Abstract
The HIV accessory protein Nef is one of the earliest and most abundantly expressed viral proteins. It is also found in the serum of infected individuals (Caby et. al, 2005). Extracellular Nef protein has deleterious effects on CD4+ T cells (James et. al, 2004), the primary targets of HIV, and can suppress immunoglobulin class switching in bystander B cells (Qiao et. al, 2006). Nevertheless, the mode of exit of Nef from infected cells remains a conundrum. We found that Nef stimulates its own export via the release of exosomes from all cells examined. Depending on its intracellular location, these Nef exosomes form at the plasma membrane, late endosomes or both compartments in Jurkat, SupT1 and primary T cells, respectively. Nef release through exosomes is conserved also during HIV-1 infection of peripheral blood lymphocytes. Released Nef exosomes cause activation-induced cell death of resting PBLs in vitro. Thus, HIV-infected cells export Nef in bioactive vesicles, which facilitate the depletion of CD4+ T cells that is a hallmark of AIDS.
Keywords: AIDS, apoptosis, exosome, HIV, multivesicular body, Nef, T cell activation
A key feature of the human immunodeficiency virus type 1 (HIV) infection is the gradual and progressive loss of CD4+ T cells, which leads to severe immunodeficiency characteristic of AIDS (1). In addition to direct cytotoxic effects on infected cells, the depletion of T cells can also result from the apoptosis of uninfected bystander cells (2). This activation-induced cell death can be due to several viral proteins, among which Nef appears to play a preeminent role (3–6).
The 27 kDa misnamed negative factor (Nef) is one of the earliest and most abundantly expressed viral proteins. Although localized primarily to cellular membranes, it is also present in the serum of infected individuals and those receiving highly active antiretroviral therapy (HAART) (7). Its pathogenic potential was demonstrated first in rhesus macaques infected with a Nef-deleted simian immunodeficiency virus (8) and later in long-term nonprogressors infected with a Nef-deleted HIV (HIVΔNef) (9). Likewise, HIVΔNef did not cause the depletion of CD4+ T cells in the SCID-hu mouse model of infection (10). To explain this phenotype in the host, several biological effects of Nef have been identified in cells. They include the internalization of critical receptors on infected cells, the activation of intracellular signaling pathways and the incorporation of Nef into progeny virions that increases their infectivity (11, 12).
In T cells, Nef forms a signaling complex with the T cell antigen receptor (TCR), thereby activating a transcriptional program that is almost identical to that triggered upon its exogenous stimulation (13). Nef further manipulates T cell activation by interacting with several proteins downstream of the TCR, which include PI3K, Vav or DOCK2-ELMO, small GTPases, Pak2 and PKC (11, 14). Whereas in some contexts, this activation leads to apoptosis (15, 16), in others, Nef protects cells by inhibiting pro-apoptotic proteins like ASK1, Bad or p53 (16–19). These differences can be explained by the presence of other co-stimulatory conditions in these assays (16, 20). In addition, Nef affects bystander T cells indirectly via the increased expression of FasL on infected cells (21) as well as directly by interacting with the CXCR4 chemokine receptor and/or via its uptake into these cells (4, 5, 22, 23).
Although Nef is present in the serum of infected individuals (22), it is not clear how it exits from infected cells (24). Most likely, this process is secondary to Nef's ability to modulate intracellular trafficking at multiple steps. By interacting with adaptor proteins (AP1, AP2 and AP3), βCOP, PACS1, vacuolar ATPase (V1H), subunits of the ESCRT complex, as well as lipid and protein kinases (11), Nef accesses many distinct intracellular organelles. Thus, it is not surprising that Nef leads to changes in endosomal morphology (25) as well as induces the proliferation of endosomes, lysosomes (26), fat granules (27) and multivesicular bodies (MVBs) (28, 29). In the degradative pathway, these latter organelles fuse with lysosomes. Alternatively, they fuse with the plasma membrane and release their intraluminal vesicles as exosomes (30). In T lymphoblastoid Jurkat cells, TCR activation induces immediate production of exosomes in discrete domains at the plasma membrane (31, 32). Substantial release of exosomes is also observed in activated primary T cells (33). This heterogeneous biogenesis prompted a new definition of exosomes as secreted membrane vesicles, which are between 40 and 100 nm in diameter and contain typical marker proteins (34). Exosomes circulate in the blood, suggesting that they play an important role in intercellular communications in the organism (35). Recent work on exosomes released by macrophages infected with various intracellular pathogens also suggests that they play a key role in immune surveillance (36). In this study, we examined the role of Nef in the release of exosomes in several cell lines and primary cells as well as in infected peripheral blood lymphocytes (PBLs). We found that Nef induces the release of exosomes from T cells, which transport extracellular Nef and cause apoptosis of bystander T cells.
Results
Nef increases the production of exosomes, thereby stimulating its own export from HeLa.CIITA cells
In previous studies, a striking increase in the number of MVBs was observed in Nef-expressing HeLa.CIITA cells (28, 29). To determine if this increase in MVBs could also lead to increased release of intraluminal vesicles, we quantified exosomes released from Nef.GFP-expressing HeLa.CIITA cells and compared them to those from control cells, which expressed only GFP or the mutant NefG2A.GFP chimera, which lacks the membrane-targeting myristoylation residue in Nef.
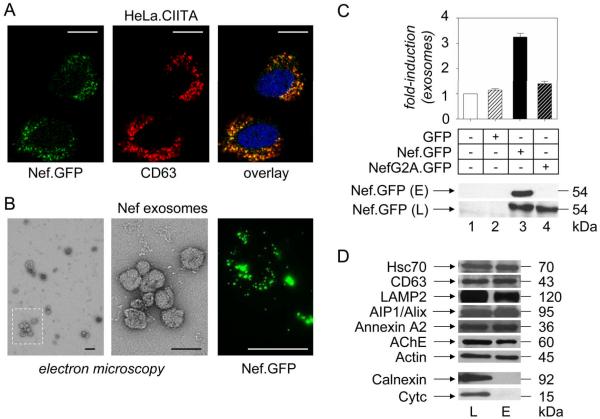
First, we expressed transiently Nef.GFP in HeLa.CIITA cells and examined them by fluorescent microscopy. Fig. 1A demonstrates that Nef was targeted to the perinuclear region (green), where it colocalized with the late endosomal marker CD63 (red) (37, 38), which indicates that Nef is present in late endosomes (yellow). Next, cellular supernatants were subjected to 0.2 μm filtration and high speed centrifugation steps, which represent established methods for preparing pure exosomes (39). Electron microscopic analyses of the purified pellet revealed small membrane vesicles of up to 100 nm in diameter, with an cup-shape structure (Fig. 1B, middle panel), which is typical for exosomes (30). Additional western blotting of the Nef.GFP-purified pellet (Fig. 1D, lane E) demonstrated the presence of several exosomal markers (CD63, AIP1/Alix and acetylcholinesterase (AChE)) and conserved exosomal proteins (Hsc70, LAMP2, annexin A2 and actin) (30). The purity of the sample was confirmed by the absence of calnexin and cytochrome c, which are markers for the endoplasmatic reticulum and mitochondria, respectively (Fig. 1D). The lysate from Nef.GFP-expressing HeLa.CIITA cells served as the control for these proteins and corresponding antibodies (Fig. 1D, lane L). From these data, we conclude that vesicles released from HeLa.CIITA cells are exosomes.
Figure 1.
Nef increases the production of exosomes in HeLa.CIITA cells. (A) Localization of Nef in HeLa.CIITA cells. Fluorescence of Nef.GFP (green) expressed in HeLa.CIITA cells was superimposed onto the indirect immunofluorescence of CD63 (red). Yellow color represents the overlay. (B) Visualization of exosomes. Exosomes from Nef.GFP-expressing cells were purified, negatively stained with uranyl formate and analyzed by transmission electron microscopy. Part of the sample was also analyzed by fluorescent confocal microscopy for the presence of Nef.GFP (green). Scale bars represent 20 μm (white) and 100 nm (black). (C) Quantification of exosomes. Amounts of exosomes released from control HeLa.CIITA cells (lane 1) and those expressing GFP (lane 2), Nef.GFP (lane 3) or NefG2A.GFP (lane 4) were assessed by normalizing the total protein content of the vesicular pellet to the number of transfected cells. They are displayed as relative values compared to those from untransfected (control) HeLa.CIITA cells. Results are presented as the mean ±SD of three independent experiments. 10 μg of above mentioned exosomal preparations were further analyzed for the presence of Nef.GFP with anti-GFP antibody by western blotting. (D) Composition of exosomes. Western blotting of Nef exosomes was performed with antibodies directed against conserved exosomal proteins (Hsc70, CD63, LAMP2, AIP1/Alix, Annexin A2, AChE, actin), markers for endoplasmatic reticulum (Calnexin) and mitochondria (Cytc). E stands for exosomes and L for cell lysate.
