Abstract
Rb/E2F is dysregulated in murine and human pituitary tumors. Pituitary tumor transforming gene (PTTG1), a securin protein, is required for pituitary tumorigenesis, and PTTG1 deletion attenuates pituitary tumor development in Rb+/− mice. E2F1 and PTTG1 were concordantly overexpressed in 29 of 46 Rb+/− murine pituitary tissues and also in 45 of 80 human pituitary tumors (P < 0.05). E2F1 specifically bound the hPTTG1 promoter as assessed by chromatin immunoprecipitation and biotin-streptavidin pull-down assay, indicating that hPTTG1 may act as a direct E2F1 target. Transfection of E2F1 and its partner DP1 dose-dependently activated hPTTG1 transcription up to 3-fold in p53-devoid H1299 cells but not in p53-replete HCT116 cells. E2F1 overexpression enhanced endogenous hPTTG1 mRNA and protein levels up to 3-fold in H1299 cells. The presence of endogenous p53/p21 constrained the induction, whereas knocking down either p53 or p21 in HCT116 cells restored E2F1-induced hPTTG1 transactivation and expression. Moreover, suppressing Rb by small interfering RNA concordantly elevated E2F1 and hPTTG1 protein levels. In contrast, transfection of E2F1 small interfering RNA lowered hPTTG1 levels 24 h later in HCT116 than in H1299 cells, indicating that p53 delays E2F1 action on hPTTG1. These results elucidate a mechanism for abundant tumor hPTTG1 expression, whereby Rb inactivation releases E2F1 to induce hPTTG1. This signaling pathway may underlie the requirement of PTTG1 for pituitary tumorigenesis.
E2F1 abundance induces pituitary tumor transforming gene expression in human pituitary tumors.
Pituitary tumors, accounting for about 15% of intracranial neoplasms, are benign adenomas secreting hormones giving rise to endocrine syndromes, or they may be clinically silent and present with compressive mass effects (1). Although disrupted growth factors, oncogene signaling, and hormonal feedback have been described (2), the proximal pathogenesis of these monoclonal tumors remains elusive. Retinoblastoma protein (Rb) controls the G1/S cell phase transition and enables cell growth by targeting key transcription factors including the E2Fs (3), comprising at least eight E2F and two DP subunits (4). These associate as E2F/DP heterodimers that bind and regulate E2F target genes whose products are required for nucleotide synthesis, DNA replication, and cell cycle progression. These targets include cyclin D1, cyclin E, c-myc, and cdc2 (5). E2F1, the index member, exhibits a yin and yang activity to induce both proliferation and apoptosis, leading potentially to both tumor-promoting as well as tumor-suppressive effects (6). Rb inactivation or phosphorylation leads to E2F release, a hallmark of experimental endocrine tumorigenesis. Rb heterozygous (Rb+/−) mice are viable but develop spontaneous pituitary tumors that originate from the intermediate (85%) or anterior (15%) lobes with almost 100% penetrance (7,8,9). A subset of these mice also develops thyroid medullary C-cell adenomas or hyperplasia (9,10). Endocrine tumorigenesis in Rb+/− mice or embryonic lethality of Rb−/− mice results from dysregulated E2F transcriptional activity. Loss of E2F1 reduces the frequency of pituitary and thyroid tumors and greatly enhances the Rb+/− E2f1−/− mouse lifespan (11). Similar to E2F1, inactivation of pituitary tumor transforming gene (PTTG1) in Rb+/− mice is a protective determinant for pituitary tumorigenesis (12). In contrast, transgenic PTTG1 overexpression facilitates pituitary tumor development (13,14). In human pituitary tumors, Rb phosphorylation is enhanced likely due to deregulated p16 or p27 (15,16,17) with increased cyclin D and E levels (18), thus enhancing E2F release to drive proliferation.
Human PTTG1 (hPTTG1), the index mammalian securin, is a vital component of the spindle checkpoint controlling faithful chromatid separation and has also been identified as a protooncogene (19,20). hPTTG1 facilitates cell cycle progression, maintains chromosomal stability, and mediates in vitro transformation and in vivo tumorigenesis (20,21). hPTTG1 abundance (22) or loss of function (21,23) both result in abnormal mitosis and chromosomal instability. In contrast to restricted normal tissue expression, hPTTG1 is abundantly expressed in pituitary (24,25), thyroid (26), breast (27), esophageal (28), and colorectal tumors (29). hPTTG1 overexpression correlates with tumor invasiveness, differentiation, tumor recurrence, and prognosis. Notably, hPTTG1 has been identified as a key signature gene associated with tumor metastases (30). hPTTG1 activates c-myc (31) and CCND3 (32) as a cotranscription factor enhancing cell proliferation. hPTTG1 interacts with Ku (33) and modulates p53 (34,) participating in DNA damage/repair and apoptosis (35,36). hPTTG1 increases basic fibroblast growth factor and vascular endothelial growth factor expression and induces tumor angiogenesis (37). Several factors induce hPTTG1, including estrogen, insulin, basic fibroblast growth factor and endothelial growth factor (38). In pituitary folliculostellate TtT/GF cells, EGF-induced PTTG1 is mediated by phosphatidylinositol 3-kinase, protein kinase C, and MAPK pathways (39). hPTTG1 is also regulated as a target gene of the β-catenin/TCF pathway in esophageal cancers (40). Nevertheless, proximal regulatory mechanisms for hPTTG1 expression are still unclear.
E2F-mediated induction of cell cycle genes such as cyclin B, cdc2, cdc20, and Bub have expanded the understanding of E2F functions to also include control of mitotic progression, not limited to the G1/S transition (41). hPTTG1 acts as a mitotic spindle checkpoint component in the surveillance of cell cycle progression, and hPTTG1 expression was reduced after E2F3 knockdown in bladder cancer cells as measured by expression cDNA microarray analysis (42). These observations imply that hPTTG1 may act as a downstream E2F target gene. In this study, potential E2F binding sites were identified in the hPTTG1 promoter, indicating a novel regulatory mechanism of hPTTG1 as a direct E2F1 target to promote cell proliferation. Immunoreactive pituitary E2F1 and PTTG1 were concordantly up-regulated in Rb+/− murine pituitary tissues and in human pituitary tumors. E2F1 overexpression induced hPTTG1 transcription and enhanced hPTTG1 mRNA and protein levels in cells devoid of p53. Suppressing endogenous Rb elevated both E2F1 and hPTTG1 protein levels, whereas attenuated E2F1 expression resulted in suppressed hPTTG1. These results elucidate a mechanism for abundant tumor hPTTG expression, whereby Rb inactivation releases E2F1 to induce PTTG1 expression. This signaling pathway may underlie the requirement of PTTG1 for pituitary tumorigenesis.
Results
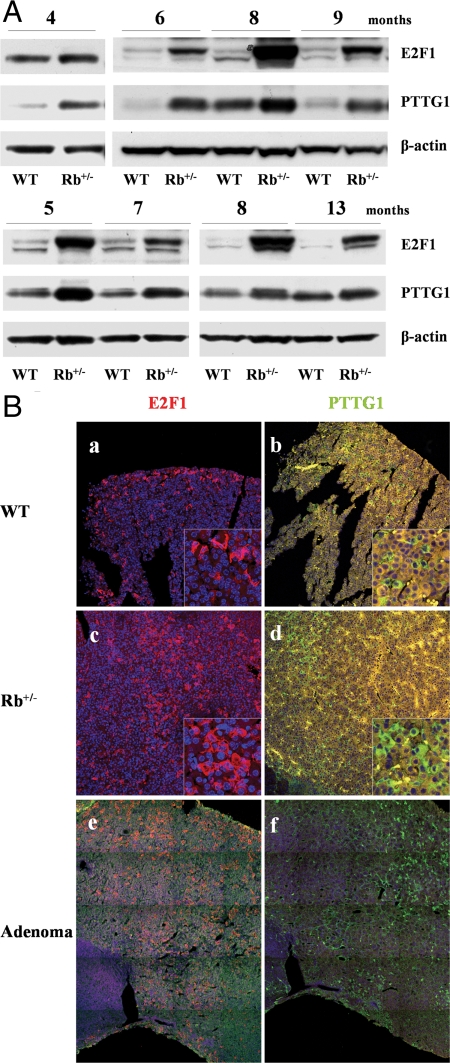
Increased pituitary E2F1 and PTTG1 expression in Rb+/− mice
Rb+/− mice develop pituitary tumors with high penetrance by 8–12 months, whereas PTTG1 inactivation attenuates Rb+/− pituitary tumor development (12). Because Rb phosphorylation or mutation leads to E2F release, we assessed E2F1 and PTTG1 expression by Western blot and confocal immunofluorescence in pituitary tissues derived from Rb+/− and age- and gender-paired wild-type (WT) mice. Increased pituitary E2F1 (∼7-fold) and PTTG1 protein levels (2.6-fold) were, respectively, observed in Rb+/− mice compared with WT. Twenty-nine of 46 Rb+/− mice (63%) exhibited concordant up-regulation of pituitary E2F1 and PTTG1. Figure 1A shows representative E2F1 and PTTG1 Western blots of Rb+/− and WT pituitary glands, and Table 1 summarizes pituitary E2F1 and PTTG1 increases at different ages. Both pituitary E2F1 and PTTG1 expression were higher than WT, whereas enhanced PTTG1 expression was attenuated in older animals. When assessed by confocal immunofluorescence, pituitary E2F1 and PTTG1 protein were found to be concordantly overexpressed in eight of 11 Rb+/− mice (73%) (Table 2). Elevated pituitary E2F1 and PTTG1 expression were observed in 6- to 10-month-old mice already harboring pituitary tumors, and also in 2- to 3-month-old mice before tumor formation, indicating early E2F1 and PTTG1 up-regulation in the pretumorous pituitary. Representative mouse pituitary specimens exhibiting immunoreactive E2F1 and PTTG1 up-regulation in 2-month-old Rb+/− mouse are shown in Fig. 1B (a–d). Twelve percent of Rb+/− pituitary cells expressed E2F1 (vs. 6% of WT cells), and 9% of Rb+/− pituitary cells expressed PTTG1 (vs. 3% of WT cells). Larger scanning fields showing abundant E2F1 and PTTG1 expression and similar distribution in a pituitary tumor derived from a 6-month-old Rb+/− mouse are depicted in Fig. 1B, e and f.
