Abstract
Due to its numerous environmental extremes, the Tibetan Plateau—the world's highest plateau—is one of the most challenging areas of modern human settlement. Archaeological evidence dates the earliest settlement on the plateau to the Late Paleolithic, while previous genetic studies have traced the colonization event(s) to no earlier than the Neolithic. To explore whether the genetic continuity on the plateau has an exclusively Neolithic time depth, we studied mitochondrial DNA (mtDNA) genome variation within 6 regional Tibetan populations sampled from Tibet and neighboring areas. Our results confirm that the vast majority of Tibetan matrilineal components can trace their ancestry to Epipaleolithic and Neolithic immigrants from northern China during the mid-Holocene. Significantly, we also identified an infrequent novel haplogroup, M16, that branched off directly from the Eurasian M founder type. Its nearly exclusive distribution in Tibetan populations and ancient age (>21 kya) suggest that M16 may represent the genetic relics of the Late Paleolithic inhabitants on the plateau. This partial genetic continuity between the Paleolithic inhabitants and the contemporary Tibetan populations bridges the results and inferences from archaeology, history, and genetics.
Keywords: mtDNA, origin
The Tibetan Plateau is characteristic of most extreme environmental conditions, with high absolute elevation, low temperature, extreme aridity, and hypoxia. Nonetheless, modern humans settled on this plateau by the Paleolithic Age. A number of Paleolithic sites excavated throughout the Tibetan Plateau have been dated to >20 thousand years ago (kya) [Fig. 1 and supporting information (SI) Table S1] (1–3), documenting the earliest human presence on the plateau well before the last glacial maximum (LGM, 22–18 kya). In contrast, evidence from classical genetic studies on the contemporary indigenous Tibetan population argues for a northern East Asian origin during the Neolithic (4), a scenario that seems compatible with the available historic records. According to the Xin Tang Shu (New Tang Annals; 11th century A.D.), proto-Tibetans (“Bo” people) can in fact trace their ancestry to the Di-Qiang, an ancient tribe that resided in northwest China about 3 kya (5). One possibility is that the Late Paleolithic settlers might have been eliminated due to exacerbated environmental conditions during the LGM or the Younger Dryas (12.8–11.6 kya), or were largely, if not completely, replaced by the Neolithic immigrants. This notion receives some support from archaeological observations; in particular, the main type of Neolithic tools excavated on the plateau, microliths, show typical features of the northern Chinese tool culture (6). However, these microliths also display some characteristics of the Tibetan paleoliths (7, 8). This mosaic feature raises another possibility that the Neolithic immigrants had received some contribution from the Paleolithic settlers through either cultural or demic contact.
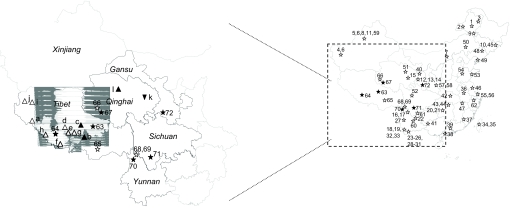
Fig. 1.
Sampling locations of the Chinese populations under study and of the excavated archaeological sites summarized from the literature. Codes 1–72 label the populations (with solid pentacles labeling locations of Tibetan populations collected in this study; see Table S2 for details). Solid triangles refer to excavation sites dating to pre-LGM, open triangles represent sites with Paleolithic age based on lithic comparison, and inverted triangles indicate post-LGM sites (see Table S1 for more details).
Based on the genetic evidence obtained so far from Y chromosome (9, 10) and mitochondrial DNA (mtDNA) (11–13) data, the majority of Tibetan genetic components can trace their origins to the Neolithic immigrants from northern East Asia. No solid genetic evidence indicates the existence of any ancient genetic relics from Paleolithic settlers. Nearly all of the Y chromosome markers in Tibetans analyzed recently (14) are indeed suggestive of more recent genetic inflow, except for the paragroup O3a5*-M134 (comprising the O3a5-M134 Y chromosomes not belonging to O3a5a-M117) which has a more ancient age of 22 kya. The high frequency of haplogroup D-M174 (the Eurasian YAP+ founder haplogroup) in Tibetans had previously led some researchers to propose an additional genetic contribution from Central Asians (9) or to infer an ancient relationship between Tibetans and Japanese (15).
One must concede that most of those genetic studies were hampered by either limited resolution of the classification tree (9, 11, 13), relatively small sample sizes (9–12), or, most importantly, potentially biased sampling coverage, in that most of the Tibetan samples came from the peripheral regions of Tibet, including Yunnan and Qinghai Provinces (9, 10, 12, 13) or from an undifferentiated “general population” (14). Consequently, phylogeographic analyses performed on Tibetans were only rudimentary and proved largely inconclusive, as fine-scale founder types could not be identified.
