Abstract
Odorant receptors are among the fastest evolving genes in animals. However, little is known about the functional changes of individual odorant receptors during evolution. We have recently demonstrated a link between the in vitro function of a human odorant receptor, OR7D4, and in vivo olfactory perception of 2 steroidal ligands—androstenone and androstadienone—chemicals that are shown to affect physiological responses in humans. In this study, we analyzed the in vitro function of OR7D4 in primate evolution. Orthologs of OR7D4 were cloned from different primate species. Ancestral reconstruction allowed us to reconstitute additional putative OR7D4 orthologs in hypothetical ancestral species. Functional analysis of these orthologs showed an extremely diverse range of OR7D4 responses to the ligands in various primate species. Functional analysis of the nonsynonymous changes in the Old World Monkey and Great Ape lineages revealed a number of sites causing increases or decreases in sensitivity. We found that the majority of the functionally important residues in OR7D4 were not predicted by the maximum likelihood analysis detecting positive Darwinian selection.
Keywords: olfaction, olfactory, molecular evolution, pheromone, GPCR
Odorant receptors (ORs) are 7-transmembrane G protein coupled receptors encoded by the largest gene family in mammalian genomes and are undergoing rapid evolutionary change, with extensive gene gains and losses (1, 2). Various lines of evidence point to the reduction of the functional OR repertoire in the human lineage (3, 4). The human genome contains a higher percentage of OR pseudogenes compared to other mammalian species, including chimpanzee, its closest primate relative (5); yet a recent study showed that the number of OR genes and the fraction of OR pseudogenes in chimpanzees are very similar to those in humans (6). Less than 400 of the approximately 900 human ORs encoded in the genome have intact open reading frames (ORFs); the rest are pseudogenes (7, 8). In contrast, mice or rats have more than 1,000 OR genes, and the majority of the ORs have intact ORFs (1).
We have previously identified a human OR, OR7D4, which selectively responds to 2 sex steroid-derived odors, androstenone and androstadienone, in a heterologous cell system (9). In the domestic pig, androstenone has been identified as a male pheromone that induces a receptive mating stance in estrous females (10). In humans, androstenone is found in saliva, sweat, and urine and olfactory exposure to androstadienone has been reported to affect physiological responses (11–13). However, their effects on human reproductive activities remain controversial (14).
In the previous study, we also found 2 linked SNPs, R88W and T133M, constituting a common variant that had severely impaired function in vitro compared to the reference OR7D4. This functional variation is well correlated with variability in human perception of these steroidal odors (9). Two rarer variants, P79L and S84N, which have severely impaired or dramatically increased function, respectively, also showed a correlation between subjects who possess these variants and respective perception to the odors (9). A cloned chimpanzee ortholog also responded to these chemicals and appeared to be more sensitive than the human S84N variant (9).
Positive Darwinian selection (positive selection) is a phenomenon whereby natural selection favors changes. As olfaction is essential for detecting food sources, avoiding toxic compounds or predators, and evaluating mates, positive selection on an OR gene, if any, can be important in modulating species-specific behaviors by changing sequences of the ORs, thereby modifying odor specificity and sensitivity. It is, however, possible that olfaction is of diminishing importance to primates and therefore the receptors are no longer under positive selection. Previous sequence analysis of a genome-wide scan on the high-coverage genome assemblies of 6 mammalian species suggested an astounding enrichment of chemosensory receptor genes with evidence for positive selection (15). Other studies suggested evidence for positive selection acting on some OR genes in mammals, including human and chimpanzee (5, 16–18), while another study found no evidence for positive selection in the human and chimpanzee lineages (19). However, sequence-based methods to predict positive selection have their limitations as various factors could cause biased sequence changes and fixation during evolution (20–22). Therefore, as a first step toward elucidating the association between sequence changes and functional changes in evolution, addressing the functional changes of OR orthologs is one of the keys to understanding the evolution of ORs.
Here we investigate the evolutionary changes in an OR in primates. We show that primate OR7D4s exhibit dramatic differences in responding to its cognate steroidal ligands. By site-directed mutagenesis using various putative common ancestors, we identified amino acid residues that are important for determining sensitivity of OR7D4 in each species.