Figure 1.
Increased pituitary E2F1 and PTTG1 expression in Rb+/− mice. A, Representative Western blots of pituitary E2F1 and PTTG1 expression in Rb+/− and paired WT mice (3–15 months old). B, E2F1 and PTTG1 immunoreactivity in Rb+/− and WT pituitary. E2F1 staining is higher in 2-month-old Rb+/− (c) than paired WT (a) mouse pituitary, and hPTTG1 expression is enhanced in 2-month-old Rb+/− (d) vs. paired WT (b) mouse pituitary. Image size, 750 × 750 μm; corner insets size, 100 × 100 μm. Abundant E2F1 (e) and PTTG1 expression (f) in 6-month-old Rb+/− pituitary tumor. Images depict a combined scan field sized approximately 1000 × 1250 μm. Red signal, E2F1 staining; green signal, PTTG1 staining; blue signal, DAPI nuclear staining; yellow signal, autofluorescence background.
Table 1.
Pituitary E2F1 and PTTG1 expression in paired Rb+/− and WT mice
| Age (months) | WT (n) | Rb+/− (n) | Concordantly increased E2F1 and PTTG1 (n) | E2F1 expression/Rb+/− pituitary (fold increase) | PTTG1 expression/Rb+/− pituitary (fold increase) |
|---|---|---|---|---|---|
| <6 | 14 | 19 | 13 | 2.7 ± 0.5 | 2.5 ± 0.6 |
| 6–7 | 5 | 9 | 7 | 5.4 ± 1.5 | 4.0 ± 1.4 |
| 8–9 | 14 | 14 | 6 | 12.7 ± 2.3 | 2.0 ± 0.3 |
| >12 | 2 | 4 | 3 | 8.9 ± 2.4 | 1.4 ± 0.1 |
| Total | 35 | 46 | 29 (63%) | 7.1 ± 1.1a | 2.6 ± 0.4a |
Pituitary E2F1 and PTTG1 expression were detected in 46 Rb+/− and 35 paired WT mice by Western blot. Concordantly increased E2F1 and PTTG1 were observed in 29 Rb+/− mice compared with WT. E2F1 and PTTG1 fold increase in Rb+/− pituitary glands were normalized to WT (as standard 1).
P < 0.001.
Table 2.
Increased pituitary E2F1 and PTTG1 expression in paired Rb+/− and WT mice
| Genotype | Age (months)
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2
|
3
|
6
|
7
|
10
|
||||||||||||
| Rb+/− | WT | Rb+/− | WT | Rb+/− | WT | Rb+/− | WT | Rb+/− | WT | Rb+/− | WT | Rb+/− | WT | Rb+/− | WT | |
| Gender | M | M | F | F | M | M | F | F | F | F | F | F | M | M | M | M |
| Pituitary tumor | No | No | No | No | Yes | Yes | No | Yes | ||||||||
| Pituitary E2F1 expression (%)a | 12 | 6 | 10 | 5 | 8 | 4 | 6 | 1 | 16 | 10 | 15 | 8 | 23 | 14 | 10 | 7 |
| Pituitary PTTG1 expression (%)a | 9 | 3 | 6 | 0.1 | 2 | 0.7 | 3 | 1 | 18 | 0.2 | 6 | 3 | 14 | 7 | 6 | 0.1 |
E2F1 and PTTG1 overexpression were observed in eight of 11 Rb+/− mice as assessed by confocal immunofluorescence. In three Rb+/− and WT mice, E2F1 was unchanged and PTTG1 was undetectable (data not shown). E2F1 expression was 12.8 ± 1.4 in Rb+/− vs. 8.8 ± 1.5 in WT (n = 11; P < 0.01); PTTG1 expression was 5.8 ± 1.8 in Rb+/− vs. 1.4 ± 0.7 in WT (n = 11; P < 0.05). F, Female; M, male.
Percentage of immunopositive pituitary cells.
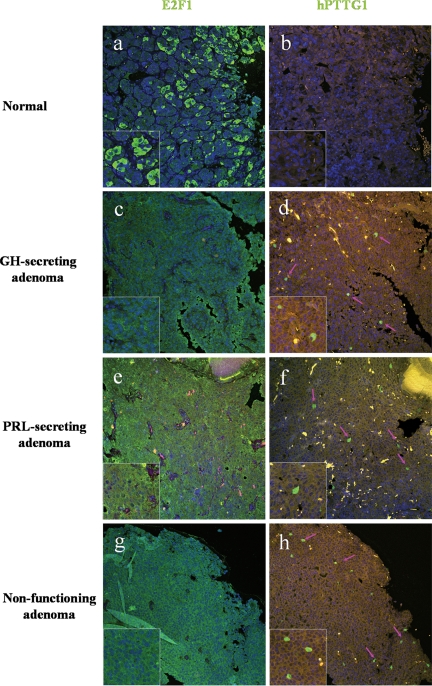
Concordant E2F1 and hPTTG1 expression in human pituitary tumors
hPTTG1 is overexpressed in human pituitary, breast, colorectal, and uterine tumors (38). We therefore assessed E2F1 and hPTTG1 expression in human pituitary tumor samples by confocal imaging. We studied three normal pituitary tissues, and as expected, normal pituitary tissue was not immunoreactive for hPTTG1. Modest cytoplasmic E2F1 staining was detected in up to 25% of normal pituitary cells, whereas both cytoplasmic and nuclear E2F1 staining was observed in most (up to 95%) pituitary tumor cells. In 80 pituitary tumors including one ACTH-secreting, 17 GH-secreting, 13 prolactin-secreting, and 49 nonfunctioning tumor specimens, 83% (66 of 80) and 63% (50 of 80) of pituitary tumors exhibited up-regulated E2F1 and hPTTG1, respectively. As indicated in Table 3, 45 tumors with high E2F1 expression were also immunoreactive for hPTTG1, and nine cases with unchanged E2F1 levels did not exhibit an enhanced hPTTG1 signal. Taken together, 68% of tumor samples (54 of 80) exhibited a concordant pattern of E2F1 and hPTTG1 expression that correlated positively (P < 0.05, Pearson χ2 test) (Table 3 and Fig. 2). hPTTG1 overexpression correlated with proliferation. Pituitary tumor hPTTG1 up-regulation was concordant with elevated Ki-67 staining (P < 0.01) (data not shown). Moreover, in 17 GH-secreting tumors, higher hPTTG1 expression was observed in macroadenomas than in microadenomas (macro, 2.66 ± 1.06, n =12, vs. micro, 0.47 ± 0.28, n = 5; P < 0.05).
Table 3.
E2F1 and hPTTG1 expression in 80 human pituitary tumors
| hPTTG1
|
Total | ||
|---|---|---|---|
| Overexpression | No change | ||
| E2F1 | |||
| Overexpression | 45 | 21 | 66 |
| No change | 5 | 9 | 14 |
| Total | 50 | 30 | 80 |
E2F1 and hPTTG1 immunoreactivity were assessed by confocal microscopy in 80 human pituitary tumor specimens (Fig. 2). Correlation was analyzed using Pearson χ 2 test; P < 0.05.
Figure 2.
E2F1 and hPTTG1 immunoreactivity in human pituitary tumor specimens. A and B, Low E2F staining and negative hPTTG1 reactivity in normal pituitary tissue; C–H, high E2F1 and hPTTG1 expression in a nonfunctioning, prolactin (PRL)-secreting and GH-secreting pituitary tumor, respectively. Pink arrows indicate positive hPTTG1 staining cells. Image size, 750 × 750 μm. Green signal, E2F1 or hPTTG1 staining; blue signal, DAPI nuclear staining; yellow signal, autofluorescence background.