Results and Discussion
To investigate at a finer scale whether any genetic relics from the Paleolithic inhabitants have survived in the modern Tibetan population, we analyzed 680 individuals, representing 6 populations sampled from all major residential regions of Tibetans across China (Fig. 1 and Table S2), for the (nearly) entire mtDNA control region sequence variation (Table S3). We then selected mtDNAs to sequence the entire genome.
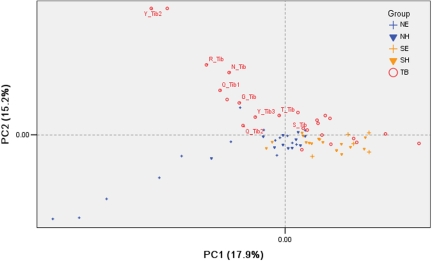
With the exception of a few subjects (classified coarsely as M*, N*, or R*), most of the samples (653/680) were unambiguously allocated to the known Eurasian mtDNA classification scheme (Table S3), among which the vast majority (637/653) belong to (a fraction of) east Asian haplogroups, whereas the remaining ones belong to haplogroups prevalent in either west Eurasians (14/653) or south Asians (2/653). Haplogroups prevalent in northern East Asia, including A, M8 (encompassing M8a, C, and Z), M9, D, and G, were found at relatively high frequencies in Tibetans (64.0% on average; Table S4), an observation consistent with previous reports (4, 11–13). This is also well expressed by a principal components analysis (PCA): most regional Tibetan populations (except for Sichuan-Tibetans) show closer relationships with the northern East Asian populations (Fig. 2).
Fig. 2.
PCA of the populations under study. The map was constructed based on the basal haplogroup frequency matrix in Table S4. N_Tib, Nakchu Tibetan; R_Tib, Shigatse Tibetan; T_Tib, Tibet Tibetan; Q_Tib, Qinghai Tibetan; Y_Tib, Yunnan Tibetan; S_Tib, Sichuan Tibetan; G_Tib, Gansu Tibetan. See Table S3 for details of the group classifications.
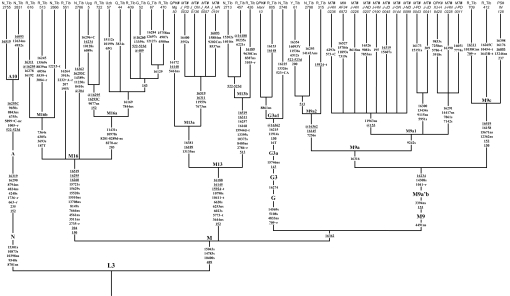
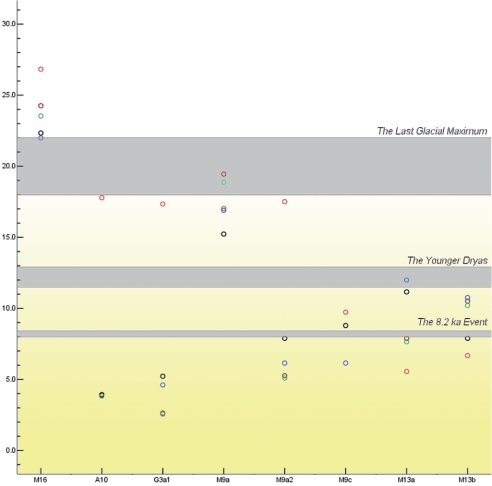
Comparison with the published East Asian mtDNA datasets revealed that most of the East Asian lineages observed in Tibetans nearly match those in other northern East Asian populations (Fig. S1). This reflects rather recent genetic contributions, in agreement with the historically recorded assimilation of ancient Di-Qiang people into the proto-Tibetans (5). Complete sequencing of representative Tibetan lineages (as judged from the control region variation motifs; Table S3) disclosed 3 novel subhaplogroups: M9c, M13b, and A10 (Fig. 3). Phylogenetic analysis showed that several haplogroups, including M9a, M9c, M13a, M13b, G3a1, and A10, are prevalent in Tibetans (Tables S3 and S4). Most of these (M9c, M13a, M13b, and G3a1 in particular) share no terminal but only root types, with their counterparts in other populations from China (Figs. S2–S6). This strongly suggests that these specific lineages have de novo origins within Tibetans. Therefore, these haplogroups could serve as optimal molecular markers for dating the start of the major migration into the Plateau. The situation for haplogroups M9a and A10 seems somewhat different. Both of these haplogroups contain several major clusters that composed nearly exclusively of non-Tibetans or a mixture of Tibetans, Han Chinese, and individuals from the other Chinese ethnic populations, suggesting that haplogroups A10 and M9a had already differentiated before their arrival in Tibet. Fig. 4 