Results
Intact Primate OR7D4/OR7D1 Orthologs Exhibit Diverse Response Levels to Androstenone and Androstadienone.
We attempted to clone the full ORF of the orthologs of OR7D4 and its closest homolog, OR7D1 from a panel of 12 primate genomes representing approximately 55 million years of evolution. This included 5 hominoids (bonobo, chimpanzee, gorilla, orangutan, and gibbon), 4 Old World Monkeys (rhesus macaque, pigtailed macaque, colobus monkey, and patas monkey), 2 New World Monkeys (spider monkey and squirrel monkey), and 1 prosimian (mongoose lemur) [supporting information (SI) Fig. S1 in SI Materials and Methods]. We verified the presence of OR7D4/OR7D1 orthologs in 8 species, namely, bonobo, chimpanzee, gorilla, orangutan, rhesus macaque, pigtailed macaque, colobus monkey, and mongoose lemur (Fig. S2). Among them, 6 species, bonobo, chimpanzee, gorilla, orangutan, rhesus macaque, and pigtailed macaque, appeared to have both OR7D4 and OR7D1 orthologs while we could identify only 1 OR7D4/OR7D1 ortholog from mongoose lemur. This result, together with previous reports (3, 5) and our data-mining, was consistent with the idea that OR7D4/OR7D1 duplication probably occurred after the divergence of the New World Monkeys, and that OR7D4/OR7D1 was lost in some branches, such as the New World Monkeys (See SI Materials and Methods for detailed information on OR7D4/OR7D1 gene gains and losses).
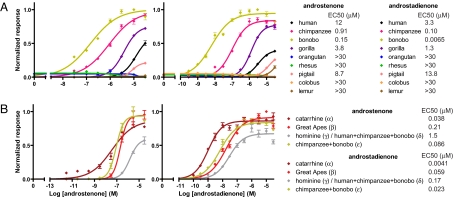
We next assessed the function of the different primate OR7D4/OR7D1 clones with a cAMP-mediated luciferase reporter gene assay in Hana3A cells, a HEK293T-derived cell line stably expressing accessory factors for OR expression, transiently expressing human receptor-transporting protein 1S (hRTP1S) to facilitate the cell-surface expression of ORs (9, 23, 24). Surprisingly, the response profile of OR7D4 orthologs exhibited an extremely broad spectrum of receptor function (Fig. 1A). The bonobo OR7D4 was the most sensitive to androstenone and androstadienone, exhibiting much lower EC50 values, thresholds, and saturation concentrations compared with the functional human OR7D4 variant, RT (9). The chimpanzee and gorilla OR7D4 were less sensitive compared with the bonobo OR7D4 but more sensitive than the human ortholog. In contrast, mongoose lemur, colobus monkey, rhesus macaque, pigtailed macaque, and orangutan OR7D4s showed little or no response to androstenone or androstadienone. This indicates a dramatic change in ligand sensitivity and/or specificity of OR7D4 during primate evolution. It is probable that OR7D4 orthologs in these species could respond to ligands other than these 2 steroidal odors. We therefore tested all of the OR7D4 orthologs against a panel of 8 additional steroidal odors (Table S1A). We found that the Great Ape (human, bonobo, chimpanzee, and gorilla) OR7D4s that responded to androstenone and androstadienone also responded to the related compound androstenone methyl ketal while the other OR7D4s that showed little or no response to androstenone or androstadienone did not respond to these additional steoridal odors (Fig. S3A). Intact OR7D1 orthologs did not respond to the 10 steroidal compounds tested (Fig. S3A). We subsequently screened all intact OR7D1 orthologs against a panel of 10 odorant mixtures, representing 169 odorants of diverse chemical structures and found no cognate ligand for OR7D1 (Fig. S3B and Table S1B).
Fig. 1.
