Abstract
An RNAi based functional screen of mitotic kinases in Drosophila recently identified a number of members of the kinome that are required for normal cell division. Depletion of these kinases resulted in a number of different mitotic abnormalities including spindle malformation, chromosome missegregation, centrosome amplification and failure of cytokinesis [1]. Since mitotic defects are commonly observed in cancer cells, these kinases may contribute to tumor development and/or progression. To investigate whether common genetic variation in the mitotic kinases are associated with breast cancer risk, we genotyped 386 single nucleotide polymorphisms (SNPs) from 44 mitotic kinase genes, in 798 breast cancer cases and 843 unaffected controls from a clinic-based study. A total of 22 SNPs from 13 kinase genes displayed significant associations with breast cancer risk (Ptrend ≤ 0.05), including two SNPs from FYN (rs6914091 and rs1465061) that remained of interest after accounting for multiple testing (q=0.06). These associations were stronger when evaluating cases with estrogen and progesterone receptor positive tumors. In addition, haplotype-based tests identified significant associations with risk for common haplotypes of the MAST2 (P = 0.04) and MAP2K4 (P = 0.006) genes. Although requiring replication, these findings suggest that genetic polymorphisms in mitotic kinases that have been implicated in chromosome instability and aneuploidy may contribute to the development of breast cancer.
Keywords: mitosis, mitotic kinase, single nucleotide polymorphism (SNP), haplotype, breast cancer risk
Introduction
Mitotic kinases exert pleiotropic effects to control and regulate all phases of mitosis including entry, progression and exit. They phosphorylate a plethora of structural and regulatory proteins involved in centrosome duplication and separation, chromosome condensation, spindle assembly and fidelity, chromosome segregation and cytokinesis, to ensure successful divisions of mammalian cells. The activity of these kinases is modulated by transcriptional regulation, post-translational modification and proteosomal degradation. Over-expression of a number of these kinases, such as members of the Aurora, PLK and NEK families, are known to induce centrosome amplification, chromosome mis-segregation and failure of cytokinesis [2, 3]. These mitotic abnormalities in turn lead to chromosomal instability (CIN), aneuploidy and polyploidy that are commonly observed in human solid tumors and hematologic malignancies.
CIN and aneuploidy are common features of invasive breast tumors and early stages of breast oncogenesis, including ductal carcinoma in-situ (DCIS), [4, 5]. The AURKA, PLK1, NEK2 and NEK8 oncogenic kinases are frequently over-expressed, both at mRNA and protein levels, in a large number of primary DCIS and invasive breast carcinomas, and are known to promote aberrant cell divisions leading to CIN, aneuploidy and polyploidy [6-8]. Inhibition of both AURKA and PLK1 kinases has been shown to reverse the oncogenic process [9, 10]. In addition, the TTK, MASTL, BUB1, BUB1B, CDC7, CDC2, AURKB, and PLK4 kinases have been identified as components of a prognostic signature in luminal A hormone receptor positive breast tumors [11].
Common genetic variation in mitotic kinase genes has been associated with breast cancer risk. A meta-analysis of rs2273535 (F31I) in the AURKA gene in 15 case-control studies of multiple types of cancers showed a modest increase in cancer risk associated with this common genetic variation [12]. Similarly, individual SNPs and haplotypes in BUB1B and TTK have been linked to a modest increase in risk in a sporadic breast cancer population in Taiwan [13]. In addition, a genome-wide association study revealed a significant association between a SNP in the MAP3K1 gene and moderately increased breast cancer risk in the general population [14]. In this study, we examined the effect of tagging SNPs in 44 mitotic kinase genes on breast cancer risk. These genes were selected based on a previous study showing that the Drosophila orthologs caused substantial mitotic disruption when down-regulated by RNAi [1]. The SNPs were genotyped in a clinic-based breast cancer case-control study of Caucasians from the Midwestern United States, and our results suggested that inherited genetic polymorphisms of novel mitotic kinase genes may be associated with breast cancer risk.
Materials and methods
Mayo Clinic Study Population
Details concerning the collection of cases and controls used in the Mayo Clinic Breast Cancer Case-Control Study have been described previously [14] [15] [16]. Briefly, this population is a clinic-based series of breast cancer cases and unaffected controls from a 6-state region (Minnesota, Iowa, Wisconsin, North and South Dakota and Illinois) who attended the Mayo Clinic between 2002 and 2005. All cancer patients included were newly diagnosed with invasive breast cancer. Controls with no history of cancer were recruited in parallel from women attending the Department of Internal Medicine at the Mayo Clinic for a prescheduled annual medical examination. Controls were frequency matched to cases by age and state of residence. Eligible women provided informed consent and a sample of blood as a source of DNA and completed a risk factor questionnare. Participation rates were 69% and 71% for cases and controls, respectively. A total of 1,741 genomic DNA samples (798 cases, 843 controls, 100 blind duplicates) were assayed at Illumina Corporation (San Diego, California) on an Illumina BeadLab using the Illumina GoldenGate genotyping assay.
GENICA Replication Study Population
Kinome SNPs that displayed strong associations with breast cancer risk in the Mayo Clinic study were further genotyped in an independent case-control population from an interdisciplinary study group on Gene Environment Interaction and Breast Cancer in Germany (GENICA). The GENICA breast cancer case-control study, including 1021 incident cases and 1015 age-matched, population-based controls from the Greater Bonn Region, Germany, has been described previously [17, 18]. In brief, breast cancer cases were included based on a histopathologic diagnosis of primary breast cancer. Cases and controls were eligible if they were of Caucasian ethnicity, currently residing in the study region, and were below age 80 years. The GENICA study was approved by the Ethic's Committee of the University of Bonn. All study participants gave written informed consent.
SNP Selection
SNPs in the genomic region from 5 kb upstream to 5 kb downstream of each of the genes with minor allele frequency (MAF) > 0.05 in the Caucasian population were selected from the Hapmap Consortium stage I and II release [19]. TagSNPs representing SNPs with pairwise correlation of r2≥0.8 were chosen by LDselect [20] analysis of genotype data from Caucasian populations. All synonymous and nonsynonymous coding SNPs, 5′ and 3′ UTR SNPs, and 5′ upstream (promoter region) SNPs within 1000 base pairs from the transcriptional start site with MAF > 0.05 in the Caucasian population were included as candidate functional SNPs.