Interestingly, aggregated and free exosomes from Nef.GFP-expressing HeLa.CIITA cells were green when analyzed by fluorescent microscopy, which suggested that Nef was incorporated into these vesicles (Fig. 1B, right panel). Western blotting confirmed this observation (Fig. 1C, sample E, lane 3). In contrast, NefG2A.GFP was absent from vesicles obtained from HeLa.CIITA cells expressing the mutant NefG2A.GFP chimera (Fig. 1C, sample E, lane 4). This finding demonstrated the importance of N-terminal myristoylation for the incorporation of Nef into exosomes. To determine levels of exosome release, we normalized the total protein content of purified preparations to the number of manipulated cells. After two days in culture, we observed a 3-fold increased release of exosomes from Nef.GFP-expressing compared to untreated or GFP-expressing HeLa.CIITA cells (Fig. 1C, top panel, compare bars 1–3). The expression of the mutant NefG2A.GFP chimera had only marginal effects on this release (Fig. 1C, top panel, bar 4). Thus, Nef not only induces the release of exosomes from HeLa.CIITA cells, but is also incorporated into them, thereby controlling its own export from cells. Since these vesicles contain Nef, we call them Nef exosomes.
Nef in exosomes enters target cells
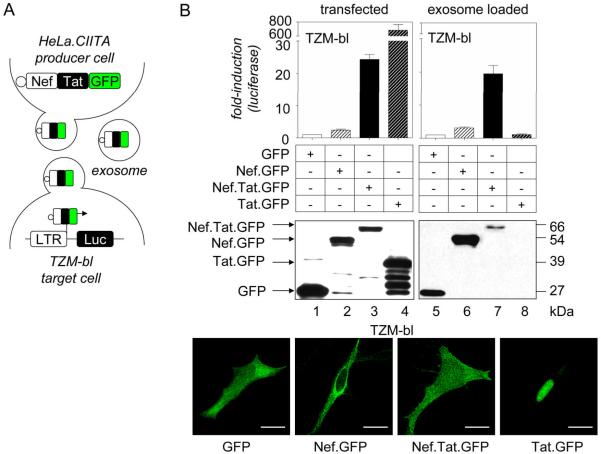
Next, we explored if exosomes are capable of shuttling Nef into target cells. To study this phenotype, we developed an exosome effector assay (Fig. 2A), based on the assumption that Nef can deliver exogenous proteins into cells (40). For this assay, Nef was first linked to the HIV transcriptional transactivator Tat fused to GFP. As expected, when expressed in HeLa.CIITA cells, Nef.Tat.GFP was released from cells in exosomes that were harvested two days later and added to the indicator TZM-bl cells. We followed the delivery of exosomal proteins into cells by measuring the luciferase activity that resulted from the induction of an integrated HIV long terminal repeat (LTR)-luciferase reporter gene. The goal of the assay was to bring Tat to the transactivation response (TAR) element in the HIV LTR, thereby increasing greatly its transcriptional activity. In addition to Nef.Tat.GFP, Nef.GFP, Tat.GFP and control GFP were also expressed in HeLa.CIITA cells and tested in this exosome effector assay.
Figure 2.
Nef chimeras in exosomes enter target cells. (A) Schematic overview of the exosome effector assay. Fusion proteins were expressed first in HeLa.CIITA cells. Two days later, exosomes were harvested as in Fig. 1 and then layered onto TZM-bl cells that harbor an integrated HIV-LTR-luciferase reporter gene. (B) Exosome effector assay. Effects of fusion proteins on the HIV LTR were determined first following their transient expression in TZM-bl cells: GFP (lane 1, white bar), Nef.GFP (lane 2, white bar with black stripes), Nef.Tat.GFP (lane 3, black bar) and Tat.GFP (lane 4, black bar with white stripes). Next, exosomes were harvested from transfected HeLa.CIITA cells and added to fresh TZM-bl cells for 48 hours: GFP (lane 5), Nef.GFP (lane 6), Nef.Tat.GFP (lane 7) and Tat.GFP (lane 8). Luciferase activity was measured at the end of the incubation. Presented are relative luciferase values to those with GFP or GFP exosome samples. Results are presented as the mean ±SD of three independent experiments. Expression of chimeras in HeLa.CIITA cells and their incorporation into exosomes were evaluated with anti-GFP antibodies by western blotting. Protein bands are labeled. Intracellular localization of the above-mentioned proteins was visualized by direct fluorescence in TZM-bl cells (green). White scale bar represents 20 μm.
First, we examined the expression of chimeras in HeLa.CIITA cells (Fig. 2B, middle panel, lanes 1–4) and their release in exosomes (Fig. 2B, middle panel, lanes 5–8) by western blotting with antibodies to GFP. Of note, whereas the incorporation of Nef.GFP into exosomes was more efficient than that of Nef.Tat.GFP, Tat.GFP was completely absent (Fig. 2B, middle panel, lanes 5–8). The localization of chimeras (Fig. 2B, bottom panels) and their effects on the HIV LTR (Fig. 2B, top panel, bars 1–4) were established in transiently transfected TZM-bl cells. Whereas the expression of GFP alone led to a diffuse cytoplasmic staining, Nef.GFP and Tat.GFP were excluded from and localized exclusively to the nucleus, respectively (Fig. 2B, bottom panels). In contrast, Nef.Tat.GFP localized simultaneously to the cytosol and nucleus (Fig. 2B, bottom panels). As observed previously (41), exogenous expression of Nef.GFP in TZM-bl cells activated the HIV promoter slightly when compared to effects of GFP alone (Fig. 2B, top left panel, bar 2). Importantly, Nef.Tat.GFP increased luciferase activity 25-fold (Fig. 2B, top left panel, bar 3). Not surprisingly, Tat.GFP had the highest activity in these cells (Fig. 2B, top left panel, bar 4). We conclude that Nef.Tat.GFP retains the abilities of Nef and Tat to associate with exosomes and to transactivate the HIV LTR, respectively.
After evaluating individual aspects of the assay, we examined the ability of various Nef exosomes to enter and affect target cells. First, we harvested exosomes produced from HeLa.CIITA cells that expressed GFP, Nef.GFP, Nef.Tat.GFP or Tat.GFP, incubated them with TZM-bl cells and measured their luciferase activity two days later. Indeed, exosomes with Nef.GFP had similar effects on the HIV LTR as Nef.GFP expressed intracellularly (Fig. 2B, top panels, bars 2 and 6). Importantly, exosomes with Nef.Tat.GFP induced the promoter up to 20-fold, which was comparable to levels observed with this chimera expressed transiently in cells (Fig. 2B, top panels, bars 3 and 7). Importantly, exosomes purified from Tat.GFP-expressing cells had no effect on the HIV LTR (Fig. 2B, top right panel, bar 8). This finding indicates that proteins transported by exosomes are internalized by and confer new properties to target cells.
Nef increases the production of exosomes in transformed T cells
CD4+ T lymphocytes are the principal cellular target of HIV infection (3) and Nef is the earliest and most abundantly expressed viral protein. In most cells, it induces the accumulation of numerous intracellular organelles (25, 26). Indeed, its effect on intracellular trafficking can contribute to the egress of viral particles from infected cells (29). Thus, it could also stimulate the production of Nef exosomes from these cells. Nevertheless, the production of Nef exosomes in T cells might also differ, as the plasma membrane is the site of activation-induced exosome biogenesis in Jurkat cells (31, 32).
