SUMMARY
Replication Protein A (RPA), the heterotrimeric SSB of eukaryotes, contains four ssDNA binding domains (DBDs) within its two largest subunits, RPA1 and RPA2. We analyzed the contribution of the four DBDs to ssDNA binding affinity by assaying recombinant heterotrimeric RPA in which a single DBD (A, B, C or D) was inactive. Inactivation was accomplished by mutating the two conserved aromatic stacking residues present in each DBD. Using a short substrate, such as (dT)12, no stable interaction could be detected with RPA containing inactive domain A (RPA-A−) while the Kafor RPA-B− or RPA-C− was approximately one third that of wild type RPA. The Kaof RPA-D− was unaffected for substrates 12 to 23 nt in length, but was one third that of wild type RPA for substrates of 40 nt or more. Protein-DNA crosslinking confirms that domain A is essential for RPA to bind substrates of 12 nt or less and that DBD-D (RPA2) requires a minimum of 40 nt to interact with ssDNA. The data support a model in which domain A makes the initial contact with ssDNA, domains A, B, and C (in RPA1) contact substrates up to 23 nt in length, and RPA2 interacts with substrates of 40 - 60 nt.
INTRODUCTION
Replication Protein A (RPA) is a single-stranded DNA (ssDNA) binding protein (SSB), that plays an essential role in DNA metabolism, including replication, repair and recombination (1). Human RPA (hsRPA) is a multimeric complex of three subunits, 70 kDa (RPA1), 34 kDa (RPA2) and 11 kDa (RPA3), that binds to ssDNA with high affinity and binds poorly to double-stranded DNA (dsDNA) and RNA (2-4). RPA has been identified in numerous species including the yeast Saccharomyces cerevisiae (scRPA) where it is a heterotrimeric complex of 69 kDa, 36 kDa and 13 kDa subunits (5,6). The genes encoding scRPA1-3 are referred to as RFA1-3, respectively, and each gene is essential for viability (5,7). Each subunit of RPA is also required for SV40 DNA replication in vitro (8,9).
The binding of RPA to ssDNA has been analyzed by a number of methods and appears to involve at least two modes as determined by its occluded binding-site size. Crosslinking of hsRPA to ssDNA revealed an initially unstable 8 nt binding mode that resolves to a stable 30 nt extended mode (10,11). A high-affinity 30 nt binding mode was also obtained for hsRPA and scRPA using fluorescence quenching and electrophoretic mobility shift assay (EMSA) (12,13). The binding site size for a number of other species of RPA have been reported, including Drosophila (22 nt) (14), calf (20 - 25 nt) (15), and yeast (20 - 30 nt) (16). An unusual 90 nt binding mode has been reported for scRPA using fluorescence quenching and electron microscopy (17).
The RPA1 subunit displays strong ssDNA binding on its own (6,18) although early structure/function analysis revealed that the N-terminal 18 kDa of RPA1 (RPA1N) is unlikely to play a role in ssDNA binding as it is dispensable for SV40 DNA replication and has no significant binding activity (19,20). The structure of the central domain of hsRPA1 has been determined and consists of two structurally similar ssDNA binding domains (DBDs), or “OB-folds” (oligonucleotide/oligosaccharide binding folds) (21). Single-stranded DNA binding by these domains (A and B) is accomplished by aromatic amino acid residues stacking with the individual bases of ssDNA and by hydrogen bonds between the protein and both the phosphate backbone and DNA bases. DBD-A and B contact 3 nt each with 2 nt between the two domains. The C-terminal domain of RPA1 (DBD-C) is a third ssDNA binding domain that requires zinc and is likely to contain another OB-fold (19,22). RPA2 contains a fourth binding domain (DBD-D) with an OB-fold structure (23-25). These four DBDs display amino acid sequence similarity particularly with respect to the aromatic residues known to stack with the ssDNA bases (19). There is currently no evidence that RPA3 binds ssDNA.
Our understanding of how these DBDs contribute to the mechanism of ssDNA binding isincomplete. RPA1 is thought to account for most if not all of the heterotrimer’s binding affinity (20,26) as the interaction of ssDNA with RPA2 is weak (23,24), and the ssDNA binding by the RPA2/3 sub-complex is difficult to detect (27). However, the binding affinity of the RPA2/3 sub-complex is stimulated 100 fold when the N- and C-termini of RPA2 are truncated to produce a “core” domain bound to RPA3 (25). A direct comparison of binding by the isolated DBDs is difficult due to their insolubility (19,23) and a systematic analysis of the role of the individual DBDs within the context of the heterotrimer is lacking. Models of ssDNA binding by the heterotrimer can be formulated based on the evidence that DBDs A and B interact with 8 nt of ssDNA although it is difficult to extrapolate the length of DNA bound by the RPA trimer based on this data. The simplest model to account for most of the data is that the four DBDs collectively interact with 18 - 20 nt and thereby occlude 30 nt of ssDNA (28).
To systematically analyze the role of the four DBDs we asked how each one contributes to the overall binding affinity of RPA. To accomplish this we inactivated a single DBD within the context of the RPA heterotrimer and compared its binding affinity to that of wild type (wt) RPA. Thus, RPA containing an inactive domain A, B, C or D was purified and bound to substrates of various size. Mutation of domain A had the most severe effect and eliminated binding of the shortest substrate (dT)12. RPA containing mutations in DBDs B and C bound to substrates (dT)12, 17, and 23 with reduced affinity compared to wt RPA. Surprisingly, mutation of DBD-D had no effect on these substrates; mutations in domain D affected the binding to (dT)40 and (dT)60 most significantly. These data suggest that RPA interacts with 23 nt, due to the binding of domains A, B and C, and that DBD-D allows RPA to interact with 40 - 60 nt. These conclusions were confirmed by in vitro crosslinking of substrate DNA to RPA.
