Abstract
PURPOSE
Historical data have indicated the potential for the histologically-normal breast to harbor pre-malignant changes at the molecular level. We postulated that a histologically-normal tissue with “tumor-like” gene expression pattern might harbor substantial risk for future cancer development. Genes associated with these high-risk tissues were considered to be “malignancy-risk genes”.
EXPERIMENTAL DESIGN
From a total of 90 breast cancer patients, we collected a set of 143 histologically-normal breast tissues derived from patients harboring breast cancer who underwent curative mastectomy, as well as a set of 42 invasive ductal carcinomas (IDC) of various histologic grades. All samples were assessed for global gene expression differences using microarray analysis. For the purpose of this study we defined normal breast tissue to include histologically normal and benign lesions.
RESULTS
Here we report the discovery of a “malignancy-risk” gene signature that may portend risk of breast cancer development in benign, but molecularly-abnormal, breast tissue. Pathway analysis showed that the malignancy-risk signature had a dramatic enrichment for genes with proliferative function, but appears to be independent of ER, PR, and HER2 status. The signature was validated by RT-PCR, with a high correlation (Pearson correlation=0.95 with p<0.0001) with microarray data.
CONCLUSION
These results suggest a predictive role for the malignancy-risk signature in normal breast tissue. Proliferative biology dominates the earliest stages of tumor development.
While breast cancer therapy has seen substantial advances over the last few decades (1, 2), predicting breast cancer risk in the apparently normal breast is still problematic (3–9). Although a few pre-malignant histologic risk factors have been identified (atypical ductal hyperplasia (ADH), lobular carcinoma in situ, microcalcifications) (10, 11), few tools exist to distinguish the normal breast from the breast at risk for cancer (3–9). Furthermore, in patients who are treated for invasive breast cancer, the risk of local recurrence remains in spite of histologically negative margins. Wapnir et al (12) observed 10-year cumulative local recurrence rates ranging from 4.8% to 10.1% across five National Surgical Adjuvant Breast and Bowel Project (NSABP) trials involving 2,669 node-positive patients treated between 1984 and 1994, and 10-year local recurrence rates of 3.5% to 6.5% were observed in node-negative patients receiving systemic treatment in NSABP trials (13) during the same time period.
Recent developments of gene signatures for breast cancer have been reported to benefit breast cancer prognosis (14–24). Despite these efforts and those of mammographic screening, it is still difficult to detect risk for malignant conversion of normal breast tissue (25). Several lines of evidence suggest that histologically-normal breast tissue may, in fact, harbor pre-malignant molecular alterations in normal breast tissue adjacent to cancer at molecular level (3–7) (4, 7–9). In this study, we developed an innovative approach to identify histologically-normal, but molecularly-abnormal “IDC-like” tissue for malignant degeneration. We postulated that a histologically-normal tissue with “tumor-like” gene expression pattern might harbor substantial risk for future cancer development. Genes associated with these high-risk tissues were referred to as “malignancy-risk genes”.
The goal of our study was to establish a malignancy-risk gene expression signature in histologically-normal breast tissues obtained from patients with ipsilateral invasive breast cancer. We have developed a gene signature to assess cancer risk by first identifying a signature for invasive ductal carcinoma (IDC), and by then refining it using IDC-like normal tissues. A set of 143 histologically-normal breast tissues and 42 IDC tissues, derived from 90 patients who underwent mastectomy for ipsilateral breast carcinoma, were assessed for global gene expression differences using microarray analysis. A signature portending tissues at risk of future malignancy was developed from this analysis of histologically-normal breast tissues. Its clinical association with cancer risk was first confirmed with RTPCR and then evaluated using two independent external datasets.
Materials and Methods
Tissues and their associated clinicopathological data
Tissues were collected in accordance with the protocols approved by the Institutional Review Board of the University of South Florida, and stored in the tissue bank of Moffitt Cancer Center. The tissues were embedded in Tissue-Tek® O.C.T., 5-µm sections cut and mounted on Mercedes Platinum StarFrost™ Adhesive slides. The slides were stained using a standard H&E protocol, and tissue boundaries marked. Using the marked slide as a “map”, tissues were microdissected. Adipose tissues were trimmed away. Both histologically-normal breast tissues and IDCs were derived from 90 patients that underwent mastectomy for various stages of breast carcinoma and were collected and frozen in liquid nitrogen. Clinico-pathological data from the patients used in the study, including the tumor ER, PR and Her2/Neu status and tumor grade, are shown in Table 1. When possible, each mastectomy specimen was prosected to yield an IDC and up to five sequentially-derived, adjacent normal tissue samples in the ipsilateral breast or from the four quadrants of the contralateral breast. As a result, we collected 42 IDCs and 143 normal breast tissues from the 90 patients for microarray analysis. Due to RNA quality issue in some IDC and normal tissues, we did not have a complete set of IDC and normal tissues for some patients. There were 11 patients (a total of 34 tissues) with at least one normal and one IDC tissue, 19 patients (a total of 28 tissues) with IDC tissue only, and 60 patients (a total of 123 tissues) with normal tissue only. Supplementary Table 1 lists number of normal and IDC tissues and their geographical locations relative to the incident tumor.
Table 1.
Pathological data of the patients used in the study, including ER, PR, Her2, and grade.
| ER/PR/Her2 status | |||
|---|---|---|---|
| ER | R P | u Her2/ne | |
| Negative | 25 | 38 | 43 |
| Positive | 55 | 42 | 12 |
| other* | 10 | 10 | 35 |
| Total cases | 90 | 90 | 90 |
| Grade | frequency |
|---|---|
| Well differentiated | 6 |
| Moderately differentiated | 27 |
| Poorly differentiated | 30 |
| Undifferentiated/anaplastic | 10 |
| No grade | 17 |
| Total cases | 90 |
Results not available
Histology
Based on the histopathologic review by one breast pathologist (AN), all of the 143 histologically normal breast tissues were confirmed to be free of atypical ductal hyperplasia (ADH) and in-situ or invasive breast carcinoma. The 42 IDC tissues were also confirmed by the histopathologic review by the same pathologist, based on the modified Bloom and Richardson grading scheme (26).
RNA Extraction
Total RNA was extracted from the breast tissues using the Trizol method. Briefly, tissues were pulverized in liquid nitrogen, resuspended in 5 ml of lysis buffer, incubated for 3 min. at room temperature, and centrifuged at 11,500 g for 15 minutes at 4°. The aqueous phase was removed and put into another tube with 2.5 ml of isopropanol, mixed well and set at −20° C for 20 minutes. The amount of RNA was quantitated by measuring A260. Microarray analysis was performed using the Affymetrix U133Plus 2.0 GeneChips (54,675 probe sets). Expression values were calculated using the robust multi-array average (RMA) algorithm (27) (Data is in the GEO repository: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE10780).
