals. The transfer function of the V. harveyi quorum-sensing circuit was recently measured using single-cell fluorescence microscopy (see Figure 1B and Long et al, 2009). In this study, we consider a class of transfer functions for the V. harveyi quorum-sensing circuit on the basis of a simple model of V. harveyi signal transduction consistent with the aforementioned experiments. We show that many features of the experimentally measured transfer function can be understood using information theory. We argue that our analysis of the V. harveyi quorum-sensing network provides insight into broader design principles applicable to many signal integration networks in cells.
Results and discussion
Overview of the information theory formalism
A central concept in information theory is mutual information (see below for a mathematical definition). The mutual information is a symmetric measure of the correlation between inputs and outputs, and measures how much one can learn on average about the input from the output, and vice versa. In the context of cellular biology, mutual information provides a measure of how much a cell can learn about external signals. Cells constantly sense their environments and adjust their gene expression accordingly. This requires cells to faithfully convey information about external signals, even in the presence of noise. The mutual information quantifies, in bits, how much bacteria learn about an external signal (e.g. autoinducer). A bit is the standard unit of information and is defined as the quantity of information required to distinguish two mutually exclusive but equally probable states from each other. Consequently, calculating the mutual information between each input signal and the output in a multi-input signaling system, such as the V. harveyi quorum-sensing circuit, allows us to quantify how much bacteria can learn on average about each input from the shared output.
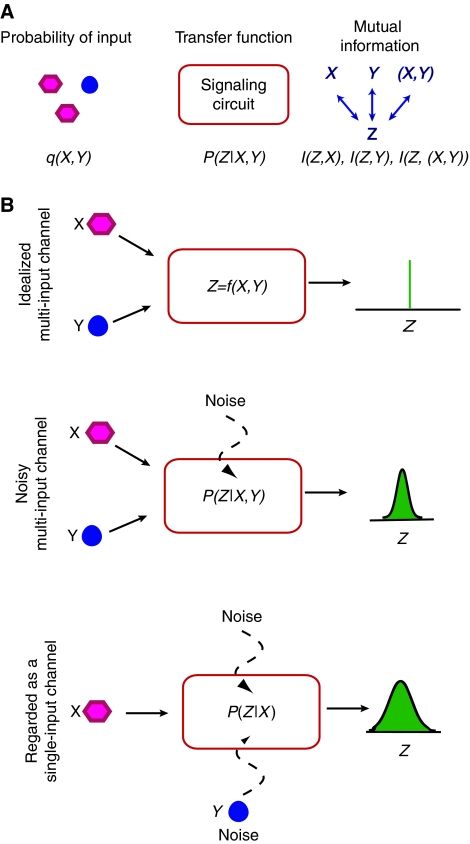
There are three basic components of our information theory formalism: (1) a model of the signaling circuit, (2) a statistical model (a ‘prior') for the likelihood of encountering a particular input signal, and (3) the various mutual information between the inputs and the output (Figure 2A). We now discuss each of these components in greater detail.
Model for the signaling circuit: Signal integration in bacteria commonly occurs through two-component signal-transduction systems, often including a phosphorelay. In addition, the time-scales on which external input signals, such as autoinducers, vary is much slower than the typical time scales for phosphorylation/dephosphorylation in the relay. This separation of time scales allows us to model the signaling circuit by its steady-state properties, with the inputs assumed to be constant in time. Furthermore, for simplicity and to facilitate comparison with experiment, we limit our considerations to multi-input circuits with two input signals, denoted X and Y, and a single output, denoted Z. The generalization to circuits with more than two inputs is straightforward. For an idealized multi-input channel without biochemical noise, an input (X, Y) gives rise to a single output Z=f(X, Y). However, signaling fidelity is generally limited by biochemical noise so that a single input can give rise to many outputs (McAdams and Arkin, 1997; Elowitz et al, 2002; Ozbudak et al, 2002; Swain et al, 2002). Noise can arise from both the stochastic nature of biochemical reactions, often called intrinsic noise, and from other cellular variability, often called extrinsic noise. Although the former can be reduced by temporal averaging, the latter often cannot. Consequently, we characterize a signaling circuit by a noisy transfer function, P(Z∣X, Y), which gives the probability of an output, Z, as a function on the inputs, X and Y (see Figure 2B).
Prior distribution on input signals: To quantify information transmission, it is necessary to define a prior distribution of input signals, q(X,Y). This prior represents the probability that a bacterium receives an input signal (X, Y). For example, q(X, Y) could be the distribution of input signals that a typical bacterium would encounter in its natural habitat (Tkacik et al, 2008a).
Mutual information: Information transmission in a signaling circuit can be quantified by the mutual information between the input and output signals. For a circuit with two inputs, X and Y, and a single output, Z, there are three distinct mutual information, I(Z, X), I(Z, Y), and I(Z,(X, Y)). They measure, respectively, how much can be learned on average about the inputs X, Y, and (X, Y), from the output Z. I(Z,(X, Y)) measures the total information transmitted about both signals X and Y, but is a poor measure of how much can be learned about individual inputs. By contrast, I(Z, X) and I(Z, Y) measure how much can be learned about the individual inputs, but these are often not reflective of total information transmission.
Figure 2.
(A) The mutual informations I(Z, X), I(Z, Y), and I(Z,(X, Y)) measure how much one can learn about the inputs, for example, autoinducer levels, X, Y, and (X, Y), respectively, from the output Z, for example, LuxR level. Mutual information is a function of the prior, q(X, Y), that is, the a priori probability of a given input (X, Y), and of the probabilistic transfer function P(Z∣X, Y) of the signaling circuit. (B) (Top) For an idealized multi-input channel without noise, an input (X, Y) gives rise to a single output Z. (Middle) In the presence of noise, a single input can give rise to many outputs with a distribution described by the noisy-transfer function, P(Z∣X, Y). (Bottom) When viewed as single-input channel with input X and output Z, the second signal, Y, effectively acts as an additional source of noise.