EXPERIMENTAL PROCEDURES
Plasmid Constructions
The plasmids used in this study are listed in Table 1. In order toexpress recombinant RPA using the T7 RNA polymerase system (29), we constructed triple expression plasmids in which each of the RFA genes is driven by it own T7 promoter. The wt RPA triple expression plasmid, pSAS106, which was used as the parent vector of all RFA1 aromatic amino acid mutants, was constructed from three separate expression plasmids. The RFA1, RFA2 and RFA3 open reading frames were ligated into pET11a (29) on NdeI/BamHI cassettes to create, pRF6, pJM223, and pJM329, respectively. The RFA1 open reading frame in pRF6 was amplified from pJM136 which lacks internal NdeI sites (23). In addition, polymerase chain reaction (PCR) amplification with Vent DNA polymerase was used to introduce unique XhoI and Asp718 sites at codons corresponding to the junctions of the single-stranded DNA binding domains A/B and B/C, respectively (Fig. 1). This resulted in the amino acid replacements S293L, N294E and I424G which were found to have no effect on ssDNA binding activity (data not shown). A unique SacII site was also engineered after the stop codon of RFA1 just upstream of the unique BamHI of pRF6. The double expression plasmid pJM332 was created by ligating the BglII-BamHI fragment of pJM223 (containing T7-RFA2) into the BamHI site of pJM329. The BglII-BamHI fragment of pJM332 was then ligated into the BamHI site of pRF6 to create the triple expression plasmid pSAS106. DNA sequencing revealed that only the intended changes were present in the final construction.
TABLE 1.
Plasmids used in this study.
| Name | Insert | Relevant amino acid changes |
Vector | Reference or Source |
|---|---|---|---|---|
| pRF6 | RFA1 | S293L, N294E, I424G | pET11a | This study |
| pJM223 | RFA2 | none | pET11a | 7 |
| pJM329 | RFA3 | none | pET11a | 7 |
| pJM332 | RFA2, 3 | none | pET11a | 45 |
| pSAS106(WT) | RFA1, 2, 3 | RFA1: S293L, N294E, I424G | pET11a | This study |
| pSAS109(A-) | RFA1 ,2 ,3 | RFA1: F238A, F269A | pET11a | This study |
| pSAS112(B-) | RFA1, 2, 3 | RFA1: W360A, F385A | pET11a | This study |
| pSAS115(C-) | RFA1, 2, 3 | RFA1: F537A, Y586A | pET11a | This study |
| pSAS103(D-) | RFA1, 2, 3 | RFA2: W101A, F143A | pET11a | This study |
| pSAS120(A-B-) | RFA1, 2, 3 | RFA1: F238A, F269A, W360A, F385A | pET11a | This study |
| pSAS105 | RFA1 | none | pET11a | This study |
| pJM136 | RFA1 | none | pRS413 | 23 |
| pJM243 | RFA2 cDNA | none | pRS415 | 23 |
| pSAS204 | RFA2 cDNA | W101A, F143A | pET11a | This study |
| pJM128 | RFA1, 3 | none | pET11a | This study |
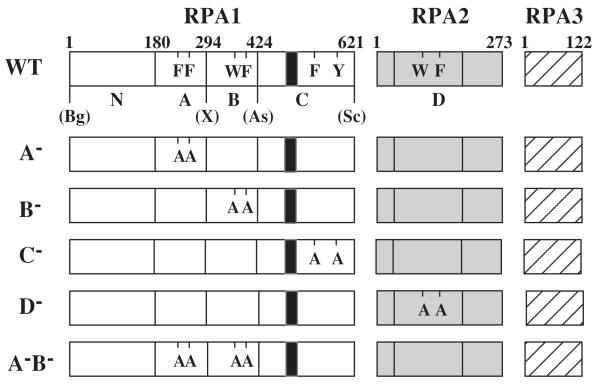
Fig. 1. Heterotrimeric RPA proteins used in this study.
The domain structure of wt RPA (Top) or the indicated mutant RPA protein is illustrated schematically. The RPA1 subunit consists of an N-terminal domain (N) and DBDs -A, -B, and -C while RPA 2 consists of DBD-D. RPA3 is wt in all RPA complexes. The amino acid residues comprising the domains of RPA1 are: N, 1 - 179; A, 180 - 294; B, 295 to 415; and C, 416 - 621. DBD-D is defined as amino acids 40-174 of RPA2. The positions of conserved aromatic residues within each DBD are indicated using single letter code. The following mutations are indicated: A−, F238A, F269A; B−, W360A, F385A; C−, F537A, Y586A; and D−, W101A, F143A. The unique restriction sites created in the wt RFA1 plasmid and their positions relative to the DBDs are presented: Bg, BglII; X, XhoI; As, Asp718; Sc, SacII; S, SalI; Bs, BsiWI. The black box located within DBD-C denotes the zinc-finger motif extending from position 486 to 508.
Point mutant derivatives of the wt RPA plasmid, pSAS106, were created by two rounds of PCR amplification with Vent DNA polymerase, mutagenic oligodeoxynucleotides that change a specific aromatic residue to alanine and the following template plasmids: pSAS105 for domain A; pJM136 for domains B and C, and pJM243 for domain D. The A−, B− and C− PCR products were digested with BglII and SalI, SalI and Asp718 or BsiWI and SacII, respectively. The digested DNA fragments were then ligated into unique BglII and XhoI, XhoI and Asp718 or Asp718 and SacII sites of pSAS106, respectively. The A−B− double mutant was made by a three-way ligation between the digested A− and B− PCR fragments and pSAS106 digested with BglII and Asp718. The D− PCR fragment was isolated on an NdeI/BamHI cassette followed by ligation into pET11a. This plasmid, pSAS204, was subsequently digested with BglII and BamHI and the released fragment was ligated into the BglII site of pJM128 to create the triple expression plasmid pSAS103.