Statistical Analysis
The quality of genotyping data was assessed by Pearson's goodness-of-fit χ2 test of deviation from Hardy-Weinberg equilibrium for all SNPs in control subjects. Unconditional logistic regression analysis was used to estimate odds ratios (OR) and 95% confidence intervals (95% CI) for risk of breast cancer associated with the genotypes of each SNP [21]. Primary tests of association between SNPs and breast cancer risk were performed assuming an ordinal (log-additive) genotypic relationship, using tests for trend within the logistic regression models. Risk factors significantly associated with case-control status were accounted for in these analyses. These covariates included age at enrollment, age at menarche, parity, age at first live birth, oral contraceptive use, menopausal status, HRT use, and body mass index. Exploratory analyses were also conducted in subgroups of women defined by menopausal status or histological subtype of tumors based on ER, PR and HER2 status, as reported in pathology records. In order to determine the degree to which the tests for association were prone to false positive findings False discovery rate (FDR) q-values were calculated [22].
Risk associations between intra-genic haplotypes and invasive breast cancer were also determined. Haplotype blocks within each gene of interest were defined in control subjects by HaploView based on the method of Gabriel et al. [23]. Pair-wise LD was estimated between SNPs by D′ and r2 values. Logistic regression analysis was performed for each haplotype to estimate haplotype frequency and to examine associations between haplotypes within the identified haplotype blocks and breast cancer status using the HaploStats analysis package [24].
Gene-gene interactions
SNP by SNP interactions were explored through the assessment of significance of all possible pair-wise combinations of SNPs [25]. Additional unconditional logistic regression analyses were performed to further explore the effect on breast cancer risk of SNP pairs selected through this comprehensive assessment. All statistical tests were two-sided, and all analyses were carried out using the SAS (SAS Institute, Inc., Cary, NC) and S-Plus (Insightful, Seattle, WA) software systems.
Results
SNPs and genes significantly associated with breast cancer risk
In a recently published study, an RNAi screen of protein kinases in Drosophila identified 80 kinases that caused gross cell cycle defects and mitotic abnormalities when depleted [1]. We selected all known kinases influencing mitosis and also those implicated in S and G2/M phase cell cycle regulation that led to subsequent mitotic defects, for a total of 44 genes. A total of 386 tagging and functional SNPs (four 5′ upstream, three 3′UTR and two non-synonymous SNPs) with MAF > 0.05 were selected from the human orthologs of these kinase genes. These SNPs were subsequently genotyped in 798 cases and 843 unaffected controls from a Mayo Clinic breast cancer case-control study. The density of the tagSNPs for each gene and percent coverage of all variation and of SNP bins (r2 > 0.8) in each gene by the selected SNPs, based on the HapMap database (NCBI36), are shown in Supplementary Table 1. A total of 381 SNPs from 44 genes displayed high call rates (> 99%), MAF > 0.05 and did not diverge significantly from Hardy-Weinberg Equilibrium (HWE) and were subjected to subsequent statistical analysis (Supplementary Table 1).
A total of 22 SNPs from 13 kinase genes displayed significant associations with breast cancer risk under the ordinal model (Ptrend ≤ 0.05) as shown in Table 1. Of those 22 SNPs, 13 were associated with increased risk and nine were associated with decreased risk. Three SNPs, rs6914091, rs1465061 and rs12910 in FYN, displayed the most significant associations (OR per allele = 1.27 to 1.33, Ptrend ≤ 0.001) (Table 1). These SNPs showed similar associations (OR = 1.31 to 1.41, Ptrend ≤ 0.004) in postmenopausal women, which account for over 70% of the case-control population (Supplementary Table 2). Interestingly, the statistical significance of these associations was substantially increased in subgroups of cases with ER positive tumors (OR = 1.38 to 1.42, Ptrend ≤ 0.0002) and PR positive tumors (OR = 1.40 to 1.45, Ptrend ≤ 0.0002) (Table 2). In contrast, the FYN SNPs were not significantly associated with ER negative breast cancer (data not shown). This finding suggested a hormone-dependent association between variation in the FYN gene and increased breast cancer risk. Similarly, the ROCK1 SNP, rs17202375, displaying a significant association with risk (OR = 1.43; Ptrend = 0.005) was associated with ER positive (OR = 1.52, Ptrend = 0.004) and PR positive (OR = 1.61, Ptrend = 0.001) breast cancer and risk of postmenopausal (OR = 1.72, Ptrend = 0.0009) but not premenopausal disease (Supplementary Table 2). Nine other SNPs from seven kinase genes (BRD4, CDC42BPA, CIT, EIF2AK4, KIAA0999, PAK6 and RYK) also displayed associations with increased breast cancer risk, although the effects were only marginally significant (Table 1). It was also noted that a single SNP from KIAA0999 exhibited an association with increased risk, whereas two others from the same gene showed protective effects (Table 1). These two SNPs remained significantly associated with decreased breast cancer risk in postmenopausal women (Supplementary Table 2). Similarly, two SNPs in both CDC42BPA and PAK6 were significantly associated with opposite effects on breast cancer risk (Table 1).
Table 1.