RT-PCR Validation
Validation of 30 selected malignancy risk signature genes (of 117 available) (Supplementary Table 2) was done using the TaqMan Low Density Arrays (Applied Biosystems, Foster City, CA, USA). Due to limitation of sample availability, 5 “IDC-like” normal tissues, 8 IDCs, and 8 normal tissues were used for validation. Single stranded cDNA was synthesized from 1 ug of total RNA using random primers in a 20 uL reaction volume using Applied Biosystem’s High Capacity cDNA Reverse Transcription kit. The 20 uL reactions were incubated in a thermal cycler for 10 min at 25°C, 120 min at 37°C, 5 sec at 85°C and then held at 4°C. Real-time PCR was carried out using sequence specific primers/probes on the Applied Biosystems 7900 HT Real-Time PCR system. cDNA was diluted 2.5-fold; 5.0 uL of diluted cDNA was mixed with 45 uL of nuclease-free water and was added to 50 uL of TaqMan Universal PCR Master Mix (Applied Biosystems). The 100 uL total reaction mixture was loaded in the corresponding ports of a TaqMan Low Density Array (TLDA) card. Each TLDA card consisted of 3 replicates (4 samples per card). Expression value (ΔCt) was calculated by first averaging replicates for each gene and then normalized (subtraction) by an endogenous control gene (18S). Since a lower value of ΔCt indicates a higher expression, a -ΔCt was used to correlate with microarray gene expression.
Signature Generation/Statistical Methods
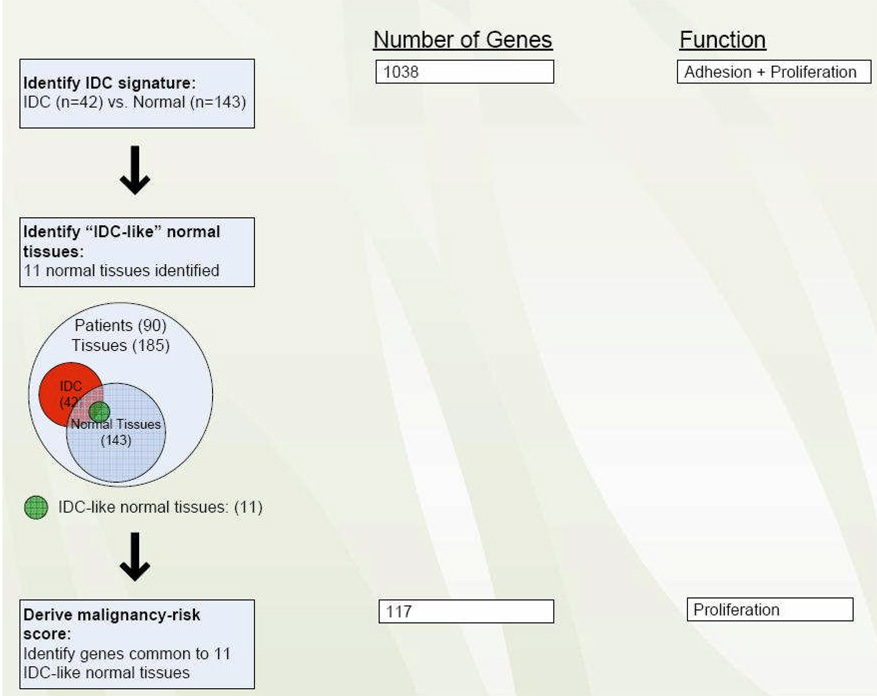
Statistical analysis included a series of steps to develop and validate the malignancy-risk gene signature (Figure 1):
Figure 1.
Flow chart to developing the malignancy-risk gene signature.
1. Identification of IDC gene signature
In this first step, a set of 1038 genes (1,554 probe sets) was identified that distinguished the IDCs (n = 42) from the histologically-normal tissues (n = 143). The IDC gene set was identified by treating IDC and normal tissues as two independent groups (although some were derived from the same patients) and using Statistical Analysis of Microarray (28) at 1% false discovery rate (FDR) with a fold change >2 (Figure 1). The study aimed to collect multiple normal and IDC tissues from the same subjects, but due the heterogeneous nature of the sample set, some patients had only normal tissues sampled while others samples were limited to IDC tissues only. This nature of unbalanced data made it difficult to adjust for subject variation. Instead, we aggregated data into normal and IDC two groups for comparison. To ensure homogeneity for data aggregation, we checked whether overall gene expression from the normal tissues in patients with normal tissues available only was similar to the normal tissues in patients with both normal and IDC tissues available. We used Kmeans approach to classify all the normal tissues into two groups based on gene expression data. Fisher exact test did not show the two types of normal tissues were statistically different (p=0.53). We found similar results for the IDC tissues (p=0.99). These results suggested homogeneity for the two types of normal tissues (also for the IDC tissues).
2. Identification of “IDC-like” normal tissues
In this step, we used the IDC gene signature to identify 11 histologically normal breast tissues that had acquired the molecular fingerprint of IDC. The method first ranked all the normal tissues for each IDC tumor gene. (e.g., A normal tissue A is ranked as the top 1% (percentile rank=100%) for tumor gene X1, top 10% (percentile rank=90%) for tumor gene X2, top 20% for tumor gene X3, and so on). As a result, for the up-regulated IDC tumor genes (e.g., k1 genes), we will have a set (k1) of the tissue percentile ranks for each tissue. If a normal tissue displayed at least half (>k1/2) of the percentile ranks over 80% (i.e., the median percentile rank>0.8), we considered it as “IDC-like” normal tissue. Similarly, a normal tissue was also considered as an IDC-like tissue if a normal tissue had the median of the percentile ranks below 20% for down-regulated IDC tumor genes. A graphical presentation of the method is included in the Supplementary Figure 1. A simulation was conducted and showed its effectiveness to identify IDC-like tissues (Supplementary Figure 2). We also compared to other approaches and results did not show these alternative approaches were as effective as our rank approach (Supplementary Figure 3).