Mutual information is a statistical quantity that measures the average amount that can be learned about an input, with the average taken over the input prior, q(X, Y). Consequently, all three mutual informations depend on both the transfer function of the signaling circuit, P(Z∣X, Y) and on the prior, q(X, Y). The relevant expressions are (Shannon, 1948; MacKay, 2003).
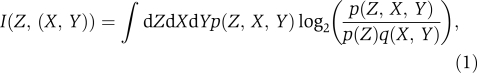 |
with the joint probability p(Z, X, Y)=P(Z∣X, Y)q(X, Y) and p(Z = ∫ dXdYP(Z|X,Y)q(X,Y), and
with p(Z,X) = ∫ dYP(Z|X,Y)q(X,Y) and p(Z) as above, and the expression for I(Z, Y) is the same as I(Z, X) except with X and Y interchanged I.
Information transmission in the V. harveyi quorum-sensing circuit
Applying information theory to the V. harveyi quorum-sensing circuit requires explicit models for the transfer function and the prior. The input–output relationship for the V. harveyi quorum-sensing circuit was experimentally quantified in genetically engineered strains lacking the CAI-1-CqsS pathway to study the integration of signals from autoinducers AI-1 and AI-2 (Long et al, 2009). In these experiments, strains were engineered with gfp fused to the promoter of qrr4, which is one of the genes encoding the quorum-sensing small RNAs activated by phospho-LuxO. As shown in Figure 1, signals from AI-1 and AI-2 are already integrated at this stage of the quorum-sensing circuit. Recent experimental and theoretical study suggest that the detection of AIs by their cognate receptors (e.g. LuxN, LuxPQ) can be understood using a simple two-state model in which receptors exists in two states: a low kinase activity state (‘off') and a high kinase activity state (‘on') (Keymer et al, 2006; Swem et al, 2008; Supplementary information). In addition to their kinase activities, the quorum-sensing receptors have a strong state-independent phosphatase activity (Long et al, 2009). AIs act by binding to a receptor and decreasing the probability that the receptor is in the high kinase activity, ‘on' state. Thus, specifying the external concentration of an AI in the environment is equivalent to specifying the probability that the corresponding receptor is in its high activity state. Hence, we take the input signals, X and Y, to be the probabilities that LuxN and LuxPQ, respectively, are in their kinase-active states. An advantage of this formulation is that input signals are bounded between 0 and 1. These probabilities are related to the autoinducer concentrations through the formulas
 |
(see Supplementary information).
Motivated by experiment (Long et al, 2009), we model the mean response of the V. harveyi quorum-sensing circuit using the expression
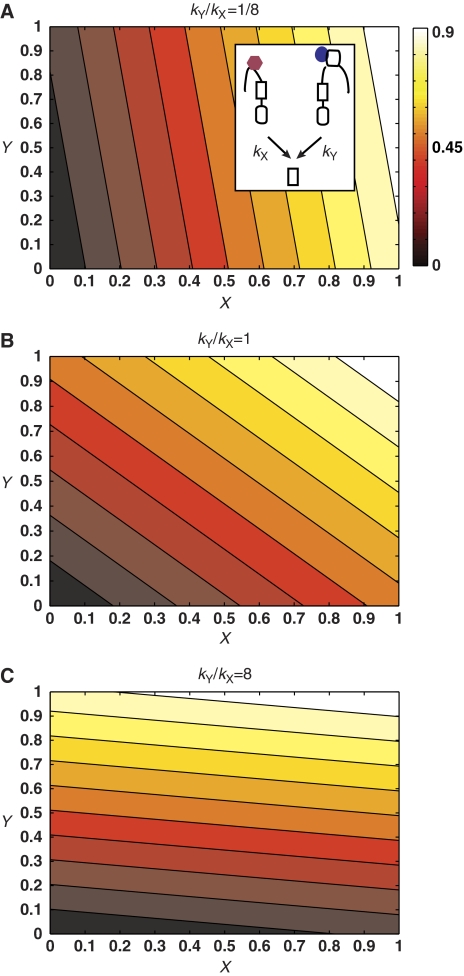
with Z the output signal, that is, the fraction of phospho-LuxO, kX the total kinase rate of active LuxN, kY the total kinase rate of active LuxPQ, and p the total phosphatase rate from both receptors. The second, approximate expression, applies because, for the quorum-sensing circuit, the total phosphatase rate is much larger than the maximal total kinase rate, p≫kX+kY. The mean transfer function f(X, Y) is plotted in Figure 3 for the cases kX≫kY, kX=kY, and kX≪kY. Experiments indicate that the actual kinase activities of the AI-1 and AI-2 pathways are nearly equal (Figure 1B, Long et al, 2009).
Figure 3.
Constant output Z contours for different relative kinase strengths of the two signaling branches carrying the inputs X and Y: (A) kY/kX=1/8, (B) kY/kX=1, (C) kY/kX=8. Output is normalized by the maximum of Z and the color bar applies to all graphs.