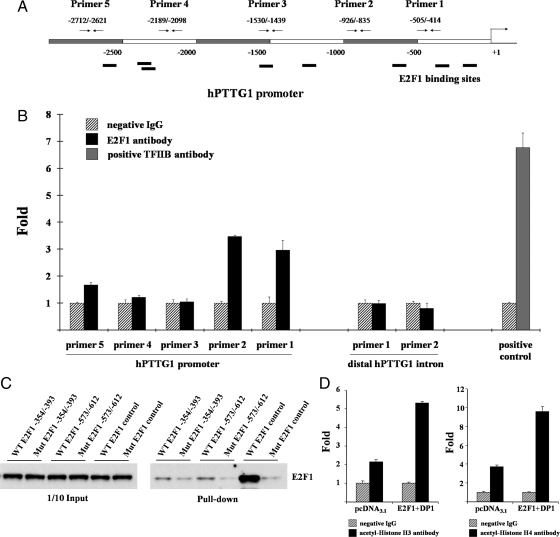
E2F1 binds the hPTTG1 promoter and increases histone acetylation on the hPTTG1 promoter
Eight potential E2F1 binding motifs were detected by screening the hPTTG1 promoter region with Genomatix MatInspector (http://www.genomatix.de/) (Fig. 3A). We performed chromatin immunoprecipitation (ChIP) in both H1299 and HEK293 cells to identify E2F1 binding to the hPTTG1 promoter in vivo. About 107 cells were fixed and sonicated into 200- to 1000-bp chromatin DNA fragments. Equal amounts of chromatin DNA (normalized inputs) were incubated separately with negative IgG control, E2F1 antibody, or positive control TFIIB antibody. Antibodies captured specific chromatin DNA fragments and then bound to subsequently added protein G beads. Chromatin DNA pulled down by protein G beads was detected as template using real-time PCR. Five hPTTG1 promoter primer pairs designed for real-time PCR included primer 1, closest to the ATG translational initial site, and primer 5 the furthest (Fig. 3A). Two negative anti-E2F1 control primers were designed to assess the contribution of nonspecific antibody binding. Both primers were located in the distal 3′ intron region and at least 1.2 kb from any E2F-binding site. As shown in Fig. 3B, anti-E2F1 immunoprecipitated DNA with the enriched E2F1 locus was amplified by hPTTG1 promoter primers 1–2 and 5 but not by primers 3–4 or negative control primers, indicating specific E2F1 binding to the hPTTG1 promoter. Amplifications obtained using primers 1 and 2 were higher than those derived from primer 5, implying that E2F1 binding motifs closer to ATG in the hPTTG1 promoter exhibit higher binding affinity.
Figure 3.
E2F1 binds the hPTTG1 promoter, and E2F1 overexpression promotes acetylation of histone H3 and H4 on the hPTTG1 promoter. A, Schematic representation of potential E2F1 binding sites on hPTTG1 promoter and five primers designed for ChIP. B, E2F1 binds the hPTTG1 promoter. Normalized inputs of H1299 chromatin DNA were pulled down by E2F1 or negative IgG antibodies, and the DNA template was amplified by real-time PCR with specific hPTTG1 promoter primers 1–5 (mentioned in Materials and Methods). Two negative control primers located in distal hPTTG1 intron were designed to assess the contribution of nonspecific binding. TFIIB antibody was used as a positive control for the ChIP assay. Hatched bars, incubation with negative control IgG; black bars, incubation with E2F1 antibody (primers 1–5); gray bar, incubation with positive control TFIIB antibody. Each negative IgG control was normalized to unit 1, each real-time PCR was performed in triplicate, and ChIP experiments were repeated twice. C, Western blot of biotin-streptavidin pull-down assay. Equal amounts of H1299 nuclear extract were incubated with one of the following oligonucleotides: WT E2F1 −354/−393 or −573/−612, intact sequences containing WT E2F1-binding motif; Mut E2F1 −354/−393 or −573/−612, devoid of vital core sequences; and consensus WT or Mut E2F1 as positive control. DNA-protein complexes precipitated by streptavidin-agarose were used to detect E2F1 by Western blot. The left 1/10 input lanes confirmed equal extract aliquots added to each reaction. D, E2F1 overexpression promotes acetylation of histone H3 and H4 on the hPTTG1 promoter. H1299 cells were transfected with pcDNA3.1 or E2F1 and DP1. Normalized inputs of chromatin DNA lysed from pcDNA3.1 or E2F1 and DP1 transfectants were pulled down by acetyl-histone H3, H4, or negative IgG antibodies, and the DNA template was amplified by real-time PCR with specific hPTTG1 promoter primers 2. Hatched bars, incubation with negative control IgG; black bars, incubation with acetyl-histone H3 antibody (left) or acetyl-histone H4 antibody (right). Each negative IgG control was normalized to unit 1, each real-time PCR was performed in triplicate, and ChIP experiments were repeated twice.
We next performed biotin-streptavidin pull-down to verify a direct interaction between E2F1 protein and E2F1 binding motifs at −364 and −585 bp on the hPTTG1 promoter. Equal amounts of H1299 nuclear extract (600 μg) were incubated with one of the following oligonucleotides: WT E2F1 −354/−393, mutant (Mut) E2F1 −354/−393, WT E2F1 −573/−612, Mut E2F1-573/−612, WT E2F1 control, or Mut E2F1 control. Oligonucleotides bound to specific proteins were subsequently precipitated with steptavidin beads by biotin-labeled 5′-ends. The pulled-down DNA-protein complex was used to detect E2F1 by Western blot (Fig. 3C). WT E2F1 −354/−393 and −573/−612 both specifically bound E2F1, and E2F1 −354/−393 binding was abrogated by about 60%, and −573/−612 binding abrogated by about 85% with mutant E2F1 oligonucleotides. The consensus E2F1 binding sequence and mutant oligonucleotides were used as controls. Inputs confirmed equal amounts of nuclear proteins added in each reaction. These results indicate that E2F1 binding motifs located at −364 and −585 bp on the hPTTG1 promoter participate in direct binding to E2F1.
E2F1–3 complexes are required for histone H3 and H4 acetylation (43). To analyze E2F1-influenced histone acetylation status on the hPTTG1 promoter in vivo, we separately transfected pcDNA3.1 control vector or E2F1 and DP1 expression plasmids to H1299 cells and subsequently subjected transfectant lysates to ChIP assays using anti-acetylated histone antibodies. H1299 transfectants were fixed and sonicated after 24 h. Equal amounts of chromatin DNA (normalized inputs) lysed from pcDNA3.1 or E2F1 and DP1 transfectants were, respectively, incubated with negative IgG control, acetyl-histone H3 or acetyl-histone H4 antibodies. Precipitated chromatin DNA was amplified with hPTTG1 promoter primer 2 by real-time PCR. As shown in Fig. 3D, compared with normalized negative IgG control (hatched bar denoted as 1), pcDNA3.1 transfectants exhibited approximately 2-fold induction of acetyl-histone H3 binding on the hPTTG1 promoter. E2F1 and DP1 overexpression enhanced acetyl-histone H3 binding, because E2F1 and DP1 transfectants showed increased binding (∼5.5-fold), higher than observed with pcDNA3.1 controls. Similar results were obtained with acetyl-histone H4 antibody (Fig. 3D); E2F1 and DP1 cotransfection promoted the acetyl-histone H4 binding of hPTTG1 promoter approximately 10-fold, higher than pcDNA3.1 control (∼4-fold). These results imply that E2F1 overexpression promotes histone acetylation on the hPTTG1 promoter, facilitating transcription factor access.
