Abstract
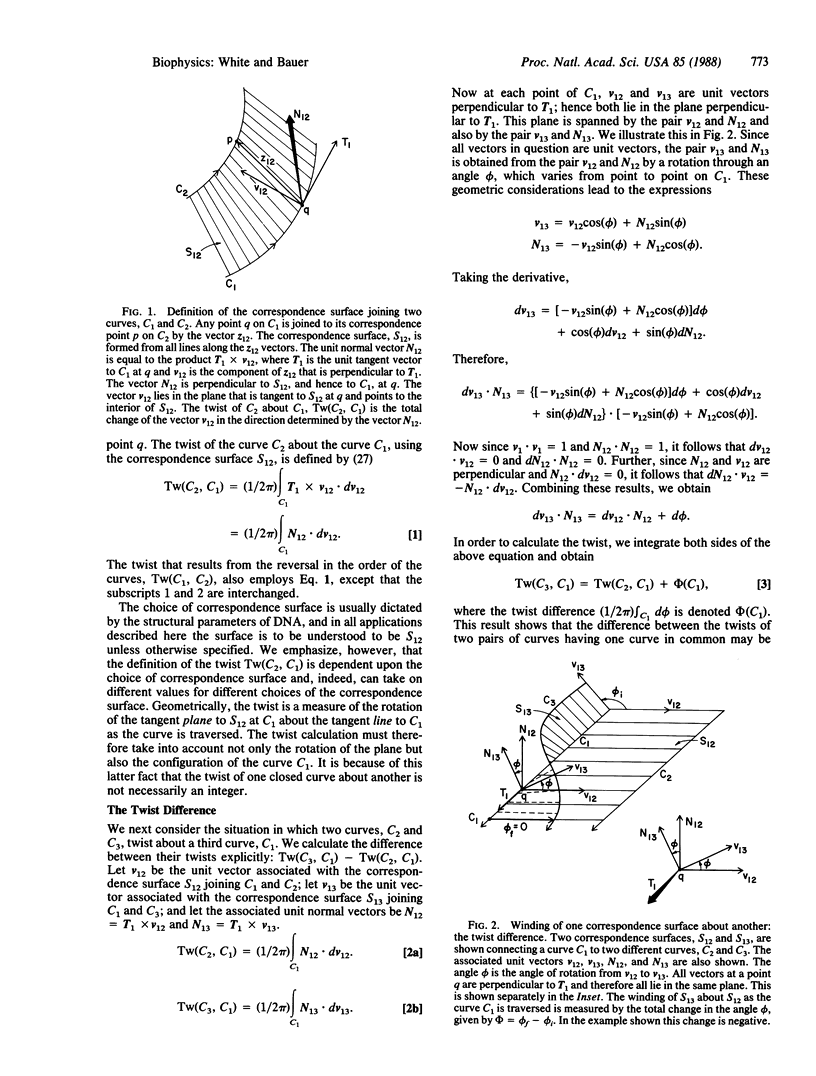
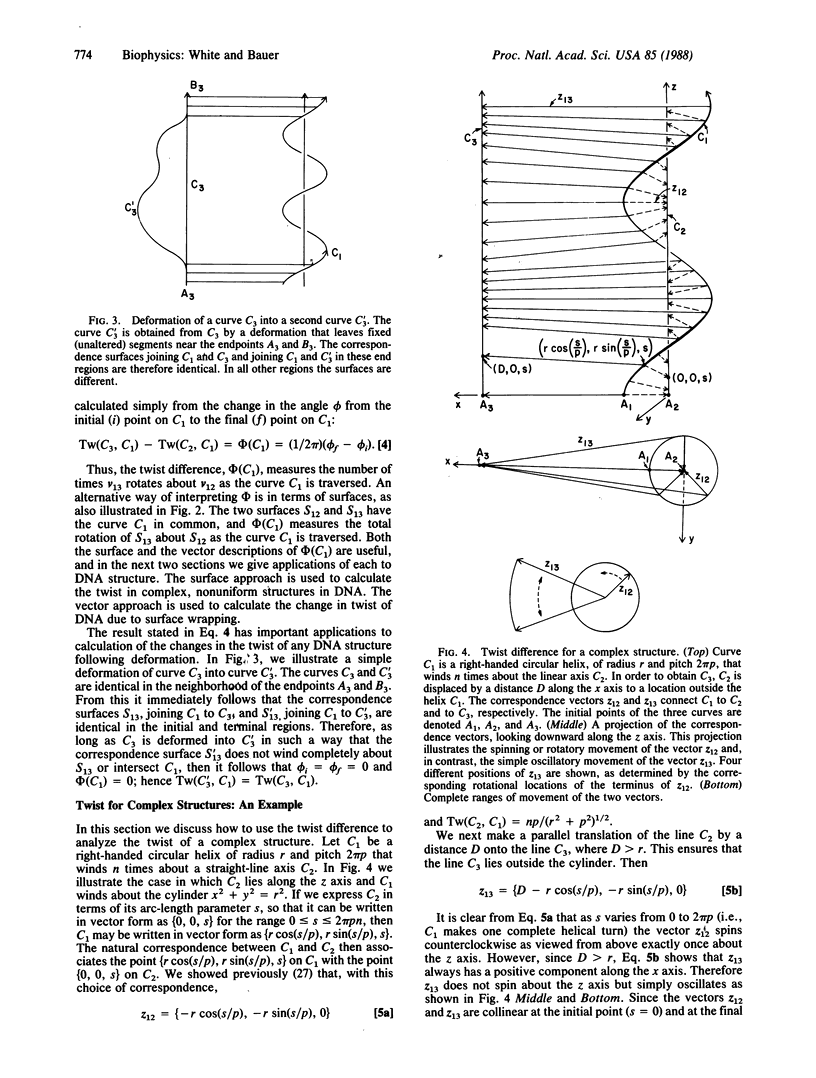
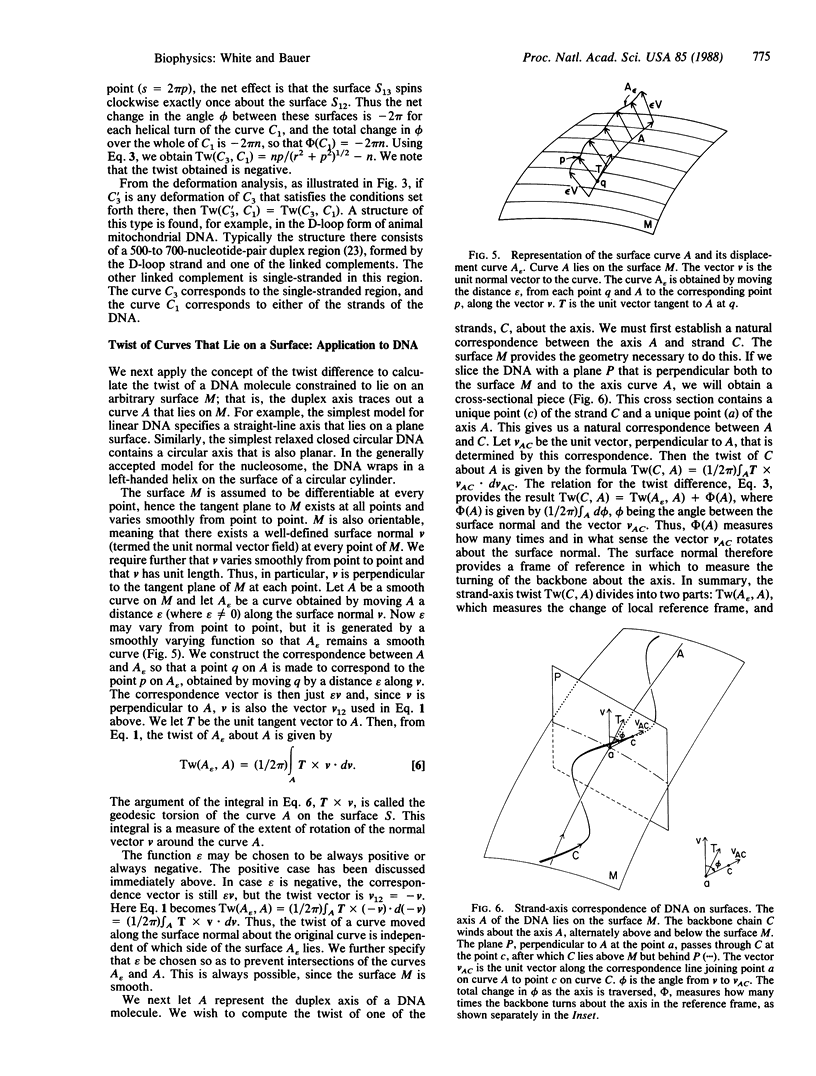
The twist is a fundamental geometric property of nucleic acids. Calculation of the twist in the most general case requires detailed specification of the three-dimensional path of each strand, but many important cases may be analyzed by considering only the twist difference. If C1, C2, and C3 are three distinct space curves, the twist difference about C1 is defined as Tw(C3, C1) - Tw(C2, C1). We show here that this difference measures the rotation of the correspondence surface joining C1 to C2 about the correspondence surface joining C1 to C3. This result has application to DNA containing local nonuniformities, such as denatured regions, cruciforms, and other altered structures. It also facilitates the calculation of twist for three-stranded structures, including D-loops in mitochondrial DNA and replication and transcription intermediates. The twist difference may also be used to simplify greatly the analysis of twist changes in duplex DNA due to winding on surfaces, such as histones and certain enzymes. In such cases the strand-axis twist of DNA divides into two independent terms. The first term arises from the twist of the local reference frame, and the second arises from the rotation of either strand about the duplex axis as measured in the local reference frame. Twist changes consequent to nucleosome winding, for example, arise from the twist of the nucleosome axis, a straight line, about the DNA axis plus the rotation of either strand of the DNA about its axis in the reference frame of the cylinder.