In a standard manner, we approximate the probabilistic transfer function, P(Z∣X, Y) as a Gaussian channel, in which the probability of observing an output for a given input is modeled by a Gaussian distribution around the mean output level for that input (Detwiler et al, 2000; MacKay, 2003; Ziv et al, 2007; Tkacik et al, 2008a, 2008b). Explicitly, we model the noisy transfer function as
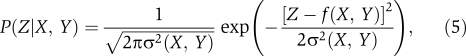 |
where f(X, Y), given by equation (4), is the deterministic transfer function describing the average output Z as a function of the inputs X and Y, and σ(X, Y) is the input-dependent standard deviation of the output signal for a given input (cf. Figure 2B). Experimentally, the standard deviation σ(X, Y) is measured by looking at cell-to-cell variations in a clonal population using single-cell fluorescence microscopy (Long et al, 2009). We expect that (5) is a good approximation for the true transfer function because, experimentally, the noise is well approximated by a Gaussian and is much smaller than the mean signal, σ(X, Y)/f(X, Y)≪1 (Long et al, 2009).
Unfortunately, little is known at a quantitative level about the natural environment of V. harveyi, making it difficult to accurately model the prior q(X, Y). Therefore, we take the approach of performing all our calculations for a variety of reasonable priors. In this report, we present results for three choices of prior: a flat prior in which all inputs are equally likely, a bimodal prior which is symmetric in the two inputs, and a non-symmetric bimodal prior (see Supplementary information). We have verified that our main conclusions are insensitive to the choice of prior.
Finally, we note that there is a one-to-one correspondence between priors on receptor kinase-active probabilities, (X, Y), and priors on autoinducer concentrations as long as fluctuations in receptor probabilities are negligible. In this case, receptor probabilities and autoinducer concentrations are related by the deterministic equation (3) and any prior on autoinducer concentrations can be transformed into a prior on receptor probabilities. Although the specific form of the transformation of the prior depends on the binding affinities of the autoinducers for the receptors, by definition, the mutual information between the input and output is independent of whether one treats the autoinducer concentrations or the receptor probabilities as the inputs.
Information about each input is limited by ‘noise' from the other input(s)
In a circuit that integrates multiple signals, information transmission about each individual signal is limited by two distinct phenomena: biochemical noise and interference from other signals (see Figure 2). Noise arises from both the stochastic nature of biochemical reactions underlying the signaling circuit and as well as other sources of cellular variability (Elowitz et al, 2002). In the presence of noise, a single input gives rise to a distribution of outputs. This type of noise limits information transmission because it introduces uncertainty about the input given the output. A second, independent phenomenon that limits information transmission about individual inputs in multi-input circuits is interference from other signals. Generally, different combinations of the input signals can give rise to the same output signal. Consequently, when a multi-input circuit is viewed as a single-input channel for a particular input, other signals introduce additional uncertainty about that input even in the absence of noise, that is, other signals act as additional noise sources (see. Figure 2C).
In the V. harveyi quorum-sensing circuit, experiments indicate that the noise is generally significantly smaller than the mean input signal, with the signal-to-noise ratio always greater than 2.5 (σ(X, Y)/f(X, Y)⩾2.5). Thus, the circuit is always in a ‘low-noise' regime. To assess whether noise or interference from other signals is the primary limitation on information transmission about individual signals, we have obtained formulas for the mutual information, I(Z, X) and I(Z, Y), in the low-noise regime using a saddle-point approximation (see Supplementary information). The saddle-point approximation, also known as the method of steepest descent, gives an asymptotic expansion for the mutual information valid in the limit of large signal-to-noise ratio. The mutual information formulas are most easily expressed in terms of transformed coordinates (f, θ) related to (X, Y) by the coordinate transformation (X, Y) → (f=f(X, Y), θ=X). Explicitly one has,
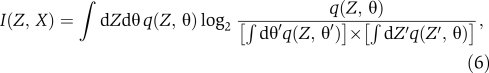 |
with
A similar formula can be derived for I(Z, X) (see Supplementary information).
Recall that I(Z, X) and I(Z, Y) measure the average amount of information that can be learned about the individual inputs X and Y from the output Z, and therefore I(Z, X) and I(Z, Y) allow us to quantify information transmission about individual inputs. We find that our approximate expressions for these quantities do not depend on the noise, indicating that information transmission about each input is primarily limited by interference from other signals.
Total information transmission is limited by biochemical noise
We can also discover how much bacteria can learn on average about all the inputs from the mutual information I(Z,(X, Y)) between the output Z and the ordered pair of inputs (Z, X). In contrast to the case of individual inputs considered above, we find that even in the low-noise regime, total information transmission is limited by noise when both signals are considered. Explicitly, one has
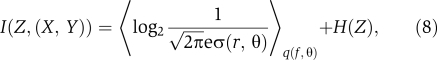 |
where 〈 〉q(f, θ) indicates the expectation value with respect to the prior distribution
and H(Z) is the entropy of the output Z (see Supplementary information). Notice that I(Z,(X, Y) is primarily limited by the noise, σ(f, θ), in the transfer function. Our approximate expressions are analogous to those obtained for a single-input, single-output biochemical network (Tkacik et al, 2008b; Supplementary information). This follows intuitively because I(Z,(X, Y)) is insensitive to the identity of the individual signals X and Y and thus the circuit effectively has a single input (X, Y) and a single output Z.
We calculated the total information transmission in the V. harveyi quorum-sensing circuit using data from Long et al, 2009 (see Supplementary information). We calculated the mutual information I(GFP, (X, Y)) between the GFP output signal and the inputs, and observed that the information is of order 1.5 bits for a variety of priors. It should be noted that by standard information theoretic inequalities, I(GFP, (X, Y)) is a lower bound on I(Phospho−LuxO, (X, Y)) the information transmitted between the inputs and LuxO, the output of the quorum-sensing phosphorelay (MacKay, 2003). Nonetheless, we stress that I(GFP, (X, Y)) is a reasonable proxy for the true information transmission because information from the inputs is eventually transmitted to the master quorum-sensing regulator LuxR through the small RNAs (see Figure 2A).
