Abstract
IGF-I is abundantly expressed in the liver under the stimulation of GH. We showed previously that expression of hepatocyte nuclear factor (HNF)-3γ, a liver-enriched transcription factor, was strongly stimulated by GH in bovine liver. In this study, we determined whether GH-increased HNF-3γ might contribute to GH stimulation of IGF-I gene expression in bovine liver and the underlying mechanism. A sequence analysis of the bovine IGF-I promoter revealed three putative HNF-3 binding sites, which all appear to be conserved in mammals. Chromatin immunoprecipitation assays showed that GH injection increased binding of HNF-3γ to the IGF-I promoter in bovine liver. Gel-shift assays indicated that one of the three putative HNF-3 binding sites, HNF-3 binding site 1, bound to the HNF-3γ protein from bovine liver with high affinity. Cotransfection analyses demonstrated that this HNF-3 binding site was essential for the transcriptional response of the IGF-I promoter to HNF-3γ in CHO cells and to GH in primary mouse hepatocytes. Using similar approaches, we found that GH increased binding of the signal transducer and activator of transcription 5 (STAT5) to the HNF-3γ promoter in bovine liver, that this binding occurred at a conserved STAT5 binding site, and that this STAT5 binding site was necessary for the HNF-3γ promoter to respond to GH. Taken together, these results suggest that in addition to direct action, GH-activated STAT5 may also indirectly stimulate IGF-I gene transcription in the liver by directly enhancing the expression of the HNF-3γ gene.
Besides direct stimulation, growth hormone-activated STAT5 may also indirectly stimulate IGF-I gene transcription in liver through upregulation of the liver-enriched transcription factor HNF-3γ.
IGF-I is a polypeptide hormone that plays important roles in growth, development, and metabolism in an endocrine, paracrine, and/or autocrine manner (1,2). In addition to its physiological roles, IGF-I has also been implicated in cancer (3,4), diabetic retinopathy (5,6,7), and aging (8). Although many organs and tissues in the body produce IGF-I (9,10), the liver is the major source of circulating IGF-I (11,12). Liver production of IGF-I is primarily controlled by the pituitary GH at the transcriptional level (10,13).
Significant insight has been recently gained into the mechanism by which GH stimulates IGF-I gene transcription in the liver. Earlier studies found that the signal transducer and activator of transcription 5 (STAT5) is essential for this regulation (14,15,16). This finding led to the subsequent identification of multiple STAT5 binding sites, with which STAT5 interacts to activate IGF-I gene transcription in response to GH (17,18,19,20). Because IGF-I mRNA is expressed in the liver at a much higher level than in other tissues (9,10), we hypothesized that GH stimulation of IGF-I gene transcription in this tissue involve liver-enriched transcription factors in addition to STAT5. The liver-enriched transcription factors include hepatocyte nuclear factor (HNF)-1α, -1β, -3α, -3β, -3γ, -4α, -4γ, -6α, and -6β, albumin D-element binding protein (DBP), and CCAAT/enhancer-binding proteins (C/EBP)-α and -β (21,22,23). The IGF-I gene promoter contains binding sites for HNF-1, C/EBP, and HNF-3 (24,25,26,27). Therefore, it is possible that HNF-1α, C/EBP-α and -β, and HNF-3α, -3β, and -3γ participate in GH regulation of IGF-I gene transcription in the liver.
In this study, we focused on the potential role of HNF-3γ, also known as Foxa3 (28,29), in GH stimulation of IGF-I gene transcription in bovine liver. We chose this focus because we previously observed that HNF-3γ expression is more potently stimulated by GH than any other liver-enriched transcription factors in bovine liver (30). In the present study, we also determined the mechanism by which GH stimulates HNF-3γ gene expression in bovine liver.
Results
The IGF-I promoter contains three putative HNF-3 binding sites
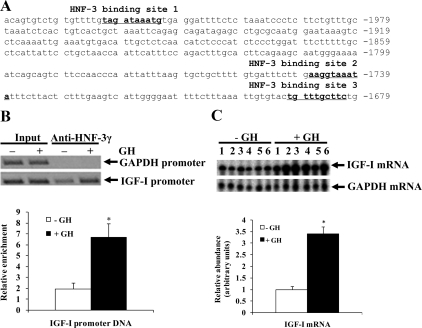
Sequence analysis of a 2-kb bovine IGF-I promoter (chromosome 5: 711195851-71198044, Bovine October 2007 Assembly) revealed three putative HNF-3 binding sites (Fig. 1A). All of them are nearly identical to the consensus HNF-3 binding sequence, WRRRYMAAYA, where W is A or T, R is A or G, Y is C or T, and M is A or C (31). The putative HNF-3 binding site 1 was located 2021 bp upstream from the major transcription start site for class 2 IGF-I mRNA (32); the putative HNF-3 binding sites 2 and 3 were located 1747 and 1690 bp upstream from this transcription start site (Fig. 1A). A sequence alignment of the corresponding DNA regions of the bovine, mouse, rat, horse, dog, human, and chimpanzee genomes revealed that all of the three putative HNF-3 binding sites are conserved among these species (data not shown).
Figure 1.
GH administration increased binding of HNF-3γ to the IGF-I promoter and IGF-I mRNA expression in bovine liver. A, Three putative HNF-3 binding sites in the bovine IGF-I promoter. Shown is the partial sequence of the bovine IGF-I promoter 2. Nucleotides of the sequence are numbered relative to the major transcription start site, numbered +1, for class 2 IGF-I mRNA. B, ChIP assay of HNF-3γ binding to the IGF-I promoter in liver of cattle before and after GH administration (indicated by −GH and +GH, respectively). In this assay, liver chromatin was precipitated with an anti-HNF-3γ antibody, and the putative HNF-3 binding sites-containing IGF-I promoter and the GAPDH promoter that does not contain a putative HNF-3 binding site in the immunoprecipitates were amplified by PCR. Shown are a representative gel image of the PCR products and average IGF-I promoter enrichment of ChIP assays on three different animals. The abundance of the IGF-I promoter DNA in the immunoprecipitates was normalized to that in the input chromatin. *, P < 0.05 (n = 3) compared with −GH. C, Ribonuclease protection assay of liver IGF-I mRNA in cattle before and after GH administration. Shown are a gel image of the assay and result of a densitometric analysis of the bands in the gel image. The ribonuclease-protected probe fragments representing IGF-I and GAPDH mRNA are indicated with arrows. The detected density of IGF-I mRNA in each sample was normalized to that of GAPDH mRNA. *, P < 0.05 (n = 6) compared with −GH.
GH increased HNF-3γ binding to the IGF-I promoter in the liver
GH is known to increase HNF-3γ expression in bovine liver (30) (also see Fig. 4D below). To determine whether GH increases HNF-3γ binding to the IGF-I promoter region that contains putative HNF-3 binding sites, we performed chromatin immunoprecipitation (ChIP) assays on liver chromatin from cattle before and after GH administration. As shown in Fig. 1B, the HNF-3γ antibody precipitated significantly more of the IGF-I promoter DNA from the liver after GH injection than before GH injection, indicating that GH injection increased HNF-3γ binding to the IGF-I promoter in the liver. The GH treatment had no effect on HNF-3γ binding to the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter (Fig. 1B), a promoter that does not contain putative HNF-3 binding sites. In the HNF-3γ antibody-immunoprecipitated chromatin from the liver before GH injection, the IGF-I promoter was enriched compared with the GAPDH promoter (Fig. 1B), indicating that there was HNF-3γ binding to the IGF-I promoter even when the animals were not injected with GH. A ribonuclease protection assay of liver IGF-I mRNA from the same animals revealed that GH-increased binding of HNF-3γ to the IGF-I promoter was associated with an approximately 3-fold increase in IGF-I mRNA expression in the liver (P < 0.05, Fig. 1C).
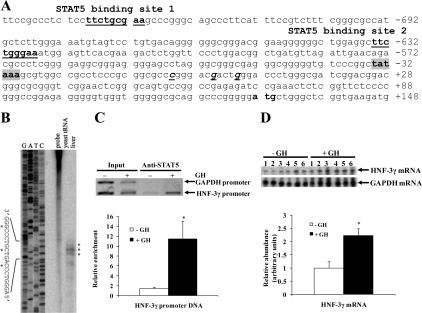
Figure 4.