summarizes the estimated ages of these haplogroups by adopting the most recent fine-tuning of the calibration rate of different segments of the mtDNA (see Table 1 for details). The ages of M9a, A10, and G3a1 fall into the period of post-LGM warming, whereas M9a2, M9c, M13a, and M13b are likely of early Holocene origin. It is noteworthy that the arrival time of these haplogroups at the Tibetan Plateau may have been somewhat more recent than their coalescent ages would indicate, because some of these haplogroups (A10 and M9a in particular) had already differentiated before their arrival on the plateau (Figs. S2 and S6). It is then conceivable that most, if not all, of these haplogroups may have actually arrived and spread on the plateau only after the 8.2 ka event (8.0–8.4 kya) at the beginning of the Holocene climatic optimum, with first Epipaleolithic and later Neolithic settlers from the upper and middle Yellow River. The distribution and frequencies of the geographically differentiated haplogroups M9a and M13 strikingly parallels that of the Y-chromosome haplogroup D-M174, which has relatively high frequencies (14.0%–72.3%) among most Tibeto-Burman populations and in Japanese (35.1%) (15). Given the limited resolution of the current set of Y-chromosome SNPs and the difficulty of identifying paternal founder types in a population, it is likely that the Y-chromosome haplogroup D-M174 is rather to be compared to the mtDNA macrohaplogroup M. Therefore, the question of whether the Tibetan-Japanese genetic link is of Pleistocene (15) or Holocene origin (as reflected by the sharing of mtDNA haplogroups M9a and M13) still awaits further investigation.
Fig. 3.
Reconstructed mtDNA tree of the completely sequenced representatives of the major Tibetan mtDNA lineages. Suffixes “A,” “C,” “G,” and “T” refer to transversions, “d” denotes deletion, and “+” indicates an insertion event (without specifying the number of inserted nucleotides). Suffixes “Y” and “R” denote heteroplasmic mutations (C/T and A/G, respectively); recurrent mutations are underlined; “@” denotes a reverse mutation; “s” means synonymous and “ns” means nonsynonymous mutation; “−nc” refers to mutations at the intergenic noncoding regions in segments 577–16023; and “∼r” and “∼t” denote mutations in rRNA genes and tRNA genes, respectively. The C stretch length polymorphism in region 303–315 was disregarded for the tree reconstruction. Suffixes “MT#,” “MI#,” “QPK#,” and “PS#” next to the sample names refer to the sources Tanaka et al. (38), Ingman et al. (39), Kong et al. (19, 24), and Soares et al. (40), respectively. Codes “N,” “R,” “Q,” “Y,” and “G” refer to sampling locations (Nakchu, Shigatse, Qinghai, Yunnan, and Gansu, respectively) of different regional Tibetan populations.
Fig. 4.
Estimated ages of the Tibetan-prevalent mtDNA lineages. Open cycles indicate the estimated ages of a haplogroup, with red, brown, green, blue, and black cycles referring to the calibration rates for segment 16051-16400 substitutions (35), coding region synonymous substitutions (35), modified coding region synonymous transitions (34), modified coding region substitutions (34), and complete genome substitutions (35), respectively.
Table 1.