Protein Expression and Purification
Recombinant RPA proteins were expressed in the E.coli strain BL21(DE3) essentially as described (29). Cells were grown in LB medium with 100 μg/ml ampicillin at 37°C until the absorbance at 600 nm was 0.5. The cultures were induced for 2 h by adding isopropyl-1-thio-ß-D-galactopyranoside (IPTG) to 0.4 mM. Cells were collected by centrifugation and resuspended in buffer B (25 mM HEPES, [pH 7.5], 0.01% NP-40, 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol [DTT], 0.1 mM phenylmethylsulfonyl fluoride [PMSF]) containing 100 mM NaCl. All subsequent steps were performed on ice or at 4°C. Samples were subjected to 3 freeze-thaw cycles and 8 sonication periods of 15 s each. The lysate was centrifuged at 12,000 × g and the supernatant applied to a 50 ml Affi-Gel blue affinity resin (Biorad). The column was washed sequentially with 3 column volumes of buffer B containing 800 mM NaCl and 0.5 M NaSCN and the protein was eluted with buffer B containing 1.5 M NaSCN. Peak fractions identified by Bradford analysis were pooled, loaded onto a 5 ml hydroxylapatite column (Biorad) and washed sequentially with 15 ml of buffer B containing 40 mM NaH2PO4 (pH7.5), 120 mM NaH2PO4 (pH7.5) or 500 mM NaH2PO4 (pH7.5). The protein eluted in the 120 mM NaH2PO4 wash. Peak fractions containing RPA were identified by sodium dodecyl sulfate-17% polyacrylamide gel electrophoresis (SDS-PAGE), pooled and dialyzed in buffer A (same as buffer B above, except that 25 mM Tris-HCl, [pH 7.5] is substituted for HEPES) containing 100 mM NaCl. The dialyzed fractions were loaded onto a 1 ml Mono Q column (Amersham Pharmacia Biotech), washed with 3 ml buffer A containing 100 mM NaCl and eluted in a 10 ml linear gradient from 100 mM to 400 mM NaCl. Peak RPA fractions were identified by SDS-PAGE, pooled and diluted with buffer A until the conductance was equivalent to buffer A plus 25 mM NaCl. The diluted sample was applied to a 2 ml phosphocellulose column (Whatman) and washed with 5 ml of buffer A containing 500 mM NaCl or 1 M NaCl. The protein was eluted in the 500 mM NaCl wash. Samples from the fractions were resolved on a SDS-PAGE and those containing highly purified RPA were pooled and dialyzed in buffer B containing 25 mM NaCl or buffer B containing no EDTA, 20 μM ZnSO4 and 25 mM NaCl. Protein concentrations were determined by the Bradford assay using bovine serum albumin as the standard.
Single-stranded DNA binding and denaturing immunoprecipitation assays
The standard DNA binding reaction was performed in a total volume of 15 μl and contained the indicated purified protein samples from E.coli, 2 fmols of 32P-labeled oligonucleotide (dT12, dT17, dT23, dT40 or dT60), 25 mM HEPES (pH 7.5), 250 mM NaCl, 0.5 mM DTT, 5% glycerol, 20 μM ZnSO4, 0.1% NP-40 and 10 mg/ml BSA. RPA titrations ranged from 0.58 pM to 35 nM. Reactions were incubated 30 min at 25°C and size separated on native 6% polyacrylamide gels (37.5:1 acrylamide:bis) containing 0.5X TBE. The band intensities of free and bound DNA were analyzed with a phosphorimager and IP-Lab Gel software. The proportions of the free and bound oligo were calculated and a reciprocal plot of the Langmuir isotherm was used to determine the dissociation constant (Kd) for each protein and oligonucleotide. Dissociation constants were then converted to their corresponding association constants (Ka).
Binding reactions used for the denaturing immunoprecipitation assay were carried out under identical conditions except for the following: equimolar amounts of purified protein and 32P-labeled oligonucleotide were incubated together in the absence of glycerol and NP-40 and in the presence of 1 mg/ml BSA. The reactions were crosslinked with ultraviolet (UV) light at a dose of 1000 J/m2, boiled for 10 min in denaturing buffer containing 40 mM Tris (pH 7.4), 1% SDS, 1 mM DTT, 5 mM EDTA, 2 mM PMSF followed by the addition of Radio Immune Precipitation Assay (RIPA) buffer lacking SDS (50 mM Tris, [pH8.0], 1 mM DTT, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.5% deoxycholate [DOC]) to dilute the SDS to 0.2%. Anti-RPA1 or -RPA2 antibody was then added to the mixture and incubated 1 h at 4°C. Samples were incubated with protein A beads at 4°C for 1 h while mixing and then centrifuged to pellet the antibody-RPA-DNA complex. SDS-PAGE loading buffer was added and the samples were boiled for 5 min. The samples were then resolved by SDS-15% PAGE and visualized with a phosphorimager.
RESULTS
Mutant forms of RPA
In order to determine contribution of the four DBDs of yeast RPA to ssDNA binding, we sought to assay RPA proteins in which a single DBD had been inactivated. The inactivation of these DBDs was accomplished by mutating residues critical for ssDNA binding. The crystal structure of hsRPA domains A and B bound to ssDNA identified a number of residues that make specific hydrophobic and hydrogen bond interactions with ssDNA (21). We focused on the two aromatic residues that make hydrophobic stacking interactions with the ssDNA bases for the following reasons. Hydrogen bonding between RPA and the DNA bases is dependent on the sequence of the substrate DNA (21) and the amino acid residues involved in these interactions are not conserved in all four DBDs (19). In contrast, the hydrophobic stacking interactions appear to be independent of DNA sequence and the positions of the aromatic residues are conserved in all four DBDs (19). Thus, mutation of the two aromatic residues would be expected to have the same effect in each DBD allowing us to compare the relative roles of the four DBDs in ssDNA binding. Effects due to DNA sequence heterogeneity was eliminated by the use of homopolymeric ssDNA oligo (dT) as substrate.
To assay mutant RPA proteins we designed an expression plasmid in which a variety of mutations could be introduced into a single RPA subunit and co-expressed with the remaining two subunits. Expression in bacteria was essential as two of the single amino acid replacements (F238A and F537A) were previously shown to be lethal in yeast (19). As illustrated in Figure 1, wt RPA was expressed from plasmid pSAS106 in which each DBD of RPA1 is encoded by a unique DNA cassette. In addition, each gene was driven by its own T7 promoter (not shown). Amino acid sequence alignment was previously used to identify the aromatic amino acids in each DBD that are homologous to the stacking residues identified in the crystal structure of hsRPA domains A and B (19,21). To express RPA with an inactive DBD these residues were mutated to alanine in pairs (Fig.1).
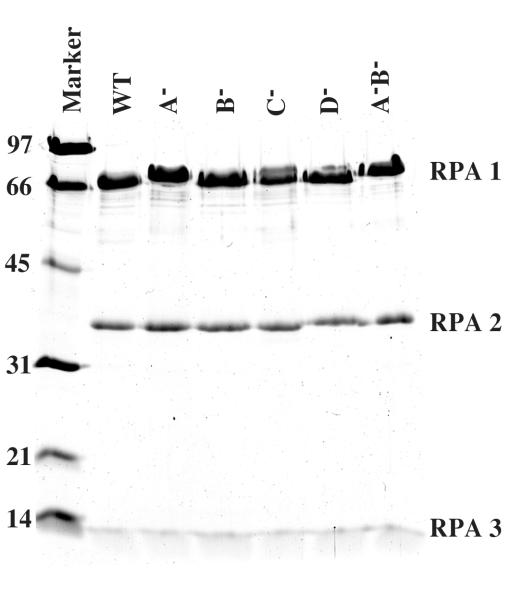
Following expression in E.coli, RPA was purified using affinity and ion exchange chromatography. SDS-PAGE analysis of the purified proteins indicated a purity of at least 95% (Fig. 2). Mutation of domain A appeared to cause a significant structural change in the protein as the bands corresponding to the RPA1 subunit in the A− and A−B− mutants migrated somewhat slower than those of wt or other RPA mutants. This behavior may be related to the significance of this domain in mediating ssDNA binding (see below).