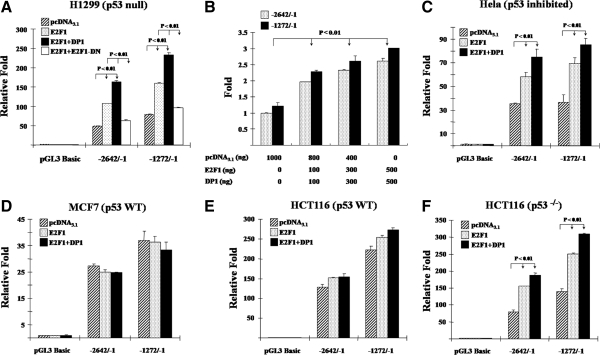
E2F1 activates hPTTG1 transcription
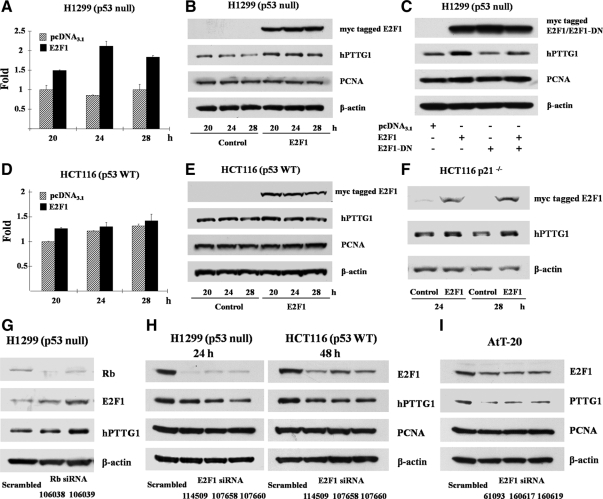
To measure hPTTG1 promoter activity in response to E2F1, we cloned two hPTTG1 promoter fragments, −2642/−1 and −1272/−1, into the pGL3-Basic luciferase reporter vector. The −2642/−1 construct contains all eight E2F1 binding motifs, whereas the shorter −1272/−1 construct contains E2F1 binding sites that exhibited higher binding affinity in ChIP assays. We cotransfected empty control, E2F1 only, E2F1 and DP1, or E2F1 and E2F1-DN (dominant negative) together with hPTTG1 promoter constructs (−2642/−1 or −1272/−1) to measure transcriptional activity by a dual-luciferase reporter system in p53-deficient H1299 cells. E2F1 overexpression or E2F1 and DP1 cotransfection transactivated both the −2642/−1 and −1272/−1 promoters approximately 3.3-fold compared with empty vector pcDNA3.1, whereas the transactivation was abolished by E2F1-DN (Fig. 4A). Similar results were confirmed in HeLa cells containing HPVE6 protein, an inhibitor of p53 (Fig. 4C). Cotransfection of E2F1 and DP1 forms more heterodimers, enhances DNA binding (44), and also leads to further hPTTG1 promoter transactivation than E2F1 transfection alone (Fig. 4, A and C). Experimental inductions were normalized to the background empty vector pGL3-Basic control. The shorter −1272/−1 fragment exhibits higher activity than the −2642/−1 promoter, and both responded dose-dependently in H1299 cells (Fig. 4B). Notably, E2F1- and DP1-induced hPTTG1 promoter activation was not observed in p53 WT MCF7 (Fig. 4D) and HCT116 cells (Fig. 4E), indicating that the presence of an intact p53/p21 pathway constrains transactivation. In p53-knockout HCT116 cells, E2F1 and DP1 induced −2642/−1 and −1272/−1 hPTTG1 promoter activity approximately 2.5-fold (Fig. 4F), similar to results obtained in HCT116 p21−/− cells (data not shown).
Figure 4.
E2F1 activates hPTTG1 transcription. E2F1 activates hPTTG1 transcriptional activity in H1299 (A) and Hela cells (C) but not in MCF7 (D) and HCT116 cells (E). Cells (each well of a 24-well plate) were cotransfected with 200 ng pGL3-Basic or either of hPTTG1 promoter −2642/−1 or −1272/−1, together with 800 ng empty vector pcDNA3.1 (hatched bar), 400 ng E2F1 and 400 ng pcDNA3.1 (dotted bar), 400 ng E2F1 and 400 ng DP1 (black bar), or 400 ng E2F1 and 400 ng E2F1-DN (white bar). B, Dose dependency of E2F1-induced hPTTG1 transcriptional activity in H1299 cells. H1299 cells were cotransfected with 200 ng hPTTG1 promoter −2642/−1 (dotted bar) or −1272/−1 (black bar), together with increasing amounts of E2F1 and DP1 (each from 100–500 ng). Empty vector pcDNA3.1 was used to adjust equivalent amounts of DNA for transfection. Results are plotted as arbitrary units with controls (transfected with empty vector pcDNA3.1) set as 1. F, Deleted p53 in HCT116 cells abolished p53 suppression of E2F1-induced hPTTG1 elevation. Relative activities were normalized by empty vector pGL3-Basic as background. Five nanograms of pRL-TK plasmid were cotransfected to normalize transfection efficiency. Assays were performed 24 h after transfection. Transfections were performed in triplicate for each experiment and repeated four times.
E2F1 up-regulates endogenous hPTTG1 expression
To assess whether E2F1 induces endogenous hPTTG1 mRNA and protein expression, we transiently transfected empty vector pcDNA3.1 or the E2F1 expression plasmid to H1299 and HCT116 cells. After 20–28 h, E2F1 transfectants exhibited enhanced hPTTG1 mRNA levels (∼2.1-fold) in H1299 (Fig. 5A) but not in HCT116 cells (Fig. 5D), as measured by real-time PCR using β-actin as an internal control. Cell lysates analyzed by Western blot verified that hPTTG1 protein expression followed a pattern similar to mRNA levels; i.e. overexpressed E2F1 enhanced hPTTG1 protein expression approximately 3-fold in H1299 cells (Fig. 5B). We did not detect obvious alterations of hPTTG1 in p53 WT HCT116 cells (Fig. 5E), similar to the unaltered mRNA levels observed. In p53-deficient H1299 cells, transfection of E2F1-DN attenuated endogenous hPTTG1 expression and also abolished WT E2F1-induced hPTTG1 expression (Fig. 5C). Although E2F1 participates in cell cycle progression and proliferation, E2F1-induced hPTTG1 was not cell cycle dependent, because no obvious cell cycle changes were evident in both H1299 and HCT116 cell controls and in E2F1 transfectants for up to 28 h (data not shown). We also used proliferating cell nuclear antigen (PCNA) as a late G1/S indicator, and PCNA expression was equally sustained in both controls and E2F1 transfectants. Thus, hPTTG1 levels may increase before aberrant cell cycle progression induced by E2F1. Furthermore, induction of hPTTG1 was sensitive to p53/p21 inhibition in HCT116 cells, where induction was not observed. As shown in Fig. 5F, E2F1 overexpression elevated hPTTG1 protein levels in HCT116 p21−/− cells. We also transfected p21 small interfering RNA (siRNA) in HCT116 cells to impair p21 expression and detected up-regulation of E2F1-induced hPTTG1 (data not shown).
Figure 5.
E2F1 induces hPTTG1 expression. E2F1-induced hPTTG1 mRNA (A) and protein expression (B) in H1299 cells but not in HCT116 cells (D and E). Cells were transfected with empty vector pcDNA3.1 or E2F1. hPTTG1 mRNA levels were detected by real-time PCR (A and D); myc-tagged E2F1 and hPTTG1 protein levels were measured by Western blot (B and E). Control is empty vector pcDNA3.1. E2F1-induced hPTTG1 was abolished by E2F1-DN in H1299 cells (C) and not observed in cells in the presence of p53 (E) but detected in HCT116 p21−/− cells (F). G, Impairing endogenous Rb expression concordantly elevated E2F1 and hPTTG1 protein in H1299 cells. H1299 cells were transfected with negative control siRNA or Rb siRNA 106038 or 106039 for 48 h. H, Suppressing endogenous E2F1 by siRNA attenuated hPTTG1 expression in both H1299 and HCT116 cells. H1299 and HCT116 cells were transfected with negative control siRNA, E2F1 siRNA 114509, 107658, or 107660 and harvested after 24 or 48 h, respectively. I, Suppressing endogenous E2F1 by siRNA attenuated hPTTG1 expression in mouse pituitary AtT-20 cells. AtT-20 cells were transfected with negative control siRNA, mouse E2F1 siRNA 61093, 160617, or 160619. Scrambled is negative control siRNA. Rb, E2F1, and hPTTG1 protein were detected by Western blot. β-Actin was used as an internal control, and proliferating cell nuclear antigen was used as a late G1/S indicator of the cell cycle.
Because inactivation of tumor Rb increases E2F1 in vivo, we transfected Rb siRNA (100 nm) to H1299 cells to inhibit endogenous Rb expression. These transfectants exhibited both E2F1 and hPTTG1 up-regulation (Fig. 5G). In contrast, impairing E2F1 expression by siRNA attenuated hPTTG1 expression in both H1299 and HCT116 cells (Fig. 5H). After suppressing E2F1 expression, hPTTG1 attenuation was observed later in HCT116 (48 h) than in H1299 cells (24 h). Because no human pituitary cell lines are available, we tested mouse pituitary AtT-20 cells and confirmed that attenuating E2F1 suppressed PTTG1 expression (Fig. 5I). Furthermore, no p53-deficient pituitary cell lines are available.