3. Derive malignancy-risk gene score
Once the IDC-like normal tissues were identified, we then formed a common set of genes, “malignancy-risk signature genes”, whose expression percentile rank was greater than 80% (or less than 20%) in most IDC-like normal tissues. Using the principal components analysis (PCA) method, we derived a “risk score” (malignancy-risk score) to represent an overall gene expression level for the malignancy-risk gene signature. First, we performed principal components analysis to reduce data dimension into a small set of uncorrelated principal components. This set of principal components was generated based on its ability to account for variation. We used the first principal component (1st PCA), as it accounts for the largest variability in the data, as a malignancy risk score to represent the overall expression level for the signature. That is, malignancy risk score=∑wixi, a weighted average expression among the malignancy-risk genes, where xi represents gene i expression level, wi is the corresponding weight with , and the wi values maximize the variance of ∑wixi . While other PCAs (e.g., the second and third principle components) may also associate with cancer risk, our experiences indicates that the 1st PCA often corresponds most effectively to cancer risk-related information for this study (see RT-PCR and DCIS validation in the Results section).
4. Cross-validation
Leave-one-out cross validation (LOOCV) was performed to evaluate robustness of the IDC and malignancy-risk gene signatures. This was done by excluding one sample at a time and repeating steps 1–3 to see how many were correctly identified (IDC genes, IDC-like normal tissues, and malignancy-risk genes).
5. Pathway analysis
Pathway analysis was done using MetaCore ™ by GeneGo for steps 1 and 3 to identify biological functions associated with IDC genes and the malignancy-risk genes. We compared pathways of the two gene sets to reveal difference of biological processes between the IDC genes and the malignancy-risk genes.
6. RT-PCR validation
Pearson correlation was used to evaluate association of the malignancy risk score between microarray and RT-PCR platforms. The malignancy-risk score was calculated using the 30 selected malignancy-risk signature genes (see Statistical Methods) for microarray and RT-PCR, respectively. Correlation analysis was also performed for each individual malignancy-risk gene. Analysis of variance was used to test the differences among the three groups (normal, IDC-like normal, and IDC) with the Tukey method (29) to adjust for p value for pair-wise comparison. We also used support vector machine (SVM) to build a classifier from the microarray platform to evaluate the prediction performance on the RT-PCR platform.
7. Evaluation of cancer risk and cancer progression
We assessed the cancer risk potential and the cancer progression of the malignancy-risk score on two independent data sets. Because each data set had a different set of available genes, we used whatever genes were in common with the malignancy risk gene signature to evaluate each data set (essentially a subset of the original malignancy-risk gene signature). The SVM was used to evaluate prediction performance. In addition, for ordinal clinical variable (e.g., from normal, ductal carcinoma in situ (DCIS), to IDC), the malignancy-risk score was used to correlate with cancer severity using Pearson correlation to evaluate the trend of the malignancy-risk gene signature with cancer progression.
Results
IDC gene signature
An IDC gene signature (1,554 probe sets: 1038 unique genes) was developed from a set of 42 IDC and 143 normal breast tissues using Statistical Analysis of Microarray (28). We found the IDC gene set remained robust in the leave-one-out cross-validation. Pathway analysis revealed two predominant cellular processes: cell adhesion and cell cycle/proliferation (Supplementary Table 3).
IDC-like normal breast tissues
We ranked the 143 normal breast tissues based on the IDC gene signature and identified 11 “IDC-like” normal breast tissues, whose gene expression profiles more closely approximated that of the IDC samples rather than the rest of the 132 normal breast tissues (i.e., these 11 IDC-like normal tissues were molecularly- abnormal, but histologically-normal) (Supplementary Figure 4).
Histology of IDC-like normal tissues
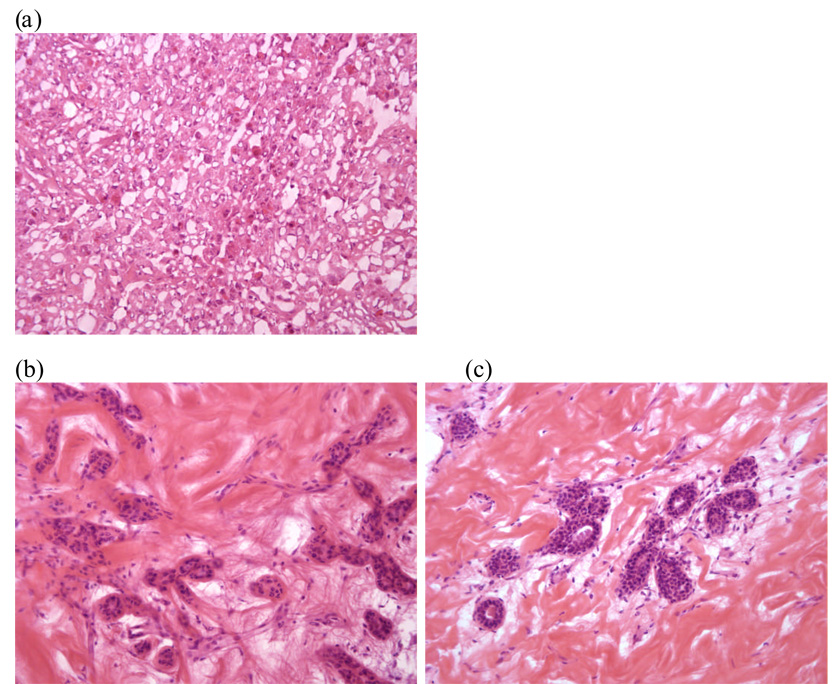
Supplementary Table 4 summarizes the normal histological findings of the 11 IDC-like normal breast tissues used in this study. All of these specimens consisted of completely unremarkable, benign breast tissues and were free from in-situ or invasive carcinoma as well as atypical ductal hyperplasia (Figure 2). Fisher exact test showed no significant association of patients harboring IDC-like normal tissues with ER/PR/Her2/grade (Supplementary Table 5).
Figure 2. Histologic images of representative frozen breast tissues (original magnification X 200).
(a) Invasive ductal carcinoma (IDC) showing sheets of tumor cells and stromal strands, (b) Normal breast lobule in a frozen breast tissue specimen that was collected at 1 cm from the tumor (IDC) shown in Figure a. This specimen was designated as ‘IDC-like normal’ based on its molecular profile, (c) Normal breast lobule in a frozen breast tissue specimen that was collected at 2 cm from the tumor (IDC) shown in Figure a.
Malignancy-risk gene signature and risk score
A malignancy-risk gene signature was developed by forming a “common set” of genes whose expression varied (up or down) at high levels in the 11 IDC-like normal tissues (see Statistical Methods). The malignancy-risk genes consisted of 109 up-regulated probe sets (96 unique genes) and 31 down-regulated probe sets (21 unique genes). Table 2 provides a selected subset of malignancy-risk genes; the entire list is presented in Supplementary Table 6. Moreover, by utilizing principal component analysis, a malignancy-risk score was derived to represent an overall gene expression level for the malignancy-risk signature (see Statistical Methods).
Table 2.