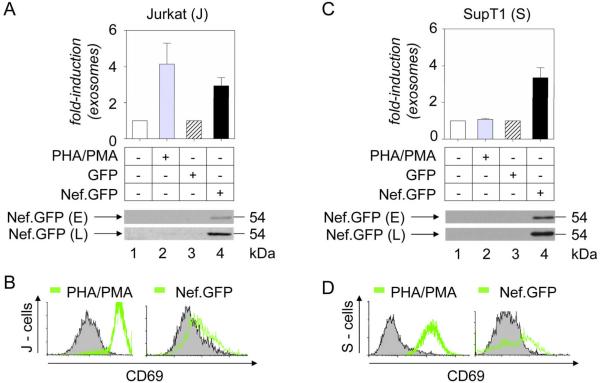
To explore the role of Nef in exosome production in T cells, we expressed Nef.GFP in Jurkat and SupT1 cells, quantified released vesicles and compared these levels to those from untreated, TCR-activated or GFP-expressing samples, as in Fig. 1. Importantly, Nef.GFP increased the production of exosomes up to 4-fold in Jurkat cells when normalized to a GFP sample, similar to effects of treating these cells with a combination of a plant lectin (phytohemagglutinin) and a phorbol ester (phorbol myristyl acetate) (PHA/PMA) (Fig. 3A, bars 1–4). Comparable effects of Nef.GFP were also observed in SupT1 cells (Fig. 3C, bars 3 and 4). In contrast, the addition of PHA/PMA to SupT1 cells, although activating them (Fig. 3D, left panel, green line), had no effect on exosome release (Fig. 3C, bar 2). Whereas PHA/PMA-treated Jurkat and SupT1 cells displayed abundant CD69 activation marker, the expression of Nef had lesser effects in both cells (Figs. 3B and 3D, right panels, green lines). Western blotting of exosomes from Jurkat and SupT1 cells confirmed that they incorporate Nef.GFP efficiently (Figs. 3A and 3C, lane 4). Thus Nef stimulates its own export via the release of exosomes from transformed T cells.
Figure 3.
Nef increases the production of exosomes in transformed T cells. (A and C) Production of exosomes. Measured were amounts of exosomes released from control Jurkat (A) or SupT1 (C) cells (lane 1, white bar), those activated with PHA/PMA (lane 2, grey bar), and those expressing GFP (lane 3, white bar with black stripes) or Nef.GFP (lane 4, black bar). Experiments were conducted and data are presented as in Fig. 1. Results are presented as the mean ±SD of three independent experiments. 10 μg of these exosomal preparations were also analyzed for the presence of Nef with anti-GFP antibodies by western blotting. Capital letters E and L stand for exosome and cell lysate, respectively. (B and D) Activation of manipulated transformed T cells. Activation states of PHA/PMA-treated and Nef-expressing Jurkat (B) or SupT1 (D) cells were analyzed with antibodies to CD69 by FACS. Representative FACS profiles from resting, untransfected (black curve, gray surface) and PHA/PMA-treated or Nef.GFP-expressing (green curve) cells are presented.
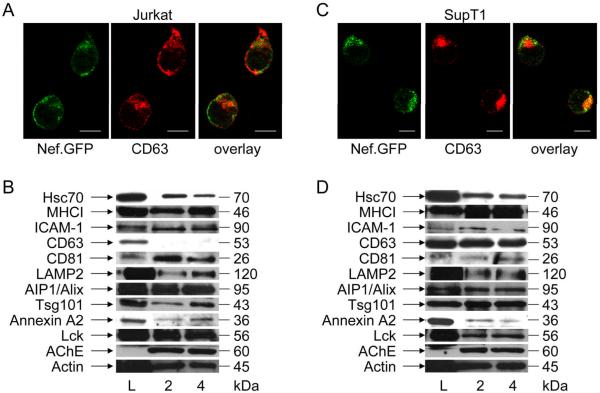
To account for differences in exosomes released from Nef-expressing or PHA/PMA-treated Jurkat and SupT1 cells, we performed additional characterization of exosomes and its producer cells. First, we examined the subcellular localization of Nef by fluorescent microscopy. In Jurkat cells, Nef.GFP localized predominantly to the plasma membrane (Fig. 4A, left panel, green) with some residual staining in the perinuclear region and did not colocalize with CD63 (Fig. 4A, middle and right panels). In contrast, Nef.GFP was found mainly in the perinuclear region in SupT1 cells (Fig. 4C, left panel, green), where it colocalized with CD63 (Fig. 4C, middle and right panels). Western blotting revealed additional differences in exosomal proteins from Jurkat (Fig. 4B) and SupT1 cells (Fig. 4D). We analyzed samples using antibodies to several conserved exosomal proteins: Hsc70, major histocompatibility complex class I (MHCI) determinants, adhesion molecules (ICAM-1), tetraspanins (CD81 and CD63), membrane transport and fusion proteins (LAMP2, AIP1/Alix, Tsg101, annexin A2); signaling proteins (Lck), enzymes (AChE) and cytoskeletal components (actin) (Figs. 4B and 4D). Although we observed similar levels for most examined proteins, Jurkat exosomes were enriched in plasma membrane-associated proteins ICAM-1, CD81 and Lck (Fig. 4B, lanes L, 2 and 4, corresponding to Fig. 4A, top panel, bars 2 and 4), whereas SupT1 exosomes were enriched in late endosomal marker CD63 (Fig. 4D, lanes L, 2 and 4, corresponding to Fig. 4C, top panel, bars 2 and 4) (38). Although both cells produce Nef exosomes, differences in their responses to activation signals and composition of secreted vesicles together with the distinct localization of Nef in producer cells indicate that their biogenesis occurs at different sites in Jurkat and SupT1 cells.
Figure 4.
Nef exosomes are formed at different sites in Jurkat and SupT1 cells. (A and C) Localization of Nef in transformed T cells. Jurkat (A) or SupT1 (C) cells expressing Nef.GFP (green) were stained with anti-CD63 antibodies by indirect immunofluorescence (red). Yellow color represents the overlay. White scale bar represents 10 μm. (B and D) Protein composition of exosomes. Exosomes, which were purified from Jurkat (B) or SupT1 (D) cultures treated with PHA/PMA (lane 2) or cells expressing Nef.GFP (lane 4) were analyzed with antibodies directed against conserved exosomal proteins (Hsc70, MHCI, ICAM-1, CD63, CD81, LAMP2, AIP1/Alix, Tsg101, Annexin A2, Lck, AChE, Actin) by western blotting. Capital L stands for cell lysate.
Nef exosomes form at the plasma membrane in Jurkat and MVBs in SupT cells
The onset of current mass spectrometry-based proteomic technologies has contributed significantly to our understanding of the composition and biogenesis of exosomes. Data from various studies suggest that subsets of cellular proteins are specifically targeted to exosomes (42), which reflect the site of their formation. Based on this information, we performed proteomic profiling of Nef exosomes originating from Jurkat and SupT1 cells.
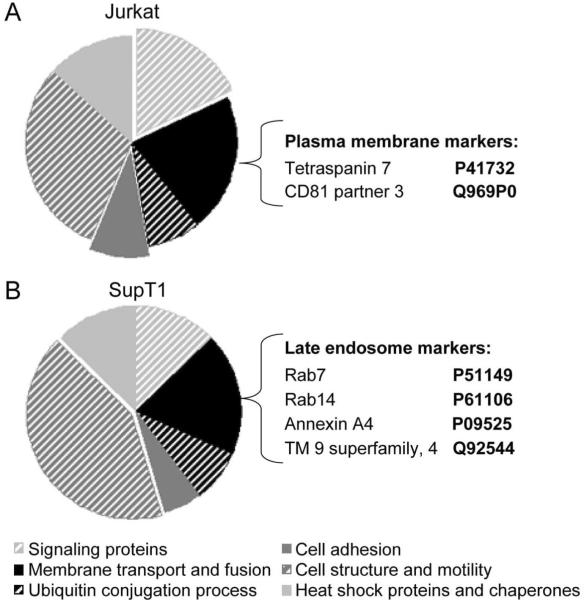
Exosomes from Nef.GFP-expressing Jurkat and SupT1 cells were separated on 12% SDS-PAGE, stained with Coomassie blue dye, excised and trypsinized for analysis by mass spectrometry. Proteins were identified by comparing tandem mass spectra with Swissprot amino acid databases. Background and non-annotated proteins were eliminated from further consideration and the remaining proteins were then analyzed by DAVID (Database for Annotation, Visualization and Integrated Discovery) program, and grouped into different classes (Supplemental Tables 1 and 2). Consistent with previous studies, the following classes of proteins were identified: cell adhesion (integrins), cell structure and motility (actin, tubulin), heat shock and chaperone (Hsc70 and proteins of T complex), ubiquitin conjugation (ubiquitin), membrane transport and fusion (annexins and Rab proteins) and signaling categories (14–33 protein and kinases) (Fig. 5). Groups of metabolic and transcription factors were also identified but were not analyzed further as they are not the focus of this study. Interestingly, about 75 % of all identified proteins were identical between Jurkat and SupT1 exosomes, likely representing a conserved set of proteins that support common exosome functions.
Figure 5.
Exosomes are formed at the plasma membrane and late endosomes in Jurkat and SupT1 cells, respectively. (A and B) Proteomic profiles of Nef exosomes from Jurkat and SupT1 cells. Exosomes were purified from Nef.GFP-expressing Jurkat (A) and SupT1 (B) cultures and analyzed by mass spectrometry. Identified proteins were annotated to different classes as indicated (bottom legend) using DAVID program. Exposed fields depict enriched categories from each sample. Markers specific for plasma membrane or late endosomes are listed beside each diagram.