GH increased binding of STAT5 to the HNF-3γ promoter and HNF-3γ mRNA expression in bovine liver. A, Two putative STAT5 binding sites in the bovine HNF-3γ promoter. Shown is the partial sequence of the bovine HNF-3γ promoter. The nucleotides are numbered relative to the major transcription start site, numbered +1, of the bovine HNF-3γ gene (see B). The two putative STAT5 binding sites are underlined. B, Mapping of the transcription start site of the bovine HNF-3γ gene. The mapping was done by ribonuclease protection assay of bovine liver total RNA using a probe covering a 239-bp HNF-3γ DNA region spanning the putative transcription start site. The ribonuclease-protected fragments were resolved in parallel with a sequencing ladder (G, A, T, C) of the same 239-bp DNA. Three ribonuclease-protected bands (indicated with asterisks) place three transcription start sites, which are 23, 29, and 33 bp downstream from a putative TATA box (see A). C, ChIP assay of STAT5 binding to the HNF-3γ promoter in bovine liver. Liver chromatin from +GH and −GH cattle was precipitated with an anti-STAT5 antibody. The abundance of the putative STAT5 binding sites-containing HNF-3γ promoter region and the GAPDH promoter region, which does not contain a STAT5 binding site, in the immunoprecipitates was quantified by PCR. Shown are a representative gel image of the PCR and average enrichment of the HNF-3γ promoter by the anti-STAT5 antibody. The density of the HNF-3γ promoter DNA in the anti-STAT5 antibody immunoprecipitates was normalized to that in the input chromatin. *, P < 0.05 (n = 3) compared with −GH. D, Ribonuclease protection assay of liver HNF-3γ mRNA in cattle before and after GH administration. This assay was done as described for Fig. 1C. Shown are a representative gel image of the ribonuclease protection assay and result of a densitometric analysis of the bands in the gel image. The detected density of HNF-3γ mRNA in each sample was normalized to that of GAPDH mRNA. *, P < 0.05 (n = 6) compared with −GH.
Two of the three putative HNF-3 binding sites in the IGF-I promoter could bind to liver HNF-3γ protein in vitro
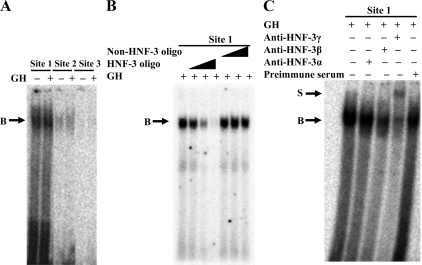
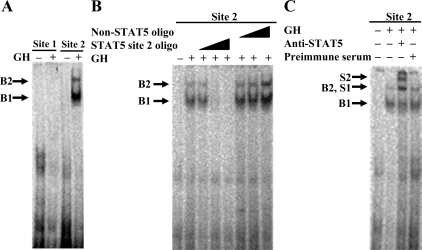
EMSA were performed to determine whether the putative HNF-3 binding sites in the IGF-I promoter can bind to the HNF-3γ protein from bovine liver. As shown in Fig. 2A, the oligonucleotides corresponding to the putative HNF-3 binding sites 1 and 2 each formed a DNA-protein complex with liver nuclear proteins from cattle both before and after GH injection, whereas the putative HNF-3 binding site 3 did not. The complex formed with the putative HNF-3 binding site 1 was much stronger than that with the binding site 2 (Fig. 2A). To determine whether the DNA-protein complex was formed with HNF-3γ, the putative HNF-3 binding site 1 oligonucleotide was further analyzed in competitive gel-shift and supershift assays. As shown in Fig. 2B, the DNA-protein complex formed with the radiolabeled HNF-3 binding site 1 oligonucleotide was increasingly diminished by an increasing molar excess of unlabeled the same oligonucleotide but was not affected by an excess of an oligonucleotide that did not contain an HNF-3 binding site (Table 1). In the supershift assay, a supershift of the DNA-protein complex was generated by the HNF-3γ antibody but not by the preimmune serum (Fig. 2C), demonstrating the presence of HNF-3γ in the DNA-protein complex. The HNF-3β antibody appeared to reduce the intensity of the DNA-protein complex (Fig. 2C). This reduction may be caused by competition of the HNF-3β antibody with the DNA oligonucleotide for binding to the same portion of the HNF-3β protein or by formation of a DNA-protein-antibody complex that was unable to enter the gel. Therefore, part of the DNA-protein complex might contain the HNF-3β protein. The HNF-3α antibody did not cause a supershift of or disrupt the DNA-protein complex (Fig. 2C), indicating that the DNA-protein complex did not contain the HNF-3α protein.
Figure 2.
Two putative HNF-3 binding sites in the bovine IGF-I promoter could bind to bovine liver HNF-3γ protein in vitro. Panel A, EMSA of the putative HNF-3 binding sites. In this assay, a 32P-labeled double-stranded oligonucleotide corresponding to each of the three putative HNF-3 binding sites (see Fig. 1A and Table 1) was incubated with liver nuclear protein extracts from cattle before (−GH) and after (+GH) GH injection, followed by gel electrophoresis. B indicates a DNA-protein complex. Panel B, Competitive gel-shift assay of the putative HNF-3 binding site 1. In this assay, the 32P-labeled oligonucleotide corresponding to the putative HNF-3 binding site was incubated with GH-treated liver nuclear protein extracts in the presence of 1×, 10×, and 100× molar excess of unlabeled the same oligonucleotide or an unrelated oligonucleotide. Panel C, Supershift assay of the putative HNF-3 binding site 1. In this assay, the 32P-labeled oligonucleotide was incubated with GH-treated liver nuclear protein extracts in the presence of an anti-HNF-3α, anti-HNF-3β, or anti-HNF-3γ antibody or an equal protein amount of goat preimmune serum. S indicates a supershift of the DNA-protein complex B. These assays were repeated at least two times; shown are representative results.
Table 1.
Gel-shift oligonucleotides used in this study
| Name | Sequence (5′–3′) | Chromosomal location |
|---|---|---|
| STAT5 binding site 1 | CCTCTCCTTCTGCGAAGCCC | chr18: 53169792–53169811 |
| STAT5 binding site 2 | GAGGCTTCTGGGAAATGGA | chr18: 53169897-53169916 |
| HNF-3 binding site 1 | TTGTAGATAAATGTGA | chr5: 71198005-71198020 |
| HNF-3 binding site 2 | TTGAAGGTAAATATTT | chr5: 71197737-71197752 |
| HNF-3 binding site 3 | GTGTACTGTTTGCTTCT | chr5: 71197683-71197699 |
| Non-STAT5 oligo | GATTATTGACTTAG | |
| Non-HNF3 oligo | CAGGGGTGGGCGGAGAGGAAGG |
All sequences correspond to the sense strand of the binding sites. The core sequences of the STAT5 and HNF-3 binding sites are indicated in bold. The chromosomal locations listed correspond to the locations in the Bovine Genome October 2007 Assembly at the UCSC Genome Browser (http://genome.ucsc.edu).
HNF-3γ activated the IGF-I promoter through the putative HNF-3 binding site 1
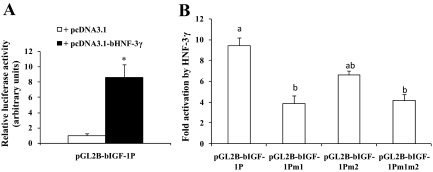
Cotransfection analyses were performed to determine whether HNF-3γ could activate the IGF-I promoter. As shown in Fig. 3A, luciferase activity expressed from the IGF-I promoter in the presence of the HNF-3γ expression plasmid was more than eight times that in the presence of the corresponding empty vector pcDNA3.1 (P < 0.05), indicating that HNF-3γ can transactivate the IGF-I promoter. We next determined whether the putative HNF-3 binding sites 1 and 2, which were demonstrated to bind to liver HNF-3γ by the gel-shift experiments (Fig. 2), are necessary for the HNF-3 response of the IGF-I promoter. As shown in Fig. 3B, mutation of the HNF-3 binding site 1 decreased the HNF-3γ response of the IGF-I promoter by 64% (P < 0.05); mutation of the HNF-3 binding site 2 did not significantly affect (P > 0.1) the response of the promoter or further reduce the HNF-3γ response of the site 1-mutated IGF-I promoter. These data indicated that most of the response of the IGF-I promoter to HNF-3γ was mediated by the HNF-3 binding site 1, which appeared to bind to HNF-3γ at much higher affinity than the HNF-3 binding site 2 (Fig. 2A). The data also showed that mutation of the HNF-3 binding site 1 or both this and the HNF-3 binding site 2 did not completely block the response of the IGF-I promoter to HNF-3γ (Fig. 3B). Mutation of the putative HNF-3 binding site 1, 2, or both did not affect luciferase activity expressed from the IGF-I promoter cotransfected with the empty vector pcDNA3.1 (data not shown), indicating that those putative HNF-3 binding sites do not mediate the basal activity of the IGF-I promoter in CHO cells.