Estimated ages of specific Tibetan mtDNA haplogroups based on different calibration rates
| Haplogroup | n | Control region substitutions |
n | Coding region substitutions |
Coding region synonymous transitions |
Coding region synonymous substitutions |
Complete genome substitution |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Soares rate for segment 16051–16400* |
Modified Mishmar rate† |
Modified Kivisild rate† |
Soares synonymous rate* |
Soares rate* |
||||||||
| ρ ± σ | T (ky) | ρ ± σ | T (ky) | ρ ± σ | T (ky) | ρ ± σ | T (ky) | ρ ± σ | T (ky) | |||
| M16 | 23 | 1.61 ± 0.53 | 26.83 | 13 | 4.77 ± 1.29 | 21.99 | 3.08 ± 1.00 | 23.54 | 3.08 ± 1.00 | 24.26 | 8.23 ± 1.61 | 22.34 |
| [18.04, 35.62] | [16.05, 27.92] | [15.87, 31.21] | [16.35, 32.17] | [17.72, 27.05] | ||||||||
| A10 | 60 | 1.07 ± 0.54 | 17.79 | 2 | 1.00 ± 0.71 | 4.61 | 0.50 ± 0.50 | 3.83 | 0.50 ± 0.50 | 3.94 | 1.50 ± 0.87 | 3.90 |
| [8.75, 26.83] | [1.35, 7.87] | [0.00, 7.65] | [0.00, 7.88] | [1.63, 6.19] | ||||||||
| G3a1 | 25 | 1.04 ± 0.39 | 17.34 | 3 | 1.00 ± 0.75 | 4.61 | 0.33 ± 0.33 | 2.55 | 0.33 ± 0.33 | 2.63 | 2.00 ± 0.94 | 5.22 |
| [10.81, 23.88] | [1.17, 8.05] | [0.00, 5.10] | [0.00, 5.26] | [2.73, 7.74] | ||||||||
| M9a | 143 | 1.02 ± 0.27 | 17.03 | 15 | 3.67 ± 1.14 | 16.90 | 2.47 ± 0.97 | 18.87 | 2.47 ± 0.97 | 19.45 | 5.67 ± 1.32 | 15.23 |
| [12.58, 21.48] | [11.64, 22.16] | [11.46, 26.28] | [11.81, 27.08] | [11.56, 18.98] | ||||||||
| M9a2 | 20 | 1.05 ± 0.42 | 17.51 | 3 | 1.33 ± 0.67 | 6.15 | 0.67 ± 0.47 | 5.10 | 0.67 ± 0.47 | 5.26 | 3.33 ± 1.15 | 7.89 |
| [10.48, 24.54] | [3.07, 9.22] | [1.49, 8.71] | [1.54, 8.97] | [4.94, 10.90] | ||||||||
| M9c | 60 | 0.58 ± 0.19 | 9.73 | 3 | 1.34 ± 0.67 | 6.15 | — | — | — | — | 3.33 ± 1.05 | 8.79 |
| [6.62, 12.84] | [3.07, 9.22] | [5.96, 11.67] | ||||||||||
| M13a | 24 | 0.33 ± 0.19 | 5.56 | 5 | 2.60 ± 1.22 | 11.99 | 1.00 ± 0.82 | 7.65 | 1.00 ± 0.82 | 7.88 | 4.20 ± 1.51 | 11.16 |
| [2.45, 8.67] | [6.38, 17.59] | [1.34, 13.96] | [1.38, 14.39] | [7.06, 15.36] | ||||||||
| M13b | 25 | 0.40 ± 0.16 | 6.67 | 3 | 2.33 ± 0.88 | 10.76 | 1.33 ± 0.67 | 10.20 | 1.33 ± 0.67 | 10.51 | 3.00 ± 1.00 | 7.89 |
| [4.00, 9.34] | [6.69, 14.82] | [5.10, 15.30] | [5.26, 15.77] | [5.22, 10.61] | ||||||||
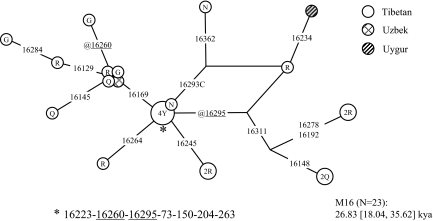
Significantly, one unclassified mtDNA lineage (designated as M* in Table S3), recognized by the control region motif 150-204-16223-16260, has a distribution restricted to Tibetans, with only 2 descendant lineages observed sporadically in Uygur and Uzbek (16); both populations were in contact with Tibetans during the Yuan Dynasty (Fig. 5). The whole mtDNA genome analysis revealed 10 diagnostic coding region mutations (Fig. 3). This lineage, named M16 here, branched off directly from the M founder type and is absent in >5,000 (nearly) complete mtDNA genomes from the worldwide mtDNA database. With hindsight, this lineage was detected in previous studies on Tibetans (11, 13, 16, 17), but has remained unrecognized due to limited information. For instance, among 54 Tibetans, there was one sample (type AS155) with salient RFLP status (−2734AluI, +8148HaeIII, and + 15520HaeIII) which clearly points to M16 status (11). The low frequency of M16 (1.9%; 1/54) in Tibetans sampled from 3 regions of Tibet (Nakchu, Tsedang, and Linchi) (11) is in line with our results (2.1%; 14/680).
Fig. 5.
Constructed median network displaying the control region information of M16 lineages. This network was constructed manually according to Bandelt et al. (31). The data used here were collected from the literature (Table S2) and the present study (Table S3). The sequence information used for network construction was confined to segment 16047-16497. Time estimation was carried out based on segment 16051-16400 as described previously (35). The asterisk denotes the ancestral node of the haplogroup defined by motif 16223-16260-16295-73-150-204-263. See the legend of Fig. 3 for more information.
