Abstract
We have analyzed the solution structure of RNA containing the branch point/3' splice site region of the first intervening sequence (IVS1) of human beta-globin pre-mRNA by nuclease mapping and NMR. Nuclease mapping indicates that there are two distinct structural domains: one contains the branch point region, and the other includes the 3' splice site and second exon sequences. In the branch point domain, the adenosine at which the RNA branch forms appears to be in the loop of a stem/loop structure. The branch point structure does not appear to interact with other parts of the RNA, since its unique nuclease digestion pattern is conserved among transcripts containing the entire intron or only the branch point region. This is confirmed by a comparison of the NMR spectra of two RNA transcripts; a distinct set of resonances appears in the spectra of the RNA containing only branch sequences or including 3' splice site/exon 2 sequences. NMR studies further show that the 3' splice site/exon 2 domain has a lower melting temperature than the branch point domain, suggesting that the two regions are distinct dynamically as well as structurally. Nuclease mapping studies of adenovirus major late IVS1 indicate that this RNA has structural features in common with the human beta-globin transcript.
Full text
PDF

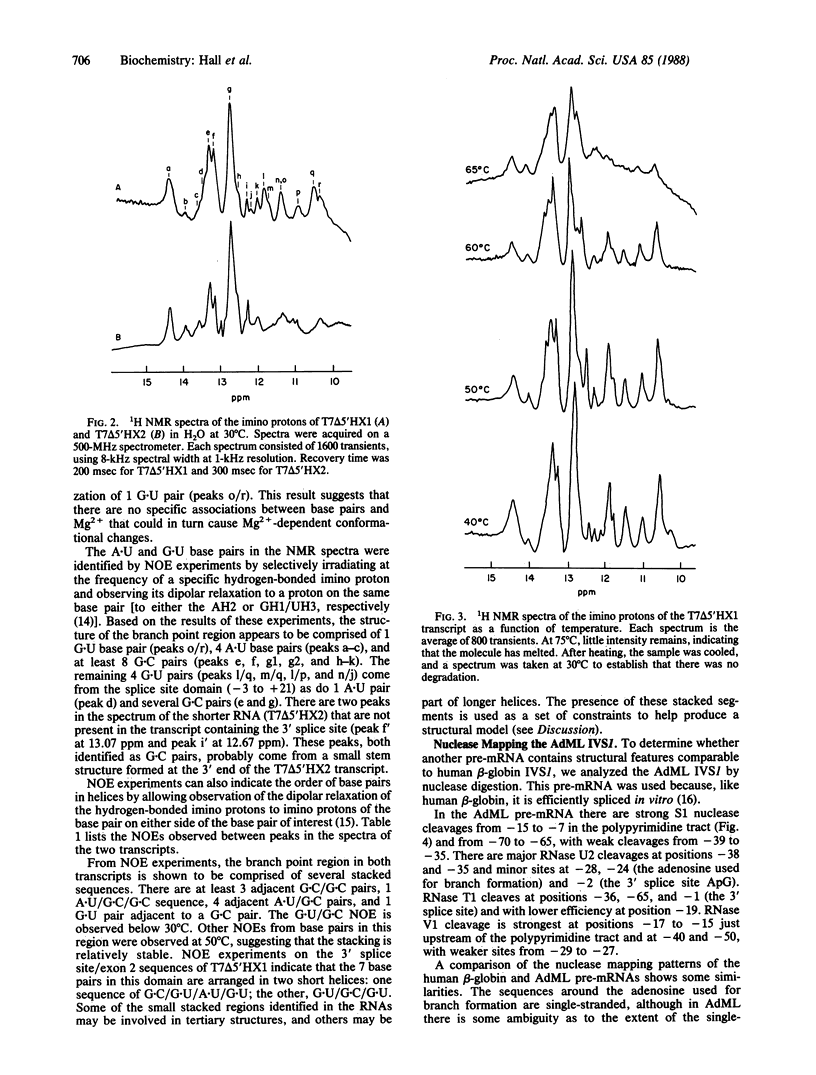
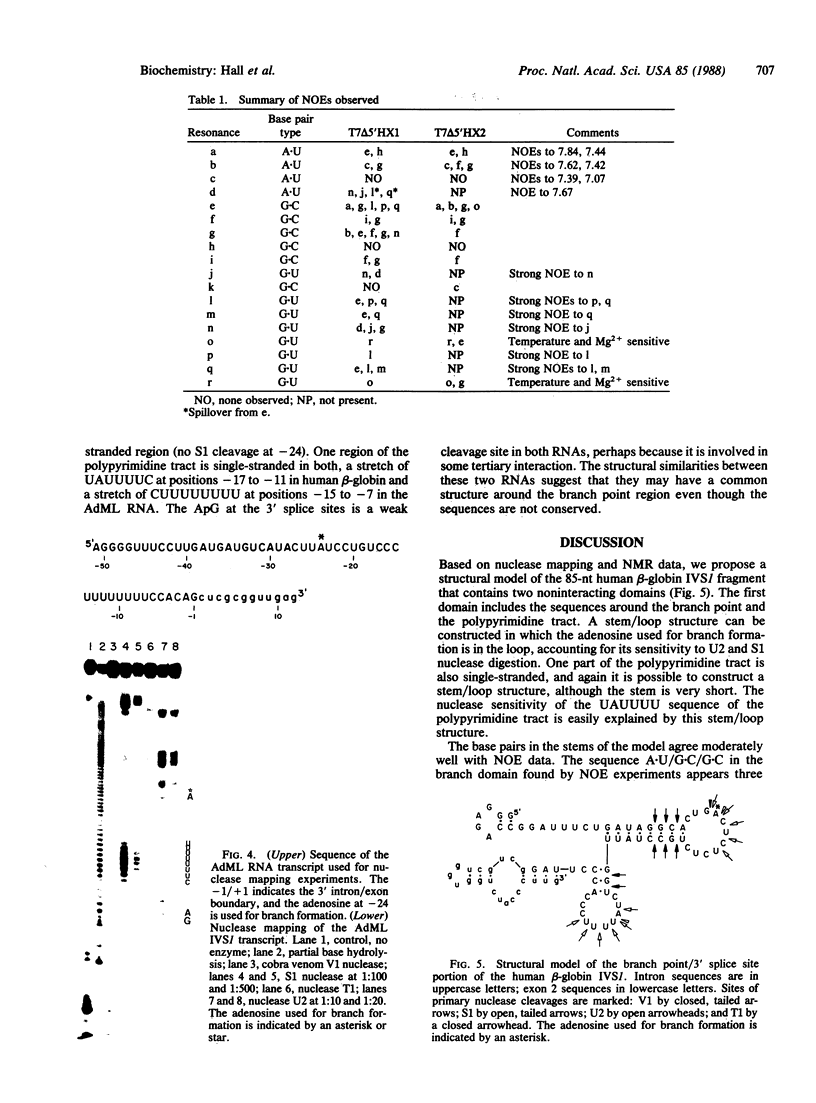

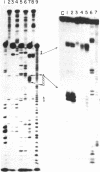
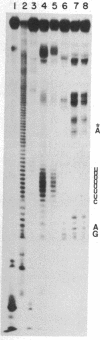
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cech T. R., Bass B. L. Biological catalysis by RNA. Annu Rev Biochem. 1986;55:599–629. doi: 10.1146/annurev.bi.55.070186.003123. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Tanner N. K., Tinoco I., Jr, Weir B. R., Zuker M., Perlman P. S. Secondary structure of the Tetrahymena ribosomal RNA intervening sequence: structural homology with fungal mitochondrial intervening sequences. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3903–3907. doi: 10.1073/pnas.80.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davanloo P., Rosenberg A. H., Dunn J. J., Studier F. W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke V., Steitz J. A. A protein associated with small nuclear ribonucleoprotein particles recognizes the 3' splice site of premessenger RNA. Cell. 1986 Dec 26;47(6):973–984. doi: 10.1016/0092-8674(86)90812-3. [DOI] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Hernandez N., Keller W. Splicing of in vitro synthesized messenger RNA precursors in HeLa cell extracts. Cell. 1983 Nov;35(1):89–99. doi: 10.1016/0092-8674(83)90211-8. [DOI] [PubMed] [Google Scholar]