Figure 3.
HNF-3γ could activate the bovine IGF-I promoter through one of the putative HNF-3 binding sites. A, Cotransfection analysis of the bovine IGF-I promoter in CHO cells. In this analysis, the IGF-I promoter-luciferase reporter plasmid pGL2B-bIGF-IP was cotransfected with the bovine HNF-3γ expression plasmid pcDNA3.1-bHNF-3γ or the empty vector pcDNA3.1 and the transfection efficiency control plasmid pRL-CMV plasmid. Luciferase activity was measured 24 h after the transfection. *, P < 0.05 (n = 4) compared with pcDNA3.1. B, Cotransfection analysis of the HNF-3 binding sites-mutated IGF-I promoter. In the pGL2B-bIGF-IP plasmid, both putative HNF-3 binding sites 1 and 2 were intact. In pGL2B-bIGF-IPm1, the HNF-3 binding site 1 was mutated. In pGL2B-bIGF-IPm2, the HNF-3 binding site 2 was mutated. In pGL2B-bIGF-IPm1m2, both HNF-3 binding sites 1 and 2 were mutated. Fold activation on the y-axis corresponds to the ratio of the luciferase activity in the presence of pcDNA3.1-bHNF-3γ to that in the presence of pcDNA3.1. Bars not labeled with the same letter are different (P < 0.05, n = 4).
The HNF-3γ promoter contains a conserved putative STAT5 binding site
We next determined the mechanism by which GH increases HNF-3γ mRNA expression in bovine liver. Unlike the IGF-I promoter, the bovine HNF-3γ promoter had not been characterized before this study. Therefore, we first mapped the transcription start site of the bovine HNF-3γ gene. A ribonuclease protection assay of bovine liver RNA using a probe expected to cover the putative transcription start site generated three protected fragments (Fig. 4B), each fragment corresponding to a possible transcription start site. Based on the sequencing ladder of the same 239-bp DNA fragment, the most abundant protected fragment corresponded to HNF-3γ mRNA transcribed from the nucleotide 29 bp downstream from the putative TATA box (Fig. 4A). The two less abundant ribonuclease-protected bands placed two additional transcription start sites, 23 and 33 bp downstream from the TATA box (Fig. 4B). These locations of the transcription start sites of the bovine HNF-3γ gene were similar to those of the mouse and human HNF-3γ genes (33,34).
A search of the bovine HNF-3γ promoter sequence revealed two putative STAT5 binding sites at approximately 720 and 620 bp upstream from the major transcription start site (Fig. 4A), and both sites conform to the consensus STAT5 binding sequences, TTCNNNGAA, where N is any nucleotide (35). A sequence alignment of the corresponding DNA regions of the bovine, mouse, rat, horse, dog, human, and chimpanzee genomes revealed that the proximal putative STAT5 binding site (i.e. the putative STAT5 binding site 2 in Fig. 4A) was conserved across these species and that the putative STAT5 binding site 1 was not conserved (data not shown).
GH increased binding of STAT5 to the HNF-3γ promoter in the liver
Because the HNF-3γ promoter contains two putative STAT5 binding sites, we next determined whether GH induced STAT5 to bind to this promoter in bovine liver, using ChIP assays. As shown in Fig. 4C, the anti-STAT5 antibody precipitated 10 times more of the HNF-3γ promoter region containing the two putative STAT5 binding sites from the liver of cattle after GH injection than before GH injection (P < 0.05). The anti-STAT5 antibody did not precipitate detectable GAPDH promoter DNA, which does not contain a putative STAT5 binding site, either before or after GH injection (Fig. 4C). These data indicated that GH increased binding of STAT5 to the HNF-3γ promoter in the liver. A ribonuclease protection assay showed that this increase was accompanied by a 2-fold increase in HNF-3γ mRNA expression in the liver (P < 0.05, Fig. 4D).
The conserved putative STAT5 binding site in the HNF-3γ promoter could bind to liver STAT5 in vitro
We performed EMSA to determine whether the two putative STAT5 binding sites in the HNF-3γ promoter can bind to the STAT5 protein from bovine liver. As shown in Fig. 5A, the oligonucleotide corresponding to the putative STAT5 binding site 2 formed two DNA-protein complexes (denoted B1 and B2 in the figure) with liver nuclear protein extracts from cattle after GH injection; these complexes were not detectable when the oligonucleotide was incubated with liver nuclear proteins from cattle before GH injection. The STAT5 protein can bind to its target DNA element in the form of a dimer or a tetramer (36). Because the B1 complex moved more quickly than the B2 complex, the B1 complex might contain a dimeric form of STAT5, and B2 a tetrameric form of STAT5, or protein(s) in addition to a STAT5 dimer. The same DNA-protein complexes were not formed by the oligonucleotide corresponding to the putative STAT5 binding site 1 (Fig. 5A), indicating that this putative STAT5 binding site is not a real STAT5 binding site. Competitive gel-shift and supershift assays were performed to confirm the specificity of and the presence of STAT5 in the DNA-protein complexes formed by the putative STAT5 binding site 2. As shown in Fig. 5B, the two DNA-protein complexes were diminished by a molar excess (1×, 10×, and 100×) of the unlabeled oligonucleotide corresponding to the putative STAT5 binding site 2 but were not affected by the same excess of an unrelated oligonucleotide (Table 1). The two DNA-protein complexes were supershifted by the anti-STAT5 antibody but not by the preimmune serum (Fig. 5C). These data demonstrated that the putative STAT5 binding site 2 of the bovine HNF-3γ promoter can form DNA-protein complexes with GH-activated STAT5 protein from bovine liver.
Figure 5.
One of the two putative STAT5 binding sites in the HNF-3γ promoter could bind to bovine liver STAT5 in vitro. A, EMSA of the two putative STAT5 binding sites. In this assay, a 32P-labeled double-stranded oligonucleotide corresponding to the putative STAT5 binding site 1 or site 2 (see Fig. 4A and Table 1) was incubated with liver nuclear protein extracts from +GH and −GH cattle followed by gel electrophoresis. B1 and B2 indicate two DNA-protein complexes. B, Competitive gel-shift assay of the putative STAT5 binding site 2. In this assay, the 32P-labeled oligonucleotide corresponding to this STAT5 binding site was incubated with liver nuclear protein extracts from +GH cattle in the presence of 1×, 10×, and 100× molar excess of the same unlabeled oligonucleotide or an unrelated oligonucleotide. C, Supershift assay of the putative STAT5 binding site 2. In this assay, the 32P-labeled oligonucleotide corresponding to this STAT5 binding site was incubated with liver nuclear protein extracts from +GH cattle in the presence of an anti-STAT5 antibody or an equal amount of rabbit preimmune serum. S1 and S2 indicate probable supershifts of the DNA-protein complexes B1 and B2, respectively. These assays were repeated at least two times; shown are representative results.
The HNF-3γ promoter was GH responsive and this response depended on the identified STAT5 binding site
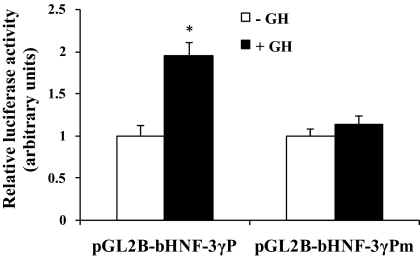
We next determined whether the HNF-3γ promoter could mediate GH-induced STAT5 activation of reporter gene expression, using a cotransfection analysis. In this analysis, a bovine HNF-3γ promoter-reporter plasmid was cotransfected with a GHR expression plasmid and a STAT5b expression plasmid into CHO cells. As shown in Fig. 6, GH treatment of the transfected CHO cells caused a 2-fold increase in luciferase activity expressed from the HNF-3γ promoter (P < 0.05). We further determined whether this GH response of the HNF-3γ promoter was dependent on the putative STAT5 binding site 2 in the promoter. As shown in Fig. 6, mutation of this STAT5 binding site completely abolished the response of the HNF-3γ promoter to GH (P < 0.05), indicating that this STAT5 binding site is essential for the HNF-3γ promoter to be activated by GH-induced STAT5.