Comparison of nonsynonymous and synonymous substitutions on the mtDNA protein-encoding genes, as well as on internal and terminal branches of the East Asian mtDNA tree, revealed no significant difference between the variations seen within haplogroup M16 and within other East Asian haplogroups of similar ages (Tables 2 and 3). This indicates that M16 might have been under similar natural selection pressure as the other haplogroups, at least since the beginning of the LGM. This finding is supported by the observation that different timing methods, including the method that considers only synonymous transitions (18), have led to coincident ages of M16 (>21 kya; Table 1 and Fig. 4). The rather long stem of M16 in the mtDNA phylogeny shows a somewhat different picture, with 5 nonsynonymous changes and 1 rRNA mutation, opposed to just 4 synonymous mutations (Fig. 3). A more extreme pattern among the basal East Asian haplogroups is seen only in the stem for haplogroup R11 (19). This could imply that the evolutionary pathway leading to the root of haplogroup M16 might have been under strong selection pressure during the millennia before the LGM, which eventually left M16 as the sole surviving mtDNA lineage among the earliest Paleolithic inhabitants of the Tibetan Plateau.
Table 2.
Comparisons of nonsynonymous and synonymous substitutions between M16 and the other East Asian M lineages
| Gene | M16 |
East Asian M lineages* |
|||
|---|---|---|---|---|---|
| NS† | S‡ | NS† | S‡ | P§ | |
| ND1 | 1 | 3 | 9 | 15 | 1.000 |
| ND2 | 2 | 1 | 8 | 15 | 0.538 |
| COX1 | 0 | 4 | 3 | 21 | 1.000 |
| COX2 | 2 | 1 | 5 | 17 | 0.180 |
| ATP8 | 0 | 1 | 2 | 5 | 1.000 |
| ATP6 | 2 | 0 | 6 | 6 | 0.473 |
| COX3 | 0 | 0 | 6 | 8 | 1.000 |
| ND3 | 0 | 2 | 1 | 3 | 1.000 |
| ND4L | 0 | 0 | 0 | 8 | 1.000 |
| ND4 | 0 | 4 | 7 | 15 | 0.546 |
| ND5 | 1 | 3 | 9 | 24 | 1.000 |
| ND6 | 0 | 1 | 4 | 11 | 1.000 |
| CytB | 2 | 4 | 14 | 13 | 0.656 |
| Totally | 10 | 24 | 74 | 161 | 1.000 |
Table 3.
Comparison of internal and terminal NS/S on the East Asian mtDNA tree
| n* | NSi/SI† | NSt/St† | P | |
|---|---|---|---|---|
| M16 | 13 | 0.78 (7/9) | 0.20 (3/15) | 0.13 |
| East Asian M lineages‡ | 38 | 0.56 (24/43) | 0.42 (50/118) | 0.44 |
| P | 0.58 | 0.29 |
*Sample size.
†Indices “i” and “t” refer to the corresponding fractions of NS/S (see Table 2) for the internal branches and terminal branches, respectively, of the East Asian mtDNA tree. P values were obtained by the 2-tailed Fisher's exact test.
Several characteristics of M16 observed so far, including its basal status on the Eurasian mtDNA tree, restricted distribution in Tibetans, and ancient age (>21 kya), strongly support that this lineage has a different origin from the other Tibetan-prevalent haplogroups of northern East Asian ancestry. The most reasonable explanation for these is that M16 represents the genetic relics of the initial Late Paleolithic settlers on the Tibetan Plateau. This inference is supported by the fact that our dating for the autochthonous haplogroup M16 is very close to the optical dating (20.6–21.7 kya) of the human handprints and footprints found at the center of the Tibetan Plateau (in the vicinity of Lhasa, 4,200 m above sea level) (1).
Our findings have significant implications for the seemingly conflicting inferences drawn from archaeology, genetics, and historic records. Essentially, the previous debate on the peopling of the Tibetan Plateau concerned the issue of whether or not the initial Late Paleolithic inhabitants on the plateau were completely replaced by the later Neolithic immigrants. In this study, the observed genetic continuity between the initial Paleolithic inhabitants and the modern populations on the Tibetan Plateau strongly suggests that modern humans did exist on the plateau before the LGM, and it is these Paleolithic people who have successfully overcome the extremely harsh climate and environments and made some genetic contribution (albeit limited) to the contemporary inhabitants. This also helps to explain why the excavated microliths on the Tibetan Plateau display mosaic features of both northern Chinese tool culture (6) and the Tibetan Paleoliths (7, 8).
In summary, although the vast majority of identified mtDNA lineages found in Tibetans can trace their origins to northern East Asia and may have entered the Tibetan Plateau in the Holocene, our study provides support for the existence of genetic relics of the Late Paleolithic settlers in Tibetans, indicating some genetic continuity between the initial Paleolithic inhabitants and the modern populations on the Tibetan Plateau. Our findings may contribute to resolving the long-standing debates among the fields of archaeology, history, and genetics.