In addition to these shared constituents, cell type-specific proteins that reflect the site of exosome biogenesis were also identified. While we observed an enrichment of cell adhesion and signaling proteins in exosomes from Jurkat cells (Fig. 5A), the cell structure and motility group was enriched in exosomes from SupT1 cells (Fig. 5B). Similarly, whereas we identified CD81 partner 3 and tetraspanin 7 (43) and other typical plasma membrane proteins in exosomes from Jurkat cells (Fig. 5A), those from SupT1 cells were enriched in proteins specific for late endosomes, which include Rab7, Rab14 (44), annexin A4 (45) and transmembrane 9 superfamily member 4 (Fig. 5B). These data support and extend the observation that the plasma membrane is the site of biogenesis of Nef exosomes as well as those from activated Jurkat cells (32). In contrast, Nef exosomes from SupT1 cells originate from MVBs, similar to what we observed with HeLa.CIITA cells.
Nef exosomes are produced from Nef-expressing and HIV-infected primary T cells
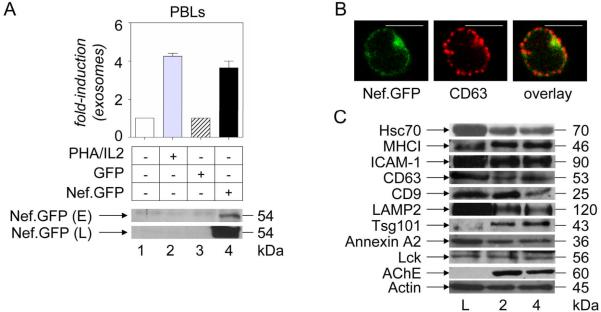
T lymphocytes represent up to 30 % of the white blood cell population. Thus, they likely contribute importantly to the pool of exosomes in the serum (35). One of the known inducers of exosome release from PBLs is T cell activation, shown to promote fusion of the limiting membrane of MVBs with the plasma membrane, resulting in the externalization of exosomes (33). Thus, to explore effects of Nef on exosome production in primary T cells, vesicles released from Nef.GFP-expressing PBLs were quantified and compared to resting, PHA and interleukin 2 (PHA/IL2)-treated or GFP-expressing samples, as in Figs. 1 and 4. Comparable to effects on Jurkat cells (Fig. 3A), the addition of PHA/IL2 or the expression of Nef increased the production of exosomes from PBLs about 4-fold (Fig. 6A, top panel, bars 1–4). Western blotting of purified exosomes again confirmed the presence of Nef.GFP in these secreted vesicles (Fig. 6A, bottom top panel E, lane 4).
Figure 6.
Nef increases the production of exosomes in primary T cells. (A) Evaluation of exosome release. Amounts of exosomes released from control primary T cells (lane 1, white bar), those treated with PHA/IL2 (lane 2, grey bar), and those expressing GFP (lane 3, white bar with black stripes) or Nef.GFP (lane 4, black bar) were assessed as in Figs. 1, 3 and 4. Presented are relative protein values in comparison to resting or untransfected cells. Results are presented as the mean ±SD of three independent experiments. 10 μg of these exosomal preparations were analyzed further for the presence of Nef with anti-GFP antibodies by western blotting. Again, capital E and L stand for exosome and cell lysate, respectively. (B) Subcellular localization of Nef. Primary T cells expressing Nef.GFP (green) were stained with anti-CD63 antibodies by indirect immunofluorescence (red). Yellow represents the overlay. White scale bar represents 10 μm. (C) Protein composition of exosomes. Exosomes from primary T cultures treated with PHA/IL2 (lane 2) or those expressing Nef.GFP (lane 4) were analyzed with antibodies directed against conserved exosomal proteins (Hsc70, MHCI, ICAM-1, CD63, CD9, LAMP2, AIP1/Alix, Tsg101, Annexin A2, Lck, AChE, Actin) by western blotting.
To characterize further exosomes and PBLs, we performed additional fluorescent microscopic and western blotting analyses. When the localization of Nef.GFP in PBLs (Fig. 6B) was compared to that in transformed T cell lines (Figs. 4A and 4C), we observed that Nef is found both at the plasma membrane and in the perinuclear region as in Jurkat and SupT1 cells, respectively. The composition of exosomal proteins from PHA/IL2-treated or Nef.GFP-expressing PBLs (Fig. 6C, lanes L, 2 and 4, corresponding to Fig. 4, top panel, bars 2 and 4) also resembled that of exosomes from transformed T cell lines (Figs. 4B and 4D). They contained several conserved exosomal (Hsc70, MHCI, LAMP2, Tsg101, Annexin A2, Lck, AChE and actin), plasma membrane (ICAM-1 and CD9) and late endosomal (CD63) proteins. Thus, Nef exosomes from PBLs represent a mixture of secreted vesicles of plasma membrane and late endosomal origin. Taken together, these data indicate that Nef increases the production of exosomes from several distinct cellular compartments.
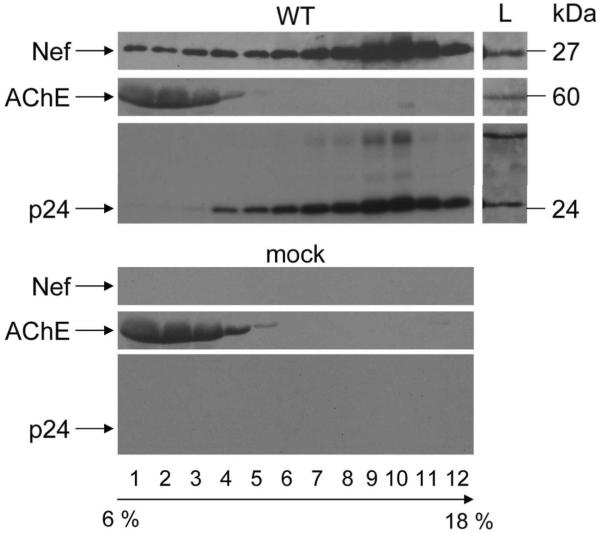
To demonstrate that Nef exosomes are also released from HIV-infected T lymphocytes, we pelleted extracellular supernatants from HIV-1NL4-3 (NL4-3) or mock- infected PBLs, which were enriched from freshly isolated PBMCs, and then subjected them to velocity gradient centrifugation in 6–18% iodixanol (Fig. 7). Optiprep TM velocity gradient centrifugation had been demonstrated to separate and purify exosomes from progeny virions produced in 293T cells, primary CD4+ T cells, macrophages and dendritic cells (46, 47). We collected thirteen fractions and analyzed them with anti-Nef, AChE and p24 (HIV capsid) antibodies by western blotting. The examination of proteins from NL4-3 sample (Fig. 7, top panel) revealed that although a major part of Nef sediments in fractions 6 to 12, corresponding to 10.8–18% iodixanol, a significant amount of Nef is also found in the top fractions, corresponding to 6–9.6% iodixanol. These top fractions also stained for AChE, indicating the presence of exosomes, whereas p24 and thus HIV was found only in 10.8–18% iodixanol fractions. The exclusion of p24 in 6–9.6% iodixanol fractions also demonstrated the purity of these Nef exosomes. Thus, in HIV-infected primary T lymphocytes, a significant amount of Nef is also released in exosomes.
Figure 7.
Nef exosomes are released from HIV-infected primary T cells. Supernatants from PBLs infected with NL4-3 (WT) or left uninfected (Mock) were first pelleted and then subjected to velocity gradient centrifugation in 6 –18% iodixanol. Fractions were collected from the top of the gradient, as indicated. Protein profiles were examined with anti-Nef, AChE and p24 antibodies by western blotting. Capital L stands for cell lysate.
Nef exosomes cause activation-induced cell death of resting CD4+ T lymphocytes
Nef not only increases the production of exosomes but is also exported in all cells examined. These numerous Nef exosomes might exert biological effects similar to those described for the free Nef protein added to cells. Among them, cellular cytotoxicity (4, 22) is the most important, as it could be involved in the extensive loss of CD4+ T cells, which is the hallmark of AIDS (1).