Primate OR7D4 orthologs and hypothetical OR7D4 ancestors exhibit functional divergence in response to androstenone and androstadienone. Dose-response curves of (A) all intact primate OR7D4 orthologs and (B) reconstructed hypothetical OR7D4 for the catarrhine ancestor (α), Great Ape ancestor (β), hominine ancestor (γ), human, bonobo, and chimpanzee ancestor (δ), and chimpanzee and bonobo ancestor (ε) to androstenone (Left) and androstadienone (Right) are assessed in a luciferase assay system. y axis denotes normalized response ± SEM. (n = 4). The EC50 value of each dose–response curve is shown.
The Amino Acid Residues That Determine OR7D4 Activity.
Given the functional variation of the OR7D4 orthologs, we attempted to trace the evolutionary history of OR7D4. A maximum likelihood calculation using the known orthologous primate sequences allowed us to infer the ancestral state for each polymorphic site at different interior nodes of the primate phylogeny (See SI Materials and Methods for detailed information on maximum likelihood analysis). We found that the putative catarrhine ancestor, or the ancestor to the Great Apes and Old World Monkeys (node α in Fig. 2), exhibited an extremely low EC50 value (i.e., high sensitivity) to both androstenone and androstadienone (Fig. 1B). The putative Great Ape ancestor (node β in Fig. 2) showed an increase in EC50 value (i.e., decrease in sensitivity) to the steroidal odorants compared with the putative catarrhine ancestor, but responded to the odorants with a high efficacy at higher concentrations (Fig. 1B). The putative hominine ancestor (node γ in Fig. 2), which had the same sequence as the ancestor of human, bonobo and chimpanzee (node δ in Fig. 2), showed a reduction in both sensitivity and efficacy compared with the putative Great Ape ancestor (Fig. 1B). In contrast, the bonobo and chimpanzee ancestor (node ε) showed greater response than the ancestor of human, bonobo, and chimpanzee (node δ) (Fig. 1B).
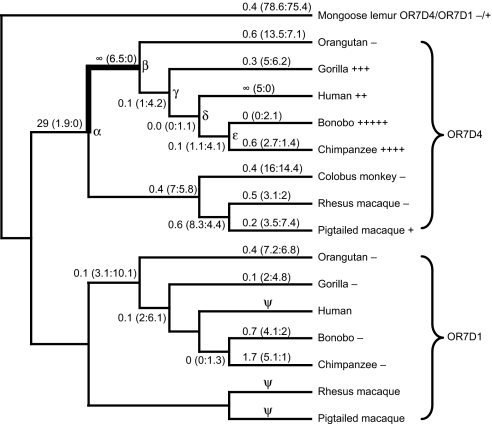
Fig. 2.
OR7D4 shows a signature for positive selection in the primate lineage. The ω values and actual numbers of nonsynonymous and synonymous changes (R:S in parentheses) along each branch are calculated by the free-ratio model of PAML using the OR7D4+OR7D1 dataset and shown on a cladogram of accepted primate phylogeny (36). Bolded line indicates accelerated evolution as computed by likelihood ratio tests shown in Table S4. Hypothetical OR7D4 ancestors, including catarrhine (α), Great Ape (β), hominine (γ), human, bonobo, and chimpanzee (δ), and bonobo and chimpanzee (ε), are indicated on the corresponding nodes on the cladogram. The functional level of the OR7D4 and OR7D1 orthologs for each species as measured in the luciferase assay (see Fig. 1A and Fig. S3A) is indicated beside the species name. The number of plus signs indicates the relative level of functional activity, and the minus signs indicate that the ortholog is nonfunctional in response to androstenone or androstadienone. Ψ denotes pseudogene lineages that are excluded from the analysis.