V. harveyi must tune kinase activities to simultaneously learn about multiple inputs
Experiments indicate that in V. harveyi, signals from two of the autoinducers, AI-1 and AI-2, are combined strictly additively in a shared phosphorelay pathway, with each autoinducer contributing very nearly equally to the total response (Long et al, 2009). In terms of the mean response (equation (4)), this means that the maximal kinase activities of the AI-1/LuxN and AI-2/LuxPQ pathways are almost identical, that is, kX≈kY. The observed transfer function seems puzzling at first—it is symmetric in the two inputs (see Figure 1B), indicating that bacteria cannot distinguish between AI-1 and AI-2 even though the two AIs encode distinct information about local species composition. This conundrum motivated us to investigate how kinase rates of the two pathways, kX and kY, and phosphatase rate, p, affect information transmission, by calculating the mutual informations, I(Z, X) and I(Z, Y), for different choices of circuit parameters.
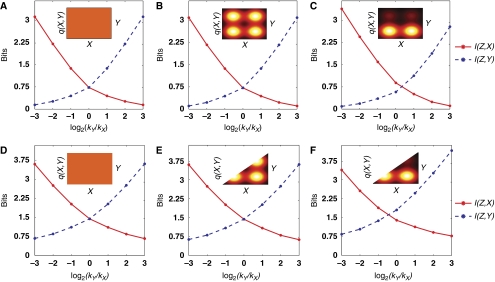
Our results indicate that the signal processing properties of the quorum-sensing circuit vary dramatically with changes in the relative strength of the kinase activities of the two pathways. In contrast, when phosphatase activity is large compared with the kinase activity and the noise is small, the net phosphatase activity, p, affects information transmission about individual inputs only modestly. The reason for this is that the phosphatase activity, p, simply rescales the output Z (see equation (4)), and mutual information is insensitive to re-scalings. Figure 4 shows plots of I(Z, X) and I(Z, Y), as functions of the ratio of kinase activities, kY/kX, for various priors. As discussed previously, I(Z, X) and I(Z, Y) are limited primarily by interference between signals, not by noise. Therefore, we used the low-noise expressions (see Supplementary information). Our results indicate that if kY/kX≫1, I(Z, Y) can be very large (≫1) but I(Z, X) is very small (≪1). However, if kY/kX≪1, I(Z, Y) is very small but I(Z, X) is very large. Thus, if the kinase activity of one pathway is much larger than the other, the cell can only learn about the stronger pathway. Only when the kinase activities of the two pathways are roughly equal, kY≈kX, can the cell learn, in an information theoretic sense, about both inputs. We conclude that V. harveyi must tune kinase activities of AI-1 and AI-2 pathways to be roughly equal to learn about both inputs. Indeed, this is what is observed in experiments (Long et al, 2009). Nonetheless, even when kinase activities are equal, small numerical values of the mutual information (see Figure 4) indicate that interference between signals is a major impediment to learning about individual inputs.
Figure 4.
Mutual information as a function of the ratio of kinase strengths, kY/kX. (A–C) Mutual information I(Z, X) between Z and X (red curves), and mutual information I(Z, X) between Z and Y, I(Z, Y) (dashed blue curves) as functions of kY/kX, for (A) a flat prior, (B) a symmetric prior, bimodal in each input, and (C) a non-symmetric prior, bimodal in each input. (Insets) Graphical representations of the corresponding priors with brighter colors representing higher probability. (D, E) Mutual information as shown in the figure, with the restriction that the inputs obey X⩾Y.
These results can be intuitively understood as follows. The architecture of prokaryotic phosphorelays is such that a single phosphate group is passed from receptors detecting the inputs to the output response regulator. Thus, in the V. harveyi quorum-sensing circuit, all information about the inputs is encoded in a single number, the number of phospho-LuxO molecules. At steady state, bacteria are limited to what is commonly referred to in the language of information theory as ‘amplitude encoding', (i.e. all information is stored in the magnitude of the output variable). Amplitude encoding places strong limitations on how signals can be integrated. If the kinase rate of the X-signaling branch is much larger than that of the Y-signaling branch, (kY/kX≪1), then the number of phospho-LuxO almost entirely reflects the magnitude of the input signal X and contains very little information about Y. However, if (kY/kX≫1), then the number of phospho-LuxO almost entirely reflects the magnitude of the input signal Y and contains very little information about X. This relation can be observed graphically from the constant output contours of Figure 3. The contours are almost vertical when kY/kX≪1, indicating that Z is highly correlated with X but largely uncorrelated with Y. The opposite is true when kY/kX≫1.
We conclude that for the number of phospho-LuxO molecules to contain information about both signaling branches, it is necessary that, on average, both signaling branches phosphorylate about equal numbers of LuxO. In terms of kinase activities, this translates into the requirement that the maximal kinase activities of the two signaling pathways be approximately equal, kX≈kY. In light of these results, we speculate that the reason that the kinase activities of the AI-1/LuxN and AI-2/LuxPQ pathways are nearly identical is to allow bacteria to learn about the concentration of both autoinducers individually.