Fig. 2. Purified RPA complexes.
ScRPA proteins were expressed in E.coli and purified as described in the Experimental Procedures. Two μg of wt RPA or the indicated mutant was resolved on by SDS-17% PAGE and visualized with Coommassie blue. The positions of RPA1 (69 kDa), RPA2 (36 kDa) and RPA3 (13 kDa) are indicated. The molecular mass standards are indicated in kDa.
RPA activity and electrophoretic mobility assay
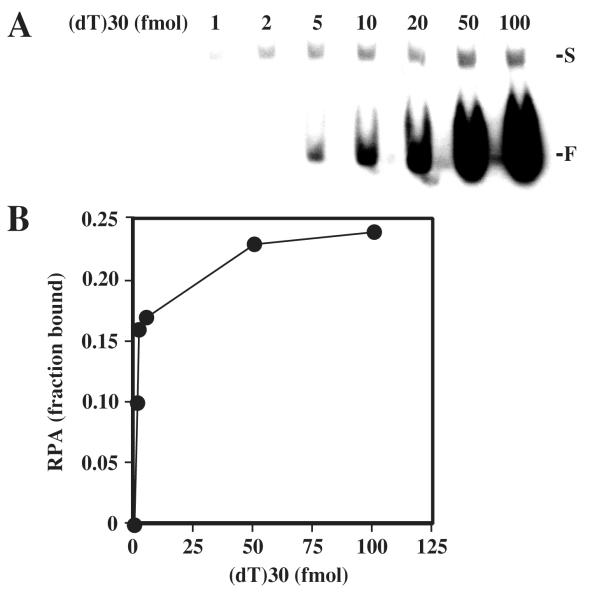
An electrophoretic mobility shift assay (EMSA) was used to determine the ssDNA binding affinity of wt and mutant RPA. This assay is a sensitive method for the analysis of RPA-DNA interactions that uses nanomolar concentrations of RPA so that equilibrium binding conditions are achieved (12,13). Prior to performing this assay on a variety of substrates we determined the percentage of purified RPA in our preparations that was able to bind ssDNA. A constant amount of RPA was incubated with increasing amounts of radiolabeled (dT)30 and then the DNA-protein complexes were separated on a nondenaturing 6% polyacrylamide gel. The radioactive signal from the free and bound DNA was visualized by phosphorimager (Fig. 3A). Only singly-liganded complexes were observed using this substrate. At low levels of input DNA binding was quantitative and no free DNA was visible. As the amount of input DNA increased, the signal for the bound complex became more intense until it remained constant. Following quantitation of these signals, the fraction of bound RPA was determined and plotted versus moles of substrate DNA. At saturation, approximately 24% of the RPA heterotrimer was bound to oligo (dT)30 (Fig. 3B). This fraction of RPA is referred to as the “active” fraction and analysis of RPA mutants revealed similar levels of activity (data not shown). Therefore, an average value of 24% active protein was used in calculating the binding constants of RPA proteins analyzed in this study.
Fig. 3. Determining the fraction of active RPA.
(A) 8.7 fmol of purified wt RPA was incubated with the indicated amount of 32P-labeled (dT)30 and the reactions were resolved on a 6% nondenaturing polyacrylamide gel. The positions of singly liganded (S) and unbound (F) substrate are indicated. (B) The radioactivity corresponding to free DNA and protein-DNA complex in (A) was quantitated using liquid scintillation counting. The fraction of RPA in the bound form was then calculated and plotted as a function of input DNA.
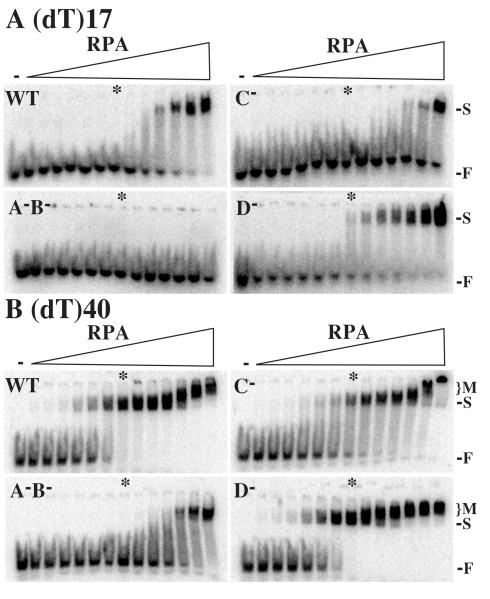
A series of electrophoretic mobility shift assays using various lengths of oligo (dT) was performed using wt and mutant RPA. We incubated increasing amounts of each RPA protein with a fixed amount of 32P-labeled oligonucleotide and resolved the bound complex from the free DNA using nondenaturing gel electrophoresis. The amounts of free and bound DNA were then analyzed using a phosphorimager. Figure 4 shows representative experiments for wt RPA as well as the C−, D−, and A−B− mutants using (dT)17 and (dT)40 as substrates. Similar experiments were performed with the A− and B− mutants and (dT)12, (dT)23 and (dT)60 substrates. In the case of (dT)17, titration with RPA resulted in a single complex migrating slower than free probe (Fig. 4A). The signal corresponding to free DNA disappears completely at high concentrations of wt RPA and the D− mutant. In contrast, the A−B− mutant yielded no DNA-protein complexes, even at high protein concentrations, and the C− mutant produced a retarded complex only at the highest protein concentrations. Based on this qualitative assay, we conclude that wt RPA binds the (dT)17 substrate as a singly-liganded form and that saturated binding requires a molar excess of RPA over substrate (asterisks in Fig. 4). Further, mutating the stacking residues appears to be an effective method of inactivating the DBDs. In the case of the C− mutant, binding of the (dT)17 substrate is compromised, while in the A-B- mutant binding of (dT)17 is eliminated. On the other hand, the D− mutant appears to bind this substrate like wt RPA.
Fig. 4. Gel mobility shift assays of wt and mutant RPA.