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benham C. J. Torsional stress and local denaturation in supercoiled DNA. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3870–3874. doi: 10.1073/pnas.76.8.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor C. R. DNA choreography. Cell. 1981 Aug;25(2):293–295. doi: 10.1016/0092-8674(81)90045-3. [DOI] [PubMed] [Google Scholar]
- Clayton D. A. Replication of animal mitochondrial DNA. Cell. 1982 Apr;28(4):693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- Dean W. W., Lebowitz J. Partial alteration of secondary structure in native superhelical DNA. Nat New Biol. 1971 May 5;231(18):5–8. [PubMed] [Google Scholar]
- Dickerson R. E. Base sequence and helix structure variation in B and A DNA. J Mol Biol. 1983 May 25;166(3):419–441. doi: 10.1016/s0022-2836(83)80093-x. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Gyurasits E. B., Wake R. G. Bidirectional chromosome replication in Bacillus subtilis. J Mol Biol. 1973 Jan;73(1):55–63. doi: 10.1016/0022-2836(73)90158-7. [DOI] [PubMed] [Google Scholar]
- Htun H., Lund E., Dahlberg J. E. Human U1 RNA genes contain an unusually sensitive nuclease S1 cleavage site within the conserved 3' flanking region. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7288–7292. doi: 10.1073/pnas.81.23.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Lee F. S., Bauer W. R. Temperature dependence of the gel electrophoretic mobility of superhelical DNA. Nucleic Acids Res. 1985 Mar 11;13(5):1665–1682. doi: 10.1093/nar/13.5.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M. How many base-pairs per turn does DNA have in solution and in chromatin? Some theoretical calculations. Proc Natl Acad Sci U S A. 1978 Feb;75(2):640–644. doi: 10.1073/pnas.75.