A subset of malignancy-risk genes associated with DNA replication, mitosis, and cancer risk *.
| Affy probe set id | Gene Symbol | Fold change | FDR | Regulation | DNA replication | Mitosis | Poola et al | Gene Title |
|---|---|---|---|---|---|---|---|---|
| 222608_s_at | ANLN | 4.01 | <0.01 | Up-Regulated | Y | anillin, actin binding protein (scraps homolog, Drosophila) | ||
| 202095_s_at | BIRC5 | 2.95 | <0.01 | Up-Regulated | Y | baculoviral IAP repeat- containing 5 (survivin) | ||
| 209642_at | BUB1 | 2.71 | <0.01 | Up-Regulated | Y | BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) | ||
| 203755_at | BUB1B | 3.05 | <0.01 | Up-Regulated | Y | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) | ||
| 214710_s_at | CCNB1 | 4.03 | <0.01 | Up-Regulated | Y | cyclin B1 | ||
| 202705_at | CCNB2 | 2.35 | <0.01 | Up-Regulated | Y | cyclin B2 | ||
| 205034_at | CCNE2 | 3.99 | <0.01 | Up-Regulated | Y | cyclin E2 | ||
| 203213_at | CDC2 | 5.5 | <0.01 | Up-Regulated | Y | Cell division cycle 2, G1 to S and G2 to M | ||
| 203214_x_at | CDC2 | 2.89 | <0.01 | Up-Regulated | Y | cell division cycle 2, G1 to S and G2 to M | ||
| 210559_s_at | CDC2 | 4.14 | <0.01 | Up-Regulated | Y | Y | cell division cycle 2, G1 to S and G2 to M | |
| 1555758_a_at | CDKN3 | 2.85 | <0.01 | Up-Regulated | cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) | |||
| 209714_s_at | CDKN3 | 2.97 | <0.01 | Up-Regulated | Y | cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) | ||
| 204962_s_at | CENPA | 2.71 | <0.01 | Up-Regulated | centromere protein A, 17kDa | |||
| 207828_s_at | CENPF | 2.6 | <0.01 | Up-Regulated | centromere protein F, 350/400ka (mitosin) | |||
| 218542_at | CEP55 | 3.46 | <0.01 | Up-Regulated | Y | chromosome 10 open reading frame 3 | ||
| 218252_at | CKAP2 | 2.72 | <0.01 | Up-Regulated | Y | Y | cytoskeleton associated protein 2 | |
| 203764_at | DLG7 | 2.84 | <0.01 | Up-Regulated | Y | Y | discs, large homolog 7 (Drosophila) | |
| 203358_s_at | EZH2 | 2.69 | <0.01 | Up-Regulated | Y | enhancer of zeste homolog 2 (Drosophila) | ||
| 213911_s_at | H2AFZ | 2.21 | <0.01 | Up-Regulated | Y | H2A histone family, member Z | ||
| 202503_s_at | KIAA0101 | 5.89 | <0.01 | Up-Regulated | KIAA0101 | |||
| 204709_s_at | KIF23 | 2.14 | <0.01 | Up-Regulated | Y | kinesin family member 23 | ||
| 202107_s_at | MCM2 | 2.08 | <0.01 | Up-Regulated | Y | MCM2 minichromosome maintenance deficient 2, mitotin (S. cerevisiae) | ||
| 204825_at | MELK | 3.76 | <0.01 | Up-Regulated | Y | Y | maternal embryonic leucine zipper kinase | |
| 204641_at | NEK2 | 5.55 | <0.01 | Up-Regulated | NIMA (never in mitosis gene a)-related kinase 2 | |||
| 201577_at | NME1 | 2.15 | <0.01 | Up-Regulated | non-metastatic cells 1, protein (NM23A) expressed in | |||
| 218039_at | NUSAP1 | 6.41 | <0.01 | Up-Regulated | Y | nucleolar and spindle associated protein 1 | ||
| 219978_s_at | NUSAP1 | 5 | <0.01 | Up-Regulated | Y | Y | nucleolar and spindle associated protein 1 | |
| 222077_s_at | RACGAP1 | 3.36 | <0.01 | Up-Regulated | Rac GTPase activating protein 1 | |||
| 201890_at | RRM2 | 8.07 | <0.01 | Up-Regulated | Y | ribonucleotide reductase M2 polypeptide | ||
| 209773_s_at | RRM2 | 6.73 | <0.01 | Up-Regulated | Y | Y | ribonucleotide reductase M2 polypeptide | |
| 209218_at | SQLE | 3.25 | <0.01 | Up-Regulated | squalene epoxidase | |||
| 1554408_a_at | TK1 | 2.72 | <0.01 | Up-Regulated | Y | thymidine kinase 1, soluble | ||
| 202338_at | TK1 | 2.86 | <0.01 | Up-Regulated | Y | thymidine kinase 1, soluble | ||
| 201291_s_at | TOP2A | 7.56 | <0.01 | Up-Regulated | Y | topoisomerase (DNA) II alpha 170kDa | ||
| 201292_at | TOP2A | 6.03 | <0.01 | Up-Regulated | Y | topoisomerase (DNA) II alpha 170kDa | ||
| 204822_at | TTK | 3.27 | <0.01 | Up-Regulated | Y | TTK protein kinase | ||
| 204026_s_at | ZWINT | 4.46 | <0.01 | Up-Regulated | ZW10 interactor |
“Y” symbol was used to indicate the association of each malignancy-risk gene with DNA replication, mitosis, and cancer risk.
Cross-validation
Analysis of the malignancy risk score by LOOCV yielded a high degree of consistency; most IDC genes (>98%), IDC-like normal tissues (>90%), and malignancy-risk genes (>90%) were identified correctly at each leave-one-out iteration (Supplementary Figure 5). Moreover, at each iteration, we calculated a predicted malignancy risk score for the sample being excluded. Correlation analysis showed a strong relationship between the predicted risk score and the disease status (i.e., by ranking normal, IDC-like normal, and IDC from 0 to 2; Pearson correlation=0.89 and Spearman correlation=0.74 with p <0.0001).
Pathway analysis of malignancy-risk genes
In contrast to the IDC gene signature, pathway analysis of the malignancy-risk gene set showed a remarkable over-expression of proliferative function genes, instead of a mixture of proliferation and adhesion genes seen with IDC. There were 11 cell cycle related pathways represented in the malignancy-risk signature (p value <0.01, Supplementary Table 7). Since the malignancy-risk gene signature was derived from the IDC gene signature, the difference in functional classes of genes would not have been expected in the absence of a selection bias. The majority of the malignancy-risk genes were classified to be primarily associated with DNA replication and mitosis, two hallmark events associated with proliferation (Supplementary Table 8). This observation may indicate the importance of these features in early stages of tumorigenesis (30).