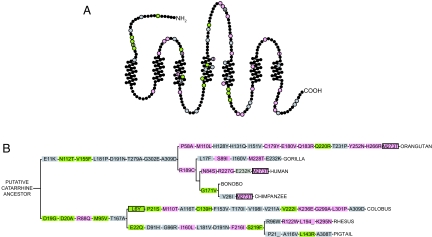
We next addressed the functional significance of amino acid changes in Old World Monkey and Great Ape OR7D4 evolution. First, using the putative OR7D4 sequence of the hypothetical hominine ancestor as a reference, we identified four nonsynonymous changes in the human reference sequence, one in bonobo, three in chimpanzee, and five in gorilla since the divergence from this ancestor (Fig. 3B and Table S2A). Second, using the putative OR7D4 sequence of the hypothetical Great Ape ancestor as a reference, we identified 13 nonsynonymous changes in the orangutan sequence and one nonsynonymous changes in the hominine ancestor since the divergence from this ancestor (Fig. 3B and Table S2B). Finally, using the putative OR7D4 sequence of the hypothetical catarrhine ancestor as a reference, we identified 35 nonsynonymous changes in the Old World Monkey species and eight nonsynonymous changes in the Great Ape ancestor since the divergence from the catarrhine ancestor (Fig. 3B and Table S2C). There are total of 58 residues out of 312 (19%) that have changed at least once in the OR7D4 evolution in catarrhines. These changes were scattered throughout the receptor (Fig. 3A), but changes in transmembrane domains were less frequent (14/156 or 9%) (P < 0.05, Fisher's exact test) than in intracellular or extracellular loops.
Fig. 3.
Nonsynonymous mutations in OR7D4 contribute to its functional divergence. (A) OR7D4 snake plot with amino acid changes from the catarrhine, Great Ape, and hominine ancestors indicated by nonblack circles. An F test that compares the best-fit values of EC50 of the ancestors with those of each of the synthetic mutants was carried out to assess whether the dose–response curves of a mutant is significantly different from those of the respective ancestors. Residues are colored in pink for decreased function, gray for no significant change, and green for increased function. (B) All of the synthetic mutations from the reconstructed hypothetical ancestors are represented on an accepted catarrhine cladogram. Positively selected sites as predicted by the M8 positive selection model of PAML using the OR7D4 only dataset (Table S5) and synthetic mutants that change function from A are plotted accordingly. Boxes are outlined in black for positive selection and are colored in pink for decreased function, gray for no significant change, and green for increased function.
To evaluate which amino acid residue could affect function, we created a panel of 65 synthetic mutants (Table S2). Each synthetic mutant was tested against androstenone in our in vitro assay, which revealed various residues critical for the function of OR7D4 (Fig. S4 and Table S2). According to the sequence of reconstructed putative ancestors, we deduced a model in which the lineage-specific amino acid changes were labeled as increasing function (green in Fig. 3), no significant functional changes (gray in Fig. 3), or decreasing function (pink in Fig. 3), as assessed in pairwise comparisons between the dose-response curves of an OR7D4 synthetic mutant and the corresponding OR7D4 putative catarrhine, Great Ape, or hominine ancestor. Unexpectedly, the majority of mutants exhibited changes in function compared with respective ancestors (39/65 or 60%); furthermore, some showed increase in function (14/65 or 22%), and others showed decrease in function (25/65 or 39%). Only a minor portion of the mutants did not change function significantly (26/65 or 40%). The residues that changed the function were scattered throughout the receptor including extracellular, transmembrane, and intracellular domains (Fig. 3A). Notably, residues that caused increase and decrease in function coexist in the same evolutionary branches in many cases (Fig. 3B). We also tested a subset of the mutants against androstadienone and found similar effects for the different site changes (Table S3 and Fig. S5).
Sites with Signatures of Positive Selection and Sites Affecting the Function of OR7D4.
Positive selection can be inferred by an excess of nonsynonymous substitutions, which lead to amino acid changes, relative to synonymous substitutions, which do not change the encoded protein. However, the validity of the statistical methods identifying sites under positive selection has been recently challenged (21, 22). Here we asked whether residues with signatures of positive selection correlate with functional changes.
We first performed phylogenetic analysis on the multiple alignment of the nucleotide sequences of OR7D4/OR7D1 cloned from different primate species using a maximum likelihood approach as implemented in the PAML software package (25). (See SI Materials and Methods for detailed information on maximum likelihood analysis.) We found evidence of positive selection in the branch leading to the Great Ape OR7D4 lineage (shown in bold in Fig. 2; see also Table S4). As expected, this branch coincided with a change in OR7D4 function since the catarrhine ancestor (Fig. 1B), and this change is a decrease in sensitivity.