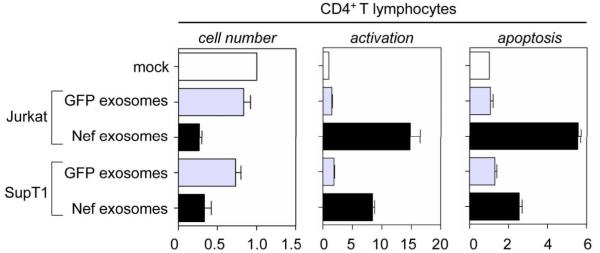
To examine this hypothesis, PBLs were incubated with equal amounts of exosomes isolated from GFP-or Nef.GFP-expressing Jurkat or SupT1 cells. Two days later, effects of exosomes on their proliferation, activation and apoptosis were examined and compared to a control sample, in which cells were co-incubated with supernatants from untransfected cells. Counts of viable cells demonstrated that Nef exosomes from Jurkat and SupT1 cells caused 70 % and 60 % reduction in cell numbers, respectively (Fig. 6, left panel, black bars). In comparison, we observed only 10 % and 25 % fall in cell counts for control exosomes (GFP) from these cells, respectively (Fig. 6, left panel, grey bars). We observed the inverse when we measured the activation of CD4+ T lymphocytes with anti-CD69 antibodies by FACS. Relative to the control, Nef exosomes from Jurkat and SupT1 cells activated these bystander cells up to 18-fold and 10-fold, respectively (Fig. 6, middle panel, black bars). Importantly control exosomes (GFP) from these cells had negligible effects on T cell activation (Fig. 6, middle panel, grey bars). Finally, we examined bystander CD4+ cells for apoptosis by measuring surface-bound annexinV-Alexa488, which generated a profile analogous to that obtained for T cell activation. Whereas Nef exosomes from Jurkat and SupT1 caused up to 6- and 3-fold increased apoptosis, respectively, control exosomes (GFP) again had only negligible effects (Fig. 6, right panel, black and grey bars). We conclude that Nef exosomes from T cells cause activation-induced cell death of target resting CD4+ T lymphocytes.
Discussion
In this study, we found that Nef stimulates its own export through the production of exosomes at the plasma membrane and from intracellular organelles. This role of Nef was conserved in all cells examined, be they transformed of primary, and was confirmed in HIV-infected T lymphocytes. Importantly, regardless of their origin within cells, as long as they contained Nef, these vesicles caused activation-induced cell death of resting CD4+ T lymphocytes, which is a hallmark of AIDS.
Although Nef is found abundantly in the serum of HIV-infected individuals (22), how it exits cells has remained largely unexplored. Nevertheless, an early study of Nef in yeast demonstrated that it could be pelleted in the culture medium, implying its association with micelles (48). This finding is supported by other studies, where Nef affected intracellular trafficking at multiple levels (11), leading to the proliferation of endosomes, lysosomes (26), fat granules (27) and multivesicular bodies (MVBs) (28, 29). Indeed, its effect on proliferation of MVBs could contribute to the egress of viral particles from infected cells (29). Thus, Nef should also lead to the release of exosomes in the absence of other viral proteins, a hypothesis, which is supported by this study.
The expression of Nef in HeLa.CIITA cells namely increased the production of secreted vesicles. They had all characteristics of exosomes, such as a diameter of up to 100 nm, a cup-shape structure and they stained positive for several exosomal markers (CD63, AIP1/Alix and AChE). Interestingly, fluorescent microscopic and western blotting analyses revealed that Nef is present in these secreted vesicles. Thus, Nef not only induces the release of exosomes from HeLa.CIITA cells, but is also incorporated in them, thereby controlling its own export from cells. Key to this function is the targeting of Nef to membranes, which was revealed by the failure of the myristoylation-defective mutant Nef protein to have any effect in these cells. This myristoylation residue is also known to be critical for all other effects of Nef in cells (49).
This phenotype of Nef was conserved in transformed and primary T lymphocytes as well as in HIV-infected PBLs. In the later scenario, a major part of Nef was still associated with virions, jet a significant share was also exported with exosomes. To separate exosomes from virions, we used an established method based on fractionation on an Optiprep TM velocity gradient (46, 47). For studying effects of Nef expression in primary cells, nucleofection was used as the gene delivery method, for which nonspecific effects on T cell activation have been excluded (50). This is important, as in addition to Nef, the release of exosomes is also induced by T cell activation in Jurkat and primary T cells (31, 32). We confirmed this by the present study, whereas only Nef but not the addition of PHA/PMA had an effect on exosome production in SupT1 cells. Thus, the effect of Nef on the production of exosomes seems to be more ubiquitous than that of T cell activation.
Importantly, Nef increased the production of exosomes from all cells examined, despite differences in their biogenesis, as implicated by the mass spectrometry data. These differences can be explained by the localization of Nef in these cells (16). At the plasma membrane, Nef interferes with signaling proteins in the TCR complex, including TCR ζ (23), and triggers a transcriptional program that mimics T cell activation (13). Additionally, Nef aggregates Lck and TCR ζ in lipid rafts, imitating a characteristic feature of activated CD4+ T cells (51). Indeed, these interactions could contribute to the biogenesis and release of exosomes in Jurkat cells, where Nef is concentrated at the plasma membrane. In contrast, in SupT1 and Hela.CIITA cells, Nef is localized to intracellular organelles and the perinuclear region. In this context, Nef interacts with trafficking intermediates, components of intracellular organelles and different PI3K isoforms (11, 14), which leads to the proliferation of MVBs, intraluminal vesicles and their egress from cells. Taken together, the multifaceted nature of Nef allows it to stimulate the biogenesis of exosomes on multiple surfaces inside cells. Importantly, regardless of their biogenesis, as long as they contain Nef, these exosomes exert the same effects on bystander cells.
How are these effects achieved? First, Nef exosomes are internalized by and confer new properties to target cells. This phenotype was first observed with our exosome effector assay where Nef.Tat.GFP transported in exosomes was released into target cells and activated the HIV LTR. Exosomes were purified extensively before they were incubated with target cells, thus excluding contributions of other contaminants. Importantly, Tat.GFP had no effect in this assay. Moreover, Nef.Tat.GFP expressed in cells or added exogenously in exosomes had similar effects on the HIV LTR. Next, when Nef exosomes were tested for its effect on resting T cells, they were able to activate resting PBLs and cause activation induced cell death. Most likely, these Nef exosomes enter cells via endocytosis, which had been reported in other systems. After acidification and fusion of these early endosomes, incorporated proteins then enter the cytosol and are targeted to appropriate compartments (52).
Most recent work on exosomes focused on elucidating the role of these bioactive vesicles in promoting intercellular communications. Studies on exosomes released from macrophages infected with various intracellular pathogens suggested a function in immune surveillance (36). Similarly, we show that Nef exosomes cause activation-induced cell death of resting CD4+ T lymphocytes in vitro. This effect depended on the presence of Nef in these vesicles, as no effect was observed for exosomes lacking Nef. Cytotoxicity likely resulted from improper T cell activation (15), which lacked appropriate co-stimulation and/or growth conditions. The involvement of soluble Nef in this process was already suggested in previous studies (22, 23), but the novelty of our observation lies in exosome association of extracellular Nef. The single membrane surrounding Nef should not only facilitate its export from and import into cells but also increase its stability in physiological fluids. Depending on specific physiological conditions, Nef exosomes could also activate directly bystander PBLs for de novo HIV infection. In any case, this bystander effect might have important implications for the gradual and progressive loss of CD4+ T cells, which is a hallmark of AIDS.
Materials and methods
Plasmid constructions
The HIV-1 SF2 Nef.GFP wild type and NefG2A.GFP mutant expression vectors are based on pEGFP-N1 plasmid (Clontech-Takara Bio Company, Madison, WI) (50). For the expression of the hybrid Nef.Tat.GFP protein, a four aminoacid linker sequence GSGM was added to the 5' end of the Tat HIV-1 SF2 ORF by PCR amplification (5'-GTACTGCAGGGCTCCGGCATGATGGAGCCAGTAGATCC TA -3', 5'-GTACCGCGGGTACCGCGGTTCCGTGGGCCCT GTCGGGT -3') and the product was inserted into the PstI/SacII restriction sites of the previously described Nef.GFP vector downstream from the nef gene. Similarly the tat gene was inserted into PstI/SacII restriction sites of the pEGFP-N1 vector for expression of Tat.GFP protein. All expression plasmids used in this study were confirmed by sequencing. For virus stock preparations, HIV-1 NL4-3 provirus plasmid was used (obtained through AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH).