Discussion
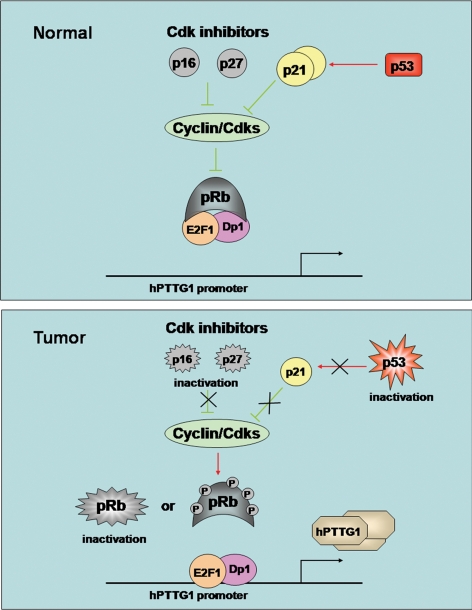
The results shown here indicate cell cycle-independent induction of hPTTG1 by E2F1. We observed eight potential E2F binding motifs in the hPTTG1 promoter and showed E2F1 binding and activation of the promoter. The results indicate that Rb inactivation indirectly increases PTTG1 expression, mediated by E2F1. This novel regulatory mechanism for intracellular hPTTG1 expression expands our understanding of E2F1 participation in the cell cycle that influences G1/S and may also control G2/M by regulating downstream genes involved in mitosis. Rb inactivation or phosphorylation leads to release of E2F transcription factors, enabling cell proliferation (3). Rb+/− mice develop pituitary tumors and thyroid medullary adenomas (7,8,9,10), which result from dysregulated E2F. Deletion of E2F1 in these animals reduces the frequency of both pituitary and thyroid tumors (11). PTTG1 inactivation in Rb+/− mice is also a protective determinant for pituitary tumor formation (12), and transgenic pituitary-directed PTTG1 overexpression facilitates pituitary tumor development (13,14). PTTG1 abundance correlates with pituitary gland plasticity and pituitary tumorigenic potential (13,14), and Pttg-null mice exhibit pituitary hypoplasia (45). Pituitary cells derived from pituitary-targeted PTTG1-overexpressing transgenic mice exhibit enlarged nuclei and marked compound chromatin redistribution. These morphological changes were even more remarkable in transgenic αGSU.PTTGxRb+/− pituitary cells (13). We show here that E2F1 and PTTG1 are concordantly up-regulated in pituitary derived from Rb+/− mice and also in human pituitary tumors. Pituitary tumors were evident in 6- to 10-month-old Rb+/− mice; however, elevated E2F1 and PTTG1 expression was observed as early as 2 months of age, indicating that E2F1 and PTTG1 increases are early events in pituitary tumor formation. By this token, up-regulated PTTG1 and disrupted G2/M checkpoint surveillance may explain, at least in part, pituitary tumor progress in this mutated genetic model. In human pituitary tumors, expression of p16 and p27 cyclin-dependent kinase (Cdk) inhibitors are low, likely due to methylation (15,16). Consequent high levels of cyclin D and cyclin E (18) enhance Rb phosphorylation, releasing E2F1. Although Rb mutations are not commonly encountered in human pituitary tumors, loss of heterozygosity at 13q14 has been reported in about 20% of invasive pituitary adenomas (46). Furthermore, high-mobility group AT-hook 2 has been shown to bind Rb and induce E2F1 activity in prolactinomas (47). Thus, the aberrantly activated pituitary Rb/E2F1 pathway may induce abundant PTTG1 expression, resulting in unfaithful chromatid separation (20), chromosomal instability (48), and loss of cell growth control (22) and subsequently triggering tumorigenesis (14) (Fig. 6).
Figure 6.
Aberrantly activated Rb/E2F1 pathway induces abundant tumor PTTG1 expression. The aberrantly activated tumor Rb pathway releases E2F1 to induce abundant hPTTG1 expression. Thus, hPTTG1 overexpression may result in chromosomal instability and proliferation and subsequently facilitate tumorigenesis.
p53, which is disrupted in about 50% of human tumors, activates p21/WAF1/CIP1, which inhibits Cdks mediating cell cycle progression (49). Cdk suppression inhibits Rb phosphorylation; thus, mutated p53 and Rb both cooperate in acceleration of tumorigenesis (10). Mice deficient in both p53 and Rb develop primarily endocrine tumors, specifically arising in the pituitary, thyroid, and pancreas. These mice, heterozygous for both mutant p53 and Rb, showed a faster rate of tumor development and decreased tumor age onset compared with singly deficient counterparts (50). Unfortunately, because no human pituitary cell lines are available, we employed WT and Mut HCT116 (kindly provided by Dr. Bert Vogelstein) and H1299 cells to further test mechanisms for our in vivo observations. We show that overexpressed E2F1-induced hPTTG1 transcriptional activity and expression of both endogenous mRNA and protein levels in p53-deficient H1299 but not in p53-replete WT HCT116 cells. Knocking down either p53 or p21 resulted in Rb phosphorylation and E2F1 release; thus, restored E2F1-induced hPTTG1 transcriptional activity and expression in HCT116 cells. This mechanism could explain the finding of abundant tumor hPTTG1 expression, whereby Rb dysfunction releases E2F leading to hPTTG1 accumulation. hPTTG1 functions to control the fidelity of chromosome separation during mitosis, and strictly balanced intracellular hPTTG1 levels are vital to maintain normal cell cycle progression and chromosome stability. Our results suggest that as a cell cycle gatekeeper, p53 exhibits a protective function on hPTTG1 expression, because p53 constrains hPTTG1 up-regulation and also delays hPTTG1 down-regulation to avoid abnormal intracellular hPTTG1 fluctuations. siRNA-mediated E2F1 suppression also attenuated hPTTG1 expression both in H1299 and HCT116 cells. We detected hPTTG1 protein expression after E2F1 siRNA transfection for up to 72 h. Decreased hPTTG1 in H1299 cells was observed after 24 h; however, reduced hPTTG1 levels in HCT116 cells were evident only after 48 h.
The p53/p21 pathway thus appears to constrain E2F1-induced PTTG1; on the other hand, Pttg-deficient mice exhibit high p21 levels (12,37). Altered pituitary PTTG abundance inversely correlates with p21 expression, and pituitary p21 levels are induced in Pttg1−/− mice (12). Targeted E2F1-3 disruption leads to p21CIP1 induction and cell cycle arrest (51). Ablation of p53 in E2F1–3-deficient cells prevented p21CIP1 induction and, surprisingly, restored both expression of E2F target genes and the capacity of these cells to proliferate (51). Thus, our results support the notion that the interactive loop between E2F1/PTTG1 and p53/p21 pathways accelerates PTTG1 accumulation in tumors associated with Rb inactivation.
E2F coordinates mitotic genes (41), affecting cell cycle progression both at the S phase and during mitosis. Importantly, the spindle checkpoint protein Mad2, also a transcriptional target of E2F, contributes to aneuploidy, unmasking a direct link for regulation of cell-cycle progression to genomic instability, implying that aneuploidy is an early event in tumor formation (52). Here, we present evidence that another spindle checkpoint protein, hPTTG1, exhibits features of an E2F1 target. Overexpressed hPTTG1 results in chromosomal instability (22), and pituitary cells in PTTG1-overexpressing transgenic mice exhibit enlarged nuclei and marked redistribution of chromatin, features more prominent in αGSU.PTTGxRb+/− mice pituitaries (13). Moreover, hPTTG1 also induces genetic instability in thyroid and colorectal tumors (48,53) and is required for DNA repair after genotoxic stress (54). The finding that hPTTG1 acts as an E2F1 target elucidates a mechanism for abundant tumor hPTTG1 expression, expands our knowledge of the Rb/E2F1 pathway playing a role in G2/M, and strengthens the link between the Rb/E2F1 pathway and aneuploidy. The Rb-E2F1-hPTTG1 signaling pathway may underlie the hPTTG1 requirement for pituitary tumorigenesis and provide a potential future antitumor target.
Materials and Methods
Animal and human specimens
Experiments were approved by the Institutional Animal Care and Use Committee. Rb+/− mice on a 129/Sv genetic background were purchased from The Jackson Laboratory (Bar Harbor, ME). Rb+/− mice and WT control mice were bred by crossing Rb+/− and Rb+/+ mice. Animals were genotyped by PCR for Rb loci as described (7). Eighty human pituitary tumor samples and three normal tissues were obtained from the Pathology Department of Cedars-Sinai Medical Center. The protocol was approved by the Institutional Review Board. All mice and human pituitary tissues were fixed in formalin, embedded in paraffin, and cut at 5 μm for immunostaining.
Immunofluorescence
Slides were deparaffinized in xylene, hydrated in graded ethanol, and heated in Target Retrieval Solution (Dako, Glostrup, Denmark) to retrieve antigen at 95 C for 40 min. Slides were permeabilized with 1% Triton X-100 in PBS for 30 min and incubated in blocking buffer (10% goat serum, 1% BSA in PBS) for 1 h. After washing with PBS, slides were hybridized with antibody against E2F1 (C-20 sc-193X, 1:500; Santa Cruz Biotechnology, Santa Cruz, CA) and hPTTG1 (Securin DCS-280, 1:50; Abcam, Cambridge, UK) at 4 C overnight. Primary antibody was omitted for negative controls. Alexa Fluor antimouse antibody with green or red fluorescence (Molecular Probes, Carlsbad, CA) was used as secondary antibody and incubated at room temperature avoiding light for 1 h. Slides were mounted with ProLong Gold Antifade Reagent with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen, Carlsbad, CA), and nuclei were dyed by DAPI with blue fluorescence.
Confocal microscopy
Samples were imaged with a Leica TCS/SP spectral confocal scanner (Leica Microsystems, Mannheim, Germany) in dual-emission mode to separate autofluorescence from specific staining. A spectral window from 500–550 nm wavelength detected emission of Alexa 488. A second window from 560–620 nm detected the autofluorescence contribution to the signal. In the final images, Alexa 488 appears green. Autofluorescence appears red. The two images were merged, so that all autofluorescence appears yellow, and any true signal appears green. Samples stained with Alexa 568 were imaged with a 540-nm HeNe laser and colored in red.