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Johnson H. N., Gartenberg M. R., Crothers D. M. The DNA binding domain and bending angle of E. coli CAP protein. Cell. 1986 Dec 26;47(6):995–1005. doi: 10.1016/0092-8674(86)90814-7. [DOI] [PubMed] [Google Scholar]
- Lomonossoff G. P., Butler P. J., Klug A. Sequence-dependent variation in the conformation of DNA. J Mol Biol. 1981 Jul 15;149(4):745–760. doi: 10.1016/0022-2836(81)90356-9. [DOI] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D. Structures of homopurine-homopyrimidine tract in superhelical DNA. J Biomol Struct Dyn. 1986 Feb;3(4):667–669. doi: 10.1080/07391102.1986.10508454. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClarin J. A., Frederick C. A., Wang B. C., Greene P., Boyer H. W., Grable J., Rosenberg J. M. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986 Dec 19;234(4783):1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- McClellan J. A., Palecek E., Lilley D. M. (A-T)n tracts embedded in random sequence DNA--formation of a structure which is chemically reactive and torsionally deformable. Nucleic Acids Res. 1986 Dec 9;14(23):9291–9309. doi: 10.1093/nar/14.23.9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., Mizuuchi M., Gellert M. Cruciform structures in palindromic DNA are favored by DNA supercoiling. J Mol Biol. 1982 Apr 5;156(2):229–243. doi: 10.1016/0022-2836(82)90325-4. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Sequence dependence of the helical repeat of DNA in solution. Nature. 1981 Jul 23;292(5821):375–378. doi: 10.1038/292375a0. [DOI] [PubMed] [Google Scholar]