Increasing amounts of wt or the indicated mutant RPA (0, 1, 2, 6, 25, 50, 100, 250, 500, 1000, 2000, 6000, 20000 or 60000 pg) were incubated with 2 fmols of radiolabeled (dT)17 (A) or (dT)40 (B). The reactions were then resolved on a 6% nondenaturing polyacrylamide gel. The first lane of each gel (−), is a negative control reaction containing no protein. The positions of RPA-DNA complexes (S, singly-liganded; M, multiply-liganded) and unbound oligonucleotide (F) are indicated. The asterisk indicates a binding reaction containing equimolar amounts of RPA and substrate.
In the case of (dT)40, titration with wt RPA revealed a retarded band that was saturated at stoichiometric levels of RPA (Fig. 4B). At the highest wt RPA concentrations a second more-slowly migrating form appeared. As previously observed, we interpret this to be a multiply-liganded complex (13). A similar response was obtained with the C− and D− mutants, although the multiply liganded complex with occurs with somewhat lower levels of RPA-D− than with wt RPA. In contrast, we observed only a singly-liganded complex with A−B− mutant even at high concentrations of protein (Fig.4B). We conclude that under these conditions (dT)40 is sufficiently large to accommodate two RPA complexes. Further, while the A-B- mutant is unable to bind a substrate of 17 nt, it retains the ability to bind a substrate of 40 nt. This suggests that RPA contains DBDs in addition to A and B that are sensitive to the length of the oligonucleotide.
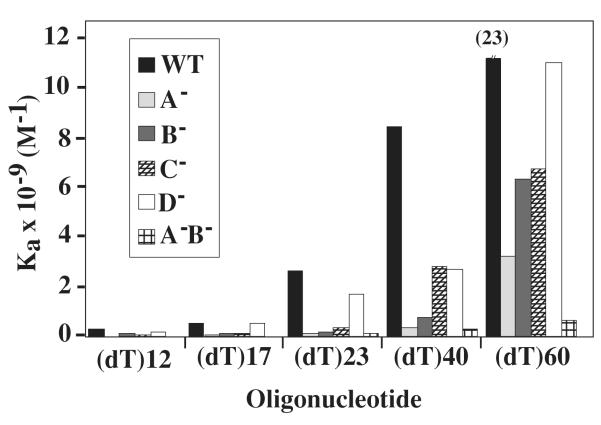
The binding affinity of wt and mutant RPA was determined by calculating an equilibrium binding constant for substrates of various lengths. This was accomplished by quantitating the intensity of the signal corresponding to the free and bound DNA and fitting the data to the Langmuir equation. The values of the binding constants determined from these and other titrations are presented in Table 2, while Figure 5 summarizes the findings. For all proteins there was an increased binding affinity for (dT)60 compared to (dT)12 (Table 2 and Figure 5). This was previously reported for hsRPA (13) and is likely the result of an increase in the number of direct interactions between the DNA and RPA protein. For example, the binding constants (Ka) for wt RPA ranged from 1.8 × 108 M−1 for (dT)12 to 2.3 × 1010 M−1 for (dT)60. These values agree closely with those obtained for hsRPA (12,13). In the case of the D− mutant, these values ranged from 1.8 × 108 M−1 to 1.1 × 1010 M−1 (Table 2). Therefore, the affinity of wt RPA to ssDNA is approximately 130-fold higher for a (dT)60 than a (dT)12, and the affinity of the D− mutant is 60-fold higher for a (dT)60 than a (dT)12. Given that the binding constants of wt and D− RPA are essentially equivalent for (dT)12 through (dT)23, this suggests that the D− mutant is compromised in its ability to bind long substrate DNAs.
TABLE 2.
Binding properties of RPA mutants determined by mobility shift assaya
| Aromatic residue mutants |
Oligonucleotide length |
||||
|---|---|---|---|---|---|
| dT12 | dT17 | dT23 | dT40 | dT60 | |
| Ka × 109 (M−1) | |||||
| WT | 0.18 ± 0.003 | 0.51 ± 0.01 | 2.6 ± 0.6 | 8.4 ± 0.7 | 23 ± 0 |
| A- | NCb | 0.088 ± 0.0008 | 0.12 ± 0.007 | 0.34 ± 0.025 | 3.2 ± 0.4 |
| B- | 0.048 ± 0.0008 | 0.11 ± 0.003 | 0.16 ± 0.01 | 0.77 ± 0.14 | 6.3 ± 0.4 |
| C- | 0.059 ± 0 | 0.087 ± 0.004 | 0.35 ± 0.05 | 2.8 ± 0.3 | 6.7 ± 0.4 |
| D- | 0.18 ± 0.003 | 0.53 ± 0 | 1.7 ± 0.01 | 2.7 ± 0 | 11 ± 0.5 |
| A-B- | NC | NC | 0.095 ± 0.005 | 0.29 ± 0.009 | 0.61 ± 0.1 |
Average apparent association constants from at least two independent titrations ± SD.
NC, no complex detected.
Fig. 5. Comparison of association constants (Ka).
The apparent binding constants (Ka) of wt or the indicated mutant RPA are presented graphically as a function of substrate size. All data is taken from Table 2.
Among the RPA proteins with singly-mutated DBDs, the most severe effect was observed with the A− mutant. No complex was detected using (dT)12 substrate and, although it bound longer substrates, its affinity was significantly reduced; binding to a 23-mer was 20-fold less than wt while binding to (dT)60 was 7-fold less than wt. This suggests that while domain A plays a critical role in binding short ssDNA the additional DBDs assist in binding longer oligos. This idea is supported when the binding affinities of the B− and C− mutants are considered. Binding of the 12-mer could be detected with the B− and C− mutants although their affinity was less than that of wt. The binding affinity of the B− and C− mutants to (dT)23 was about one tenth that of wt while their binding to (dT)60 was 1/4 that of wt. This suggests that, in contrast to domain D, domains B and C play a significant role in binding 12 through 23-mer oligonucleotides.
Lastly, the A−B− mutant was severely affected in binding ssDNA. It showed no complex formation with (dT)12 and (dT)17, and a 30- to 40-fold decrease in binding affinity for (dT)40 and (dT)60 compared to wt. Taken together, these results suggest that domain A is important for all binding events and is essential for (dT)12. Domains A and B are essential for binding dt17 and likely cooperate with domain C for binding to (dT)23. DBD-D is likely to play a role in binding oligos of between 40 and 60 nt.