Subjects and Methods
Sampling.
Blood samples from 680 unrelated individuals of 6 Tibetan populations were collected with informed consent. Total DNA was extracted by the standard phenol/chloroform method. The populations were labeled as follows: Nakchu-Tibetans, 168 Tibetans from Nakchu Prefecture of Tibet; Shigatse-Tibetans, 220 Tibetans from Shigatse Prefecture of Tibet; Yunnan-Tibetans, 71 Tibetans from Diqing Tibetan Autonomous Prefecture of Yunnan Province; Qinghai-Tibetans, 76 Tibetans from Qinghai Province; Sichuan-Tibetans, 62 Tibetans from Liangshan Yi Autonomous Prefecture of Sichuan Province; and Gansu-Tibetans, 83 Tibetans from Gannan Tibetan Autonomous Prefecture of Gansu Province.
Sequencing and RFLP Typing.
With the exception of 37 Qinghai-Tibetans, for which only the segment spanning from position 16001 to position 16497 (relative to the revised Cambridge reference sequence, rCRS) (20, 21) was amplified and sequenced as described elsewhere (22), the entire mtDNA control region for the other 643 samples was amplified, sequenced, and dealt with as described previously (23), with minor modifications in the reverse primers [i.e., replacement of the previous reverse primer H408 by H902 (5′-GACTTGGGTTAATCGTGTGAC-3′) or H575 (5′-TGAGGAGGTAAGCTACATAAACTG-3′), to cover more informative sites, such as position 489]. To confirm the haplogroup status inferred from the control region motifs, the following coding region sites were selected for typing by either RFLP or DNA sequencing [according to the reconstructed East Asian mtDNA tree (19, 24)]: 10397AluI (for macrohaplogroup M), 5176AluI/4883 (D), 3008TaqI/3010 (D4), 4831HhaI/4833 (G), 9820HinfI/6455 (M7), 6680 (M7b), 4715 (M8), 14465AccI (M8a), 13262AluI (C), 3391HaeIII/3394 (M9a), 12549 (M10), 10644RsaI (M10a), 7641AluI (M11), 12030 (M12), 6023/6253 (M13), 663HaeIII/663 (A), 5417 (N9), 10310 (F), 12406HincI (F1), 14766 (HV), 7025AluI (H), and 8281–8289del (B). For some Tibetan samples [i.e, 32 Yunnan-Tibetans and 8 Qinghai-Tibetans, for which segments from 16001 to 16497 have been reported by Yao et al. (12)], the corresponding HVS-II segments and some further coding region sites were sequenced and/or screened as well.
Data Analyses.
Based on the combined control region and coding region information, the majority of the samples were unambiguously assigned to haplogroups under the guidance of the reconstructed mtDNA trees of East Asian (19, 24) and South Asian (25, 26) mtDNA lineages. For those Tibetan mtDNAs that remained unassignable, complete mtDNA genome sequencing was performed, as described previously (27), to fully determine their exact phylogenetic status. Specifically, the whole mtDNA genome was amplified in 4 overlapping fragments by using 4 pairs of primers (L13894/H2187, L1677/H6505, L5868/H10718, and L9877/H14676), then each fragment was sequenced using a set of inner primers. (See table 1 in ref. 27 for detailed information on the primers.) With this approach, the previously reported problems in mtDNA genome datasets, such as artificial recombination (28) and amplification of pseudomitochondrial gene (29), could be minimized. Two samples (Uzb57 and Uyg5) reported with M16 control region variation motif (16) (GenBank accession numbers AY678062 and AY678009) were also selected for complete sequencing.
The haplogroup allocations of the reported mtDNA data from the literature (Table S2) were reevaluated by the near-matching strategy (30). The reduced median network for each haplogroup was constructed manually [as described by Bandelt et al. (31)] and then confirmed using Network 4.510 (http://www.fluxus-engineering.com/sharenet.htm). The time to the most recent common ancestor of a haplogroup was estimated as described previously (32–35). PCA was conducted as described previously (30).
The Chinese M sequences used in our comparative analysis of nonsynonymous and synonymous substitution were obtained from the literature (19, 24). Three sequences (GenBank accession numbers AY255153, DQ272115, and DQ272108) belonging to M9a, M13, and G3a1, respectively, were disregarded, because these haplogroups are very frequent in Tibetans and thus might have suffered similar high-altitude selection pressure as M16 did. Mutations were classified into nonsynonymous and synonymous substitutions for each gene using mtDNA-GeneSyn software (36). Each mutation was classified as internal or terminal on the mtDNA tree, as described previously (37). Fisher's exact test was used to examine the difference in each gene between M16 and the other East Asian M lineages.