Multivariate analysis of polymorphic variants of the mitotic kinase genes with Ptrend ≤ 0.05
| Gene Name | rsID | Major Allele Homozygotes |
Heterozygotes |
Minor Allele Homozygotes |
OR (95%CI) Per Allele |
Ptrend | Q value | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | |||||
| BRD4 | rs4808278 | 613 | 676 | 166 | 157 | 18 | 10 | 1.27 (1.02-1.58) | 0.03 | 0.77 |
| BUB1B | rs7182070 | 602 | 590 | 180 | 234 | 16 | 19 | 0.81 (0.66-1.00) | 0.05 | 0.77 |
| CDC42BPA | rs2802269 | 659 | 666 | 134 | 168 | 5 | 9 | 0.77 (0.61-0.98) | 0.04 | 0.77 |
| CDC42BPA | rs1929860 | 345 | 403 | 358 | 353 | 95 | 87 | 1.17 (1.00-1.36) | 0.05 | 0.77 |
| CIT | rs17496224 | 694 | 770 | 102 | 72 | 1 | 1 | 1.49 (1.08-2.05) | 0.02 | 0.77 |
| DBF4 | rs9655955 | 706 | 718 | 89 | 119 | 3 | 6 | 0.73 (0.55-0.97) | 0.03 | 0.77 |
| EIF2AK4 | rs2291627 | 646 | 715 | 143 | 124 | 9 | 4 | 1.34 (1.04-1.73) | 0.02 | 0.77 |
| FYN | rs6914091 | 221 | 297 | 412 | 414 | 165 | 131 | 1.33 (1.14-1.54) | 0.0002 | 0.06 |
| FYN | rs1465061 | 290 | 360 | 377 | 388 | 129 | 92 | 1.32 (1.13-1.53) | 0.0003 | 0.06 |
| FYN | rs12910 | 182 | 247 | 416 | 421 | 200 | 175 | 1.27 (1.10-1.46) | 0.001 | 0.19 |
| KIAA0999 | rs10047459 | 544 | 612 | 227 | 208 | 27 | 23 | 1.22 (1.01-1.48) | 0.04 | 0.77 |
| KIAA0999 | rs1473177 | 644 | 647 | 147 | 185 | 7 | 11 | 0.79 (0.63-0.99) | 0.04 | 0.77 |
| KIAA0999 | rs17120241 | 625 | 614 | 156 | 213 | 17 | 15 | 0.81 (0.65-0.99) | 0.04 | 0.77 |
| PAK6 | rs4924445 | 263 | 328 | 390 | 386 | 145 | 129 | 1.17 (1.01-1.35) | 0.03 | 0.77 |
| PAK6 | rs7169803 | 471 | 451 | 284 | 335 | 43 | 57 | 0.84 (0.71-0.99) | 0.04 | 0.77 |
| PRKACB | rs12728744 | 734 | 750 | 63 | 89 | 1 | 3 | 0.70 (0.50-0.98) | 0.04 | 0.77 |
| ROCK1 | rs17202375 | 638 | 714 | 153 | 124 | 7 | 5 | 1.43 (1.11-1.84) | 0.005 | 0.53 |
| RYK | rs10935104 | 568 | 639 | 204 | 190 | 26 | 14 | 1.29 (1.05-1.58) | 0.01 | 0.77 |
| RYK | rs9283588 | 595 | 661 | 182 | 174 | 21 | 8 | 1.30 (1.05-1.61) | 0.02 | 0.77 |
| RYK | rs1131262 | 595 | 661 | 182 | 173 | 21 | 9 | 1.28 (1.04-1.59) | 0.02 | 0.77 |
| STK4 | rs17420378 | 420 | 393 | 313 | 362 | 64 | 86 | 0.85 (0.73-0.99) | 0.04 | 0.77 |
| STK4 | rs6073636 | 420 | 394 | 313 | 364 | 64 | 85 | 0.85 (0.73-1.00) | 0.05 | 0.77 |
NOTE: rsID: SNP identification number; OR: odds ratio; CI: confidence interval
Logistic regression analysis adjusted for age, geographical region, menopausal status, age at menarche, oral contraceptive use, age first birth, hormone replacement therapy and pack-years cigarette smoking
Table 2.
Association of SNPs in mitotic kinase genes with breast cancer by ER, PR and HER2 status of tumors
| Gene Name | rsID | ER positive |
PR positive |
HER2 positive |
|||
|---|---|---|---|---|---|---|---|
| OR (95% CI) Per Allele | Ptrend | OR (95% CI) Per Allele | Ptrend | OR (95% CI) Per Allele | Ptrend | ||
| BRD4 | rs4808278 | 1.25 (0.97-1.61) | 0.09 | 0.98 (0.82-1.16) | 0.82 | 1.40 (0.94-2.08) | 0.10 |
| BUB1B | rs7182070 | 0.74 (0.58-0.94) | 0.01 | 0.72 (0.56-0.93) | 0.01 | 0.72 (0.48-1.09) | 0.13 |
| BUB1B* | rs3759843 | 0.82 (0.67-1.00) | 0.06 | 0.80 (0.65-1.00) | 0.05 | 0.95 (0.68-1.33) | 0.76 |
| CDC42BPA | rs2802269 | 0.76 (0.57-1.01) | 0.06 | 0.77 (0.57-1.03) | 0.08 | 0.70 (0.43-1.14) | 0.15 |
| CDC42BPA | rs1929860 | 1.24 (1.04-1.47) | 0.02 | 1.17 (0.98-1.40) | 0.09 | 1.36 (1.02-1.82) | 0.04 |
| CIT | rs17496224 | 1.44 (0.99-2.08) | 0.06 | 1.51 (1.03-2.20) | 0.03 | 1.12 (0.59-2.15) | 0.73 |
| DBF4 | rs9655955 | 0.73 (0.52-1.03) | 0.07 | 0.75 (0.53-1.05) | 0.10 | 0.89 (0.53-1.48) | 0.65 |
| EIF2AK4 | rs2291627 | 127. (0.95-1.70) | 0.11 | 1.38 (1.02-1.85) | 0.03 | 1.14 (0.70-1.86) | 0.60 |
| EIF2AK4* | rs17720604 | 1.00 (0.76-1.31) | 0.98 | 0.94 (0.70-1.24) | 0.65 | 1.64 (1.12-2.40) | 0.01 |
| FYN | rs6914091 | 1.42 (1.20-1.68) | 0.00006 | 1.45 (1.22-1.73) | 0.00003 | 1.19 (0.90-1.58) | 0.22 |
| FYN | rs1465061 | 1.40 (1.18-1.67) | 0.0001 | 1.40 (1.18-1.67) | 0.0002 | 1.24 (0.92-1.66) | 0.16 |
| FYN | rs12910 | 1.38 (1.16-1.63) | 0.0002 | 1.42 (1.19-1.69) | 0.00008 | 1.19 (0.90-1.56) | 0.23 |
| KIAA0999 | rs10047459 | 1.32 (1.06-1.64) | 0.01 | 1.31 (1.04-1.64) | 0.02 | 1.75 (1.24-2.46) | 0.001 |
| KIAA0999 | rs1473177 | 0.75 (0.57-0.98) | 0.04 | 0.76 (0.58-1.01) | 0.06 | 0.66 (0.41-1.08) | 0.10 |
| KIAA0999 | rs17120241 | 0.81 (0.63-1.03) | 0.08 | 0.81 (0.62-1.04) | 0.10 | 0.77 (0.50-1.17) | 0.22 |
| KIAA0999* | rs11216257 | 1.28 (1.03-1.58) | 0.03 | 1.25 (1.00-1.56) | 0.05 | 1.73 (1.23-2.42) | 0.001 |
| KIAA0999* | rs499910 | 1.14 (0.94-1.39) | 0.19 | 1.12 (0.92-1.38) | 0.26 | 1.40 (1.02-1.90) | 0.03 |
| KIAA0999* | rs2000615 | 1.17 (0.91-1.51) | 0.23 | 1.15 (0.88-1.51) | 0.30 | 1.54 (1.04-2.29) | 0.03 |
| PAK6 | rs4924445 | 1.20 (1.02-1.43) | 0.03 | 1.17 (0.98-1.39) | 0.09 | 1.26 (0.96-1.65) | 0.10 |
| PAK6 | rs7169803 | 0.86 (0.71-1.05) | 0.14 | 0.88 (0.72-1.08) | 0.22 | 0.94 (0.69-1.30) | 0.72 |
| PAK6* | rs2277562 | 1.13 (0.92-1.39) | 0.26 | 1.13 (0.91-1.40) | 0.26 | 1.56 (1.13-2.16) | 0.007 |
| PRKACB | rs12728744 | 0.77 (0.53-1.13) | 0.19 | 0.68 (0.45-1.03) | 0.07 | 1.14 (0.64-2.01) | 0.66 |