In vitro crosslinking of ssDNA to RPA
We have previously described a UV crosslinking assay to detect the interaction of ssDNA with RPA. In this assay RPA is incubated with an equimolar amount of 32P-labeled ssDNA, crosslinked with UV light, and analyzed by SDS-PAGE (19,23). In this study we searched for interactions with specific RPA subunits by denaturing the crosslinked product in the presence of 2% SDS and immunoprecipitating RPA1 or RPA2 with specific antiserum. The resulting antibody-RPA-DNA complex was collected on Protein-A beads, resolved by SDS-PAGE and visualized with a phosphorimager.
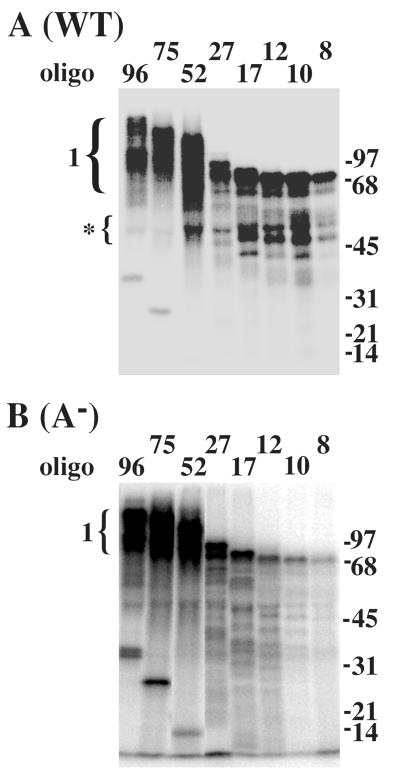
As shown in Figure 6A, when RPA was crosslinked to small substrates such as (dT)8 a 70 kDa protein corresponding to the RPA1 subunit was labeled. In this experiment we also detected binding by RPA1 breakdown fragments that migrated at approximately 50 kDa. As the substrate size was increased from 8 to 96 nt the intensity and size of this band increased. This increase in intensity reflects the increase in the Kaof RPA as substrate size increases (Table 2). At 75 to 96 nt the signal splits into two species that likely correspond to the substrate bound to multiple RPA1 subunits. To observe the contribution of domain A to this reaction we repeated the experiment with RPA-A− (Fig. 6B). While the profile of signal is roughly the same, there was a great reduction in the signal obtained with the 8, 10, and 12 nt substrates. Thus, domain A is essential for binding small substrates as previously observed (Table 2). The binding of RPA-A− to substrates 17 nt or greater is reduced somewhat compared to wt, however the increase in signal obtained with the 52-mer and the two species of RPA1 bound to the 96-mer was identical to wt RPA.
Fig. 6. DBD-A is required for binding short oligonucleotides.
One pmol of wild type (A) or mutant (B) RPA was incubated with one pmol of the indicated radiolabeled oligonucleotide. Following UV-crosslinking, the reactions were denatured and incubated with antiserum against RPA1. The reactions were then incubated with protein A beads to precipitate the RPA1-DNA complexes which were resolved by SDS-PAGE and analyzed with a phosphorimager. The numbered bracket indicates the position of RPA1 crosslinked to the indicated oligonucleotide. The bracket with an asterisk indicates binding by RPA1 break-down products. Oligonucleotides of 17 - 96 nt are of random sequence while those of 8 - 12 nt are oligo(dT). Unbound oligonucleotide can be observed in the 96, 75 and 52 lanes at molecular weights of 32, 25 and 14 kDa, respectively. Molecular mass standards are indicated in kDa.
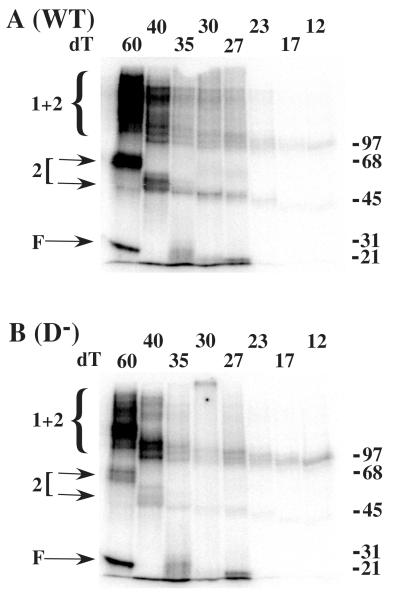
We next tested the interaction of RPA2 with ssDNA by UV-crosslinking. When wt RPA was incubated with substrates of 35 nt or less, we observed no interaction between RPA2 and ssDNA (Fig. 7A). In contrast, when incubated with larger oligos, such as (dT)60, a robust band migrating at 56 kDa was detected. This size of this band is consistent with that of RPA2 (36 kDa) bound to the oligonucleotide (~20 kDa). When incubated with (dT)40 we observed a much weaker band with a mobility of ~49 kDa consistent with a contribution of ~13 kDa from the oligo. In both cases we also observed bands >97 kDa in size that likely represent RPA1 crosslinked to ssDNA and RPA2. These species may include both direct protein-protein crosslinks as well as indirect tethering of RPA1 and RPA2 via ssDNA. To confirm that these RPA2-labeled bands represent authentic interactions between ssDNA and RPA2 we repeated the experiment with D− mutant RPA (Fig. 7B). In this case the intensity of the bands corresponding to RPA2 bound to (dT)60 or (dT)40 is dramatically reduced, as is the signal migrating at >97 kDA. Thus mutation of the aromatic residues in RPA2 directly reduces the interaction of RPA2 bound to (dT)40 and (dT60).
Fig. 7. Optimal binding by RPA2 requires 40 to 60 nt of ssDNA.
0.4 pmol of wild type (A) or mutant (B) RPA was incubated with an equimolar amount of radiolabeled oligo(dT) of the indicated length. The reactions were UV-crosslinked, denatured and incubated with antiserum against RPA2. The reactions were then incubated with protein A beads to precipitate the RPA2-DNA complexes which were then resolved by SDS-PAGE and visualized with a phosphorimager. Arrows (2) indicate the position of RPA2 singly crosslinked to DNA. The brackets (2 + 1) indicate the position of RPA2 bound to RPA1 and DNA. Unbound oligonucleotide (F) and molecular mass standards (kDa) are indicated.