Evaluation and statistical analysis
Mouse pituitary Western blot results were quantified by ImageJ according to densitometry and relative E2F1 and PTTG1 protein expression levels (E2F1/β-actin and PTTG1/β-actin) calculated. Immunofluorescence slides were independently examined by two observers who were blinded to the pathological results and to each other’s records. Mouse pituitary specimens staining PTTG1 and E2F1 were scaled as percentage of positive cells (counted by ImageJ). Human pituitary tumor specimens staining hPTTG1 were calculated as a ratio of immunoreactive cell numbers per field (×200 magnification). Three to 30 different fields were counted on each slide according to the tissue size. The average score of all three normal tissues (scaled as 0.02) was used as basal control and compared with all tumors. Tumor hPTTG1 expression was evaluated as follows: 1) no change if score was 0.2 or lower or 2) overexpression if score was higher than 0.2. Samples detected with E2F1 were scaled as a percentage of positively stained cells. The average score of all three normal tissues (scaled as 25%) was used as basal control and compared with all tumors. Tumor E2F1 expression was evaluated as follows: 1) no change if score was 50% or lower or 2) overexpression if score was higher than 50%. E2F1 and hPTTG1 expression were correlated using Pearson χ2 test with statistical software SPSS version 10.0. P < 0.05 was considered significant.
Plasmids and siRNA
hPTTG1 promoter fragments were amplified from human genomic DNA using TaKaRa LA Taq and inserted into the pGL3-Basic luciferase reporter vector (Promega, Madison, WI). The following primers were used for PCR of hPTTG1 promoters −2642/−1 and −1272/−1: forward 1 (from −2642), 5′-GGG GTA CCA TCT ATG CTA TCC CAT CCC T-3′; forward 2 (from −1272), 5′-GGG GTA CCA ACA GGA AAA GGT CGT CAA C-3′; and reverse (to −1), 5′-GAA GAT CTT CTG GAT TAT TCT AAG AAT G-3′. E2F1 and DP1 expression plasmids were amplified separately from MHS1010-9205161 clone (Open Biosystems, Huntsville, AL) and 6715577 clone [American Type Culture Collection (ATCC), Manassas, VA] by using TaKaRa LA Taq and cloned into the pcDNA3.1/myc-his vector (Invitrogen). The following primers were used for PCR of E2F1 and DP1: E2F1 forward, 5′-CGG GAT CCA CCA TGG CCT TGG CCG GGG CCC CT-3′; E2F1 reverse, 5′-GGA ATT CGA AAT CCA GGG GGG TGA GGT CCC CAA A-3′; DP1 forward, 5′-CGG GAT CCA CCA TGG CAA AAG ATG CCG GTC TAA T-3′; and DP1 reverse, 5′-CCG CTC GAG GTC GTC CTC GTC ATT CTC GTT-3′. To create the DN E2F1-E132 plasmid (designated as E2F1-DN), the sequence at E2F1 codons L132 and N133 (CTGAAT) was replaced with an EcoRI restriction site by QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) using WT E2 F1 plasmid as template. The following primers were designed for mutation: sense, 5′-GGG GGA GAA GTC ACG CTA TGA GAC CTC AGA ATT CCT GAC CAC CAA GCG C-3′, and antisense, 5′ GCG CTT GGT GGT CAG GAA TTC TGA GGT CTC ATA GCG TGA CTT CTC CCC C-3′. Plasmids were sequenced by Sequetech (Mountain View, CA). Predesigned Rb siRNA (ID 106038 and 106039), p21 siRNA (ID 1531), human E2F1 siRNA (ID 114509, 107658, and 107660), mouse E2F1 siRNA (ID 61093, 160617, or 160619) and negative control siRNA (catalog item 4611) were obtained from Ambion (Austin, TX).
Cell culture and transfections
HEK293, H1299, HeLa, MCF7, HCT116, and AtT-20 cells were obtained from the ATCC and cultured according to conditions of the ATCC manual. Transfection was performed in 70–80% confluent cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol.
ChIP
Using a ChIP kit (Active Motif, Carlsbad, CA), about 107 cells were cross-linked and lysed. Chromatin was sonicated to 200- to 1000-bp fragments with four rounds of 10-sec pulses using 25% power. Normalized inputs of sheared chromatin DNA were incubated with 3 μg E2F1 antibody (C-20 sc-193X; Santa Cruz) overnight at 4 C. Negative control IgG and positive control TFIIB antibody were added at 10 μl (4 μg) per ChIP reaction. Real-time PCR were amplified using precipitated immunocomplexes as template and the following hPTTG1 promoter primers: primer 1 forward, 5′-TTG GGC CGC GAG TTG T-3′; primer 1 reverse, 5′-CAC TCA CGC AGG TCT TAA CAG C-3′; primer 2 forward, 5′-GAG AAT GAC TCA AAC GCT GCT G-3′; primer 2 reverse, 5′-TTG GGT CTA AAG AAT ACT AGC AGA GAA G-3′; primer 3 forward, 5′-CAA TAT TGG TCC TGA AAT GCC A-3′; primer 3 reverse, 5′-AAA GGC TTG ACA CTA TAC CTG ACA TAG A-3′; primer 4 forward, 5′-GGA ATC AGA AAG CGC AGG AC-3′; primer 4 reverse, 5′-CCC GGC TTT CCT AGA CCT TC-3′; primer 5 forward, 5′-GGT GCT GTG AGA ACA TAA GGC AT-3′; and primer 5 reverse, 5′-AAG GGA TGG GAT AGC ATA GAT ACT GT-3′. These five primer pairs were designed in different promoter regions, with primer 1 the closest to ATG and primer 5 the furthest. Two negative control primers were designed to assess the contribution of nonspecific antibody binding. Both primers were designed in PTTG genomic intron 4 and distant from E2F-binding sites: negative control primer 1 forward, 5′-TCC TCT GGA GTC TTC TGT CTA TGG-3′; negative control primer 1 reverse, 5′-TTG TCC GTA GGT GTG AAA GTT CTC-3′; negative control primer 2 forward, 5′-AGT CTA AGC ACC TGT CAA G-3′; and negative control primer 2 reverse, 5′-TTG AAC TCG TGA CCT CAG-3′. The TFIIB-positive control was amplified with GAPDH primers. Negative primers, not expected to be enriched, were used as negative controls. ChIP of acetylated histone H3 and H4 were similarly performed as described above. H1299 cells transfected with pcDNA3.1 or E2F1 and DP1 were fixed and sonicated separately. Chromatin DNA were incubated with acetyl-histone H3 (3 μg, catalog item 06-599; Millipore, Billerica, MA), acetyl-histone H4 (3 μg, catalog item 06-866; Millipore), or negative IgG (10 μl), respectively. Precipitated immunocomplexes were amplified as template with the hPTTG1 promoter primer 2.
Biotin-streptavidin pull-down assay
Six different 5′ ends of biotin-labeled sense oligonucleotides were synthesized: 1)WT E2F1 −354/−393, 5′-TAGTT GAGCC GGCTC CGGCG GGGAA GGAGG CGGGC TGCGG-3′, corresponding to positions −354 to −393 of the hPTTG1 promoter; 2) Mut E2F1 −354/−393, 5′-TAGTT GAGCC GGCTC CGGGA AGGAG GCGGG CTGCG G-3′, devoid of core GGCG nucleotides; 3) WT E2F1 −573/−612, 5′-TTTGC AAGAA AGAGC TTTTG GGGCG GAGCC CTGGC TGCTT-3′, corresponding to positions −573 to −612 of the hPTTG1 promoter; 4) Mut E2F1 −573/−612, 5′-TTTGC AAGAA AGAGC TGGGC GGAGC CCTGG CTGCT T-3′, devoid of core TTTG nucleotides; 5) WT E2F1 control, 5′-ATTTA AGTTT CGCGC CCTTT CTCAA-3′, a consensus E2F1 binding sequence; and 6) Mut E2F1 control, 5′-ATTTA AGTTT CGATC CCTTT CTCAA-3′, in which AT nucleotides replaced CG. Underlined text emphasizes WT and Mut differences. All six oligonucleotides were annealed to their respective complementary antisense oligonucleotides. H1299 nuclear protein was extracted using nuclear extract kit (Active Motif). Protein concentration was measured by Coomassie Plus Assay Kit (Pierce, Rockford, IL). Two micrograms of each double-stranded oligonucleotide were incubated with 600 μg nuclear protein for 20 min at room temperature in binding buffer containing 12% glycerol, 12 mm HEPES (pH 7.9), 4 mm Tris (pH 7.9), 150 mm KCl, 1 mm EDTA, 1 mm dithiothreitol, and 10 mg poly(deoxyinosine-deoxycytosine) competitor. Subsequently, 50 μl streptavidin-agarose (Sigma Chemical Co., St. Louis, MO) was added and incubated at 4 C overnight. Before this step, 300 μl original streptavidin-agarose beads were preabsorbed with 500 mg BSA, 50 mg poly(deoxyinosine-deoxycytosine) (Sigma), and 50 mg sheared salmon sperm DNA (Sigma) for 20 min at room temperature; streptavidin-agarose beads were washed three times and resuspended in 300 μl binding buffer. The protein-DNA-streptavidin-agarose complex was gently washed three times with binding buffer and loaded onto NuPAGE Novex Bis-Tris Gel (Invitrogen) for E2F1 detection using Western blot.