| PRKACB* | rs7546625 | 0.80 (0.64-1.00) | 0.05 | 0.78 (0.62-0.99) | 0.04 | 0.86 (0.59-1.25) | 0.43 |
| PRKACB* | rs1565823 | 1.20 (1.02-1.42) | 0.01 | 1.21 (1.02-1.43) | 0.03 | 0.98 (0.75-1.28) | 0.87 |
| PRKACB* | rs10782824 | 1.19 (1.01-1.40) | 0.03 | 1.21 (1.02-1.43) | 0.03 | 0.94 (0.72-1.24) | 0.66 |
| PRKACB* | rs1057738 | 1.20 (1.02-1.41) | 0.03 | 1.21 (1.03-1.44) | 0.02 | 0.95 (0.73-1.25) | 0.73 |
| PRKACB* | rs787858 | 1.19 (1.00-1.42) | 0.05 | 1.17 (0.97-1.40) | 0.10 | 1.06 (0.79-1.41) | 0.72 |
| PRKACB* | rs957828 | 0.97 (0.81-1.16) | 0.74 | 0.97 (0.81-1.16) | 0.72 | 0.72 (0.53-0.97) | 0.03 |
| ROCK1 | rs17202375 | 1.52 (1.14-2.02) | 0.004 | 1.61 (1.21-2.15) | 0.001 | 1.15 (0.72-1.85) | 0.55 |
| RYK | rs10935104 | 1.35 (1.07-1.70) | 0.01 | 1.33 (1.04-1.68) | 0.02 | 1.43 (0.98-2.08) | 0.06 |
| RYK | rs9283588 | 1.35 (1.06-1.73) | 0.02 | 1.34 (1.04-1.73) | 0.02 | 1.45 (0.97-2.16) | 0.07 |
| RYK | rs1131262 | 1.33 (1.05-1.70) | 0.02 | 1.33 (1.03-1.71) | 0.03 | 1.48 (1.00-2.21) | 0.05 |
| RYK* | rs12186098 | 1.39 (0.96-2.01) | 0.08 | 1.47 (1.01-2.15) | 0.05 | 1.29 (0.71-2.35) | 0.41 |
| RYK* | rs13067800 | 1.39 (0.97-1.99) | 0.07 | 1.42 (0.99-2.04) | 0.06 | 1.96 (1.16-3.30) | 0.01 |
| STK4 | rs17420378 | 0.83 (0.69-0.99) | 0.04 | 0.87 (0.72-1.05) | 0.33 | 0.90 (0.67-1.22) | 0.50 |
| STK4 | rs6073636 | 0.83 (0.69-1.00) | 0.05 | 0.87 (0.72-1.05) | 0.16 | 0.90 (0.67-1.22) | 0.50 |
NOTE: ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; OR, odds ratio; CI, confidence interval
Logistic regression analysis adjusted for age, geographical region, menopausal status, age at menarche, oral contraceptive use, age first birth, hormone replacement therapy and cigarette smoking in packyears
Non-significant association in the complete case-control study population
Six of the 22 SNPs that displayed strong associations with breast cancer risk in the Mayo Clinic study were further genotyped in the GENICA study of 1021 cases and 1015 controls of German Caucasian women. These SNPs were selected based on the significance of the associations with risk in the primary analyses. The number of SNPs in the replication study was limited by cost. None were significantly associated with breast cancer risk (Table 3). Further stratified analyses of associations with risk in histological subgroups within the GENICA study were not performed.
Table 3.
Multivariate analysis of SNPs genotyped in GENICA replication study
| Gene Name | rsID | Major Allele Homozygotes |
Heterozygotes |
Minor Allele Homozygotes |
OR (95%CI) Per Allele |
Ptrend | |||
|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | ||||
| CIT | rs4766950* | 271 | 260 | 499 | 502 | 232 | 228 | 1.02 (0.90-1.16) | 0.79 |
| FYN | rs6914091 | 317 | 337 | 488 | 480 | 167 | 170 | 1.03 (0.90-1.17) | 0.68 |
| KIAA0999 | rs7928320* | 598 | 604 | 342 | 334 | 65 | 52 | 1.07 (0.92-1.24) | 0.38 |
| PAK6 | rs16970458* | 497 | 492 | 400 | 423 | 102 | 80 | 1.04 (0.90-1.19) | 0.62 |
| ROCK1 | rs17202375 | 805 | 807 | 180 | 181 | 8 | 12 | 0.96 (0.77-1.18) | 0.69 |
| RYK | rs9283588 | 781 | 789 | 222 | 212 | 14 | 11 | 1.07 (0.89-1.30) | 0.46 |
NOTE: rsID: SNP identification number; OR: odds ratio; CI: confidence interval
Logistic regression analysis adjusted for age, geographical region, menopausal status, age at menarche, oral contraceptive use, age first birth, hormone replacement therapy and pack-years cigarette smoking
rs7928320 was significant in the general model in Mayo study (Ptrend = 0.0004)
rs4766950 and rs1697045 were significant in the general model in the pre-menopausal women
Haplotypes and haplotype blocks significantly associated with breast cancer risk
To examine the associations of specific haplotypes with breast cancer risk, haplotype blocks were defined for each gene based on the model reported by Gabriel et al [23]. In each haplotype block, haplotypes with frequency > 2% were subsequently analyzed by multiple logistic regression. A total of nine haplotypes in specific blocks displayed significant associations with breast cancer risk (Table 4 and Supplementary Table 3). In particular, the combined “rare” haplotypes in haplotype block 1 of MAP2K4 were significantly associated with risk (global P = 0.04), as was the specific haplotype 1211111 (Frequency = 17.8%, Score = 2.76, P = 0.006). Similarly, the specific haplotype 12112122 of MAST2 was associated with a decreased risk of breast cancer (Frequency = 6.6%, Score = −2.08, P = 0.04). None of the individual SNPs in these two genes displayed significant associations (Table 1 and 3), indicating that the associations of these specific combinations of alleles with risk were not due to the effects of individual SNPs. Haplotypes exhibiting significant associations with risk (P ≤ 0.05) were also identified in BRD4 (21112), BUB1B (1122), DBF4 (21122), KIAA0999 (11121), ROCK1 (1122), RYK (1221121), and STK4 (1122111122) (Table 3). Each of these specific haplotypes contained single SNPs displaying significant associations with risk suggesting that these rare alleles account for the haplotype associations (Table 2 and Table 4)
Table 4.