DISCUSSION
Although RPA is well studied, the functions of its individual subunits and multiple DBDs remain obscure. For example, it is not known what combination of DBDs account for the major ssDNA binding mode. Estimates of the occluded binding-site size of several species of RPA range between 22 and 30 nt (14-16), and experiments with both yeast and human RPA indicate that this 30 nt binding mode is achieved by RPA directly interacting with 20-30 nt of ssDNA (12,13,30). It is known that this binding mode involves an initially unstable interaction with 8 nt of ssDNA that resolves into a stable elongated complex covering 30 nt (10,11). Despite the consistency of these values, we have considered the possibility that RPA binds ssDNA in a second stable mode. This idea is proposed to reconcile the following facts: the prokaryotic cellular SSB of E. coli is a homotetramer that binds ssDNA in at least two modes (35 nt and 65 nt) (31); both RPA and the prokaryotic SSB contain multiple DBDs (19,28); RPA2 is an essential ssDNA binding subunit whose function is unexplained (23,32); and a 90 nt binding mode was previously reported for yeast RPA (17).
Recent structural analysis of RPA has provided sufficient details on the mechanism of ssDNA binding to allow us to test this hypothesis. The crystal structure of domains A and B has been determined in the presence (21) and absence of ssDNA (28). These domains, each comprising an OB-fold, reorient upon binding ssDNA and interact with a total of 8 nt. The solution structure of human RPA1N also revealed an OB-fold-like structure (22,33), but this domain is not known to bind ssDNA and may be required to mediate interactions with other proteins due to its interaction with other proteins (34-38) as it fails to bind ssDNA on its own (19) and deletion of RPA1N does not affect its activity in vitro (20). The C-terminal portion of RPA1 is a third ssDNA binding domain that binds zinc and appears to contain another OB-fold (DBD-C) (19,22). Finally, the structure of a sub-complex consisting of the RPA2 core bound to RPA3 revealed OB-folds in each of these domains (32). However, only the fold in RPA2 (DBD-D) resembles domains A and B and only RPA2 is known to bind ssDNA in vitro (23,25,32). Thus, RPA consists of six potential ssDNA binding domains of which four are known to bind ssDNA.
To explain the role of the multiple DBDs in the mechanism of ssDNA binding by RPA we considered the following two models. The simplest idea, that all six DBDs are required for stable ssDNA binding, is difficult to support as RPA1N and RPA3 are not known to bind ssDNA. A second model proposes that the four known DBDs are required for the stable 30 nt binding mode while the remaining two domains mediate protein-protein interactions. Bochkarev and colleagues have recently proposed a detailed version of this model (28). In this model domains A, B, and C, align in a linear fashion and contact 13 - 15 nt. DBD-D is then proposed to align with these domains such that a total of 18 - 20 nt of ssDNA is contacted by RPA. Domains RPA1N and RPA3 are proposed to account for the observed occlusion of 30 nt (28).
Some, but not all, of our data are compatible with this model. By inactivating each DBD of scRPA and measuring the apparent association constant of the resulting complex we have determined that DBD-A is essential for RPA to interact with (dT)12 and that DBD-B and -C are required for full binding affinity to this substrate. Mutation of DBD-D had no effect on the affinity of RPA for (dT)12. Consistent with the above view, we interpret this to mean that domains A, B, and C make contact with (dT)12 while DBD-D does not. In contrast to the predictions of this model, mutation of DBD-D had no significant effect on RPA’s binding affinity to 17 or 23 nt substrates. The binding affinity of RPA-D− was significantly impaired only when the substrate size was increased to 40 or 60 nt. These data indicate that DBD-D binds ssDNA in a 40 or 60 nt mode and is unlikely to be involved in the well-characterized 30 nt binding mode.
Our results, as well as the earlier report of a 90 nt binding mode (17), were obtained with scRPA not hsRPA. We do not believe, however, that this larger binding mode is unique to scRPA. The contribution of RPA2 to ssDNA binding may have gone undetected because it interacts only with long substrates. As a result, more sensitive assays are required to detect RPA2 binding as it does not significantly affect the overall binding affinity of RPA to these substrates. This idea can explain the discrepancy in ssDNA binding site size and earlier evidence that RPA1 possesses all the ssDNA binding activity of the trimer. In this model, the occlusion of 30 nt arises from the interaction of DBDs A, B, and C with 23 nt. Binding by RPA2 (DBD-D) does not occur until the substrate is 40 nt or more which could account for the occlusion of 60 nt or more. The binding by RPA1 alone is expected to be sufficient for the stable 30 nt binding mode. Indeed, the affinity of RPA-D− for a 23 nt substrate (Ka= 1.7 × 10E9) is in the same order of magnitude as that obtained with wt RPA and a 30 nt substrate (Ka= 4.6 × 10E9) (13).
We previously described a simple UV crosslinking assay to identify interactions between ssDNA and the RPA subunits. This assay suggested that the binding of ssDNA by RPA2 occurred with low efficiency and that it could be stimulated by increased concentrations of NaCl (23). By including an immunoprecipitation step in the experiments described here we have found that the interaction between ssDNA and RPA2 is more efficient than originally thought. This result is consistent with the fact that dimeric (DBD-D/RPA3) or trimeric (DBD-C/-D/RPA3) subcomplexes of hsRPA bind ssDNA with relatively high affinity (25). We suggest that it is inherently difficult to identify an interaction between RPA2 and ssDNA in the context of wt RPA because binding by RPA2 requires prior binding by the potent RPA1 subunit. This idea is confirmed by crosslinking studies; immunoprecipitated RPA2 was associated with a significant amount of RPA1 that was itself bound to labeled ssDNA. As above, a second difficulty in identifying this interaction is that substrate must be at least 40 nt in length before DBD-D is able to contact it. If the stable 30 nt binding mode required RPA2, then significant crosslinking would be expected with this substrate. However, crosslinking was not obtained with 30 or 35 nt substrates. We observed only weak interactions with the 40 nt substrate and strong interactions with the 60 nt substrate. These data support the model in which RPA1 is exclusively responsible for the 30 nt mode and that DBD-D promotes a second, larger binding mode. An alternative explanation for the improved crosslinking of RPA2 to the 60 nt substrate is that two RPA trimers bind this substrate and undergo a conformational change that results in the interaction between ssDNA and RPA2. However, this explanation can be excluded as we fail to see doubly-occupied 60-mers under the stoichiometric conditions used in the crosslinking experiment.