Weak correlation of malignancy risk score with ER, PR, and Her2
Since ER, PR, and Her2 are key markers in cancer development, we examined their correlation with the malignancy risk score. Results showed only a weak correlation for ER and PR (r=−0.2~0.3) and a moderate correlation with Her2 (r=0.37~0.47 by spearman correlation and r=0.43~0.63 by Pearson correlation), suggesting relative independence of the risk score from these biomarkers (Supplementary Figure 6).
Higher malignancy risk score of IDC-like normal tissues
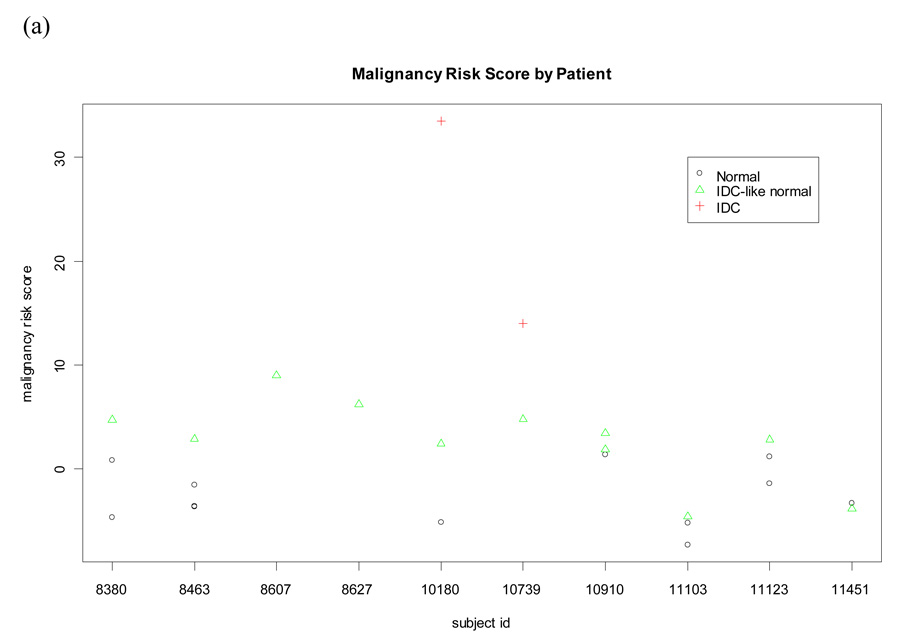
We identified 11 IDC-like normal tissues from 10 patients. There were another 12 normal tissues collected from the same 10 patients. These 12 normal tissues were molecularly and histologically normal and labeled as matched normal tissues to reflect they were derived from the same subject. The other normal tissues (n=120) from subjects without IDC-like normal tissues (i.e., not from the 10 subjects) were also molecularly and histologically normal and labeled as unmatched normal tissues for distinction. Interestingly, we found the malignancy risk score was higher in the IDC-like normal tissues and the matched normal tissues than in the unmatched normal tissues. Difference of the risk score was statistically significant for (a) IDC-like normal tissues versus the matched normal tissues (adjusted p value<0.0001 using the Tukey method) and (b) matched versus unmatched normal tissues (adjusted p value=0.0054). An increasing trend of the malignancy risk score was also seen from the unmatched normal tissues, the matched normal tissue, to the IDC-like normal tissues at the pooled data level (Pearson correlation=0.63 with p<0.0001; Figure 3). Moreover, among the 10 patients with IDC-like normal tissues, analysis results showed a higher malignancy risk score in the IDC-like normal tissues than in the matched normal tissues at subject level (p=0.01 using the random effect model; Figure 3). Since the malignancy risk score was derived without knowing subject information, a trend of the risk score decreasing from the IDC-like normal tissues, to the matched normal tissue, to the unmatched normal tissues would not be expected.
Figure 3. Comparison of malignancy-risk score between IDC-like normal tissues, their matched normal tissues, and unmatched normal tissues.
Malignancy risk score was compared among the three groups in both subject level and pooled data level (ignore subject information). (a) Subject level: A higher malignancy risk score was seen in the IDC-like normal tissues than in the matched normal tissues for most subjects (p=0.01 using the random effect model); (b) Pooled data level: An increasing trend of the malignancy risk score was seen from the unmatched normal tissues, the matched normal tissue, to the IDC-like normal tissues (Pearson correlation=0.63 with p<0.0001); (c) Pair-wise comparison of the malignancy risk score among the unmatched normal tissues, the matched normal tissue, and the IDC-like normal tissues using the Tukey method to control for the type I error.
RT-PCR validation of malignancy-risk genes
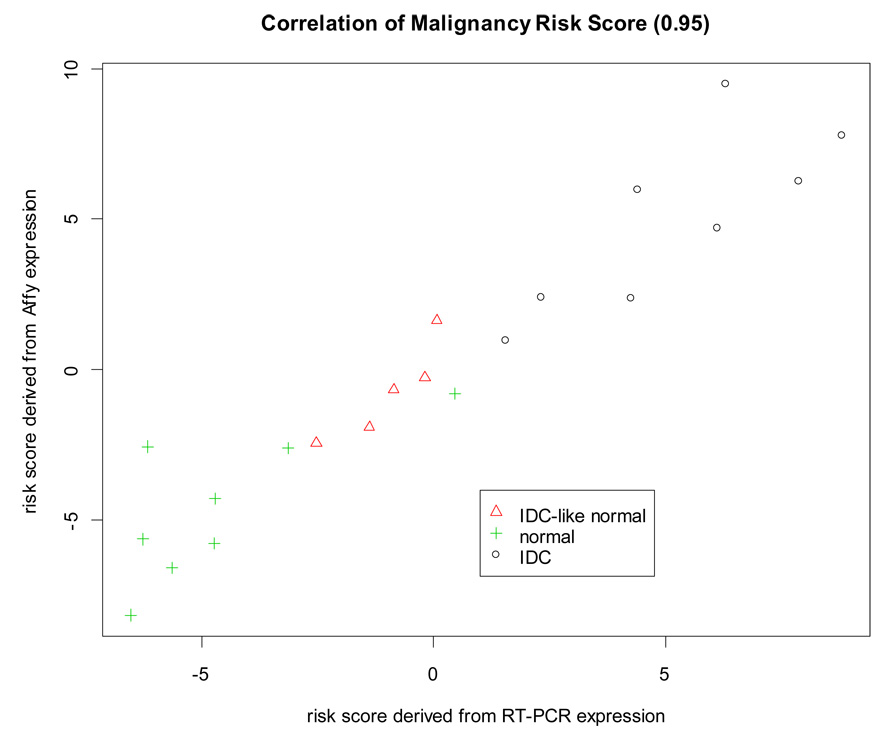
Expression of the 30 selected malignancy risk signature genes identified by microarray profiling was successfully validated by RT-PCR. There were 27 genes showing a strong Pearson correlation >0.7 (correlation>0.9: 12 genes, 0.8–0.9: 13, and 0.7–0.8: 2; the p values were <0.0001) (Supplementary Figure 7). The composite malignancy risk score (based on microarray data from 30 genes) also demonstrated a very high correlation (0.95) with RT-PCR results. The risk score for the IDC-like normal tissues fell in the middle between the IDC and normal samples (Figure 4). In addition, we used support vector machine (SVM) to build a classifier for the 30 genes from microarray (accuracy rate=86% using LOOCV) and predicted on the RT-PCR expression with 90% accuracy (Supplementary Figure 7). In comparing the malignancy risk score generated by various PCAs, the use of the 1st PCA as the risk score showed a very high correlation (r=0.95) between microarray and RT-PCR and an increasing trend of the risk score from normal to IDC samples. The other PCAs had a weak correlation (r<0.5) and did not yield their association with cancer progression.