Association of specific haplotypes with breast cancer risk
| Haplotypes and Blocks | Haplotype | Hap-Frequency | Hap-Score | P value |
|---|---|---|---|---|
| BRD4 block 1 | 21112 | 0.115 | 2.04 | 0.04 |
| BUB1B block 1 | 1122 | 0.147 | −2.04 | 0.04 |
| DBF4 block 1 | 21122 | 0.068 | −2.31 | 0.02 |
| KIAA0999 block 1 | 11121 | 0.110 | −2.10 | 0.04 |
| MAST2 block 1 | 12112122 | 0.066 | −2.08 | 0.04 |
| MAP2K4 block 1 | 1211111 | 0.178 | 2.76 | 0.006 |
| ROCK1 block 1 | 1122 | 0.091 | 2.70 | 0.007 |
| RYK block 1 | 1221121 | 0.125 | 2.47 | 0.01 |
| STK4 block 1 | 1122111122 | 0.139 | −2.41 | 0.02 |
NOTE: 1=major allele, 2=minor allele; Hap-Frequency: frequency of each haplotype in the controls; Hap-Score: statistical measurement of association of each specific haplotype with breast cancer risk
BRD4 block 1: rs4808272 rs11880801 rs8104223 rs4809130 rs4808278
BUB1B block 1: rs1801376 rs1047130 rs2305653 rs7182070
DBF4 block 1: rs9655955 rs13238458 rs11764107 rs4728713 rs6977687
KIAA0999 block 1: rs687172 rs11216164 rs681524 rs1473177 rs499910
MAP2K4 block 1: rs8067785 rs7216812 rs12603036 rs17614045 rs2322123 rs9907196 rs10432016. The individual SNPs in this block displayed no significant associations with breast cancer risk (in bold and underlined). *The global P value for this block is also significant at 0.04
MAST2 block 1: rs4660891 rs12732188 rs4660318 rs12141934 rs10789481 rs12759880 rs2275426 rs1707336. The individual SNPs in this block displayed no significant associations with breast cancer risk (in bold and underlined)
ROCK1 block 1: rs7227454 rs288989 rs17202375 rs1481280
RYK block 1: rs13067800 rs9283588 rs10935104 rs6795658 rs9859680 rs1131262 rs4339087
STK4 block 1: rs17322289 rs2284271 rs6017460 rs17420378 rs6073604 rs10485454 rs6073627 rs6073629 rs8000 rs607363
Pair-wise interactions of SNPs significantly associated with breast cancer risk
Pair-wise SNPs from 44 kinase genes were subjected to linear logistic regression analysis to identify significant interactions. A total of 19 combinations were found to have highly significant associations with breast cancer risk (P ≤ 0.0001) (Supplementary Table 4). These 19 interactions, together with an additional 89, produced a common FDR q-value of 0.329, suggesting that only one-third of these interactions are expected to be false positives. Of the 19 interactions with the smallest p-values, two were composed of SNPs that independently displayed significant associations with risk (Table 1). For the other 17 combinations, either one or both SNPs in each pair were not significantly associated with cancer risk. Six in 17 of these pair-wise interactions included one of the three SNPs in FYN displaying significant associations with risk. Notably, interactions between FYN and IRAK1 and FYN and MAST2 displayed dose-dependent associations with breast cancer risk (OR1/1 copy = 1.6 to 1.7, P ≤ 0.007; OR2/2 copies = 2.6 to 3.7, P ≤ 0.03) (Supplementary Table 4). Combinations of rs7928320 in KIAA0999 and SNPs in ROCK1, PAK4 and MAST2 were significantly associated with increased breast cancer risk when minor alleles were present at each locus (OR2/2 copies = 3.3 to 13.4; P = 0.02 to 0.004; data not shown). Interestingly, none of these SNPs individually displayed significant associations with breast cancer risk.