Theoretical analysis of possible strategies for increasing information transmission
Setting kX≈kY has the consequence of introducing symmetry in the input–output relation such that bacteria cannot distinguish the input (X, Y) from the input (Y, X). For example, bacteria cannot distinguish saturating AI-1 and no AI-2 condition from saturating AI-2 and no AI-1 condition (Long et al, 2009). This constraint limits how much cells can learn about either input. Quantitatively, from our calculations, for kX≈kY, cells can learn only about 0.6–0.8 bits about each input signal, with the exact number depending on the choice of prior. This is significantly less information than that contained in the input distribution. For comparison, bacteria would have to learn 1 bit to distinguish between two states of the autoinducers (e.g. high and low concentration) and the Bicoid–Hunchback system in early Drosophila development was recently measured to transmit 1.5 bits (Tkacik et al, 2008a). The low mutual information between inputs and outputs indicates that interference between signals greatly limits information transmission in these systems and serves as a major impediment to learning about multiple signals. This observation led us to consider possible mechanisms that would allow bacteria to increase how much they learned about individual inputs. Below, we consider two such strategies: manipulation of individual input signals and feedbacks on receptors.
Bacteria can increase information transmission by manipulating the inputs
In quorum sensing, bacteria both produce and detect autoinducers. This led us to consider whether bacteria could increase the information they obtain about their environments by manipulating relative autoinducer production rates. As discussed, the primary limit on information transmission when the kinase rates of AI-1 and AI-2 signaling pathways are equal is the symmetry in the input–output relation. We hypothesized that bacteria may distinctly manipulate the different autoinducer production rates to remove the ambiguity between the two input signals, and thereby increase the information the AIs provide. For example, bacteria could temporally segregate signals by first producing one autoinducer and then the other. Alternatively, V. harveyi could produce AI-1 and AI-2 at the same rate. This solution would ensure that there was always more AI-2 than AI-1 in the environment because AI-1 is produced only by V. harveyi, whereas AI-2 is produced by almost all bacteria. Within our model, this arrangement corresponds to limiting the input signaling space to X⩾Y. We calculated I(Z, X) and I(Z, Y) for the latter scenario and the results are shown in Figure 4. We found that when kX≈kY and X⩾Y bacteria could learn ≈1.5 bits about each signal, double of what they can learn when the input space is unrestricted. The additional information gained by the cell stems from the elimination of the degeneracy between AI-1 and AI-2. Namely, for intermediate levels of output, bacteria now know that there is high amount of AI-1 and low amounts of AI-2 in the environment and not high AI-2 and low AI-1. This result confirms that in principle, bacteria can increase information transmission by manipulating autoinducer production rates.
Feedback on receptor number allows bacteria to focus attention on individual inputs
The information theory analysis presented above shows that the signaling properties of the V. harveyi quorum-sensing circuit are sensitive to changes in the kinase rates of the inputs. This raises the intriguing possibility that bacteria might implement more sophisticated signal detection strategies by varying kinase activities as a function of inputs. A simple architecture for achieving this goal would be a feedback on receptor number. Indeed, preliminary experiments indicate such a feedback may exist in the quorum-sensing circuit of V. harveyi, with sRNAs negatively regulating production of LuxN (L Schaffer and B Bassler, unpublished data). We show below that such feedbacks on receptor number potentially allow bacteria to ‘focus attention' on different inputs depending on their external environments.
The maximal kinase activity of each autoinducer pathway in V. harveyi depends on two separate quantities: (1) the total number of receptors, and (2) the maximal kinase activity of each individual receptor. Explicitly, maximal kinase rates of the X (AI-1) and Y (AI-2) pathways obey kX=kX0NX and kY=kY0NY, with NX and NY the number of receptors in the X and Y pathways, respectively, and kX0 and kY0 the maximal kinase activities of the individual receptors. Thus, in principle, bacteria can modulate the ratio of maximal kinase rates between the two pathways, kY/kX, as a function of the output, Z, through feedback on receptor number (see Figure 5 and Supplementary information).
Figure 5.
Graphical representation of input–output relations in the presence of a positive feedback on receptor number restricted to the domain X>Y. Equally spaced, constant output Z contours for a signaling circuit with positive feedback on receptor number (see inset). The output is normalized by the maximum of Z. Parameters are K=6, C=1/8, δC=8.
We consider here two simple feedback architectures: (1) positive feedback on NY and (2) negative feedback on NX (see Figure 5). In the main text, we restrict our discussion to the case of positive feedback, assumed to act on the receptors in the Y pathway. We consider a positive feedback on the receptors in the Y pathway. In this case, the transfer function, Z=ffb(X, Y), describing the output signal (the fraction of phosphorylated output regulators), as a function of the inputs X and Y (the probability that the corresponding receptors are in their on states) is obtained by solving for the steady state of the differential equations
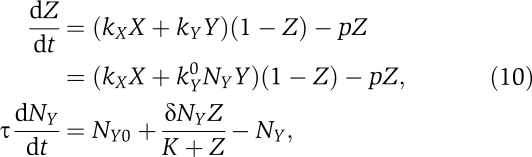 |
where NY0 is the number of receptors in the absence of feedback, δNY measures the strength of the feedback, K is value of Z, which is half-maximal for the feedback. When δNy≫NY0, this implies that at high Z, δNY≫NY0. For simplicity, we have assumed a Hill coefficient of 1 for the feedback and that the phosphatase rate is independent of the receptor number. We also assume, as above, that the phosphatase rate is much larger than the maximal kinase rate p≫kX, kY for all choices of inputs. We obtain the steady-state solution by setting the left hand sides of the above equations to zero, which yields
where, for simplicity, we denote the steady-state output by Z. This equation can be solved for Z to obtain the transfer function, ffb(X, Y), in the presence of feedback.