Using site-specific models for positive selection along with their nested null models, we carried out additional likelihood ratio tests asking if there was a subset of amino acid residues of OR7D4/OR7D1 under positive selection. All likelihood ratio tests supported positive selection on OR7D4 in the site models (Table S5A). We then used the Bayes empirical Bayes (BEB) calculations under all positive selection site models to identify specific residues that have been subject to positive selection in primates. We found that only 2 residues (6 and 273) in OR7D4 appeared to be under positive selection with a high posterior probability (P > 0.95) (Fig. S7 and Table S5).
Finally, to assess the validity of sites with signatures of positive selection in affecting the dynamic functional evolution of OR7D4, we computed the success rate of the maximum likelihood analysis used to infer the codon sites that change OR gene functions. Intriguingly, we found the vast majority of the sites (37/39) affecting the function of OR7D4 were not predicted by this method, that is, only 2 sites (2/39, 5%) that changed receptor function were putative positively selected sites as detected by BEB with M8 in PAML (Table S5B). We note, however, that these results are highly dependent on the model and computational assumption used. For example, naïve empirical Bayes (NEB) with M8 predicted 5 sites (5/39, 13%) that changed receptor function; yet, this method falsely detected 5 more sites as although they were positively selected. These results stress the importance of verifying computational predictions with experimental analysis.
Discussion
Evolution of the odorant receptor genes has been a focal point in the field of evolutionary genomics since the surfacing of genome sequence data for various model species. However, the functional assessment of the mammalian OR orthologs are limited (26, 27). As more primate genome sequences become available, it is also increasingly feasible to compare orthologous genes in different primate species. In this study, we investigated the functional evolution of an OR for 2 sex steroid-derived odors in primates. OR7D4 is an ideal model for such an assessment since it is the first OR shown to have a direct link to olfactory perception (9). Although results of in vitro assays must be interpreted with caution as there is no information on the in vivo function of this OR in nonhuman primates, previous studies suggested that in vitro responses of ORs predict in vivo chemosensory perception in primates (9, 28).
Functional evaluation of the cloned OR7D4 orthologs suggested various episodes of dramatic gain- and loss-of-function events in the primate lineage as a consequence of sequence variation among different species. For example, while bonobo OR7D4 shows extreme sensitivity to the steroid ligands, orangutan OR7D4 shows no response to these chemicals. In the future, it will be interesting to know whether the insensitive OR7D4s respond to other untested odorants. In addition, future studies involving additional ORs in multiple species can reveal whether the dynamic functional evolution shown here is restricted to specific ORs or a more general phenomenon.
In the case of visual opsins, the combination of comparing gene sequences in various species and testing the function of each variant in a heterologous system has proven to be a powerful way to elucidate their molecular mechanisms (29). Here we traced the evolutionary history of OR7D4 at the amino acid level and identified critical residues that alter receptor function by performing a comprehensive mutational analysis based on the interspecific functional differences among the primate species. This allowed us to gain critical information on whether species-specific amino acid changes are responsible for the increase or decrease in OR activity. We found a number of sites that change receptor function. We propose this strategy of structure-function analysis based on evolutionary changes as a powerful way to identify critical amino acid residues of OR function. Since we found changes in function in different domains of the receptor, in the future, it will be important to investigate how these changes affect function of the receptor. Testing double and triple mutants with a subset of the residues in the hominine lineage is consistent with the idea that the effect could be additive in some cases (Fig. S6). However, the sensitivity to androstenone and androstadienone decreased from the putative catarrhine ancestor to the putative Great Ape ancestor (Fig. 1B), even though 2 amino acid changes (N112T and V155F) increase the sensitivity and other changes do not affect the function in this lineage (Fig. 3B). Further analyses involving making combinations of point mutations will reveal how multiple amino acid changes in a given lineage might affect the function of the receptor.