Reporter assay
Cells were split into 24-well plates, and each well was cotransfected with 1) 200 ng luciferase promoter, pGL3-Basic as control, and hPTTG1 promoter −2642/−1 or −1272/−1 and 2) 800 ng empty pcDNA3.1 vector as control, 400 ng E2F1 plus 400 ng pcDNA3.1 empty vector, 400 ng E2F1 plus 400 ng DP1, or 400 ng E2F1 plus 400 ng E2F1-DN plasmids. pRL-Tk (Promega) encoding renilla luciferase was used as an internal control (5 ng/well) to assess transfection efficiency. After 24 h, whole-cell lysates were collected for reporter detection by the dual-luciferase reporter system (Promega) according to the protocol. Reactions were measured using an Orion microplate luminometer (Berthold Detection System, Huntsville, AL). Transfections were performed in triplicate and repeated three times to assure reproducibility.
RNA extraction and real-time PCR
Total RNA was isolated using Trizol reagent (Invitrogen). Three micrograms of total RNA were used to synthesize cDNA with SuperScript II reverse transcriptase (Invitrogen). Real-time PCR was amplified in 20-μl reaction mixtures [100 ng template, 0.5 μm of each primer, 10 μl 2× Sybr Green Master Mix (Applied Biosystems, Foster City, CA)] using the following parameters: 95 C for 1 min followed by 40 cycles of 95 C for 20 sec and 60 C for 40 sec. β-Actin was used as internal control. Real-time PCR primers were designed as follows: hPTTG1 forward, 5′-TGA TCC TTG ACG AGG AGA GAG-3′; hPTTG1 reverse, 5′-GGT GGC AAT TCA ACA TCC AGG-3′; β-actin forward, 5′-CAT GTA CGT TGC TAT CCA GGC-3′; and β-actin reverse, 5′-CTC CTT AAT GTC ACG CAC GAT-3′.
Cell lysis and Western blot analysis
Total cell lysate was prepared in RIPA buffer (Sigma) containing protease inhibitor cocktail (Sigma). Protein concentrations were measured by Coomassie Plus assay Kit (Pierce) using BSA as standard. Equal amounts (100 μg) of proteins were separated by NuPAGE Novex Bis-Tris Gels (Invitrogen) and transferred onto polyvinylidene difluoride membrane (Millipore). The membrane was incubated in blocking solution (TBS buffer containing 5% nonfat dry milk; Bio-Rad, Hercules, CA) for 1 h at room temperature, followed by incubation with primary antibody [Rb IF8 sc-102, 1:500 (Santa Cruz); E2F-1(KH95) sc-251, 1:500 (Santa Cruz); PCNA (PC10) sc-56, 1:1000 (Santa Cruz); p21 Waf1/Cip1 (DCS60), 1:1000 (Cell Signaling, Beverly, MA); Securin DCS-280, 1:200 (Abcam); β-actin, 1:5000 (Sigma); and anti-myc tag clone 9E10, 1:1000 (Millipore)] at 4 C overnight. After washes with 0.5% Tween 20 in TBS, membranes were subsequently incubated with horseradish peroxidase-linked secondary antibody (GE Healthcare, Piscataway, NJ) for 1 h at room temperature and developed using enhanced chemiluminescence Western blotting detection reagents (GE Healthcare).
HCT116 p21−/− and HCT116 p53−/− cells were kindly provided by Dr. Bert Vogelstein, Johns Hopkins University (Baltimore, MD).
Footnotes
This work was supported by National Institutes of Health Grant CA75979 (to S.M.) and the Doris Factor Molecular Endocrinology Laboratory.
Disclosure Summary: The authors have nothing to disclose
First Published Online October 16, 2009
Abbreviations: Cdk, Cyclin-dependent kinase; ChIP, chromatin immunoprecipitation; DAPI, 4′,6-diamidino-2-phenylindole; DN, dominant negative; hPTTG, human pituitary tumor transforming gene; Mut, mutant; PCNA, proliferating cell nuclear antigen; Rb, retinoblastoma protein; siRNA, small interfering RNA; WT, wild type.
References
- Melmed S 2003 Mechanisms for pituitary tumorigenesis: the plastic pituitary. J Clin Invest 112:1603–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Pereda M, Giacomini D, Echenique C, Stalla GK, Holsboer F, Arzt E 2005 Signaling processes in tumoral neuroendocrine pituitary cells as potential targets for therapeutic drugs. Curr Drug Targets Immune Endocr Metabol Disord 5:259–267 [DOI] [PubMed] [Google Scholar]
- Weinberg RA 1995 The retinoblastoma protein and cell cycle control. Cell 81:323–330 [DOI] [PubMed] [Google Scholar]
- Attwooll C, Lazzerini Denchi E, Helin K 2004 The E2F family: specific functions and overlapping interests. EMBO J 23:4709–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Ciro M, Cocito A, Helin K 2004 E2F target genes: unraveling the biology. Trends Biochem Sci 29:409–417 [DOI] [PubMed] [Google Scholar]
- La Thangue NB 2003 The yin and yang of E2F-1: balancing life and death. Nat Cell Biol 5:587–589 [DOI] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA 1992 Effects of an Rb mutation in the mouse. Nature 359:295–300 [DOI] [PubMed] [Google Scholar]
- Hu N, Gutsmann A, Herbert DC, Bradley A, Lee WH, Lee EY 1994 Heterozygous Rb-1δ20/+ mice are predisposed to tumors of the pituitary gland with a nearly complete penetrance. Oncogene 9:1021–1027 [PubMed] [Google Scholar]
- Harrison DJ, Hooper ML, Armstrong JF, Clarke AR 1995 Effects of heterozygosity for the Rb-1t19neo allele in the mouse. Oncogene 10:1615–1620 [PubMed] [Google Scholar]
- Williams BO, Remington L, Albert DM, Mukai S, Bronson RT, Jacks T 1994 Cooperative tumorigenic effects of germline mutations in Rb and p53. Nat Genet 7:480–484 [DOI] [PubMed] [Google Scholar]
- Yamasaki L, Bronson R, Williams BO, Dyson NJ, Harlow E, Jacks T 1998 Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1+/− mice. Nat Genet 18:360–364 [DOI] [PubMed] [Google Scholar]
- Chesnokova V, Kovacs K, Castro AV, Zonis S, Melmed S 2005 Pituitary hypoplasia in Pttg−/− mice is protective for Rb+/− pituitary tumorigenesis. Mol Endocrinol 19:2371–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donangelo I, Gutman S, Horvath E, Kovacs K, Wawrowsky K, Mount M, Melmed S 2006 Pituitary tumor transforming gene overexpression facilitates pituitary tumor development. Endocrinology 147:4781–4791 [DOI] [PubMed] [Google Scholar]
- Abbud RA, Takumi I, Barker EM, Ren SG, Chen DY, Wawrowsky K, Melmed S 2005 Early multipotential pituitary focal hyperplasia in the α-subunit of glycoprotein hormone-driven pituitary tumor-transforming gene transgenic mice. Mol Endocrinol 19:1383–1391 [DOI] [PubMed] [Google Scholar]
- Woloschak M, Yu A, Post KD 1997 Frequent inactivation of the p16 gene in human pituitary tumors by gene methylation. Mol Carcinog 19:221–224 [DOI] [PubMed] [Google Scholar]
- Komatsubara K, Tahara S, Umeoka K, Sanno N, Teramoto A, Osamura RY 2001 Immunohistochemical analysis of p27 (Kip1) in human pituitary glands and in various types of pituitary adenomas. Endocr Pathol 12:181–188 [DOI] [PubMed] [Google Scholar]
- Asa SL, Ezzat S 2005 Genetics and proteomics of pituitary tumors. Endocrine 28:43–47 [DOI] [PubMed] [Google Scholar]
- Jordan S, Lidhar K, Korbonits M, Lowe DG, Grossman AB 2000 Cyclin D and cyclin E expression in normal and adenomatous pituitary. Eur J Endocrinol 143:R1–R6 [DOI] [PubMed] [Google Scholar]
- Zhang X, Horwitz GA, Prezant TR, Valentini A, Nakashima M, Bronstein MD, Melmed S 1999 Structure, expression, and function of human pituitary tumor-transforming gene (PTTG). Mol Endocrinol 13:156–166 [DOI] [PubMed] [Google Scholar]
- Zou H, McGarry TJ, Bernal T, Kirschner MW 1999 Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 285:418–422 [DOI] [PubMed] [Google Scholar]
- Jallepalli PV, Waizenegger IC, Bunz F, Langer S, Speicher MR, Peters JM, Kinzler KW, Vogelstein B, Lengauer C 2001 Securin is required for chromosomal stability in human cells. Cell 105:445–457 [DOI] [PubMed] [Google Scholar]
- Yu R, Lu W, Chen J, McCabe CJ, Melmed S 2003 Overexpressed pituitary tumor-transforming gene causes aneuploidy in live human cells. Endocrinology 144:4991–4998 [DOI] [PubMed] [Google Scholar]