Figure 6.
GH induced reporter gene expression from the bovine HNF-3γ promoter through the identified STAT5 binding site. This was demonstrated by a cotransfection analysis. In this analysis, the plasmid pGL2B-bHNF3γP, in which the putative STAT5 binding site 2 was intact, or pGL2B-bHNF3γPm, in which the binding site was mutated, was cotransfected with a GHR expression plasmid and a STAT5b expression plasmid into CHO cells. Twenty-four hours after the transfection, the cells were serum starved for 16 h and then treated with 500 ng/ml GH or PBS (indicated as −GH) for 8 h before luciferase assay. Variation in transfection efficiency was controlled by cotransfecting the pRL-CMV plasmid. *, P < 0.05 (n =4) compared with −GH.
GH could activate the IGF-I promoter through the HNF-3 binding sites in primary hepatocytes
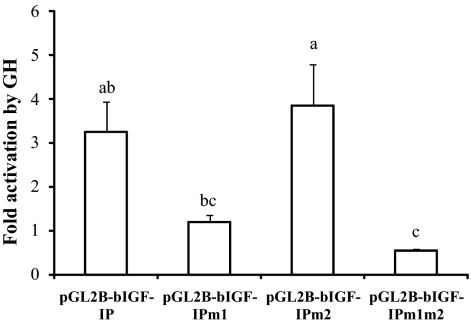
The above results suggest that in addition to directly stimulating IGF-I gene transcription, GH-activated STAT5 might indirectly stimulate IGF-I gene transcription in bovine liver through up-regulation of HNF-3γ gene expression. To further study this possibility, we determined the effect of the two HNF-3 binding sites on the transcriptional response of the bovine IGF-I promoter to GH in primary mouse hepatocytes. We chose to use primary mouse hepatocytes over bovine primary hepatocytes because the latter are difficult to isolate and culture. As shown in Fig. 7, GH stimulated a 3-fold increase in luciferase activity expressed from the transfected bovine IGF-I promoter in mouse hepatocytes (P < 0.05). Deletion of the HNF-3 binding site 1 completely blocked this increase (P = 0.1), deletion of the second HNF-3 binding site had no effect on this increase, and deletion of both HNF-3 binding sites was not different from deletion of the HNF-3 binding site 1 in terms of blocking the transcriptional response of the IGF-I promoter to GH. Deletion of either HNF-3 binding site did not affect the transcriptional activity of the IGF-I promoter in mouse hepatocytes not treated with GH (data not shown). These data further supported the indication of earlier experiments that GH might indirectly stimulate IGF-I gene transcription in bovine liver through up-regulation of HNF-3γ gene expression and binding to the IGF-I promoter.
Figure 7.
GH could activate the bovine IGF-I promoter in primary mouse hepatocytes through the HNF-3 binding site 1. The hepatocytes from male mice were cotransfected with the plasmid pGL2B-bIGF-IP, pGL2B-bIGF-IPm1, pGL2B-bIGF-IPm2, or pGL2B-bIGF-IPm1m2 and the transfection efficiency control plasmid pRL-CMV. The cells were cultured in serum-containing medium for 24 h followed by serum-free medium for 8 h and then treated with 500 ng/ml GH or an equal volume of PBS for 24 h before dual-luciferase assay. Fold activation on the y-axis corresponds to the ratio of the luciferase activity in the presence of GH to that in the presence of PBS. Bars not labeled with the same letter are different (P < 0.05, n = 3). The difference between pGL2B-bIGF-IP and pGL2B-bIGF-IPm1 tends to be significant (P = 0.1).
Discussion
Based on the facts that GH increases HNF-3γ mRNA expression in the liver (30) and that the IGF-I promoter contains conserved putative HNF-3 binding sites, we hypothesized that GH-increased HNF-3γ might contribute to GH stimulation of IGF-I gene transcription in the liver. This hypothesis is supported by the following results of this study; 1) There was binding of HNF-3γ to the IGF-I promoter in the liver of pituitary-intact cattle, and this binding is increased by exogenous GH; 2) two of the three putative HNF-3 binding sites in the IGF-I promoter bound to HNF-3γ from bovine liver in vitro; 3) overexpression of HNF-3γ activated the IGF-I promoter, and this activation was dependent on one of the identified HNF-3 binding sites; and 4) deletion of the HNF-3 binding sites blocked the transcriptional response of the IGF-I promoter to GH in primary cultured mouse hepatocytes.
The IGF-I promoter contains three HNF-3 binding sites, but only two (i.e. the putative HNF-3 binding sites 1 and 2) of them bind to HNF-3γ in vitro, and one of them (i.e. the putative HNF-3 binding site 1) is essential for the IGF-I promoter to be activated by HNF-3γ. The HNF-3 binding site 1 does not match the HNF-3 binding consensus sequence more than the other two (31) but is identical to one of the two HNF-3 binding sites in the human IGF-I promoter (37). In the EMSA (Fig. 2C), this HNF-3 binding site bound to HNF-3γ and perhaps also HNF-3β but not to HNF-3α from bovine liver (Fig. 2C), suggesting that this HNF-3 binding site might be a HNF-3γ and HNF-3β-specific binding site. However, because the same site from the human IGF-I promoter can bind to both overexpressed HNF-3β and HNF-3α (37), an alternative explanation of the EMSA result is that the HNF-3 binding site did not form a detectable DNA-protein complex with HNF-3α in bovine liver nuclear extracts because HNF-3α is expressed at a lower level than HNF-3γ and HNF-3β in bovine liver (30). Even if HNF-3α and HNF-3β bind to the IGF-I promoter, HNF-3γ is probably the major HNF-3 protein that mediates GH regulation of IGF-I gene expression in bovine liver, because HNF-3α expression is not affected by GH and HNF-3β expression is only transiently increased by GH in bovine liver (HNF-3β mRNA was increased at 6 h but not at 24 h or a week after injection of GH formulated for a 2-week release) (30). In rat, GH increases liver expression of both HNF-3β and HNF-3γ (38,39). GH seems to also stimulate liver expression of HNF-3β in mouse because mice with an intracellular domain-truncated GH receptor had reduced expression of HNF-3β in the liver (40). However, transgenic or adenoviral overexpression of HNF-3β reduces liver IGF-I mRNA expression in mice (41,42), suggesting that the GH-increased HNF-3β might inhibit rather than stimulate IGF-I gene expression in the liver.
In the cotransfection analysis using CHO cells, mutation of the two confirmed HNF-3 binding sites did not completely abolish the response of the IGF-I promoter to HNF-3γ (Fig. 3B). This suggests that the IGF-I promoter may contain additional binding sites for HNF-3γ or another HNF-3γ-dependent factor. One such additional HNF-3 binding site might be located 16 bp downstream from the HNF-3γ binding site 2 identified in this study, because the corresponding region of the human IGF-I promoter is a HNF-3 binding site (37). This potential HNF-3 binding site was not investigated in this study because it was not identified as a putative HNF-3 binding site by the TF search program we used to search for the putative HNF-3 binding sites (43). However, in the transfection analysis using mouse hepatocytes, mutation of the confirmed HNF-3 binding site 1 completely blocked the transcriptional response of the IGF-I promoter to GH. This suggests that even though the IGF-I promoter contains additional HNF-3 binding sites, they may be dispensable for the IGF-I promoter to respond to GH. We further speculate that this may be because these HNF-3 binding sites are less competitive than the identified HNF-3 binding site in binding to the GH-induced HNF-3γ, which is likely limited in amount compared with the overexpressed HNF-3γ.
Based on the prediction that the bovine HNF-3γ promoter contains two putative STAT5 binding sites and the fact that STAT5 is an important component of the signaling pathway from the GH receptor (44), we thought that GH stimulation of HNF-3γ expression in bovine liver might be mediated by direct interaction of STAT5 with the HNF-3γ promoter. In this study, we found that one (i.e. the proximal one) of the two putative STAT5 binding sites in the bovine HNF-3γ promoter was able to bind to liver STAT5 protein in vitro and to mediate GH-induced STAT5 activation of the HNF-3γ promoter and that GH increased binding of STAT5 to the HNF-3γ promoter and HNF-3γ mRNA expression in the liver. These results support the hypothesis that GH stimulates HNF-3γ mRNA expression in bovine liver through direct interaction of STAT5 with the HNF-3γ promoter. The distal putative STAT5 binding site in the HNF-3γ promoter did not bind to GH-activated STAT5 from the liver, although it is identical to the STAT5 binding consensus sequence (45,46). Unlike the proximal one, the distal putative STAT5 binding site is not conserved among other mammals. In a previous study, we also found that some putative STAT5 binding sites, despite conforming to the consensus sequences, did not bind to STAT5 (18). Thus, the consensus sequence for STAT5 binding sites (as well as that for HNF-3 binding sites) needs to be refined, and this requires the identification of many additional in vivo STAT5 (and HNF-3) binding sites.