Figure 4. RT-PCR validation.
Pearson correlation was used to evaluate association of the malignancy risk score between microarray and RT-PCR. The malignancy-risk score was calculated using the 30 selected malignancy risk genes (Supplementary table 2) for microarray and RT-PCR, respectively.
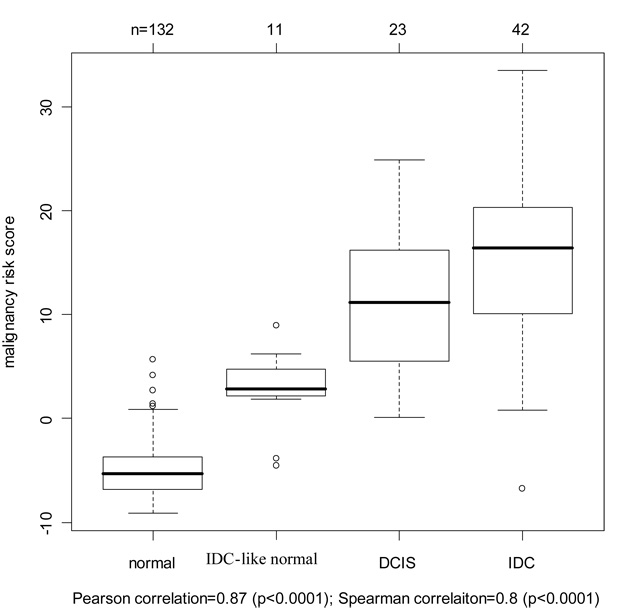
Validation of Moffitt ductal carcinoma in situ (DCIS) samples for cancer progression
A set of 23 DCIS samples (from 11 patients: 8 from the 90 patients and 3 new patients) were collected from the Moffitt Cancer Center to evaluate the cancer progression feature of the malignancy-risk gene signature. We compared the malignancy risk score among the four groups: normal breast, IDC-like normal, DCIS, and IDC. Data showed an increasing risk score pattern with progression from normal, to IDC-like normal, to DCIS, and to IDC. Pearson and Spearman correlation was 0.87 and 0.8, respectively, with a significant p value<0.0001 (by ranking the disease status from 0 to 3 for normal breast to IDC). Moreover, the malignancy-risk score of DCIS was lower than IDC, but higher than normal tissue (p =0.0005) within each patient (Figure 5). In addition, we evaluated the prediction performance on the DCIS samples. A SVM classifier was built with an accuracy rate of 92% by 10-fold cross validation. The classifier predicted most of the 23 DCIS samples into the IDC category (18/23) and two samples favoring to the “IDC-like normal” group (Supplementary Figure 8). We also compared the malignancy risk score generated by PCA1 (1st PCA) versus other PCAs (PCA2 and PCA3). In contrast to PCA1 showing a cancer progression pattern, PCA 2 and PCA3 did not demonstrate a cancer progression from non-IDC like normal to IDC (Supplementary Figure 8).
Figure 5. Validation of Moffitt ductal carcinoma in situ (DCIS) samples for cancer progression.
The malignancy-risk score was generated by the 1st PCA and showed an increasing trend from non-IDC-like normal, IDC-like normal, to IDC. Additional analysis results were in Supplementary Figure 7.
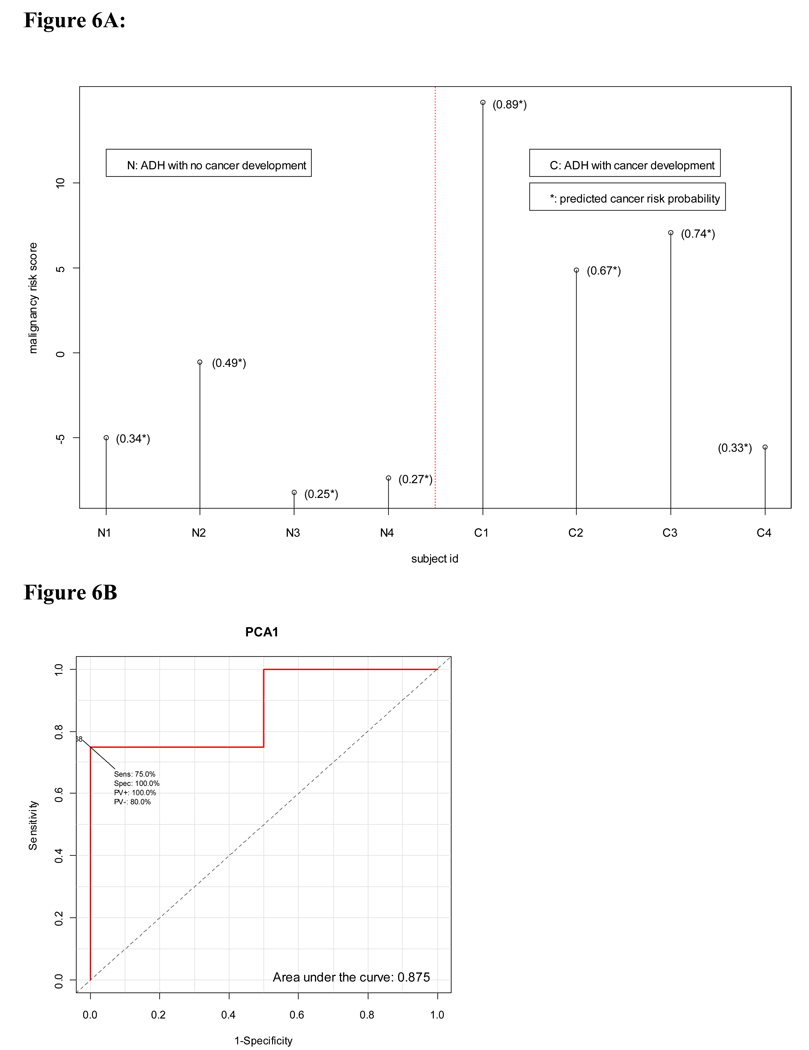
Evaluation of cancer risk in Poola et al’s ADH study (31)
This study was selected in order to assess the potential of the malignancy-risk score to predict the risk of future cancer development in the breast associated with ADH. The study collected 4 ADH tissues from patients without breast cancer development (labeled as ADH-N) and 4 ADH samples with cancer developed (labeled as ADH-C). We used 102 probe sets from their platform (in common with the malignancy-risk gene signature) to calculate the malignancy risk score by the 1st PCA and the 2nd PCA, respectively. SVM was then used to classify the 8 patients based on the malignancy-risk score. Data analysis showed the use of the 1st PCA as the malignancy risk score yielding a higher risk score in the ADH-C group than in the ADH-N group (Figure 6). The SVM correctly classified 7 out 8 patients (87.5%) although it was not statistically significant (p= 0.14 based on the fisher exact test) due to a very limited sample size (n=4 per group). Notably, three out the four ADH-C patients had a risk score above 5 with a higher cancer-risk probability, in contrast to most ADH-N patients with negative scores and a lower cancer-risk probability. As the 2nd PCA was incorporated with the 1st PCA in the model, SVM correctly classified all the 8 patients (Supplementary Figure 9), suggesting the malignancy-risk gene signature was able to differentiate ADH patients between with and without cancer development, and indicating its ability to assessing cancer risk. We also calculated area under curve (AUC) for response operating characteristic curve. The results were similar to the ones by SVM: AUC was higher by the use of the first PCAs (AUC=1) than by the 1st PCA (AUC=0.875) (Supplementary Figure 9).
Figure 6. Evaluation for cancer risk in Poola et al’s ADH study.