Supplementary Material
Acknowledgments.
We thank the volunteers for participating in the project. This work was supported by grants from the National Natural Science Foundation of China (30900797 and 30621092), the Chinese Academy of Sciences (Special Grant for the President Scholarship Winner), the Natural Science Foundation of Yunnan Province, and the Kunming Institute of Zoology, CAS (Special Grant for Young Researchers).
Footnotes
The authors declare no conflicts of interest.
This article is a PNAS Direct Submission.
Data deposition: All of the sequences obtained in the present study have been deposited into GenBank, with accession numbers FJ544230-FJ544243, FJ968772-FJ968775, and GU014563-GU014569 (for whole mtDNA genomes) and FJ543469-FJ544148 (for control region sequences).
This article contains supporting information online at www.pnas.org/cgi/content/full/0907844106/DCSupplemental.
References
- 1.Zhang DD, Li SH. Optical dating of Tibetan human hand- and footprints: An implication for the palaeoenvironment of the last glaciation of the Tibetan Plateau. Geophys Res Lett. 2002;29:1072–1074. [Google Scholar]
- 2.Aldenderfer M, Zhang Y. The prehistory of the Tibetan Plateau to the seventh century AD: Perspectives and research from China and the West since 1950. J World Prehist. 2004;18:1–55. [Google Scholar]
- 3.Yuan B, Huang W, Zhang D. New evidence for human occupation of the northern Tibetan Plateau, China during the Late Pleistocene. Chin Sci Bull. 2007;52:2675–2679. [Google Scholar]
- 4.Cavalli-Sforza LL, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton, NJ: Princeton Univ Press; 1994. p. 206. [Google Scholar]
- 5.Wang F-R. In: History of Chinese Ethnic Groups. Wang Z-H, editor. Vol 4. Beijing: China Social Sciences Press; 1994. p. 363. [Google Scholar]
- 6.Huang W-W. In: Early Humankind in China. Wu R-K, Wu X-Z, Zhang S-S, editors. Beijing: Science Press; 1989. pp. 234–235. [Google Scholar]
- 7.Huo W. Archaeological discoveries and research in Tibet in the last decade. Cultural Relics. 2000:85–95. [Google Scholar]
- 8.Li Y-X. Original Art in Tibet. Hebei, China: Hebei Education Press; 2000. pp. 10–37. [Google Scholar]
- 9.Qian Y-P, et al. Multiple origins of Tibetan Y chromosomes. Hum Genet. 2000;106:453–454. doi: 10.1007/s004390000259. [DOI] [PubMed] [Google Scholar]
- 10.Su B, et al. Y chromosome haplotypes reveal prehistorical migrations to the Himalayas. Hum Genet. 2000;107:582–590. doi: 10.1007/s004390000406. [DOI] [PubMed] [Google Scholar]
- 11.Torroni A, et al. Mitochondrial DNA analysis in Tibet: Implications for the origin of the Tibetan population and its adaptation to high altitude. Am J Phys Anthropol. 1994;93:189–199. doi: 10.1002/ajpa.1330930204. [DOI] [PubMed] [Google Scholar]
- 12.Yao Y-G, et al. Genetic relationship of Chinese ethnic populations revealed by mtDNA sequence diversity. Am J Phys Anthropol. 2002;118:63–76. doi: 10.1002/ajpa.10052. [DOI] [PubMed] [Google Scholar]
- 13.Wen B, et al. Analyses of genetic structure of Tibeto-Burman populations reveals sex-biased admixture in southern Tibeto-Burmans. Am J Hum Genet. 2004;74:856–865. doi: 10.1086/386292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gayden T, et al. The Himalayas as a directional barrier to gene flow. Am J Hum Genet. 2007;80:884–894. doi: 10.1086/516757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi H, et al. Y chromosome evidence of earliest modern human settlement in East Asia and multiple origins of Tibetan and Japanese populations. BMC Biol. 2008;6:45. doi: 10.1186/1741-7007-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao Y-G, Kong Q-P, Wang C-Y, Zhu C-L, Zhang Y-P. Different matrilineal contributions to genetic structure of ethnic groups in the Silk Road region in China. Mol Biol Evol. 2004;21:2265–2280. doi: 10.1093/molbev/msh238. [DOI] [PubMed] [Google Scholar]
- 17.Qian Y-P, et al. Mitochondrial DNA polymorphisms in Yunnan nationalities in China. J Hum Genet. 2001;46:211–220. doi: 10.1007/s100380170091. [DOI] [PubMed] [Google Scholar]