The HNF-3 proteins affect gene transcription by opening the highly compacted chromatin in a manner not requiring the SWI/SNF chromatin remodeling complex (47,48,49) and thereby enhancing the binding of RNA polymerase II to the gene promoters (49) and/or promoting binding of other transcription factors (47). The HNF-3γ protein is known to induce DNA bending, which may facilitate interaction between proteins bound to distant sites (50). STAT5 mediates GH regulation of IGF-I gene expression by binding to multiple DNA regions distantly located from the IGF-I promoter (17,18,19,20). Therefore, it is tempting to speculate that binding of HNF-3γ to the IGF-I promoter may induce bending of the IGF-I DNA in a way to facilitate binding of STAT5 to the IGF-I gene and/or interaction of distantly bound STAT5, resulting in sustained IGF-I gene transcription. STAT5 and HNF-3β cooperate in mediating GH regulation of cytochrome P-450 gene expression in the liver (51). Therefore, it is also tempting to speculate that HNF-3γ and STAT5 might cooperate in activating IGF-I gene transcription. However, HNF-3γ and STAT5 did not appear to do so in stimulating reporter gene expression from the IGF-I promoter in CHO cells (data not shown). It remains to be determined whether such cooperation exists in the liver.
In summary, this study shows that GH increases gene transcription of the liver-enriched transcription factor HNF-3γ in bovine liver through STAT5 interaction with a conserved STAT5 binding site in the HNF-3γ promoter and that GH-increased HNF-3γ may increase gene transcription of IGF-I by binding to a conserved HNF-3 binding site in the IGF-I promoter. These results suggest that in addition to direct stimulation, GH-activated STAT5 may also indirectly stimulate IGF-I gene transcription by enhancing gene expression of the liver-enriched transcription factor HNF-3γ in the liver. Such an indirect mechanism may explain in part why IGF-I mRNA is expressed at a much higher level in liver than in other tissues and why IGF-I mRNA expression continues to increase, whereas STAT5 activation is temporary in response to GH (52). Because both the HNF-3 binding site and the STAT5 binding site identified in this study are conserved in various mammals, we speculate that the GH-STAT5-HNF-3γ-IGF-I relationship identified in this study may exist in animals in addition to cattle.
Materials and Methods
Animal experiments
Nonlactating and nonpregnant Angus crossbred cows (3–5 yr of age, 510–550 kg weight) were used in this study. Liver biopsy samples were taken from each cow 1 wk before and 24 h after injection of 500 mg recombinant bovine GH formulated for sustained (2 wk) release (Monsanto Co., St. Louis, MO). The liver biopsy procedure was performed as previously described (30). Upon collection, the tissue samples were immediately processed for chromatin isolation or nuclear protein extraction or stored at −80 C for RNA isolation. These as well as the following animal-related procedures were approved by the Virginia Tech Institutional Animal Care and Use Committee.
Plasmid construction
A 2185-bp bovine IGF-I promoter, between nucleotide −2058 and nucleotide +146 relative to the major transcription start site, numbered +1, for class 2 IGF-I mRNA (32), was amplified from bovine genomic DNA by standard PCR with sequence-specific primers bIGF-IP1921F1 and bIGF-IPR2 (Table 2) and cloned into the promoterless luciferase reporter vector pGL2-basic (Promega, Madison, WI) at the restriction sites KpnI and XhoI to generate the plasmid pGL2B-bIGF-IP. A 239-bp bovine HNF-3γ DNA region, predicted to cover the putative transcription start site based on the 5′ end sequences of the human, mouse, and rat HNF-3γ mRNA (GenBank accession numbers NM_004497, NM_008260, and NM_017077, respectively), was amplified by a standard PCR with primers bHNF3gRPAF1 and bHNF3gRPAR1 (Table 2). This PCR product was cloned into the pGEM-T Easy vector (Promega) to generate the plasmid, pGEM-TEbHNF-3γ239. A standard PCR was used to amplify a 961-bp promoter region, between nucleotide −811 and nucleotide +149 relative to the major transcription start site (+1) of HNF-3γ mRNA, of the bovine HNF-3γ gene with primers bHNF3gPF1 and bHNF3gPR1 (Table 2). This PCR product was cloned into the pGL2B vector between the NheI and HindIII sites to generate the plasmid pGL2B-bHNF-3γP. A 1063-bp bovine HFN-3γ cDNA, which was expected to encode the full-length HNF-3γ protein, was amplified from bovine liver total RNA by standard RT-PCR using primers HNF-3gF and HNF-3gR (Table 2). This PCR product was cloned into the expression vector pcDNA3.1 (Invitrogen, Carlsbad, CA) at the EcoRI and NotI sites to generate the plasmid pcDNA3.1-bHNF-3γ.
Table 2.
PCR primers used in this study
| Name | Sequence (5′–3′) | Application | Amplicon size (bp) |
|---|---|---|---|
| bHNF3gRPAF1 | GAGCGGGCGGGATCCGAGG | PCR | 239 |
| bHNF3gRPAPR1 | CATCTTCACTGAGCCCAGCAT | ||
| bHNF3gPF1 | ATGCTAGCCGCCGGGAAATGGAGTC | PCR | 961 |
| bHNF3gPR1 | GCAAGCTTCATCTTCACTGAGCCCAGCAT | ||
| bHNF3gPChIPF1 | AGCCCTTCATTTCCGTCTTT | ChIP | 120 |
| bHNF3gPChIPR1 | AGGGAGCAGAGTCTTCGTGA | ||
| HNF3gF | GAGAATTCATGCTGGGCTCAGTGAAGAT3 | PCR | 1082 |
| HNF3gR | CCAGCGGCCGCACCCCTGCTAGGATGCATTA | ||
| bIGF-IP1921F1 | TTCGGTACCACAGTGTCTGTGTTTTGTA | PCR | 2185 |
| bIGF-IPR2 | AAACTCGAGCAGCAAAATTTGAGGGCAAT | ||
| bIGF-IPChIPF1 | TTTGCCAGAAGAGGGAGAGA | ChIP | 161 |
| bIGF-IP1ChIPR1 | GCAGGCTCTATCTGCTCTGAA | ||
| bGAPDHPF1 | ACTACTCTCCCGCAGTGCTC | ChIP | 185 |
| bGAPDHPR1 | AGTAGTCGGCCTACCGCTTT | ||
| bHNF3gPm2F | TGGAGGCTGCGGCCGCATGGAGTTCA | STAT5 site 2 mutagenesis | |
| bHNF3gPm2R | TGAACTCCATGCGGCCGCAGCCTCCA | STAT5 site 2 mutagenesis | |
| bIGF-IP1921m1F1 | TTCGGTACCACAGTGTCTGTGTTTTGTGCGGCCGCGTGAGGATTTTCTCTAAAT | HNF-3 site 1 mutagenesis | |
| bIGF-IP1921m2F1 | GTGATTTCTTGAGCGGCCGCGCGATTTCTTACTC | HNF-3 site 2 mutagenesis | |
| bIGF-IP1921m2R1 | GAGTAAGAAATCGCGGCCGCGCTCAAGAAATCAC | HNF-3 site 2 mutagenesis |
The top sequence of a pair of primers is the forward primer and the bottom sequence the reverse primer. Underlined are restriction enzyme recognition sites added for cloning or mutagenesis.
The putative HNF-3 binding site 1, site 2, or both, in the IGF-I promoter insert of the pGL2B-bIGF-IP plasmid were mutated into NotI restriction sites to generate the plasmids pGL2B-bIGF-Ipm1, pGL2B-bIGF-Ipm2, and pGL2B-bIGF-Ipm1m2, respectively. The mutations were made by PCR-based site-directed mutagenesis as described previously (19), using primers bIGF-IP1921m1F1, bIGF-IP1921m2F1, and bIGF-IP1921m2R1 (Table 2). Similarly, the putative STAT5 binding site in the HNF-3γ promoter insert in pGL2B-bHNF-3γP was mutated to generate the plasmid pGL2B-bHNF-3γPm, using primers bHNF3gPm2F and bHNF3gPm2R (Table 2).