A particularly interesting parameter range is the regime when kY0(NY0+δNY)≫kX≫kY0NY0. In this case, the maximal kinase activity of the X-pathway is much greater than the maximal kinase activity of the Y-pathway at low Z, and the opposite is true at large Z. Thus, the positive feedback on receptor number, NY, allows the bacteria to access information preferentially about input X at low Z (i.e. at high cell density) and learn preferentially about Y at high Z (i.e. at low cell density). In the low-noise limit, the mutual informations, I(Z, X) and I(Z, Y), only depend on three combinations of parameters, ratios of the maximal kinase activities in the presence and absence of feedback, and the half-maximal value of the feedback K (data not shown). Thus, by rescaling X and Y, we can consider the equivalent transfer function
with C=kY0NY0/kX, δC=kY0δNY/kX.
Both of these feedback architectures allow bacteria to tune kinase rates of the two pathways so that kY/kX≫1 at large (X, Y) and kY/kX≪1 at small (X, Y). This can be understood graphically in Figure 5A that shows contour lines of constant output, Z, for different values of the inputs X and Y in the presence of a positive feedback from Z on NY. It is noteworthy that for X and Y near 1 (i.e. at low cell density), the constant-Z contours are more horizontal indicating that kY/kX≫1, whereas for X and Y close to zero (i.e. high cell density), the contour lines are much more vertical indicating that kY/kX≪1. Therefore, for the positive feedback shown in Figure 5, bacteria preferentially learn about Y (AI-2) at low cell densities and about X (AI-1) at high cell densities. Though less pronounced, analogous results can be achieved using a negative feedback on NX (see Supplementary information).
To quantify information transmission for such feedback architectures, we calculated the mutual informations, I(Z, X) and I(Z, Y), in the presence of feedbacks for various choices of kinetic parameters and a flat prior (see Supplementary information). In all our calculations, we assumed the signal-to-noise ratio to be large, σ(X, Y)/f(X, Y)≪1, and used the low-noise expressions derived in the Supplementary information. In this limit, I(Z, X) and I(Z, Y) are limited primarily by interference between signals and are independent of any specific choice of noise, σ(X, Y). We found that mutual informations in the presence of either a positive feedback on the receptors for input Y or a negative feedback on the receptors for input X are comparable to those in the absence of feedback. This finding indicates that bacteria can preferentially detect AI-2 (X) at low cell densities and AI-1 (X) at high cell densities without sacrificing how much they learn on average about both inputs. For example, for both the feedback transfer functions shown in Figure 5, I(Z, X) and I(Z, Y) are both ≈1.5 bits for the case when X⩾Y, comparable with their values in the absence of feedback (see Figure 4).
Discussion
Cells constantly sense their environments and adjust their behavior accordingly. Specifically, cells often integrate temporally coincident information from multiple environmental inputs to modulate their gene expression states. However, mechanisms and logic by which cells integrate multiple signals remain by and large poorly understood. We developed a new, mathematical framework for analyzing information processing in cells based on information theory, and used it to study the integration of multiple autoinducer signals using the model quorum-sensing bacterium Vibrio harveyi. Our studies revealed that there are two distinct mechanisms that limit information transmission when bacteria integrate multiple signals, biochemical noise and interference between different signals. Although the former limits the total information that bacteria can learn about all the inputs, signal interference is the primary impediment to learning about individual input signals. Furthermore, we showed that because of signal interference, V. harveyi cells must precisely tune the kinase activity of each input branch of the quorum-sensing pathway to simultaneously learn about individual autoinducer inputs. These theoretically motivated conclusions are consistent with recent quantitative experiments on V. harveyi showing that the maximal kinase activities of AI-1 (LuxN) and AI-2 (LuxPQ) pathways are nearly equal (Long et al, 2009). Our information theory analysis also indicates that bacteria can increase how much they learn about individual inputs by manipulating the different autoinducer production rates. Finally, we have shown that bacteria can learn preferentially about a particular signal in a particular environment, even with a single-output pathway, by using simple feedback loops to control receptor numbers.
Our theory not only explains the puzzling experimental observation of nearly equal kinase activities of the LuxN and LuxPQ pathways (Long et al, 2009), but also makes several testable predictions about the V. harveyi quorum-sensing circuit. First, we predict that the maximal kinase activity of the CAI-1/CqsS branch, when measured, will prove to be similar to that of the AI-1/LuxN and AI-2/LuxPQ pathways (see Figure 2). This prediction follows directly from our information theory analysis, which indicates that the three signaling branches must phosphorylate about equal numbers of LuxO for cells to simultaneously learn about all three input signals. Second, the theoretical work presented here suggests that V. harveyi may manipulate both autoinducer production and receptor numbers to reduce interference between signals, and thereby increase information transmission. Preliminary evidence suggests that this is the case.
An as yet unanswered question is why V. harveyi and related species use multiple autoinducers (AIs) and then funnel all the information from these autoinducers into a single-output pathway. We speculate that different concentrations of multiple autoinducers may represent different stages of community development such as the stages of growth in a biofilm. Unlike eukaryotic development, for example, embryogenesis, in which the rate of development is fixed and driven by a clock (Nieuwkoop and Faber, 1994) and the input signal is often stereotyped (Gregor et al, 2007a, 2007b), the rate of development of a bacterial community depends on unpredictable environmental conditions, such as nutrient availability and population composition and density. To compensate for such variability, quorum sensing could allow bacteria to monitor stages of community development and act accordingly. The architecture of the V. harveyi quorum-sensing circuit, with multiple inputs and a single output, is consistent with the idea that V. harveyi uses quorum sensing to implement a single, multi-stage developmental program, with different genes activated at different levels of LuxR expression. Indeed, Long et al (2009) showed that V. harveyi can ‘count' the number of autoinducer signals present. Thus, if AIs accumulate in a defined sequential order, the number of autoinducers present at saturating concentration could signal different stages of development. For example, models of biofilm growth suggest that the universal autoinducer AI-2 may be more informative at early stages of biofilm growth in which communities are expected to be mixed, whereas the species specific autoinducer AI-1 may be more informative at later stages when mostly progeny are nearby (Nadell et al, 2008).