The use of the 1st PCA as the malignancy risk score showed that the ADH-C group had a higher risk score than the ADH-N group (Figure 6A) with AUC=0.875 (Figure 6B) . Additional analysis results were in Supplementary Figure 8.
Discussion
Identification of normal tissue at risk for malignant conversion has great potential application in clinical practice, in both evaluating the malignancy risk measured by routine breast biopsies as well as the risk of local recurrence following lumpectomy. Detecting these high-risk normal, appearing tissues, however, remains a challenging task. In this study, we utilized the IDC data information to develop a new concept of "IDC-like normal tissue". The "IDC-like normal tissue" could be histologically normal tissue with a molecular proclivity towards IDC. Based on this hypothesis, we identified 11 “IDC-like” normal tissues (out of 143 normal breast tissues) and developed the malignancy-risk gene signature and risk score.
A careful re-examination of all the IDC-like normal tissues showed that they were histologically-normal, with no evidence of in-situ or invasive carcinoma of the breast, and no atypia (Figure 2 and Supplementary Table 4). However, these IDC-like normal tissues showed gene expression profiles resembling invasive carcinomas, indicating that these tissues had already acquired the molecular fingerprint of cancer and, therefore, may be at increased risk for subsequent cancer development. Moreover, from these IDC-like normal tissues, we developed a “malignancy-risk” gene signature that may serve as a marker of subsequent risk of breast cancer development. The malignancy-risk gene signature was internally validated by RT-PCR and leave-one-out cross validation as well as by two additional datasets. Further analysis of external datasets also demonstrated its clinical relevance to cancer-risk and cancer progression. While this gene signature requires further clinical validation, this is an intriguing finding with substantive clinical implications. Several studies have suggested that cell cycle/proliferation are key hallmarks of existing cancer (22, 34–36). This is the first study, however, to suggest the proliferative program of gene expression may be the earliest detectable event in normal breast tissues at risk for developing breast cancer. Moreover, this is the largest molecular analysis of histologically benign breast tissues. A recently reported study of 14 normal breast tissues from breast cancer cases identified genes differentially expressed in these tissues versus normal breast reduction mammoplasties, but did not decipher a predominantly proliferative gene function (37).
The large preponderance of proliferative genes in the malignancy-risk gene set was not expected. By comparison, IDC associated genes were biased towards both proliferative and adhesive gene sets. These findings suggest a temporal relationship between proliferative and adhesive gene expression programs, with the former being precursors to histological alterations and responsible for malignancy risk. Interestingly, there was also no statistical association of the IDC-like normal tissues with ER/PR, Her2/neu, and grade suggesting the malignancy risk signature may be not be dependent on these factors. The lack of association of the IDC-like normal tissues with the triple negative (ER/PR/Her2Neu) phenotype also suggests no link to BRCA1 and BRCA2.
Evaluation on two external independent datasets demonstrated the clinical relevance of the malignancy-risk gene signature to cancer risk. While further validation of the malignancy-risk signature is warranted, the signature has promise for impacting clinical decisions. These include altering strategies for follow-up of histologically-normal, but molecularly abnormal breast biopsies, determining which patients might benefit from radiotherapy following lumpectomy, or determining which patients might benefit from mastectomy due to multifocal disease risk.