Other plasmids used in this study included the GH receptor expression plasmid pcDNA3.1-bGHR (19), the wild-type STAT5b expression plasmid pMX-STAT5b (53), and cDNA plasmids to generate antisense riboprobes for ribonuclease protection assay of HNF-3γ, IGF-I, and GAPDH mRNA (30,54).
All inserts and mutations of the plasmids made in this study were verified by DNA sequencing. The ability of pcDNA3.1-bHNF-3γ to express HNF-3γ protein was confirmed by gel electrophoresis of products of in vitro transcription and translation of this plasmid in the presence of [35S]Met, and this was done essentially as previously described (19).
Ribonuclease protection assay
Total RNA from liver samples was isolated using TRI reagent (Molecular Research Center, Cincinnati, OH), essentially according to the manufacturer’s instructions. Ribonuclease protection assays were performed to analyze IGF-I, HNF-3γ, and GAPDH mRNA levels and also to determine the transcription start site of the HNF-3γ gene in bovine liver. The antisense riboprobes for the assays were synthesized as described previously (18). The assays were carried out using the RPA II kit (Ambion, Austin, TX). Briefly, 20 μg of total RNA were mixed with 1 × 105 dpm of HNF-3γ or IGF-I antisense probe in a total volume of 20 μl hybridization buffer. In the assay of IGF-I and HNF-3γ mRNA, 1 × 104 dpm GAPDH antisense probe synthesized at 10 times lower specific activity was included as a loading control. The mixture was incubated at 42 C for 16 h and then digested with ribonucleases A and T1 at 37 C for 45 min. The protected RNA fragments were precipitated and resolved on 6% polyacrylamide gels containing 7 m urea and were visualized by phosphorimaging. The densities of the protected bands were measured using the ImageJ software (http://rsb.info.nih.gov/ij).
A sequencing ladder of the antisense strand of the 239-bp HNF-3γ DNA fragment was generated by using the fmol DNA sequencing system (Promega) and primer 5′-CATCTTCACTGAGCCCAGCAT-3′. The primer was labeled with [32P]γ-ATP and T4 polynucleotide kinase as described previously (55). This sequencing ladder served as a reference in the ribonuclease protection assay of the HNF-3γ mRNA start site.
ChIP assay
The nuclei from fresh liver samples were prepared as described previously (18). The nuclei were subsequently sheared on ice with 10 pulses of 20 sec sonication using a sonic dismembrator model 100 at setting 3 (Fisher Scientific, Pittsburgh, PA). Under these conditions, the chromatin was sheared to fragments 200–500 bp long. The sheared chromatin was either immediately immunoprecipitated or stored at −80 C. Immunoprecipitation of the sheared chromatin was performed using a ChIP-IT kit (Active Motif, Carlsbad, CA), following the manufacturer’s directions. Briefly, the sheared chromatin from approximately 200 mg tissue was mixed with 25 μl protein G-Dynal magnetic beads, 3 μg STAT5 antibody (sc-835; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), or 3 μg HNF-3γ antibody (sc-5361; Santa Cruz Biotechnology) in a final volume of 200 μl ChIP buffer 1 (from the kit) and was incubated overnight at 4 C. The immunocomplexes binding to the protein G-Dynal magnetic beads were collected using a magnetic stand. The magnetic beads were washed, and the chromatin was eluted using the reagents from the ChIP-IT kit, according to the manufacturer’s instructions. The eluted chromatin was reverse cross-linked by overnight incubation at 65 C. Meanwhile, 10% of the sonicated chromatin before immunoprecipitation (i.e. the input chromatin) was also reverse cross-linked. The DNA from the reverse cross-linked samples was extracted by proteinase K digestion followed by phenol-chloroform extraction.
Abundance of the IGF-I or HNF-3γ promoter DNA in the HNF-3γ antibody or STAT5 antibody-precipitated chromatin or input chromatin was determined by semiquantitative PCR using 2× PCR Master Mix (Promega) and sequence-specific primers (Table 2). The abundance of the GAPDH promoter in these chromatin samples was also analyzed by PCR as a loading control. The GAPDH promoter was not expected to be bound by STAT5 or HNF-3 because it does not contain a putative STAT5 or HNF-3 binding site and is not regulated by GH (30). The PCR conditions were 30 cycles of 94 C for 30 sec, 60 C for 1 min, and 72 C for 2 min. The PCR products were resolved through standard agarose gels. Densities of DNA bands were measured using the ImageJ software. The density of a DNA fragment in an antibody-precipitated chromatin sample was normalized to that of the same fragment in the corresponding input chromatin to control for variation in PCR efficiency.
EMSA
Nuclear proteins from liver samples were prepared as described previously (18). Double-stranded oligonucleotides corresponding to the putative HNF-3 binding sites in the IGF-I promoter or the putative STAT5 binding sites in the HNF-3γ promoter were end-labeled with 32P using T4 polynucleotide kinase (Promega). The sequences of these oligonucleotides are shown in Table 1. Ten micrograms of bovine liver nuclear proteins were incubated with 1 × 105 dpm 32P-labeled oligonucleotide probe in a reaction buffer containing 20% glycerol, 20 mm Tris-HCl (pH 7.5), 100 mm KCl, 1 mm dithiothreitol, 1 mm EDTA, and 2 μg poly(deoxyinosine-deoxycytosine) for 90 min at 4 C. After the incubation, the DNA-protein mixtures were resolved on native 6% polyacrylamide gels. After electrophoresis, the gels were dried and visualized by phosphorimaging. For supershift assays of the STAT5 binding site, the nuclear protein extracts were incubated with 2 μg anti-STAT5 antibody (sc-835; Santa Cruz) or 2 μg rabbit preimmune serum in the reaction buffer for 1 h at 4 C before being incubated with the radiolabeled oligonucleotide. For supershift assays of the HNF-3 binding sites, the nuclear protein extracts were incubated with 2 μg anti-HNF-3α (sc-6553), anti-HNF-3β (sc-6554), anti-HNF-3γ (sc-5361) (Santa Cruz), or goat preimmune serum at 4 C overnight before the labeled oligonucleotide was added. For the competitive gel-shift assays, the 32P-labeled oligonucleotide was incubated with the nuclear protein extracts in the presence of 1×, 10×, and 100× molar excess of the same unlabeled oligonucleotide or an unrelated oligonucleotide (Table 1) before gel electrophoresis.
Transfection of CHO cells and luciferase assay
The Chinese hamster ovary cell line CHO cells were grown in MEM supplemented with 1 mm sodium pyruvate, 2 mm l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% fetal bovine serum. All reagents used in cell culture were purchased from Sigma-Aldrich, Inc. (St. Louis, MO). The cells were cultured at 37 C in a humidified 5% CO2 atmosphere. Approximately 24 h before transfection, the cells were plated in 24-well plates at a density of 5 × 104 per well. In the transfection analyses to determine the ability of HNF-3γ to activate the IGF-I promoter, CHO cells in each well were transfected with 0.5 μg of the IGF-I promoter construct pGL2B-bIGF-IP, pGL2B-bIGF-Ipm1, pGL2B-bIGF-Ipm2, or pGL2B-bIGF-Ipm1m2, 0.5 μg pcDNA3.1-bHNF-3γ or pcDNA3.1 (empty vector), and 1 ng of the transfection efficiency control plasmid pRL-CMV (Promega), using FuGENE 6 as the transfection reagent (Roche Applied Science, Indianapolis, IN). The cells were lysed 48 h after the transfection. In the transfection analyses to determine GH response of the HNF-3γ promoter, the cells in each well were transfected with 0.5 μg of the HNF-3γ promoter construct pGL2B-bHNF-3γP or pGL2B-bHNF-3γPm, 0.5 μg pcDNA3.1-bGHR, 0.5 μg pMX-STAT5b, and 1 ng pRL-CMV. About 24 h after the transfection, the medium was replaced with serum-free MEM, and the cells were further cultured for 16 h. Subsequently, the cells were treated with 500 ng/ml recombinant bovine GH (National Hormone and Peptide Program, Torrance, CA) or an equal volume of PBS (the vehicle for GH) for 8 h. Cell lysis and dual-luciferase assay were performed using the dual-luciferase reporter assay system (Promega), according to the manufacturer’s instructions. The luciferase activity expressed from a promoter construct was divided by that from pRL-CMV in the same well to normalize the variation in transfection efficiency.