- Rau D. C., Gellert M., Thoma F., Maxwell A. Structure of the DNA gyrase-DNA complex as revealed by transient electric dichroism. J Mol Biol. 1987 Feb 5;193(3):555–569. doi: 10.1016/0022-2836(87)90266-x. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Sequence-dependent helical periodicity of DNA. Nature. 1981 Jul 23;292(5821):378–380. doi: 10.1038/292378a0. [DOI] [PubMed] [Google Scholar]
- Richmond T. J. Protein--DNA interaction. Another folding problem? Nature. 1987 Mar 5;326(6108):18–19. doi: 10.1038/326018a0. [DOI] [PubMed] [Google Scholar]
- Shen C. K. Superhelicity induces hypersensitivity of a human polypyrimidine . polypurine DNA sequence in the human alpha 2-alpha 1 globin intergenic region to S1 nuclease digestion--high resolution mapping of the clustered cleavage sites. Nucleic Acids Res. 1983 Nov 25;11(22):7899–7910. doi: 10.1093/nar/11.22.7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D., Baldwin R. L. Energetics of DNA twisting. II. Topoisomer analysis. J Mol Biol. 1983 Nov 15;170(4):983–1007. doi: 10.1016/s0022-2836(83)80199-5. [DOI] [PubMed] [Google Scholar]
- Sullivan K. M., Lilley D. M. Influence of cation size and charge on the extrusion of a salt-dependent cruciform. J Mol Biol. 1987 Jan 20;193(2):397–404. doi: 10.1016/0022-2836(87)90227-0. [DOI] [PubMed] [Google Scholar]
- Travers A., Klug A. Nucleoprotein complexes. DNA wrapping and writhing. 1987 May 28-Jun 3Nature. 327(6120):280–281. doi: 10.1038/327280a0. [DOI] [PubMed] [Google Scholar]
- White J. H., Bauer W. R. Calculation of the twist and the writhe for representative models of DNA. J Mol Biol. 1986 May 20;189(2):329–341. doi: 10.1016/0022-2836(86)90513-9. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]