Cells and transfections
Hela.CIITA and TZM-bl cells were grown in DMEM with 10% FBS and antibiotics and transfections performed using FuGENE 6 (Roche, Basel, Switzerland). SupT1 cells were grown in RPMI 1640 medium with 10% FBS, antibiotics and L-glutamine. Cells were electroporated using a BioRad electroporator (BioRad USA Life Sciences, Hercules, CA) as follows: 1 × 107 cells in the presence of 10 μg of pDNA, electroporated at 950 μ-F and 250 or 230 V, respectively. When appropriate, Jurkat and SupT1 cells were activated with 5 μg /ml phytohaemagglutinin P (PHA) and 10 ng /ml or 50 ng /ml PMA for 16 hours, respectively (both Sigma; Sigma-Aldrich, St. Louis, MO). PBMCs were isolated from buffy coats of healthy HIV negative donors in a ficollhypaque density gradient (Pharmacia; GE Healthcare, Uppsala, Sweden). PBMCs were then plated at 5 × 106 cells per ml in complete RPMI 1640 (containing penicillin (100 IU/ml), streptomycin (100 μg /ml) and 10% FBS) and after overnight incubation, non-adherent cells were isolated. Washed, freshly isolated cells were transiently transfected using the Human T cell Nucleofector kit (Amaxa; Lonza Company, Cologne, Germany), according to the manufacturer's instructions: 107 cells in the presence of 5 μg plasmid DNA, nucleofection program V-24. Immediately after nucleofection, cells were transferred into pre-warmed complete RPMI 1640. For activation, PBMCs were grown in complete RPMI 1640 with 25 U/ml IL2 and incubated with 2 μg /ml PHA for 16 h.
Virus stocks for infection experiments were generated by transfection of proviral HIV plasmids into 293T cells as described (29). Briefly, three days after transfection, precleared culture supernatants were filtered through a 0.22 μm-pore-size filter (Millipore, Bedford, MA), pelleted by ultracentrifugation through a 20% sucrose cushion at 100000 × g for 1.5 h and resuspended in PBS overnight at 4 °C. The HIV-1 p24 content of concentrated stocks was determined by a p24 antigen enzyme-linked immunosorbent assay (PerkinElmer, Waltham, MA).
Exosome purification and immunoblotting
Exosomes were purified as described previously (39). Briefly, cells were cultured in complete medium depleted of contaminating vesicles and protein aggregates by overnight centrifugation at 100,000 × g. Untreated, transfected or activated cells were removed by centrifugation at 2,000 × g for 10 min followed by passage of the supernatant through a 0, 22 μm filter. The filtrate was then ultracentrifuged at 100,000 × g for 1 h at 4 °C, pellet collected and washed in PBS by several repeated ultracentrifugation steps. Final pellet was resuspended in 50 μl of lysis buffer with protease inhibitors (Sigma) and protein concentration measured with BCA Protein Assay Kit (Pierce, Thermo Fisher Scientific, Waltham, MA). The quantity of the released exosomes was assessed by measuring total amount of exosomal proteins per 1 × 107 manipulated cells. For use in biological assays, protocol for exosome preparation was modified by exchanging the pelleting of supernatants with concentrating the supernatants in amicon tubes (53).
For immunoblotting, ten μg of total exosomal proteins or 30 μg of cell lysates were separated by 12 % SDS-PAGE and transferred to nitrocellulose membrane (Amersham; GE Healthcare). Goat polyclonal antibodies to Hsc70 (sc-1059, Santa Cruz Biotechnology, Santa Cruz, CA.), annexin A2 (sc-1924) and actin (sc-1615); or rabbit polyclonal to AIP1/Alix (kindly provided by W. Sundquist), calnexin (sc-11397), CD63 (sc-15363), ICAM-1 (sc-7891), Lck (sc-28882) and MHC I (sc-25619); or mouse monoclonal to acetylcholinesterase (AChE, MAB303, Millipore), CD9 (sc-13118), CD81 (sc-7637), cytochrome c (Pharmingen; BD Biosciences, Franklin Lakes, NJ), GFP (sc-9996), HIV-1 p24 (ab9071, Abcam, Cambridge, MA), LAMP2 (sc-18822), HIV-1 Nef (3689, AIDS Research and Reference Reagent Program), and tsg101 (sc-7964) were used as the primary; and appropriate HRP-conjugated anti-goat, anti-rabbit or anti-mouse (Santa Cruz Biotechnology) were used as the secondary antibodies. Membranes were developed by ECL Western Detection reagent (Amersham).
Separation and purification of exosomes and HIV-1 virions
Exosomes and HIV-1 virions, produced by acute infection of primary human CD4+ T cells, were separated as described previously (46). Briefly, freshly isolated PBMCs were activated with 5 μg/ml PHA-P and 10U/ml IL2 for 72 h and later inoculated with HIV-1 virus stocks (500 ng p24/1 ×107 cells) for 2 h at 1200 × g and 24 °C. After removing excess virus by several washing steps, cells were resusupended in fresh complete RPMI with 10U/ml IL2 at concentration 1 × 106 cells/ml. Supernatant was collected after 7 days, filtered through 0, 22 μm pore filter and pelleted at 100 000 × g for 1 h at 4 °C. Pellet containing exosomes and virions was resuspended in 500 μl of PBS and centrifuged for 1.5h at 250 000 × g and 4 °C through a 6–18% OptiprepTM velocity (60% [wt/vol] iodixanol, Sigma Aldrich) gradient, as described in Dettenhofer and Yu (47). Fractions collected from the top of the gradient were precipitated with trichloroacetic acid, resuspended in 1 × loading dye and subjected to immunoblot analysis.
Mass spectrometry (MS) analysis
50 μg of exosomal proteins purified from Nef.GFP-expressing Jurkat or SupT1 cultures were separated on 12% SDS-PAGE, the Coomassie-stained protein bands excised from the gel, trypsin digested and analysed on an LTQOrbitrap (Thermo Fisher Scientific). Fractions were separated on a 100μm × 10cm C18 column at flow rate 350 nl/min over a 60 min HPLC run. MS spectra were captured using one survey scan in FT at 30,000 resolution and 6 MS/MS events in the ion trap. Proteins were identified using Protein Prospector (54) against the SwissProt database (SwissProt.2007.04.19; 264492 entries). The database search parameters were as follows: number of missed cleavages 1; variable modifications included oxidation of methionine, N-terminal protein acetylation, pyroglutamate formation from N-terminal glutamine residues, modification of cysteine by carbamidomethylation; mass tolerance for precursor ions 20 ppm; mass tolerance for fragment ions 0.8 Da; threshold for peptide score 15; maximum E-value for accepting individual MS/MS spectra 0.01. The identified proteins were grouped into different protein classes by a DAVID (Database for Annotation, Visualization and Integrated Discovery) program (http://david.abcc.ncifcrf.gov/home.jsp).
Microscopy
For fluorescence microscopy, cells were fixed with 4% PFA in 1× PBS (pH 7.2), for 20 min and washed twice in PBS, all at 20–25 °C. Then cells were permeabilized with 1% Triton X-100 in 1× PBS for 20 min, followed by three washes in 1× PBS with 1 mg/ml BSA. Next, cells were sequentially incubated with diluted mouse antibodies to GFP (sc-9996), CD63 (Pharmingen), and appropriate anti-mouse Alexa Fluor 488 and 568 or anti-goat Alexa Fluor 488 labeled secondary antibodies (Molecular Probes, Invitrogen, Carlsbad, California). Samples were washed extensively in PBS and placed in the mounting medium with DAPI (Molecular probes). Images were obtained using a LCA ZEISS LSM 510 confocal fluorescence microscope, with a 63× NA 1.4 lens and LSM image browser imaging software (Carl Zeiss, Inc., Oberkochen, Germany). Images presented are representative of three independent experiments. For fluorescence microscopy of exosomes, the 100 000 × g pellet was resuspended in PBS, applied to poly-L-lysine coated coverslips and imaged as described, using a direct fluorescence of the fusion protein. Digital images were adjusted for brightness and contrast using Photoshop.
To image exosomes by electron microscope, pellets obtained after 100,000g centrifugation were fixed in 2% PFA and negatively stained by uranyl formate following the established protocol (55). All samples were imaged at room temperature in a Tecnai T12 microscope (FEI Company, USA) and images recorded on a 4K × 4K UltraScan CCD camera (Gatan Inc, USA).
Luciferase activity measurements
TZM-bl cells were untreated, transfected with GFP, Nef.GFP, Nef.Tat.GFP and Tat.GFP plasmids or incubated with exosomes purified from GFP, Nef.GFP, Nef.Tat.GFP and Tat.GFP transfected HeLa.CIITA cultures, respectively. After 48 hours they were lysed in 200 μl luciferase reporter lysis buffer (Promega, Madison, WI). After centrifugation, cell debris was discarded and 20 μl of the lysis solution was used for analysis using a luminometer (EG&G Berthold; Berthold Technologies, Bad Wilbad, Germany). Luciferase activity is expressed as relative light units per 10 μg of total cell protein, normalized to the mock sample.