Positive selection implies the paramount importance of a certain gene to the overall fitness of an animal. At present, it is not clear whether functional changes of an OR such as OR7D4 is adaptive for each species. The functional change of a protein is not necessarily the same as the fitness change. Actually, it is very difficult to estimate even the relative fitness of different genotypes because fitness is determined by many other genes controlling morphology and physiology as well as by environmental factors (30). Our analysis on OR7D4 represents one of the first steps toward understanding the functional changes of ORs as a whole in each species. Although our computational analysis is consistent with the idea that OR7D4 is under positive selection in a subset of branches, the results should be interpreted cautiously: We showed that the vast majority of the residues that caused functional changes were not predicted by the statistical methods. Consistent with our analysis, Yokoyama et al. (21) recently questioned the validity of the statistical methods to predict positive selection in visual opsins, in which they show no correlation between the signature of positive selection and functional changes. In addition, Nozawa et al. (22) have suggested that the current statistical methods often produce false positives.
Many genes involved in mate choice and mating behavior, including chemosensory receptors, are suggested to be under positive selection (31). Various studies point to the potential effect of the steroidal odorants on human physiology and behavior (11–13), although there is no evidence for a behavioral role of these odorants in a primate species other than human. As an OR for sex-steroid derived compounds, OR7D4 is not likely to be involved in food detection or toxicity avoidance. If functional evolution of OR7D4 is adaptive, it is tempting to speculate that sensitivity to androstenone and androstadienone, which is at least partly determined by OR7D4 in humans, could play a role in the reproductive fitness in some primate species. Nonetheless, it is also likely that functional ORs for androstenone other than OR7D4 exist in primate species. For example, behavioral data from 1 Old World Monkey, pigtailed macaque, and 2 New World Monkey species, spider monkey and squirrel monkey, which all appeared to be sensitive to androstenone at similar concentrations to humans who are sensitive to androstenone (32, 33), although we show that OR7D4/OR7D1 orthologs are probably lost in New World Monkeys. Taken together to address the natural selection more directly, future studies involving identifying additional ORs that control the level of perception to the sex-steroid derived odors and showing associations between the phenotype and a certain fitness component will be required.
Materials and Methods
Cloning and Sequencing of OR7D4 and OR7D1 Orthologs.
OR7D4 and OR7D1 orthologs were amplified from various primate genomic DNA. For 4 species, squirrel monkey, spider monkey, patas monkey, and gibbon, no amplicon corresponding to a potential OR7D4/OR7D1 gene or pseudogene was obtained using the ORF-flanking primers or degenerate primers, or of expected size using the internal primers, supporting the potential absence or significant modification of OR7D4/OR7D1 gene(s) in these species. The PCR products were sequenced with 3130 or 3730 Genetic Analyzers (Applied Biosystems). See SI Materials and Methods for detailed procedures and primer information.
Sequence Analysis.
Nucleotide and peptide sequences of OR7D4 from 8 species (colobus monkey, pigtailed macaque, rhesus macaque, orangutan, gorilla, bonobo, chimpanzee, and human), OR7D1 from 4 species (orangutan, gorilla, bonobo, and chimpanzee), and OR7D4/OR7D1 from mongoose lemur were aligned using ClustalW as implemented in MEGA4 (34). OR7D1 sequences from 3 species (pigtailed macaque, rhesus macaque, and human) were excluded due to pseudogenizations. Phylogenetic trees for primate OR7D4/OR7D1 were constructed using a neighbor-joining method in MEGA4 with support for branches assessed by bootstrap analyses of 1,000 replicates. Maximum likelihood analysis was performed with OR7D4/OR7D1 sequences with CODEML in the PAML4a or 3.15 software package (25). When the individual of a certain species was heterozygous for OR7D4/OR7D1, the intact, functional, and/or the most common allele was used in PAML analysis. Detailed methods for PAML analysis is described in SI Materials and Methods.
Luciferase Assay and Data Analysis.
Rho-tagged ORs were transfected into the Hana3A cell line (23) along with a short form of human RTP1, hRTP1S, which enhances functional expression of the ORs (24). Plasmid DNA was transfected using Lipofectamine2000 (Invitrogen). cAMP-response element luciferase reporter was used to measure the OR activation as previously described (23, 35). Data were analyzed with Microsoft Excel and GraphPad Prism 4. We note that the EC50 value of a given receptor can vary between experiments due to a range of varying assay conditions, but the relative sensitivity of the receptor variants remains the same. See SI Materials and Methods for detailed information.