Discussion
Our study systematically evaluated associations between genetic polymorphisms in 44 genes encoding kinases required for normal cell division and breast cancer risk. The common genetic variation in these genes was represented by tagging and candidate functional SNPs. The strongest associations observed in our study were individual SNPs in the FYN kinase gene, especially in women with ER-positive and PR-positive tumors, which represent the majority of the sporadic breast cancer cases. In addition, we identified two haplotypes in MAP2K4 and MAST2 that were significantly associated with breast cancer risk. Interestingly, the individual SNPs designating these haplotypes showed no significant associations with risk. Furthermore, we identified marginally significant associations with risk for polymorphisms in 12 other genes (BRD4, BUB1B, CDC42BPA, CIT, DBF4, EIF2AK4, KIAA0999, PAK6, PRKACB, ROCK1, RYK
As noted above, SNPs in FYN displayed the most significant associations with increased breast cancer risk in our clinic-based case-control series. FYN is a membrane associated non-receptor tyrosine kinase member of the src-family, implicated in diverse biological functions such as neuronal development, T-cell receptor signaling and cell-cell adhesion [26, 27]. FYN binds the p85 subunit of PI3-kinase and other FYN-binding proteins via the Src homology domains (SH) of its cytoplasmic tails and phosphorylates these targets to control cell proliferation and differentiation. FYN has been proposed as a tumor suppressor due to down-regulation in prostate cancers and inactivation in neuroblastoma [28, 29]. Disease-association studies have linked common genetic variations of FYN to alcoholism and Alzheimer's disease [30, 31]. In our study, we showed for the first time that genetic polymorphisms of FYN may also be linked to increased breast cancer risk, especially in women with expression of ER and PR in their breast tumors. This is consistent with the finding that FYN is an upstream regulator of the PI3K/Akt-signaling pathway, a critical cell survival, proliferation and motility pathway in cancer, that is activated predominantly in ER and PR positive breast tumors [32]. The three SNPs in FYN that displayed significant associations with breast cancer risk were located in the 5′ upstream region of isoforms b and c (NM_153048 and NM_153047), and the 5′ UTR (rs12910), and third intron of isoform a (rs1465061 and rs6914091). Further studies are needed to determine whether these alterations influence promoter activity or mRNA stability of FYN transcripts. Similarly to FYN, ROCK1 is a multifunctional kinase involved in focal adhesion formation, cell motility andapoptosis. ROCK1 binds the Rho small GTPase and regulates the dynamics of actin cytoskeleton and stress fiber formation. In cancer cells, Rho/ROCK signaling promotes migration and invasion by phosphorylating caveolin-1 at cell protrusions to regulate focal adhesion dynamics that are both ROCK-dependent and src-dependent [33]. In addition, ROCK1 can also bind and phosphorylate PTEN, a well-known tumor suppressor, to activate its function in chemotaxis and potential anti-tumorigenic activities [34]. Significantly increased expression of ROCK1 has been correlated with higher grade and poor overall survival of patients with breast cancers, possibly due to increased cell motility and invasiveness [35]. In keeping with these observations, we found that ROCK1 rs17202375 was associated with a substantially increased risk of breast cancer in post-menopausal women and women with ER and PR positive tumors.
Experimental data from studies of breast, ovarian, pancreatic and biliary cancers suggest that MAP2K4 is a tumor suppressor gene involved in both tumorigenesis and metastasis [36-40]. MAP2K4 has been reported to link MEKK1 to stress-activated protein kinase/JNK1 and p38 mitogen-activated protein kinase. This mitogen-activated protein kinase pathway has been implicated in the signal transduction of cytokine- and stress-induced apoptosis in a variety of cell types. None of the SNPs of the MAP2K4 gene were significantly associated with breast cancer risk in any of the genetic models tested above. However, a combination of seven SNPs forming haplotype block 1 was significantly associated with increased breast cancer risk. All seven SNPs are located in the first three introns of the MAP2K4 gene (Figure 1). It will be interesting to examine whether downregulation of the MAP2K4 promoter is correlated with this haplotype.
Fig. 1.
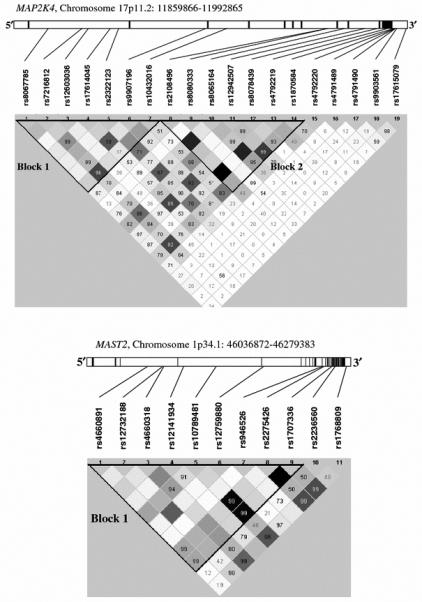
Haplotype blocks for the MAP2K4 and MAST2 genes. Haplotypes blocks were defined using the Gabriel model in Haploview. Each haplotype block is shown as a triangle. Shaded regions indicate strength of linkage disequilibrium (LD) between pairwise combinations of polymorphism (white: r2=low LD, black: r2=near perfect LD). Numbers in squares indicate estimate of pairwise D-prime, expressed in percentages. Pairs with unreported values have D-prime values of 1.0 (100%). MAP2K4 has 2 successive haplotype blocks from 5′ to 3′. Physical locations of each SNP, exons (dark bars) and introns are also shown for the gene. Exon 1 starts from the left side for both MAP2K4 and MAST2
Similarly to MAP2K4, we identified a specific haplotype of nine SNPs in MAST2 that displayed a significant association with breast cancer risk. MAST2 encodes a microtubule-associated serine/threonine kinase with largely unknown function. However, MAST2 has been shown tobind the C-terminus of PTEN through a unique PDZ domain, target the C-terminus of PTEN for phosphorylation and modulate PTEN activity [41, 42]. Interestingly, the phosphatase activity of PTEN can also be modulated by ROCK1 and it has recently been shown to control FYN function in glioma cells regulating migration. These findings suggest that FYN, ROCK1 and MAST2 may function together in the same signaling pathway to regulate the tumor suppressor function of PTEN [43]. In support of this observation, we identified significant pairwise interactions between SNPs in FYN and ROCK1 (Supplementary Table 3) and FYN and MAST2 (Supplementary Table 4).
The major strength of our association study is the comprehensive assessment of associations between genetic polymorphisms of mitotic kinase genes and breast cancer risk. The systematic approach increased the likelihood of identifying significant associations in these candidate genes that were selected because of their critical functions in breast cancer biology. However, the large number of statistical tests performed raised the possibility of false positive observations. FDR based assessment of multiple testing yielded q values of 0.06 for the most significantly associated FYN SNPs (Table 1). When accounting for the high LD between a number of the SNPs in the study, this suggests that these FYN SNPs maintain significance even after accounting for multiple testing. This is further emphasized when assessing the significance of the associations between these SNPs and risk of ER positive and/or PR positive breast cancer (Ptrend = 0.00006). However, six SNPs including rs6914091 from FYN, that were further evaluated in the GENICA case-control series, failed to replicate these significant associations, although it should be noted that these SNPs were not evaluated for associations with risk in menopausal or histological subgroups in GENICA. These findings suggest that additional studies that focus on replication of these results in independent case-control series and in histological subgroups of tumors are merited.