Supplementary Material
Acknowledgments
This Research is funded by National Cancer Institute grant R01CA112215 (PI: TJY) and National Cancer Institute Grant R01 CA 098522(PI: TJY)
References
- 1.Berry DA, Cronin KA, Plevritis SK, Fryback DG, Clarke L, Zelen M, et al. Effect of screening and adjuvant therapy on mortality from breast cancer. N Engl J Med. 2005;353(17):1784–1792. doi: 10.1056/NEJMoa050518. [DOI] [PubMed] [Google Scholar]
- 2.Giordano SH, Buzdar AU, Smith TL, Kau SW, Yang Y, Hortobagyi GN. Is breast cancer survival improving? Cancer. 2004;100(1):44–52. doi: 10.1002/cncr.11859. [DOI] [PubMed] [Google Scholar]
- 3.Lewis CM, Cler LR, Bu DW, Zochbauer-Muller S, Milchgrub S, Naftalis EZ, et al. Promoter hypermethylation in benign breast epithelium in relation to predicted breast cancer risk. Clinical Cancer Research. 2005;11(1):166–172. [PubMed] [Google Scholar]
- 4.Deng GR, Lu Y, Zlotnikov G, Thor AD, Smith HS. Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science. 1996;274(5295):2057–2059. doi: 10.1126/science.274.5295.2057. [DOI] [PubMed] [Google Scholar]
- 5.Fredriksson I, Liljegren G, Palm-Sjovall M, Arnesson LG, Emdin SO, Fornander T, et al. Risk factors for local recurrence after breast-conserving surgery. British Journal of Surgery. 2003;90(9):1093–1102. doi: 10.1002/bjs.4206. [DOI] [PubMed] [Google Scholar]
- 6.Ellsworth DL, Ellsworth RE, Love B, Deyarmin B, Lubert SM, Mittal V, et al. Outer breast quadrants demonstrate increased levels of genomic instability. Annals of Surgical Oncology. 2004;11(9):861–868. doi: 10.1245/ASO.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 7.Botti C, Pescatore B, Mottolese M, Sciarretta F, Greco C, Di Filippo F, et al. Incidence of chromosomes 1 and 17 aneusomy in breast cancer and adjacent tissue: An interphase cytogenetic study. Journal of the American College of Surgeons. 2000;190(5):530–539. doi: 10.1016/s1072-7515(00)00252-0. [DOI] [PubMed] [Google Scholar]
- 8.Larson PS, de las Morenas A, Bennett SR, Cupples LA, Rosenberg CL. Loss of heterozygosity or allele imbalance in histologically normal breast epithelium is distinct from loss of heterozygosity or allele imbalance in co-existing carcinomas. American Journal of Pathology. 2002;161(1):283–290. doi: 10.1016/S0002-9440(10)64180-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Moore DH, Meng ZH, Ljung BM, Gray JW, Dairkee SH. Increased risk of local recurrence is associated with allelic loss in normal lobules of breast cancer patients. Cancer Research. 2002;62(4):1000–1003. [PubMed] [Google Scholar]
- 10.Schnitt SJ, Morrow M. Lobular carcinoma in situ: current concepts and controversies. Semin Diagn Pathol. 1999;16(3):209–223. [PubMed] [Google Scholar]
- 11.Fitzgibbons PL, Henson DE, Hutter RV. Benign breast changes and the risk for subsequent breast cancer: an update of the 1985 consensus statement. Cancer Committee of the College of American Pathologists. Arch Pathol Lab Med. 1998;122(12):1053–1055. [PubMed] [Google Scholar]
- 12.Wapnir IL, Anderson SJ, Mamounas EP, Geyer CE, Jr, Jeong JH, Tan-Chiu E, et al. Prognosis after ipsilateral breast tumor recurrence and locoregional recurrences in five National Surgical Adjuvant Breast and Bowel Project node-positive adjuvant breast cancer trials. J Clin Oncol. 2006;24(13):2028–2037. doi: 10.1200/JCO.2005.04.3273. [DOI] [PubMed] [Google Scholar]
- 13.Wapnir SA, Mamounas E, Geyer C, Hyeon-Jeong J, Costantino J, Fisher B, Wolmark N. Survival after IBTR in NSABP Node Negative Protocols B-13, B-14, B-19, B-20 and B-23. Journal of Clinical Oncology. 2005;23(No 16S) June 1 Supplement:517. [Google Scholar]
- 14.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nature Genetics. 2006;38(9):1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 15.Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sorlie T, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005;102(10):3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14(5):518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 17.Glas AM, Floore A, Delahaye LJ, Witteveen AT, Pover RC, Bakx N, et al. Converting a breast cancer microarray signature into a high-throughput diagnostic test. BMC Genomics. 2006;7:278. doi: 10.1186/1471-2164-7-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang E, Cheng SH, Dressman H, Pittman J, Tsou MH, Horng CF, et al. Gene expression predictors of breast cancer outcomes. Lancet. 2003;361(9369):1590–1596. doi: 10.1016/S0140-6736(03)13308-9. [DOI] [PubMed] [Google Scholar]
- 19.Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, et al. Gene expression profiles of human breast cancer progression. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(10):5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 21.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 22.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van de Vijver MJ, He YD, van 't Veer LJ, Dai H, Hart AAM, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. New England Journal of Medicine. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 25.Shah VI, Raju U, Chitale D, Deshpande V, Gregory N, Strand V. False-negative core needle biopsies of the breast - An analysis of clinical, radiologic, and pathologic findings in 27 consecutive cases of missed breast cancer. Cancer. 2003;97(8):1824–1831. doi: 10.1002/cncr.11278. [DOI] [PubMed] [Google Scholar]
- 26.Robbins P, Pinder S, Deklerk N, Dawkins H, Harvey J, Sterrett G, et al. Histological grading of breast carcinomas - a study of interobserver agreement. Human Pathology. 1995;26(8):873–879. doi: 10.1016/0046-8177(95)90010-1. [DOI] [PubMed] [Google Scholar]
- 27.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller RG. Simultaneous Statistical Inference. Springer; 1981. [Google Scholar]
- 30.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Molecular Biology of the Cell. 2002;13(6):1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poola I, DeWitty RL, Marshalleck JJ, Bhatnagar R, Abraham J, Leffall LD. Identification of MMP-1 as a putative breast cancer predictive marker by global gene expression analysis. Nature Medicine. 2005;11(5):481–483. doi: 10.1038/nm1243. [DOI] [PubMed] [Google Scholar]
- 32.Turashvili G, Bouchal J, Baumforth K, Wei W, Dziechciarkova M, Ehrmann J, et al. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. Bmc Cancer. 2007:7. doi: 10.1186/1471-2407-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chanrion M, Negre V, Fontaine H, Salvetat N, Bibeau F, Mac Grogan G, et al. A gene expression signature that can predict the recurrence of tamoxifen-treated primary breast cancer. Clin Cancer Res. 2008;14(6):1744–1752. doi: 10.1158/1078-0432.CCR-07-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3(2):185–197. doi: 10.1016/s1535-6108(03)00028-x. [DOI] [PubMed] [Google Scholar]
- 35.Whitfield ML, George LK, Grant GD, Perou CM. Common markers of proliferation. Nat Rev Cancer. 2006;6(2):99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- 36.Chung CH, Bernard PS, Perou CM. Molecular portraits and the family tree of cancer. Nat Genet. 2002;(32 Suppl):533–540. doi: 10.1038/ng1038. [DOI] [PubMed] [Google Scholar]
- 37.Tripathi A, King C, de la Morenas A, Perry VK, Burke B, Antoine GA, et al. Gene expression abnormalities in histologically normal breast epithelium of breast cancer patients. Int J Cancer. 2008;122(7):1557–1566. doi: 10.1002/ijc.23267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.