Isolation and transfection of mouse hepatocytes
Mouse hepatocytes were isolated from livers of male C56BL/6 mice 6–8 wk of age using the two-step perfusion procedure (56). Briefly, the mouse was anesthetized with 0.5 mg/g body weight of Avertin, and a catheter was inserted and advanced into the inferior vena cava for perfusion. The liver was first perfused with the Krebs Ringer buffer containing 0.1 mm EGTA. The liver was subsequently perfused and digested with Ca2+-activated collagenase D at 1 mg/ml (Roche) in Krebs Ringer buffer. The hepatocytes were collected from the perfused liver and washed with precooled MEM supplemented with 10% fetal bovine serum. The hepatocytes were pelleted by centrifugation at 50 × g for 2 min. Viability of the hepatocytes was determined by trypan blue staining. Only the isolations containing more than 85% viable hepatocytes were used in transfection experiments. The hepatocytes were plated into 24-well plates at 2 × 105 per well. Twenty-four hours later, the hepatocytes in each well were transfected with 0.5 μg of the IGF-I promoter construct pGL2B-bIGF-IP, pGL2B-bIGF-IPm1, pGL2B-bIGF-IPm2, or pGL2B-bIGF-IPm1m2, and 5 ng pRL-CMV, using GeneJet as the transfection reagent (SignaGen Laboratories, Gaithersburg, MD). About 24 h after the transfection, the hepatocytes were serum starved for 8 h, followed by 24 h treatment with 500 ng/ml recombinant bovine GH or an equal volume of PBS. Cell lysis and dual-luciferase assay were performed as described above.
Statistical analyses
The statistical significance of the difference between two means in a given experiment was determined using Student’s t test. Means of more than two groups were compared using ANOVA followed by Tukey’s test. These analyses were performed using the general linear model of SAS (SAS Institute, Inc., Cary, NC). All data are expressed as mean ± sem.
Footnotes
This work was supported by National Institutes of Health Grant DK67961 and United States Department of Agriculture Cooperative State Research, Education, and Extension Service Grant no. 2009-35205-05221 (to H.J.).
Current address for S.E.: Division of Experimental Hematology and Cancer Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio 45229.
Current address for Y.W.: Eton Bioscience, Inc., Research Triangle Park, North Carolina 27709.
J.Y. is a visiting scholar from Institute of Animal Nutrition, Sichuan Agricultural University, China.
Disclosure Summary: The authors have nothing to disclose.
First Published Online October 9, 2009
Abbreviations: C/EBP, CCAAT/enhancer-binding protein; ChIP, chromatin immunoprecipitation; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HNF, hepatocyte nuclear factor; STAT5, signal transducer and activator of transcription 5.
References
- Stewart CE, Rotwein P 1996 Growth, differentiation, and survival: multiple physiological functions for insulin-like growth factors. Physiol Rev 76:1005–1026 [DOI] [PubMed] [Google Scholar]
- LeRoith D 2008 Clinical relevance of systemic and local IGF-I: lessons from animal models. Pediatr Endocrinol Rev 5(Suppl 2):739–743 [PubMed] [Google Scholar]
- Pollak M 2008 Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer 8:915–928 [DOI] [PubMed] [Google Scholar]
- Samani AA, Yakar S, LeRoith D, Brodt P 2007 The role of the IGF system in cancer growth and metastasis: overview and recent insights. Endocr Rev 28:20–47 [DOI] [PubMed] [Google Scholar]
- Meyer-Schwickerath R, Pfeiffer A, Blum WF, Freyberger H, Klein M, Lösche C, Röllmann R, Schatz H 1993 Vitreous levels of the insulin-like growth factors I and II, and the insulin-like growth factor binding proteins 2 and 3, increase in neovascular eye disease. Studies in nondiabetic and diabetic subjects. J Clin Invest 92:2620–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson-Berka JL, Wraight C, Werther G 2006 The role of growth hormone, insulin-like growth factor and somatostatin in diabetic retinopathy. Curr Med Chem 13:3307–3317 [DOI] [PubMed] [Google Scholar]
- Clemmons DR 2007 Modifying IGF1 activity: an approach to treat endocrine disorders, atherosclerosis and cancer. Nat Rev Drug Discov 6:821–833 [DOI] [PubMed] [Google Scholar]
- Bartke A 2005 Role of the growth hormone/insulin-like growth factor system in mammalian aging. Endocrinology 146:3718–3723 [DOI] [PubMed] [Google Scholar]
- D'Ercole AJ, Stiles AD, Underwood LE 1984 Tissue concentrations of somatomedin C: further evidence for multiple sites of synthesis and paracrine or autocrine mechanisms of action. Proc Natl Acad Sci USA 81:935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughaday WH, Rotwein P 1989 Insulin-like growth factors I and II. Peptide, messenger ribonucleic acid and gene structures, serum, and tissue concentrations. Endocr Rev 10:68–91 [DOI] [PubMed] [Google Scholar]
- Yakar S, Liu JL, Stannard B, Butler A, Accili D, Sauer B, LeRoith D 1999 Normal growth and development in the absence of hepatic insulin-like growth factor I. Proc Natl Acad Sci USA 96:7324–7329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren K, Liu JL, Blad K, Skrtic S, Vidal O, Wallenius V, LeRoith D, Törnell J, Isaksson OG, Jansson JO, Ohlsson C 1999 Liver-derived insulin-like growth factor I (IGF-I) is the principal source of IGF-I in blood but is not required for postnatal body growth in mice. Proc Natl Acad Sci USA 96:7088–7092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichell DP, Kikuchi K, Rotwein P 1992 Growth hormone rapidly activates insulin-like growth factor I gene transcription in vivo. Mol Endocrinol 6:1899–1908 [DOI] [PubMed] [Google Scholar]
- Woelfle J, Billiard J, Rotwein P 2003 Acute control of insulin-like growth factor-I gene transcription by growth hormone through Stat5b. J Biol Chem 278:22696–22702 [DOI] [PubMed] [Google Scholar]
- Davey HW, Xie T, McLachlan MJ, Wilkins RJ, Waxman DJ, Grattan DR 2001 STAT5b is required for GH-induced liver IGF-I gene expression. Endocrinology 142:3836–3841 [DOI] [PubMed] [Google Scholar]
- Udy GB, Towers RP, Snell RG, Wilkins RJ, Park SH, Ram PA, Waxman DJ, Davey HW 1997 Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci USA 94:7239–7244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia DJ, Ono M, Woelfle J, Schlesinger-Massart M, Jiang H, Rotwein P 2006 Characterization of distinct Stat5b binding sites that mediate growth hormone-stimulated IGF-I gene transcription. J Biol Chem 281:3190–3197 [DOI] [PubMed] [Google Scholar]
- Eleswarapu S, Gu Z, Jiang H 2008 Growth hormone regulation of insulin-like growth factor-I gene expression may be mediated by multiple distal signal transducer and activator of transcription 5 binding sites. Endocrinology 149:2230–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jiang H 2005 Identification of a distal STAT5-binding DNA region that may mediate growth hormone regulation of insulin-like growth factor-I gene expression. J Biol Chem 280:10955–10963 [DOI] [PubMed] [Google Scholar]
- Woelfle J, Chia DJ, Rotwein P 2003 Mechanisms of growth hormone (GH) action. Identification of conserved Stat5 binding sites that mediate GH-induced insulin-like growth factor-I gene activation. J Biol Chem 278:51261–51266 [DOI] [PubMed] [Google Scholar]
- Schrem H, Klempnauer J, Borlak J 2004 Liver-enriched transcription factors in liver function and development. Part II: the C/EBPs and D site-binding protein in cell cycle control, carcinogenesis, circadian gene regulation, liver regeneration, apoptosis, and liver-specific gene regulation. Pharmacol Rev 56:291–330 [DOI] [PubMed] [Google Scholar]