Supplementary Material
Acknowledgments.
We thank D. Marchuk for access to sequencing instrumentation, critical reading, and advice; R. Valdivia for microscope access; R. Molday for anti-rhodopsin antibody; J. Mainland for advice on statistics; Q. Chi, M. Kubota, A. Toyama, and W. L. Liu for expert technical assistance; and L. Vosshall, A. Keller, W. Grus, A. Moehring, H. Amrein, M. Caron, D. Tracey, M. Noor, K. Adipietro, R. Roberts, and X. F. Duan for critical comments on the manuscript. We also thank A. Yoder and S. Zehr for access to lemur samples. This is DLC publication #1167. This research was supported by grants to H.M. from the National Institutes of Health and Human Frontier Science Program and to H.Z. from an National Institutes of Health National Research Service Award Predoctoral fellowship.
Footnotes
The authors declare a potential conflict of interest. The authors plan to submit a patent application relevant to the work.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession nos. GU139530–GU139544).
This article contains supporting information online at www.pnas.org/cgi/content/full/0808378106/DCSupplemental.
References
- 1.Niimura Y, Nei M. Extensive gains and losses of olfactory receptor genes in Mammalian evolution. PLoS ONE. 2007;2:e708. doi: 10.1371/journal.pone.0000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young JM, Trask BJ. The sense of smell: Genomics of vertebrate odorant receptors. Hum Mol Genet. 2002;11:1153–1160. doi: 10.1093/hmg/11.10.1153. [DOI] [PubMed] [Google Scholar]
- 3.Rouquier S, Blancher A, Giorgi D. The olfactory receptor gene repertoire in primates and mouse: Evidence for reduction of the functional fraction in primates. Proc Natl Acad Sci USA. 2000;97:2870–2874. doi: 10.1073/pnas.040580197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilad Y, Man O, Paabo S, Lancet D. Human specific loss of olfactory receptor genes. Proc Natl Acad Sci USA. 2003;100:3324–3327. doi: 10.1073/pnas.0535697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilad Y, Man O, Glusman G. A comparison of the human and chimpanzee olfactory receptor gene repertoires. Genome Res. 2005;15:224–230. doi: 10.1101/gr.2846405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Go Y, Niimura Y. Similar numbers but different repertoires of olfactory receptor genes in humans and chimpanzees. Mol Biol Evol. 2008;25:1897–1907. doi: 10.1093/molbev/msn135. [DOI] [PubMed] [Google Scholar]
- 7.Glusman G, Yanai I, Rubin I, Lancet D. The complete human olfactory subgenome. Genome Res. 2001;11:685–702. doi: 10.1101/gr.171001. [DOI] [PubMed] [Google Scholar]
- 8.Zozulya S, Echeverri F, Nguyen T. The human olfactory receptor repertoire. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-6-research0018. 0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keller A, Zhuang H, Chi Q, Vosshall LB, Matsunami H. Genetic variation in a human odorant receptor alters odour perception. Nature. 2007;449:468–472. doi: 10.1038/nature06162. [DOI] [PubMed] [Google Scholar]
- 10.Dorries K, Adkins-Regan E, Halpern B. Sensitivity and behavioral responses to the pheromone androstenone are not mediated by the vomeronasal organ in domestic pigs. Brain Behav Evol. 1997;49:53–62. doi: 10.1159/000112981. [DOI] [PubMed] [Google Scholar]
- 11.Jacob S, Kinnunen LH, Metz J, Cooper M, McClintock MK. Sustained human chemosignal unconsciously alters brain function. NeuroReport. 2001;12:2391–2394. doi: 10.1097/00001756-200108080-00021. [DOI] [PubMed] [Google Scholar]