Flow cytometry and viable cell count
For measuring the transfection efficiency, cells were analysed by FACS (Facs Calibur, BD Biosciences) using direct fluorescence of the GFP fusion protein as a signal. Jurkat and SupT1 activation status was assessed by analyzing anti-CD69-PE (Pharmingen) stained cells, while in the case of primary lymphocytes, cells were first gated for CD4-APC (Pharmingen) signal. For measuring apoptosis, CD4-APC gated cells were stained with propidium iodide and annexin V conjugated to Alexa Fluor 488 (both Molecular Probes), according to the manufacturer's instructions. Cell viability was determined by trypan blue (Sigma) exclusion and hematocytometer cell density measurement.
Supplementary Material
Figure 8.
Nef exosomes cause activation-induced cell death of resting CD4+ T lymphocytes. Freshly isolated PBLs were incubated with exosomes isolated from Jurkat or SupT1 cells expressing GFP (grey bars, lane 2 and 4) or Nef.GFP (black bars, lane 3 and 5). PBLs, which were incubated with supernatants of untransfected cells served as the control (white bar, lane 1). After 40 h incubation, numbers of viable cells were determined for each sample. CD4+ T cells were then examined with anti-CD69 antibodies and Alexa488-conjugated antibodies to AnnexinV for activation and apoptosis, respectively. Presented are relative cell numbers compared to untreated cells. Results are presented as the mean ±SD of three independent experiments.
Acknowledgements
We thank members of the Peterlin laboratory for helpful advice and discussions. Mass spectrometry was provided by the Bio-Organic Biomedical Mass Spectrometry Resource at UCSF (A.L. Burlingame, Director) supported by the Biomedical Research Technology Program of the NIH National Center for Research Resources, NIH NCRR P41RR001614. The work was supported with funds from HARC Center (NIH P50 GMO82250).
Abbreviations
- AChE
acetylcholinesterase
- AIDS
acquired immunodeficiency syndrome
- AP
adaptor protein
- FACS
fluorescence- activated cell sorter
- HAART
highly active antiretroviral therapy
- HIV
human immunodeficiency virus
- LTR
long terminal repeat
- MS
mass spectrometry
- MVB
multivesicular bodies
- Nef
negative factor
- PBL
peripheral blood lymphocyte
- PHA
phytohaemagglutinin
- TCR
T cell antigen receptor
Footnotes
Online supplemental material Tables S1 and S2 list all Jurkat or SupT1 mass spectrometry identified exosomal proteins included in the data analysis, respectively.
References
- 1.Shearer GM. HIV-induced immunopathogenesis. Immunit. 1998;9(5):587–593. doi: 10.1016/s1074-7613(00)80656-1. [DOI] [PubMed] [Google Scholar]
- 2.Finkel TH, Tudor-Williams G, Banda NK, Cotton MF, Curiel T, Monks C, Baba TW, Ruprecht RM, Kupfer A. Apoptosis occurs predominantly in bystander cells and not in productively infected cells of HIV- and SIV-infected lymph nodes. Nat Med. 1995;1(2):129–134. doi: 10.1038/nm0295-129. [DOI] [PubMed] [Google Scholar]
- 3.Alimonti JB, Ball TB, Fowke KR. Mechanisms of CD4+ T lymphocyte cell death in human immunodeficiency virus infection and AIDS. J Gen Virol. 2003;84(Pt 7):1649–1661. doi: 10.1099/vir.0.19110-0. [DOI] [PubMed] [Google Scholar]
- 4.James CO, Huang MB, Khan M, Garcia-Barrio M, Powell MD, Bond VC. Extracellular Nef protein targets CD4+ T cells for apoptosis by interacting with CXCR4 surface receptors. J Virol. 2004;78(6):3099–3109. doi: 10.1128/JVI.78.6.3099-3109.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Homann S, Tibroni N, Baumann I, Sertel S, Keppler OT, Fackler OT. Determinants in HIV-1 Nef for enhancement of virus replication and depletion of CD4+ T lymphocytes in human lymphoid tissue ex vivo. Retrovirology. 2009;6:6. doi: 10.1186/1742-4690-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schindler M, Munch J, Kutsch O, Li H, Santiago ML, Bibollet-Ruche F, Muller-Trutwin MC, Novembre FJ, Peeters M, Courgnaud V, Bailes E, Roques P, Sodora DL, Silvestri G, Sharp PM, et al. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell. 2006;125(6):1055–1067. doi: 10.1016/j.cell.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, Crumpacker CS. Human immunodeficiency virus type 1 RNA in peripheral blood mononuclear cells of patients receiving prolonged highly active antiretroviral therapy. J Infect Dis. 2001;184(10):1341–1344. doi: 10.1086/324002. [DOI] [PubMed] [Google Scholar]
- 8.Kestler HW, 3rd, Ringler DJ, Mori K, Panicali DL, Sehgal PK, Daniel MD, Desrosiers RC. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65(4):651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 9.Dyer WB, Geczy AF, Kent SJ, McIntyre LB, Blasdall SA, Learmont JC, Sullivan JS. Lymphoproliferative immune function in the Sydney Blood Bank Cohort, infected with natural nef/long terminal repeat mutants, and in other long-term survivors of transfusion-acquired HIV-1 infection. Aids. 1997;11(13):1565–1574. doi: 10.1097/00002030-199713000-00004. [DOI] [PubMed] [Google Scholar]
- 10.Jamieson BD, Aldrovandi GM, Planelles V, Jowett JB, Gao L, Bloch LM, Chen IS, Zack JA. Requirement of human immunodeficiency virus type 1 nef for in vivo replication and pathogenicity. J Virol. 1994;68(6):3478–3485. doi: 10.1128/jvi.68.6.3478-3485.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roeth JF, Collins KL. Human immunodeficiency virus type 1 Nef: adapting to intracellular trafficking pathways. Microbiol Mol Biol Rev. 2006;70(2):548–563. doi: 10.1128/MMBR.00042-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi M, Aiken C. Nef enhances HIV-1 infectivity via association with the virus assembly complex. Virology. 2008;373(2):287–297. doi: 10.1016/j.virol.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simmons A, Aluvihare V, McMichael A. Nef triggers a transcriptional program in T cells imitating single-signal T cell activation and inducing HIV virulence mediators. Immunity. 2001;14(6):763–777. doi: 10.1016/s1074-7613(01)00158-3. [DOI] [PubMed] [Google Scholar]
- 14.Geyer M, Fackler OT, Peterlin BM. Structure--function relationships in HIV-1 Nef. EMBO Rep. 2001;2(7):580–585. doi: 10.1093/embo-reports/kve141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tai XG, Toyooka K, Yashiro Y, Abe R, Park CS, Hamaoka T, Kobayashi M, Neben S, Fujiwara H. CD9-mediated costimulation of TCR-triggered naive T cells leads to activation followed by apoptosis. J Immunol. 1997;159(8):3799–3807. [PubMed] [Google Scholar]
- 16.Baur AS, Sawai ET, Dazin P, Fantl WJ, Cheng-Mayer C, Peterlin BM. HIV-1 Nef leads to inhibition or activation of T cells depending on its intracellular localization. Immunity. 1994;1(5):373–384. doi: 10.1016/1074-7613(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 17.Geleziunas R, Xu W, Takeda K, Ichijo H, Greene WC. HIV-1 Nef inhibits ASK1-dependent death signalling providing a potential mechanism for protecting the infected host cell. Nature. 2001;410(6830):834–838. doi: 10.1038/35071111. [DOI] [PubMed] [Google Scholar]
- 18.Wolf D, Witte V, Laffert B, Blume K, Stromer E, Trapp S, d'Aloja P, Schurmann A, Baur AS. HIV-1 Nef associated PAK and PI3-kinases stimulate Akt-independent Bad-phosphorylation to induce anti-apoptotic signals. Nat Med. 2001;7(11):1217–1224. doi: 10.1038/nm1101-1217. [DOI] [PubMed] [Google Scholar]
- 19.Greenway AL, McPhee DA, Allen K, Johnstone R, Holloway G, Mills J, Azad A, Sankovich S, Lambert P. Human immunodeficiency virus type 1 Nef binds to tumor suppressor p53 and protects cells against p53-mediated apoptosis. J Virol. 2002;76(6):2692–2702. doi: 10.1128/JVI.76.6.2692-2702.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexander L, Du Z, Rosenzweig M, Jung JU, Desrosiers RC. A role for natural simian immunodeficiency virus and human immunodeficiency virus type 1 nef alleles in lymphocyte activation. J Virol. 1997;71(8):6094–6099. doi: 10.1128/jvi.71.8.6094-6099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu XN, Laffert B, Screaton GR, Kraft M, Wolf D, Kolanus W, Mongkolsapay J, McMichael AJ, Baur AS. Induction of Fas ligand expression by HIV involves the interaction of Nef with the T cell receptor zeta chain. J Exp Med. 1999;189(9):1489–1496. doi: 10.1084/jem.189.9.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujii Y, Otake K, Tashiro M, Adachi A. Soluble Nef antigen of HIV-1 is cytotoxic for human CD4+ T cells. FEBS Lett. 1996;393(1):93–96. doi: 10.1016/0014-5793(96)00859-9. [DOI] [PubMed] [Google Scholar]