Our detailed analysis of the V. harveyi quorum-sensing network has implications for other prokaryotic signal-integration networks. Signal integration is a common feature of many organisms, and bacteria have developed sophisticated molecular mechanisms for integrating signals from a broad range of inputs using two-component systems and phosphorelays (Perego, 1998; Bassler and Losick, 2006; Kato et al, 2007; Mitrophanov and Groisman, 2008). For example, the sporulation and competence circuits of the soil-dwelling bacterium Bacillus subtilis integrate signals from the environment, cell cycle, and metabolism using a network design based on competition between various protein kinases and phosphatases (Perego, 1998; Veening et al, 2008). Our information theory analysis suggests that the need to minimize interference between signals probably places strong constraints on the design of such signal-integration networks. In particular, our work indicates that the information transmission properties are likely to be extremely sensitive to changes in kinase and phosphatase rates, and that bacteria may have evolved strategies for minimizing interference. One possible strategy for learning about individual input signals is to temporally coordinate signals (Mitrophanov and Groisman, 2008). For example, recent experiments on the B. subtilis sporulation and competence networks indicate that bacteria probably temporally separate input signals (Smits et al, 2007; Veening et al, 2008).
Bacteria may use a range of mechanisms to minimize signal interference by actively controlling both signal production and detection. A simple way to achieve such control is through feedbacks on synthases/receptors. For example, feedbacks on AI production are a common feature of many quorum-sensing systems (Waters and Bassler, 2005), suggesting bacteria actively manipulate the temporal profile of AI production. In V. harveyi, recent experimental evidence suggests such a feedback may act on the AI-1 synthase LuxM (L Schaffer and B Bassler, unpublished data). Moreover, the gene encoding LuxM is located in an operon with the gene encoding the AI-1 receptor LuxN, indicating the feedback also acts on receptor numbers, potentially allowing V. harveyi to focus on different autoinducers at different stages of development. Recent experiments also indicate that Escherichia coli cells manipulate chemoreceptor numbers using feedbacks. When starving, E. coli cells change the ratio of Tar to Tsr receptors, resulting in a change of behavior from heat seeking to cold seeking (Salman and Libchaber, 2007).
The use of signaling pathways with multiple inputs and a single output necessarily entails a loss of information about input signals. This raises the natural question of why such pathways are used by bacteria. For example, one can imagine alternative architectures in which each input is detected by a dedicated signaling pathway and information about multiple inputs is integrated at the promoters of regulated genes through combinatorial gene regulation (Kato et al, 2007; Mitrophanov and Groisman, 2008). We have argued that for V. harveyi such a multi-input, single-output architecture facilitates the implementation of a linear, multi-stage, developmental program. The architecture of signal integration networks may also reflect evolutionary constraints. For example, such networks may have evolved from a single pathway by gene duplication. In addition, when the output of a signal integration network is a master transcription factor regulating the expression of many genes (e.g. LuxR in V. harveyi), the use of a single-output pathway may be more efficient with regard to use of space on the genome than a competing architectures consisting of individual signaling pathways, one for each input, culminating in combinatorial gene regulation. Recent experiments indicate that the sporulation network in B. subtilis may have a similar role in regulating biofilm formation (Vlamakis et al, 2008). In light of the accumulating evidence that bacterial populations behave similarly as multicellular organisms (Shapiro, 1998), we suspect that the use signal integration networks to coordinate development programs may be widespread in prokaryotes.
The study presented here focuses on signal integration in bacteria. The architecture of prokaryotic phosphorelays, in which a single phosphate group is transferred sequentially to downstream components, constrains bacteria to encode information using amplitude encoding, that is, all information about input signals is contained in the number of active response regulator molecules. The use of amplitude encoding places strong constraints on network architecture and limits the amount of information that bacteria can transmit. This contrasts with neural networks, in which spike timing allows neurons to encode information using more sophisticated schemes (Rieke et al, 1997). Signaling in bacteria also differs from signaling in eukaryotes, which often uses multiple phosphorylation sites and kinase cascades that permit temporal encoding schemes, such as dose–duration encoding (Detwiler et al, 2000; Behar et al, 2008). It may prove fruitful to generalize our information theoretic formalism to these more complicated intracellular circuits.
Finally, our results suggest that information theory may prove to be a powerful general tool for analyzing biological signaling networks. Information theory provides a natural language for formulating questions about information processing and signaling integration. An additional advantage of an information theoretic analysis is that no detailed knowledge of the signaling circuit is required. All quantities are calculated using the input–output relationship of a signaling circuit, often an experimentally accessible quantity, even for large signaling networks. For these reasons, we expect the application of information theory to yield new biological insights into cellular signaling in the future.