Supplementary Material
Acknowledgments
This study was funded in part by a grant (5R01CA122340-02) and a Breast Cancer Specialized Program of Research Excellence (SPORE) grant P50CA166201 from the National Cancer Institute. J. C. and H. B. are supported by the Federal Ministry of Education and Research Germany grants 01KW9975/5, 01KW9976/8, 01KW9977/0, and 01KW0114 and the Robert Bosch Foundation of Medical Research, Stuttgart, Deutsches Krebsforschungszentrum, Heidelberg, Berufsgenossenschaftliches Forschungsinstitut für Arbeitsmedizin Bochum, and Medizinische Universitäts-und Poliklinik, Bonn, Germany (GENICA study); and the Deutsche Krebshilfe (GESBC study).
References
- 1.Bettencourt-Dias M, Giet R, Sinka R, Mazumdar A, Lock WG, Balloux F, Zafiropoulos PJ, Yamaguchi S, Winter S, Carthew RW, et al. Genome-wide survey of protein kinases required for cell cycle progression. Nature. 2004;432:980–987. doi: 10.1038/nature03160. [DOI] [PubMed] [Google Scholar]
- 2.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 3.Mundt KE, Golsteyn RM, Lane HA, Nigg EA. On the regulation and function of human polo-like kinase 1 (PLK1): effects of overexpression on cell cycle progression. Biochem Biophys Res Commun. 1997;239:377–385. doi: 10.1006/bbrc.1997.7378. [DOI] [PubMed] [Google Scholar]
- 4.Lingle WL, Barrett SL, Negron VC, D'Assoro AB, Boeneman K, Liu W, Whitehead CM, Reynolds C, Salisbury JL. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci U S A. 2002;99:1978–1983. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li JJ, Li SA. Mitotic kinases: the key to duplication, segregation, and cytokinesis errors, chromosomal instability, and oncogenesis. Pharmacol Ther. 2006;111:974–984. doi: 10.1016/j.pharmthera.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka T, Kimura M, Matsunaga K, Fukada D, Mori H, Okano Y. Centrosomal kinase AIK1 is overexpressed in invasive ductal carcinoma of the breast. Cancer Res. 1999;59:2041–2044. [PubMed] [Google Scholar]
- 7.Yuan J, Horlin A, Hock B, Stutte HJ, Rubsamen-Waigmann H, Strebhardt K. Polo-like kinase, a novel marker for cellular proliferation. Am J Pathol. 1997;150:1165–1172. [PMC free article] [PubMed] [Google Scholar]
- 8.Hayward DG, Clarke RB, Faragher AJ, Pillai MR, Hagan IM, Fry AM. The centrosomal kinase Nek2 displays elevated levels of protein expression in human breast cancer. Cancer Res. 2004;64:7370–7376. doi: 10.1158/0008-5472.CAN-04-0960. [DOI] [PubMed] [Google Scholar]
- 9.Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, Graham JA, Demur C, Hercend T, Diu-Hercend A, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 10.Spankuch B, Heim S, Kurunci-Csacsko E, Lindenau C, Yuan J, Kaufmann M, Strebhardt K. Down-regulation of Polo-like kinase 1 elevates drug sensitivity of breast cancer cells in vitro and in vivo. Cancer Res. 2006;66:5836–5846. doi: 10.1158/0008-5472.CAN-06-0343. [DOI] [PubMed] [Google Scholar]
- 11.Finetti P, Cervera N, Charafe-Jauffret E, Chabannon C, Charpin C, Chaffanet M, Jacquemier J, Viens P, Birnbaum D, Bertucci F. Sixteen-kinase gene expression identifies luminal breast cancers with poor prognosis. Cancer Res. 2008;68:767–776. doi: 10.1158/0008-5472.CAN-07-5516. [DOI] [PubMed] [Google Scholar]
- 12.Ewart-Toland A, Dai Q, Gao YT, Nagase H, Dunlop MG, Farrington SM, Barnetson RA, Anton-Culver H, Peel D, Ziogas A, et al. Aurora-A/STK15 T+91A is a general low penetrance cancer susceptibility gene: a meta-analysis of multiple cancer types. Carcinogenesis. 2005;26:1368–1373. doi: 10.1093/carcin/bgi085. [DOI] [PubMed] [Google Scholar]
- 13.Lo YL, Yu JC, Chen ST, Hsu GC, Mau YC, Yang SL, Wu PE, Shen CY. Breast cancer risk associated with genotypic polymorphism of the mitotic checkpoint genes: a multigenic study on cancer susceptibility. Carcinogenesis. 2007;28:1079–1086. doi: 10.1093/carcin/bgl256. [DOI] [PubMed] [Google Scholar]
- 14.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Goode EL, Fredericksen ZS, Vierkant RA, Pankratz VS, Liu-Mares W, Rider DN, Vachon CM, Cerhan JR, Olson JE, Couch FJ. Association of genetic variation in genes implicated in the beta-catenin destruction complex with risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:2101–2108. doi: 10.1158/1055-9965.EPI-08-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelemen LE, Couch FJ, Ahmed S, Dunning AM, Pharoah PD, Easton DF, Fredericksen ZS, Vierkant RA, Pankratz VS, Goode EL, Scott CG, Rider DN, Wang X, Cerhan JR, Vachon CM. Genetic variation in stromal proteins decorin and lumican with breast cancer: investigations in two case-control studies. Breast Cancer Res. 2008;10:R98. doi: 10.1186/bcr2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pesch B, Ko Y, Brauch H, Hamann U, Harth V, Rabstein S, Pierl C, Fischer HP, Baisch C, Justenhoven C, et al. Factors modifying the association between hormone-replacement therapy and breast cancer risk. Eur J Epidemiol. 2005;20:699–711. doi: 10.1007/s10654-005-0032-0. [DOI] [PubMed] [Google Scholar]
- 18.Justenhoven C, Pierl CB, Haas S, Fischer HP, Baisch C, Hamann U, Harth V, Pesch B, Bruning T, Vollmert C, et al. The CYP1B1_1358_GG genotype is associated with estrogen receptor-negative breast cancer. Breast Cancer Res Treat. 2008;111:171–177. doi: 10.1007/s10549-007-9762-x. [DOI] [PubMed] [Google Scholar]