- Schrem H, Klempnauer J, Borlak J 2002 Liver-enriched transcription factors in liver function and development. I. The hepatocyte nuclear factor network and liver-specific gene expression. Pharmacol Rev 54:129–158 [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Wang W, Ninomiya T, Nagano H, Ohta K, Itoh H 1999 Liver enriched transcription factors and differentiation of hepatocellular carcinoma. Mol Pathol 52:19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulik VP, Kavsan VM, van Schaik FM, Nolten LA, Steenbergh PH, Sussenbach JS 1995 The promoter of the salmon insulin-like growth factor I gene is activated by hepatocyte nuclear factor 1. J Biol Chem 270:1068–1073 [DOI] [PubMed] [Google Scholar]
- Nolten LA, Steenbergh PH, Sussenbach JS 1995 Hepatocyte nuclear factor 1α activates promoter 1 of the human insulin-like growth factor I gene via two distinct binding sites. Mol Endocrinol 9:1488–1499 [DOI] [PubMed] [Google Scholar]
- Nolten LA, van Schaik FM, Steenbergh PH, Sussenbach JS 1994 Expression of the insulin-like growth factor I gene is stimulated by the liver-enriched transcription factors C/EBPα and LAP. Mol Endocrinol 8:1636–1645 [DOI] [PubMed] [Google Scholar]
- Umayahara Y, Ji C, Centrella M, Rotwein P, McCarthy TL 1997 CCAAT/enhancer-binding protein δ activates insulin-like growth factor-I gene transcription in osteoblasts. Identification of a novel cyclic AMP signaling pathway in bone. J Biol Chem 272:31793–31800 [DOI] [PubMed] [Google Scholar]
- Kaestner KH 2000 The hepatocyte nuclear factor 3 (HNF3 or FOXA) family in metabolism. Trends Endocrinol Metab 11:281–285 [DOI] [PubMed] [Google Scholar]
- Clevidence DE, Overdier DG, Tao W, Qian X, Pani L, Lai E, Costa RH 1993 Identification of nine tissue-specific transcription factors of the hepatocyte nuclear factor 3/forkhead DNA-binding-domain family. Proc Natl Acad Sci USA 90:3948–3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleswarapu S, Jiang H 2005 Growth hormone regulates the expression of hepatocyte nuclear factor-3γ and other liver-enriched transcription factors in bovine liver. J Endocrinol 184:95–105 [DOI] [PubMed] [Google Scholar]
- Roux J, Pictet R, Grange T 1995 Hepatocyte nuclear factor 3 determines the amplitude of the glucocorticoid response of the rat tyrosine aminotransferase gene. DNA Cell Biol 14:385–396 [DOI] [PubMed] [Google Scholar]
- Wang Y, Price SE, Jiang H 2003 Cloning and characterization of the bovine class 1 and class 2 insulin-like growth factor-I mRNAs. Domest Anim Endocrinol 25:315–328 [DOI] [PubMed] [Google Scholar]
- Kaestner KH, Hiemisch H, Luckow B, Schütz G 1994 The HNF-3 gene family of transcription factors in mice: gene structure, cDNA sequence, and mRNA distribution. Genomics 20:377–385 [DOI] [PubMed] [Google Scholar]
- Navas MA, Vaisse C, Boger S, Heimesaat M, Kollee LA, Stoffel M 2000 The human HNF-3 genes: cloning, partial sequence and mutation screening in patients with impaired glucose homeostasis. Hum Hered 50:370–381 [DOI] [PubMed] [Google Scholar]
- Darnell JE Jr 1997 STATs and gene regulation. Science 277:1630–1635 [DOI] [PubMed] [Google Scholar]
- John S, Vinkemeier U, Soldaini E, Darnell Jr JE, Leonard WJ 1999 The significance of tetramerization in promoter recruitment by Stat5. Mol Cell Biol 19:1910–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolten LA, Steenbergh PH, Sussenbach JS 1996 The hepatocyte nuclear factor 3β stimulates the transcription of the human insulin-like growth factor I gene in a direct and indirect manner. J Biol Chem 271:31846–31854 [DOI] [PubMed] [Google Scholar]
- Lahuna O, Rastegar M, Maiter D, Thissen JP, Lemaigre FP, Rousseau GG 2000 Involvement of STAT5 (signal transducer and activator of transcription 5) and HNF-4 (hepatocyte nuclear factor 4) in the transcriptional control of the hnf6 gene by growth hormone. Mol Endocrinol 14:285–294 [DOI] [PubMed] [Google Scholar]
- Lahuna O, Fernandez L, Karlsson H, Maiter D, Lemaigre FP, Rousseau GG, Gustafsson J, Mode A 1997 Expression of hepatocyte nuclear factor 6 in rat liver is sex-dependent and regulated by growth hormone. Proc Natl Acad Sci USA 94:12309–12313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland JE, Lichanska AM, Kerr LM, White M, d'Aniello EM, Maher SL, Brown R, Teasdale RD, Noakes PG, Waters MJ 2005 In vivo analysis of growth hormone receptor signaling domains and their associated transcripts. Mol Cell Biol 25:66–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Hughes D, Wang X, Costa RH 2002 Adenovirus-mediated increase in HNF-3β or HNF-3α shows differences in levels of liver glycogen and gene expression. Hepatology 35:30–39 [DOI] [PubMed] [Google Scholar]
- Rausa FM, Tan Y, Zhou H, Yoo KW, Stolz DB, Watkins SC, Franks RR, Unterman TG, Costa RH 2000 Elevated levels of hepatocyte nuclear factor 3β in mouse hepatocytes influence expression of genes involved in bile acid and glucose homeostasis. Mol Cell Biol 20:8264–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer T, Chen X, Karas H, Kel AE, Kel OV, Liebich I, Meinhardt T, Reuter I, Schacherer F, Wingender E 1999 Expanding the TRANSFAC database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Res 27:318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichanska AM, Waters MJ 2008 How growth hormone controls growth, obesity and sexual dimorphism. Trends Genet 24:41–47 [DOI] [PubMed] [Google Scholar]
- Ehret GB, Reichenbach P, Schindler U, Horvath CM, Fritz S, Nabholz M, Bucher P 2001 DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J Biol Chem 276:6675–6688 [DOI] [PubMed] [Google Scholar]
- Verdier F, Rabionet R, Gouilleux F, Beisenherz-Huss C, Varlet P, Muller O, Mayeux P, Lacombe C, Gisselbrecht S, Chretien S 1998 A sequence of the CIS gene promoter interacts preferentially with two associated STAT5A dimers: a distinct biochemical difference between STAT5A and STAT5B. Mol Cell Biol 18:5852–5860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS 2002 Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 9:279–289 [DOI] [PubMed] [Google Scholar]
- Holmqvist PH, Belikov S, Zaret KS, Wrange O 2005 FoxA1 binding to the MMTV LTR modulates chromatin structure and transcription. Exp Cell Res 304:593–603 [DOI] [PubMed] [Google Scholar]
- Zhao H, Friedman RD, Fournier RE 2007 The locus control region activates serpin gene expression through recruitment of liver-specific transcription factors and RNA polymerase II. Mol Cell Biol 27:5286–5295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrou S, Hellqvist M, Samuelsson L, Enerbäck S, Carlsson P 1994 Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. EMBO J 13:5002–5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Waxman DJ 2001 Inhibitory cross-talk between STAT5b and liver nuclear factor HNF3β: impact on the regulation of growth hormone pulse-stimulated, male-specific liver cytochrome P-450 gene expression. J Biol Chem 276:43031–43039 [DOI] [PubMed] [Google Scholar]
- Ram PA, Park SH, Choi HK, Waxman DJ 1996 Growth hormone activation of Stat 1, Stat 3, and Stat 5 in rat liver. Differential kinetics of hormone desensitization and growth hormone stimulation of both tyrosine phosphorylation and serine/threonine phosphorylation. J Biol Chem 271:5929–5940 [DOI] [PubMed] [Google Scholar]
- Ariyoshi K, Nosaka T, Yamada K, Onishi M, Oka Y, Miyajima A, Kitamura T 2000 Constitutive activation of STAT5 by a point mutation in the SH2 domain. J Biol Chem 275:24407–24413 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Boyd CK, Bracken CJ, Lamberson WR, Keisler DH, Lucy MC 1999 Reduced growth hormone receptor (GHR) messenger ribonucleic acid in liver of periparturient cattle is caused by a specific down-regulation of GHR 1A that is associated with decreased insulin-like growth factor I. Endocrinology 140:3947–3954 [DOI] [PubMed] [Google Scholar]
- Xu Q, Walther N, Jiang H 2004 Chicken ovalbumin upstream promoter transcription factor II (COUP-TFII) and hepatocyte nuclear factor 4γ (HNF-4γ) and HNF-4α regulate the bovine growth hormone receptor 1A promoter through a common DNA element. J Mol Endocrinol 32:947–961 [DOI] [PubMed] [Google Scholar]
- Seglen PO 1976 Preparation of isolated rat liver cells. Methods Cell Biol 13:29–83 [DOI] [PubMed] [Google Scholar]