- Johnston P. D., Redfield A. G. Pulsed FT-NMR double resonance studies of yeast tRNAPhe: specific nuclear Overhauser effects and reinterpretation of low temperature relaxation data. Nucleic Acids Res. 1978 Oct;5(10):3913–3927. doi: 10.1093/nar/5.10.3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühne T., Wieringa B., Reiser J., Weissmann C. Evidence against a scanning model of RNA splicing. EMBO J. 1983;2(5):727–733. doi: 10.1002/j.1460-2075.1983.tb01492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987 Feb 19;325(6106):673–678. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe S. H., Duthie R. S. Splice site consensus sequences are preferentially accessible to nucleases in isolated adenovirus RNA. Nucleic Acids Res. 1986 Nov 11;14(21):8447–8465. doi: 10.1093/nar/14.21.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Parker R., Siliciano P. G., Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987 Apr 24;49(2):229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Roy S., Redfield A. G. Nuclear Overhauser effect study and assignment of D stem and reverse-Hoogsteen base pair proton resonances in yeast tRNAAsp. Nucleic Acids Res. 1981 Dec 21;9(24):7073–7083. doi: 10.1093/nar/9.24.7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. Specific and stable intron-factor interactions are established early during in vitro pre-mRNA splicing. Cell. 1985 Nov;43(1):131–142. doi: 10.1016/0092-8674(85)90018-2. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Greene J. M., Green M. R. Cryptic branch point activation allows accurate in vitro splicing of human beta-globin intron mutants. Cell. 1985 Jul;41(3):833–844. doi: 10.1016/s0092-8674(85)80064-7. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. Splicing of messenger RNA precursors. Science. 1987 Feb 13;235(4790):766–771. doi: 10.1126/science.3544217. [DOI] [PubMed] [Google Scholar]
- Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985 Dec;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- Solnick D., Lee S. I. Amount of RNA secondary structure required to induce an alternative splice. Mol Cell Biol. 1987 Sep;7(9):3194–3198. doi: 10.1128/mcb.7.9.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi J., Alibert C., Temsamani J., Reveillaud I., Cathala G., Brunel C., Jeanteur P. A protein that specifically recognizes the 3' splice site of mammalian pre-mRNA introns is associated with a small nuclear ribonucleoprotein. Cell. 1986 Dec 5;47(5):755–766. doi: 10.1016/0092-8674(86)90518-0. [DOI] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]