- 19.International HapMap Consortium et al. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng Z, Zaykin DV, Xu CF, Wagner M, Ehm MG. Selection of genetic markers for association analyses, using linkage disequilibrium and haplotypes. Am J Hum Genet. 2003;73:115–130. doi: 10.1086/376561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breslow NE, Day NE. Statistical methods in cancer research. IARC Workshop IARC Sci Publ. 1983;1987:1–406. [PubMed] [Google Scholar]
- 22.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 24.Lake SL, Lyon H, Tantisira K, Silverman EK, Weiss ST, Laird NM, Schaid DJ. Estimation and tests of haplotype-environment interaction when linkage phase is ambiguous. Hum Hered. 2003;55:56–65. doi: 10.1159/000071811. [DOI] [PubMed] [Google Scholar]
- 25.Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet. 2005;37:413–417. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- 26.Cooke MP, Abraham KM, Forbush KA, Perlmutter RM. Regulation of T cell receptor signaling by a src family protein-tyrosine kinase (p59fyn) Cell. 1991;65:281–291. doi: 10.1016/0092-8674(91)90162-r. [DOI] [PubMed] [Google Scholar]
- 27.Calautti E, Grossi M, Mammucari C, Aoyama Y, Pirro M, Ono Y, Li J, Dotto GP. Fyn tyrosine kinase is a downstream mediator of Rho/PRK2 function in keratinocyte cell-cell adhesion. J Cell Biol. 2002;156:137–148. doi: 10.1083/jcb.200105140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorensen KD, Borre M, Orntoft TF, Dyrskjot L, Torring N. Chromosomal deletion, promoter hypermethylation and downregulation of FYN in prostate cancer. Int J Cancer. 2008;122:509–519. doi: 10.1002/ijc.23136. [DOI] [PubMed] [Google Scholar]
- 29.Berwanger B, Hartmann O, Bergmann E, Bernard S, Nielsen D, Krause M, Kartal A, Flynn D, Wiedemeyer R, Schwab M, et al. Loss of a FYN-regulated differentiation and growth arrest pathway in advanced stage neuroblastoma. Cancer Cell. 2002;2:377–386. doi: 10.1016/s1535-6108(02)00179-4. [DOI] [PubMed] [Google Scholar]
- 30.Ishiguro H, Saito T, Shibuya H, Toru M, Arinami T. Mutation and association analysis of the Fyn kinase gene with alcoholism and schizophrenia. Am J Med Genet. 2000;96:716–720. [PubMed] [Google Scholar]
- 31.Watanabe T, Ohnuma T, Shibata N, Ohtsuka M, Ueki A, Nagao M, Arai H. No genetic association between Fyn kinase gene polymorphisms (−93A/G, IVS10+37T/C and Ex12+894T/G) and Japanese sporadic Alzheimer's disease. Neurosci Lett. 2004;360:109–111. doi: 10.1016/j.neulet.2004.02.046. [DOI] [PubMed] [Google Scholar]
- 32.Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmstrom PO, Mansukhani M, Enoksson J, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 33.Joshi B, Strugnell SS, Goetz JG, Kojic LD, Cox ME, Griffith OL, Chan SK, Jones SJ, Leung SP, Masoudi H, et al. Phosphorylated caveolin-1 regulates Rho/ROCK-dependent focal adhesion dynamics and tumor cell migration and invasion. Cancer Res. 2008;68:8210–8220. doi: 10.1158/0008-5472.CAN-08-0343. [DOI] [PubMed] [Google Scholar]
- 34.Li Z, Dong X, Wang Z, Liu W, Deng N, Ding Y, Tang L, Hla T, Zeng R, Li L, et al. Regulation of PTEN by Rho small GTPases. Nat Cell Biol. 2005;7:399–404. doi: 10.1038/ncb1236. [DOI] [PubMed] [Google Scholar]
- 35.Lane J, Martin TA, Watkins G, Mansel RE, Jiang WG. The expression and prognostic value of ROCK I and ROCK II and their role in human breast cancer. Int J Oncol. 2008;33:585–593. [PubMed] [Google Scholar]
- 36.Teng DH, Perry WL, 3rd, Hogan JK, Baumgard M, Bell R, Berry S, Davis T, Frank D, Frye C, Hattier T, et al. Human mitogen-activated protein kinase kinase 4 as a candidate tumor suppressor. Cancer Res. 1997;57:4177–4182. [PubMed] [Google Scholar]
- 37.Su GH, Hilgers W, Shekher MC, Tang DJ, Yeo CJ, Hruban RH, Kern SE. Alterations in pancreatic, biliary, and breast carcinomas support MKK4 as a genetically targeted tumor suppressor gene. Cancer Res. 1998;58:2339–2342. [PubMed] [Google Scholar]
- 38.Su GH, Song JJ, Repasky EA, Schutte M, Kern SE. Mutation rate of MAP2K4/MKK4 in breast carcinoma. Hum Mutat. 2002;19:81. doi: 10.1002/humu.9002. [DOI] [PubMed] [Google Scholar]
- 39.Couvelard A, Hu J, Steers G, O'Toole D, Sauvanet A, Belghiti J, Bedossa P, Gatter K, Ruszniewski P, Pezzella F. Identification of potential therapeutic targets by gene-expression profiling in pancreatic endocrine tumors. Gastroenterology. 2006;131:1597–1610. doi: 10.1053/j.gastro.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 40.Yamada SD, Hickson JA, Hrobowski Y, Vander Griend DJ, Benson D, Montag A, Karrison T, Huo D, Rutgers J, Adams S, et al. Mitogen-activated protein kinase kinase 4 (MKK4) acts as a metastasis suppressor gene in human ovarian carcinoma. Cancer Res. 2002;62:6717–6723. [PubMed] [Google Scholar]
- 41.Adey NB, Huang L, Ormonde PA, Baumgard ML, Pero R, Byreddy DV, Tavtigian SV, Bartel PL. Threonine phosphorylation of the MMAC1/PTEN PDZ binding domain both inhibits and stimulates PDZ binding. Cancer Res. 2000;60:35–37. [PubMed] [Google Scholar]
- 42.Valiente M, Andres-Pons A, Gomar B, Torres J, Gil A, Tapparel C, Antonarakis SE, Pulido R. Binding of PTEN to specific PDZ domains contributes to PTEN protein stability and phosphorylation by microtubule-associated serine/threonine kinases. J Biol Chem. 2005;280:28936–28943. doi: 10.1074/jbc.M504761200. [DOI] [PubMed] [Google Scholar]
- 43.Dey N, Crosswell HE, De P, Parsons R, Peng Q, Su JD, Durden DL. The protein phosphatase activity of PTEN regulates SRC family kinases and controls glioma migration. Cancer Res. 2008;68:1862–1871. doi: 10.1158/0008-5472.CAN-07-1182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