- 18.Kivisild T, et al. The role of selection in the evolution of human mitochondrial genomes. Genetics. 2006;172:373–387. doi: 10.1534/genetics.105.043901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong Q-P, et al. Phylogeny of East Asian mitochondrial DNA lineages inferred from complete sequences. Am J Hum Genet. 2003;73:671–676. doi: 10.1086/377718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 21.Andrews RM, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 22.Yao Y-G, Lu X-M, Luo H-R, Li W-H, Zhang Y-P. Gene admixture in the silk road region of China: Evidence from mtDNA and melanocortin 1 receptor polymorphism. Genes Genet Syst. 2000;75:173–178. doi: 10.1266/ggs.75.173. [DOI] [PubMed] [Google Scholar]
- 23.Yao Y-G, Kong Q-P, Man X-Y, Bandelt H-J, Zhang Y-P. Reconstructing the evolutionary history of China: A caveat about inferences drawn from ancient DNA. Mol Biol Evol. 2003;20:214–219. doi: 10.1093/molbev/msg026. [DOI] [PubMed] [Google Scholar]
- 24.Kong Q-P, et al. Updating the East Asian mtDNA phylogeny: A prerequisite for the identification of pathogenic mutations. Hum Mol Genet. 2006;15:2076–2086. doi: 10.1093/hmg/ddl130. [DOI] [PubMed] [Google Scholar]
- 25.Palanichamy M, et al. Phylogeny of mitochondrial DNA macrohaplogroup N in India, based on complete sequencing: Implications for the peopling of South Asia. Am J Hum Genet. 2004;75:966–978. doi: 10.1086/425871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun C, et al. The dazzling array of basal branches in the mtDNA macrohaplogroup M from India as inferred from complete genomes. Mol Biol Evol. 2006;23:683–690. doi: 10.1093/molbev/msj078. [DOI] [PubMed] [Google Scholar]
- 27.Wang H-W, et al. Strikingly different penetrance of LHON in two Chinese families with primary mutation G11778A is independent of mtDNA haplogroup background and secondary mutation G13708A. Mutat Res. 2008;643:48–53. doi: 10.1016/j.mrfmmm.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Kong Q-P, et al. Distilling artificial recombinants from large sets of complete mtDNA genomes. PLoS ONE. 2008;3:e3016. doi: 10.1371/journal.pone.0003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao Y-G, Kong Q-P, Salas A, Bandelt H-J. Pseudomitochondrial genome haunts disease studies. J Med Genet. 2008;45:769–772. doi: 10.1136/jmg.2008.059782. [DOI] [PubMed] [Google Scholar]
- 30.Yao Y-G, Kong Q-P, Bandelt H-J, Kivisild T, Zhang Y-P. Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am J Hum Genet. 2002;70:635–651. doi: 10.1086/338999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bandelt H-J, Macaulay V, Richards M. Median networks: Speedy construction and greedy reduction, one simulation, and two case studies from human mtDNA. Mol Phylogenet Evol. 2000;16:8–28. doi: 10.1006/mpev.2000.0792. [DOI] [PubMed] [Google Scholar]
- 32.Forster P, Harding R, Torroni A, Bandelt H-J. Origin and evolution of Native American mtDNA variation: A reappraisal. Am J Hum Genet. 1996;59:935–945. [PMC free article] [PubMed] [Google Scholar]
- 33.Saillard J, Forster P, Lynnerup N, Bandelt H-J, Norby S. MtDNA variation among Greenland Eskimos: The edge of the Beringian expansion. Am J Hum Genet. 2000;67:718–726. doi: 10.1086/303038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perego UA, et al. Distinctive Paleo-Indian migration routes from Beringia marked by two rare mtDNA haplogroups. Curr Biol. 2009;19:1–8. doi: 10.1016/j.cub.2008.11.058. [DOI] [PubMed] [Google Scholar]
- 35.Soares P, et al. Correcting for purifying selection: An improved human mitochondrial molecular clock. Am J Hum Genet. 2009;84:740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pereira L, et al. The diversity present in 5140 human mitochondrial genomes. Am J Hum Genet. 2009;84:628–640. doi: 10.1016/j.ajhg.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun C, Kong Q-P, Zhang Y-P. The role of climate in human mitochondrial DNA evolution: A reappraisal. Genomics. 2007;89:338–342. doi: 10.1016/j.ygeno.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka M, et al. Mitochondrial genome variation in eastern Asia and the peopling of Japan. Genome Res. 2004;14:1832–1850. doi: 10.1101/gr.2286304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ingman M, Kaessmann H, Pääbo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 40.Soares P, et al. Climate change and postglacial human dispersals in southeast Asia. Mol Biol Evol. 2008;25:1209–1218. doi: 10.1093/molbev/msn068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.