- 23.Okada H, Takei R, Tashiro M. HIV-1 Nef protein-induced apoptotic cytolysis of a broad spectrum of uninfected human blood cells independently of CD95(Fas) FEBS Lett. 1997;414(3):603–606. doi: 10.1016/s0014-5793(97)01080-6. [DOI] [PubMed] [Google Scholar]
- 24.Qiao X, He B, Chiu A, Knowles DM, Chadburn A, Cerutti A. Human immunodeficiency virus 1 Nef suppresses CD40-dependent immunoglobulin class switching in bystander B cells. Nat Immunol. 2006;7(3):302–310. doi: 10.1038/ni1302. [DOI] [PubMed] [Google Scholar]
- 25.Madrid R, Janvier K, Hitchin D, Day J, Coleman S, Noviello C, Bouchet J, Benmerah A, Guatelli J, Benichou S. Nef-induced alteration of the early/recycling endosomal compartment correlates with enhancement of HIV-1 infectivity. J Biol Chem. 2005;280(6):5032–5044. doi: 10.1074/jbc.M401202200. [DOI] [PubMed] [Google Scholar]
- 26.Sanfridson A, Hester S, Doyle C. Nef proteins encoded by human and simian immunodeficiency viruses induce the accumulation of endosomes and lysosomes in human T cells. Proc Natl Acad Sci U S A. 1997;94(3):873–878. doi: 10.1073/pnas.94.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mujawar Z, Rose H, Morrow MP, Pushkarsky T, Dubrovsky L, Mukhamedova N, Fu Y, Dart A, Orenstein JM, Bobryshev YV, Bukrinsky M, Sviridov D. Human immunodeficiency virus impairs reverse cholesterol transport from macrophages. PLoS Biol. 2006;4(11):e365. doi: 10.1371/journal.pbio.0040365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stumptner-Cuvelette P, Jouve M, Helft J, Dugast M, Glouzman AS, Jooss K, Raposo G, Benaroch P. Human immunodeficiency virus-1 Nef expression induces intracellular accumulation of multivesicular bodies and major histocompatibility complex class II complexes: potential role of phosphatidylinositol 3-kinase. Mol Biol Cell. 2003;14(12):4857–4870. doi: 10.1091/mbc.E03-04-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costa LJ, Chen N, Lopes A, Aguiar RS, Tanuri A, Plemenitas A, Peterlin BM. Interactions between Nef and AIP1 proliferate multivesicular bodies and facilitate egress of HIV-1. Retrovirology. 2006;3:33. doi: 10.1186/1742-4690-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16(4):415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 31.Blanchard N, Lankar D, Faure F, Regnault A, Dumont C, Raposo G, Hivroz C. TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/zeta complex. J Immunol. 2002;168(7):3235–3241. doi: 10.4049/jimmunol.168.7.3235. [DOI] [PubMed] [Google Scholar]
- 32.Booth AM, Fang Y, Fallon JK, Yang JM, Hildreth JE, Gould SJ. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J Cell Biol. 2006;172(6):923–935. doi: 10.1083/jcb.200508014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fomina AF, Deerinck TJ, Ellisman MH, Cahalan MD. Regulation of membrane trafficking and subcellular organization of endocytic compartments revealed with FM1-43 in resting and activated human T cells. Exp Cell Res. 2003;291(1):150–166. doi: 10.1016/s0014-4827(03)00372-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simons M, Raposo G. Exosomes - vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009 doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 35.Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17(7):879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 36.Schorey JS, Bhatnagar S. Exosome function: from tumor immunology to pathogen biology. Traffic. 2008;9(6):871–881. doi: 10.1111/j.1600-0854.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolf D, Giese SI, Witte V, Krautkramer E, Trapp S, Sass G, Haller C, Blume K, Fackler OT, Baur AS. Novel (n)PKC kinases phosphorylate Nef for increased HIV transcription, replication and perinuclear targeting. Virology. 2008;370(1):45–54. doi: 10.1016/j.virol.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 38.Pols MS, Klumperman J. Trafficking and function of the tetraspanin CD63. Exp Cell Res. 2008 doi: 10.1016/j.yexcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 39.Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166(12):7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 40.Peretti S, Schiavoni I, Pugliese K, Federico M. Cell death induced by the herpes simplex virus-1 thymidine kinase delivered by human immunodeficiency virus-1-based virus-like particles. Mol Ther. 2005;12(6):1185–1196. doi: 10.1016/j.ymthe.2005.06.474. [DOI] [PubMed] [Google Scholar]
- 41.Varin A, Manna SK, Quivy V, Decrion AZ, Van Lint C, Herbein G, Aggarwal BB. Exogenous Nef protein activates NF-kappa B, AP-1, and c-Jun N-terminal kinase and stimulates HIV transcription in promonocytic cells. Role in AIDS pathogenesis. J Biol Chem. 2003;278(4):2219–2227. doi: 10.1074/jbc.M209622200. [DOI] [PubMed] [Google Scholar]
- 42.Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8(19):4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 43.Tarrant JM, Robb L, van Spriel AB, Wright MD. Tetraspanins: molecular organisers of the leukocyte surface. Trends Immunol. 2003;24(11):610–617. doi: 10.1016/j.it.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 44.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2(2):107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 45.Gruenberg J. The endocytic pathway: a mosaic of domains. Nat Rev Mol Cell Biol. 2001;2(10):721–730. doi: 10.1038/35096054. [DOI] [PubMed] [Google Scholar]
- 46.Cantin R, Diou J, Belanger D, Tremblay AM, Gilbert C. Discrimination between exosomes and HIV-1: purification of both vesicles from cell-free supernatants. J Immunol Methods. 2008;338(1–2):21–30. doi: 10.1016/j.jim.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 47.Dettenhofer M, Yu XF. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J Virol. 1999;73(2):1460–1467. doi: 10.1128/jvi.73.2.1460-1467.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macreadie IG, Castelli LA, Lucantoni A, Azad AA. Stress- and sequence-dependent release into the culture medium of HIV-1 Nef produced in Saccharomyces cerevisiae. Gene. 1995;162(2):239–243. doi: 10.1016/0378-1119(95)00316-x. [DOI] [PubMed] [Google Scholar]
- 49.Giese SI, Woerz I, Homann S, Tibroni N, Geyer M, Fackler OT. Specific and distinct determinants mediate membrane binding and lipid raft incorporation of HIV-1(SF2) Nef. Virology. 2006;355(2):175–191. doi: 10.1016/j.virol.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 50.Keppler OT, Tibroni N, Venzke S, Rauch S, Fackler OT. Modulation of specific surface receptors and activation sensitization in primary resting CD4+ T lymphocytes by the Nef protein of HIV-1. J Leukoc Biol. 2006;79(3):616–627. doi: 10.1189/jlb.0805461. [DOI] [PubMed] [Google Scholar]
- 51.Djordjevic JT, Schibeci SD, Stewart GJ, Williamson P. HIV type 1 Nef increases the association of T cell receptor (TCR)-signaling molecules with T cell rafts and promotes activation-induced raft fusion. AIDS Res Hum Retroviruses. 2004;20(5):547–555. doi: 10.1089/088922204323087804. [DOI] [PubMed] [Google Scholar]
- 52.Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009;137(3):433–444. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lamparski HG, Metha-Damani A, Yao JY, Patel S, Hsu DH, Ruegg C, Le Pecq JB. Production and characterization of clinical grade exosomes derived from dendritic cells. J Immunol Methods. 2002;270(2):211–226. doi: 10.1016/s0022-1759(02)00330-7. [DOI] [PubMed] [Google Scholar]
- 54.Chalkley RJ, Baker PR, Huang L, Hansen KC, Allen NP, Rexach M, Burlingame AL. Comprehensive analysis of a multidimensional liquid chromatography mass spectrometry dataset acquired on a quadrupole selecting, quadrupole collision cell, time-of-flight mass spectrometer: II. New developments in Protein Prospector allow for reliable and comprehensive automatic analysis of large datasets. Mol Cell Proteomics. 2005;4(8):1194–1204. doi: 10.1074/mcp.D500002-MCP200. [DOI] [PubMed] [Google Scholar]
- 55.Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.