Supplementary Material
Supplementary Information, Supplementary figures S1–3
Acknowledgments
We would like to thank Thierry Mora, Anirvan Senputa, the Wingreen and Bassler labs, and the Princeton biophysics theory group for useful discussions. This study was supported by US National Institute of Health (NIH) grant K25 GM086909-01 (PM); partially supported by the Burroughs Wellcome Fund Graduate Training Program (SG and TL); partially supported by the Defense Advanced Research Projects Agency (DARPA) under grant HR0011-05-1-0057 (SG and NW). This work was also partially supported by National Science Foundation (NSF) Grant Phys-0650617.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bassler BL, Losick R (2006) Bacterially speaking. Cell 125: 237–246 [DOI] [PubMed] [Google Scholar]
- Behar M, Hao N, Dohlman HG, Elston TC (2008) Dose-to-duration encoding and signaling beyond saturation in intracellular signaling networks. PLoS Comput Biol 4: e1000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst A, Theunissen FE (1999) Information theory and neural coding. Nat neurosci 2: 11. [DOI] [PubMed] [Google Scholar]
- Detwiler PB, Ramanathan S, Sengupta A, Shraiman BI (2000) Engineering aspects of enzymatic signal transduction: photoreceptors in the retina. Biophys J 79: 2801–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS (2002) Stochastic gene expression in a single cell. Science 297: 1183–1186 [DOI] [PubMed] [Google Scholar]
- Gregor T, Tank DW, Wieschaus EF, Bialek W (2007a) Probing the limits to positional information. Cell 130: 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T, Wieschaus EF, McGregor AP, Bialek W, Tank DW (2007b) Stability and nuclear dynamics of the bicoid morphogen gradient. Cell 130: 141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke JM, Bassler BL (2004) Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J Bacteriol 186: 6902–6914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Mitrophanov AY, Groisman EA (2007) A connector of two-component regulatory systems promotes signal amplification and persistence of expression. Proc Natl Acad Sci USA 104: 12063–12068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keymer JE, Andres RG, Skoge M, Meir Y, WIngreen NS (2006) Chemosensing in Escherichia coli: two regimes of two-state receptors. Proc Natl Acad Sci USA 103: 1786–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long T, Tu KC, Wang Y, Mehta P, Ong NP, Bassler BL, Wingreen NS (2009) Quantifying the integration of quorum-sensing signals with single-cell resolution. PLOS Biol 78: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay D (2003) Information Theory, Inference, and Learning Algorithms. London: Cambridge University Press [Google Scholar]
- McAdams HH, Arkin A (1997) Stochastic mechanisms in gene expression. Proc Natl Acad Sci USA 94: 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrophanov AY, Groisman EA (2008) Signal integration in bacterial two-component regulatory systems. Genes Dev 22: 2601–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadell CD, Xavier JB, Levin SA, Foster KR (2008) The evolution of quorum sensing in bacterial biofilms. PLoS Biol 6: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J (1994) Normal Table of Xenopus laevis (Daudin). New York and London: Garland Publishing [Google Scholar]
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A (2002) Regulation of noise in the expression of a single gene. Nat Genet 31: 69–73 [DOI] [PubMed] [Google Scholar]
- Perego M (1998) Kinase-phosphatase competition regulates Bacillus subtilis development. Trends Microbiol 6: 366–370 [DOI] [PubMed] [Google Scholar]
- Rieke F, Warland D, van Steveninck RR, Bialek W (1997) Spikes: Exploring the Neural Code. Cambridge, MA: MIT Press [Google Scholar]
- Salman H, Libchaber A (2007) A concentration-dependent switch in the bacterial response to temperature. Nat Cell Biol 9: 1098–1100 [DOI] [PubMed] [Google Scholar]
- Shannon CE (1948) A mathematical theory of communication. Bell Syst Technical J 27: 379–423 [Google Scholar]
- Shapiro J (1998) Thinking about bacterial populations as mutlicellular organisms. Annu Rev Microbiol 52: 81–104 [DOI] [PubMed] [Google Scholar]
- Smits WK, Bongiorni C, Veening JW, Hamoen LW, Kuipers OP, Perego M (2007) Temporal separation of distinct differentiation pathways by a dual specificity Rap-Phr system in Bacillus subtilis. Mol Microbiol 65: 103–120 [DOI] [PubMed] [Google Scholar]
- Swain PS, Elowitz MB, Siggia ED (2002) Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA 99: 12795–12800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swem LR, Swem DL, Wingreen NS, Bassler BL (2008) Deducing receptor signaling parameters from in vivo analysis: LuxN/AI-1 quorum sensing in Vibrio harveyi. Cell 134: 461–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkacik G, Callan CG Jr, Bialek W (2008a) Information flow and optimization in transcriptional regulation. Proc Natl Acad Sci USA 105: 12265–12270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkacik G, Callan CG Jr, Bialek W (2008b) Information capacity of genetic regulatory elements. Phys Rev E 78: 011910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tostevin F, ten Wolde PR (2009) Mutual information between in- and output trajectories of biochemical networks. Phys Rev Lett 102: 218101. [DOI] [PubMed] [Google Scholar]
- Tu KC, Bassler BL (2007) Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev 21: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Stewart EJ, Berngruber TW, Taddei F, Kuipers OP, Hamoen LW (2008) Bet-hedging and epigenetic inheritance in bacterial cell development. Proc Natl Acad Sci USA 105: 4393–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H, Aguilar C, Losick R, Kolter R (2008) Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev 22: 945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak AM, Mugler A, Wiggins CH (2009) A stochastic spectral analysis of transcriptional regulatory cascades. Proc Natl Acad Sci USA 106: 6529–6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Bassler BL (2005) Quorum sensing: Cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21: 319–346 [DOI] [PubMed] [Google Scholar]
- Waters CM, Bassler BL (2006) The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev 20: 2754–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv E, Nemenman I, Wiggins CH (2007) Optimal signal processing in small stochastic biochemical networks. PLoS ONE 2: e1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information, Supplementary figures S1–3