- Wang Z, Moro E, Kovacs K, Yu R, Melmed S 2003 Pituitary tumor transforming gene-null male mice exhibit impaired pancreatic β-cell proliferation and diabetes. Proc Natl Acad Sci USA 100:3428–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Horwitz GA, Heaney AP, Nakashima M, Prezant TR, Bronstein MD, Melmed S 1999 Pituitary tumor transforming gene (PTTG) expression in pituitary adenomas. J Clin Endocrinol Metab 84:761–767 [DOI] [PubMed] [Google Scholar]
- Filippella M, Galland F, Kujas M, Young J, Faggiano A, Lombardi G, Colao A, Meduri G, Chanson P 2006 Pituitary tumour transforming gene (PTTG) expression correlates with the proliferative activity and recurrence status of pituitary adenomas: a clinical and immunohistochemical study. Clin Endocrinol (Oxf) 65:536–543 [DOI] [PubMed] [Google Scholar]
- Boelaert K, McCabe CJ, Tannahill LA, Gittoes NJ, Holder RL, Watkinson JC, Bradwell AR, Sheppard MC, Franklyn JA 2003 Pituitary tumor transforming gene and fibroblast growth factor-2 expression: potential prognostic indicators in differentiated thyroid cancer. J Clin Endocrinol Metab 88:2341–2347 [DOI] [PubMed] [Google Scholar]
- Ogbagabriel S, Fernando M, Waldman FM, Bose S, Heaney AP 2005 Securin is overexpressed in breast cancer. Mod Pathol 18:985–990 [DOI] [PubMed] [Google Scholar]
- Shibata Y, Haruki N, Kuwabara Y, Nishiwaki T, Kato J, Shinoda N, Sato A, Kimura M, Koyama H, Toyama T, Ishiguro H, Kudo J, Terashita Y, Konishi S, Fujii Y 2002 Expression of PTTG (pituitary tumor transforming gene) in esophageal cancer. Jpn J Clin Oncol 32:233–237 [DOI] [PubMed] [Google Scholar]
- Heaney AP, Singson R, McCabe CJ, Nelson V, Nakashima M, Melmed S 2000 Expression of pituitary-tumour transforming gene in colorectal tumours. Lancet 355:716–719 [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Ross KN, Lander ES, Golub TR 2003 A molecular signature of metastasis in primary solid tumors. Nat Genet 33:49–54 [DOI] [PubMed] [Google Scholar]
- Pei L 2001 Identification of c-myc as a down-stream target for pituitary tumor-transforming gene. J Biol Chem 276:8484–8491 [DOI] [PubMed] [Google Scholar]
- Tong Y, Tan Y, Zhou C, Melmed S 2007 Pituitary tumor transforming gene interacts with Sp1 to modulate G1/S cell phase transition. Oncogene 26:5596–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero F, Multon MC, Ramos-Morales F, Domínguez A, Bernal JA, Pintor-Toro JA, Tortolero M 2001 Human securin, hPTTG, is associated with Ku heterodimer, the regulatory subunit of the DNA-dependent protein kinase. Nucleic Acids Res 29:1300–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal JA, Luna R, Espina A, Lázaro I, Ramos-Morales F, Romero F, Arias C, Silva A, Tortolero M, Pintor-Toro JA 2002 Human securin interacts with p53 and modulates p53-mediated transcriptional activity and apoptosis. Nat Genet 32:306–311 [DOI] [PubMed] [Google Scholar]
- Yu R, Heaney AP, Lu W, Chen J, Melmed S 2000 Pituitary tumor transforming gene causes aneuploidy and p53-dependent and p53-independent apoptosis. J Biol Chem 275:36502–36505 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Mehta KR, Choi AP, Scolavino S, Zhang X 2003 DNA damage-induced inhibition of securin expression is mediated by p53. J Biol Chem 278:462–470 [DOI] [PubMed] [Google Scholar]
- Kim CS, Ying H, Willingham MC, Cheng SY 2007 The pituitary tumor-transforming gene promotes angiogenesis in a mouse model of follicular thyroid cancer. Carcinogenesis 28:932–939 [DOI] [PubMed] [Google Scholar]
- Vlotides G, Eigler T, Melmed S 2007 Pituitary tumor-transforming gene: physiology and implications for tumorigenesis. Endocr Rev 28:165–186 [DOI] [PubMed] [Google Scholar]
- Vlotides G, Cruz-Soto M, Rubinek T, Eigler T, Auernhammer CJ, Melmed S 2006 Mechanisms for growth factor-induced pituitary tumor transforming gene-1 expression in pituitary folliculostellate TtT/GF cells. Mol Endocrinol 20:3321–3335 [DOI] [PubMed] [Google Scholar]
- Zhou C, Liu S, Zhou X, Xue L, Quan L, Lu N, Zhang G, Bai J, Wang Y, Liu Z, Zhan Q, Zhu H, Xu N 2005 Overexpression of human pituitary tumor transforming gene (hPTTG), is regulated by β-catenin /TCF pathway in human esophageal squamous cell carcinoma. Int J Cancer 113:891–898 [DOI] [PubMed] [Google Scholar]
- Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins JR 2001 Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol 21:4684–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson AY, Feber A, Edwards S, Te Poele R, Giddings I, Merson S, Cooper CS 2007 Role of E2F3 expression in modulating cellular proliferation rate in human bladder and prostate cancer cells. Oncogene 26:1028–1037 [DOI] [PubMed] [Google Scholar]
- Taubert S, Gorrini C, Frank SR, Parisi T, Fuchs M, Chan HM, Livingston DM, Amati B 2004 E2F-dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol Cell Biol 24:4546–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helin K, Wu CL, Fattaey AR, Lees JA, Dynlacht BD, Ngwu C, Harlow E 1993 Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev 7:1850–1861 [DOI] [PubMed] [Google Scholar]
- Wang Z, Yu R, Melmed S 2001 Mice lacking pituitary tumor transforming gene show testicular and splenic hypoplasia, thymic hyperplasia, thrombocytopenia, aberrant cell cycle progression, and premature centromere division. Mol Endocrinol 15:1870–1879 [DOI] [PubMed] [Google Scholar]
- Pei L, Melmed S, Scheithauer B, Kovacs K, Benedict WF, Prager D 1995 Frequent loss of heterozygosity at the retinoblastoma susceptibility gene (RB) locus in aggressive pituitary tumors: evidence for a chromosome 13 tumor suppressor gene other than RB. Cancer Res 55:1613–1616 [PubMed] [Google Scholar]
- Fedele M, Visone R, De Martino I, Troncone G, Palmieri D, Battista S, Ciarmiello A, Pallante P, Arra C, Melillo RM, Helin K, Croce CM, Fusco A 2006 HMGA2 induces pituitary tumorigenesis by enhancing E2F1 activity. Cancer Cell 9:459–471 [DOI] [PubMed] [Google Scholar]
- Kim D, Pemberton H, Stratford AL, Buelaert K, Watkinson JC, Lopes V, Franklyn JA, McCabe CJ 2005 Pituitary tumour transforming gene (PTTG) induces genetic instability in thyroid cells. Oncogene 24:4861–4866 [DOI] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ 1993 The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805–816 [DOI] [PubMed] [Google Scholar]
- Harvey M, Vogel H, Lee EY, Bradley A, Donehower LA 1995 Mice deficient in both p53 and Rb develop tumors primarily of endocrine origin. Cancer Res 55:1146–1151 [PubMed] [Google Scholar]
- Timmers C, Sharma N, Opavsky R, Maiti B, Wu L, Wu J, Orringer D, Trikha P, Saavedra HI, Leone G 2007 E2f1, E2f2, and E2f3 control E2F target expression and cellular proliferation via a p53-dependent negative feedback loop. Mol Cell Biol 27:65–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando E, Nahlé Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R, Lowe SW, Cordon-Cardo C 2004 Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature 430:797–802 [DOI] [PubMed] [Google Scholar]
- Kim DS, Franklyn JA, Smith VE, Stratford AL, Pemberton HN, Warfield A, Watkinson JC, Ishmail T, Wakelam MJ, McCabe CJ 2007 Securin induces genetic instability in colorectal cancer by inhibiting double-stranded DNA repair activity. Carcinogenesis 28:749–759 [DOI] [PubMed] [Google Scholar]
- Bernal JA, Roche M, Méndez-Vidal C, Espina A, Tortolero M, Pintor-Toro JA 2008 Proliferative potential after DNA damage and non-homologous end joining are affected by loss of securin. Cell Death Differ 15:202–212 [DOI] [PubMed] [Google Scholar]