- 12.Bensafi M, Brown WM, Khan R, Levenson B, Sobel N. Sniffing human sex-steroid derived compounds modulates mood, memory and autonomic nervous system function in specific behavioral contexts. Behav Brain Res. 2004;152:11–22. doi: 10.1016/j.bbr.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Wyart C, Webster WW, Chen JH, Wilson SR, McClary A, Khan RM, Sobel N. Smelling a single component of male sweat alters levels of cortisol in women. J Neurosci. 2007;27:1261–1265. doi: 10.1523/JNEUROSCI.4430-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pause BM. Are androgen steroids acting as pheromones in humans? Physiol Behav. 2004;83:21–29. doi: 10.1016/j.physbeh.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 15.Kosiol C, et al. Patterns of positive selection in six Mammalian genomes. PLoS Genet. 2008;4:e1000144. doi: 10.1371/journal.pgen.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moreno-Estrada A, et al. Signatures of Selection in the Human Olfactory Receptor OR5I1 Gene. Mol Biol Evol. 2007;25:144–154. doi: 10.1093/molbev/msm240. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen R, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabeti PC, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gimelbrant AA, Skaletsky H, Chess A. Selective pressures on the olfactory receptor repertoire since the human-chimpanzee divergence. Proc Natl Acad Sci USA. 2004;101:9019–9022. doi: 10.1073/pnas.0401566101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurst LD. Evolutionary genomics: A positive becomes a negative. Nature. 2009;457:543–544. doi: 10.1038/457543a. [DOI] [PubMed] [Google Scholar]
- 21.Yokoyama S, Tada T, Zhang H, Britt L. Elucidation of phenotypic adaptations: Molecular analyses of dim-light vision proteins in vertebrates. Proc Natl Acad Sci USA. 2008;105:13480–13485. doi: 10.1073/pnas.0802426105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nozawa M, Suzuki Y, Nei M. Reliabilities of identifying positive selection by the branch-site and the site-prediction methods. Proc Natl Acad Sci USA. 2009;106:6700–6705. doi: 10.1073/pnas.0901855106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 24.Zhuang H, Matsunami H. Synergism of accessory factors in functional expression of mammalian odorant receptors. J Biol Chem. 2007;282:15284–15293. doi: 10.1074/jbc.M700386200. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 26.Gaillard I, Rouquier S, Chavanieu A, Mollard P, Giorgi D. Amino-acid changes acquired during evolution by olfactory receptor 912–93 modify the specificity of odorant recognition. Hum Mol Genet. 2004;13:771–780. doi: 10.1093/hmg/ddh086. [DOI] [PubMed] [Google Scholar]
- 27.Krautwurst D, Yau KW, Reed RR. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell. 1998;95:917–926. doi: 10.1016/s0092-8674(00)81716-x. [DOI] [PubMed] [Google Scholar]
- 28.Menashe I, et al. Genetic elucidation of human hyperosmia to isovaleric acid. PLoS Biol. 2007;5:e284. doi: 10.1371/journal.pbio.0050284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yokoyama S. Phylogenetic analysis and experimental approaches to study color vision in vertebrates. Methods Enzymol. 2000;315:312–325. doi: 10.1016/s0076-6879(00)15851-3. [DOI] [PubMed] [Google Scholar]
- 30.Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: Roles of chance and necessity. Nat Rev Genet. 2008;9:951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- 31.Horth L. Sensory genes and mate choice: evidence that duplications, mutations, and adaptive evolution alter variation in mating cue genes and their receptors. Genomics. 2007;90:159–175. doi: 10.1016/j.ygeno.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 32.Laska M, Wieser A, Hernandez Salazar LT. Olfactory responsiveness to two odorous steroids in three species of nonhuman primates. Chem Senses. 2005;30:505–511. doi: 10.1093/chemse/bji043. [DOI] [PubMed] [Google Scholar]
- 33.Laska M, Wieser A, Salazar LT. Sex-specific differences in olfactory sensitivity for putative human pheromones in nonhuman primates. J Comp Psychol. 2006;120:106–112. doi: 10.1037/0735-7036.120.2.106. [DOI] [PubMed] [Google Scholar]
- 34.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 35.Zhuang H, Matsunami H. Evaluating cell-surface expression and measuring activation of mammalian odorant receptors in heterologous cells. Nat Protoc. 2008;3:1402–1413. doi: 10.1038/nprot.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purvis A. A composite estimate of primate phylogeny. Philos Trans R Soc London B. 1995;348:405–421. doi: 10.1098/rstb.1995.0078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.