A mutational approach has previously been used to study the role of DBDs A and B in hsRPA (39). Walther and colleagues concluded that mutating a single aromatic residue of DBD-A or B had a minimal effect on the overall ssDNA binding affinity of hsRPA to (dT)30 (39). Our results are in partial agreement with this study as we found that RPA-A− and RPA-B− were equally defective in binding (dT)23. In addition, both studies revealed a synergistic effect in simultaneously mutating domains A and B. In contrast, we found that the double aromatic mutation had a more profound effect than the single point mutation. The affinity of RPA-A− for (dT)23 or (dT)40 was 1/20 that of wt RPA (Table 2) while the affinity of RPA containing a single aromatic mutation in domain A (F238A) for (dT)30 was 2/3 that of wt RPA (39). This defect was amplified when binding to smaller substrates was examined; binding of RPA-A− to substrates such as (dT)12 was not detectable (Table 2). We conclude that mutating both aromatic residues significantly reduces the activity of a single DBD. In addition, it is important to consider substrate size when determining the effects of these mutations as some effects are masked by the activity of additional DBDs within the RPA complex.
Crosslinking of labeled ssDNA to RPA results in a 97 kDa complex that represents RPA2 crosslinked to RPA1 (Fig. 7A). This species is under-represented when using (dT)12 - (dT)35, even though these oligos bind well to RPA1. Thus, RPA1 and RPA2 are poorly crosslinked to each other under these conditions. The dramatic increase in signal that occurs with substrates of 40 to 60 nt suggests that the interaction of RPA2 with ssDNA increases the probability of a crosslink between RPA1 and RPA2. This is consistent with previously observed rearrangements that occur upon ssDNA binding. The binding of ssDNA to RPA has previously been shown to result in the alignment of domains A and B (28), increased proteolysis of RPA2 (40), and increased phosphorylation of RPA2 by DNA-PK (11). Interestingly, Blackwell and colleagues observed that while phosphorylation of RPA by DNA-PK was dependent on ssDNA binding, there was a significant increase in modification as the ssDNA template was increased from 30 to 45 nt (11). We suggest that this modification, like the crosslinking of RPA1 and RPA2, is due to the interaction of RPA2 with ssDNA and rearrangement of the heterotrimer.
The crosslinking assay described here will allow us to further examine the role of NaCl and other factors in modulating ssDNA binding by RPA2. The function of this binding is still unclear. Unlike domain A, which has a significant effect on ssDNA binding affinity, RPA2 can contribute only a small amount to the overall binding affinity of RPA. One possibility is that this small degree of binding affinity is significant in vivo given that RPA2 is essential for viability in yeast. On the other hand, ssDNA binding by RPA2 might control RPA’s cooperativity or its interaction with other proteins. This function may in turn be regulated by the cell-cycle and DNA damage-dependent phosphorylation of RPA2 (41,42). In light of the present results, it is not surprising that phosphorylation of RPA2 did not significantly affect the ssDNA binding activity of RPA in vitro (43). Changes in this activity would be expected to have a small affect with large substrates and no effect with small substrates. An alternative role for ssDNA binding by RPA2 could be to mediate the compaction of RPA-ssDNA complexes that has been observed by electron microscopy at high salt (44). Further experimentation will be required to determine whether RPA2 or its modifications affect these activities.
Acknowledgements
The authors thank Alexey Bochkarev for communicating results prior to publication. We also thank lab members for comments on the manuscript.
The abreviations used are
- wt
wild type
- OB-fold
oligonucleotide/oligosaccharide binding fold
- RIPA
radio-immuno-precipitation assay
- DTT
dithiothreitol
- PAGE
polyacrylamide gel electrophoresis
- PMSF
phenylmethylsulfonyl fluoride
- RPA
replication protein A
- RPA1N
N-terminal 18 kDa of RPA1
- IP
immunoprecipitation
- PCR
polymerase chain reaction
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
- nt
nucleotide
- hsRPA
human RPA
- scRPA
Saccharomyces cerevisiae RPA
Footnotes
This work was supported by National Institutes of Health Grant GM55583.
REFERENCES
- 1.Wold MS. Annu. Rev. Biochem. 1997;66:61–91. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 2.Wold MS, Kelly T. Proc. Natl. Acad. Sci. USA. 1988;85:2523–2527. doi: 10.1073/pnas.85.8.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fairman MP, Stillman B. EMBO J. 1988;7:1211–1218. doi: 10.1002/j.1460-2075.1988.tb02933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wobbe CR, Weissbach L, Borowiec JA, Dean FB, Murakami Y, Bullock P, Hurwitz J. Proc. Natl. Acad. Sci. USA. 1987;84:1834–1838. doi: 10.1073/pnas.84.7.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heyer WD, Rao MR, Erdile LF, Kelly TJ, Kolodner RD. EMBO J. 1990;9:2321–2329. doi: 10.1002/j.1460-2075.1990.tb07404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brill SJ, Stillman B. Nature. 1989;342:92–95. doi: 10.1038/342092a0. [DOI] [PubMed] [Google Scholar]
- 7.Brill SJ, Stillman B. Genes Dev. 1991;5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- 8.Erdile LF, Heyer WD, Kolodner R, Kelly TJ. J. Biol. Chem. 1991;266:12090–12098. [PubMed] [Google Scholar]
- 9.Kenny MK, Schlegel U, Furneaux H, Hurwitz J. J. Biol. Chem. 1990;265:7693–7700. [PubMed] [Google Scholar]
- 10.Blackwell LJ, Borowiec JA. Mol. Cell. Biol. 1994;14:3993–4001. doi: 10.1128/mcb.14.6.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blackwell LJ, Borowiec JA, Masrangelo IA. Mol. Cell. Biol. 1996;16:4798–4807. doi: 10.1128/mcb.16.9.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim C, Snyder RO, Wold MS. Mol. Cell. Biol. 1992;12:3050–3059. doi: 10.1128/mcb.12.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim C, Paulus BF, Wold MS. Biochemistry. 1994;33:14197–14206. doi: 10.1021/bi00251a031. [DOI] [PubMed] [Google Scholar]
- 14.Mitsis PG, Kowalczykowski SC, Lehman IR. Biochemistry. 1993;32:5257–5266. doi: 10.1021/bi00070a038. [DOI] [PubMed] [Google Scholar]
- 15.Atrazhev A, Zhang S, Grosse F. European Journal of Biochemistry. 1992;210:855–865. doi: 10.1111/j.1432-1033.1992.tb17489.x. [DOI] [PubMed] [Google Scholar]
- 16.Sugiyama T, Zaitseva EM, Kowalczykowski SC. J. Biol. Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- 17.Alani E, Thresher R, Griffith JD, Kolodner RD. J. Mol. Biol. 1992;227:54–71. doi: 10.1016/0022-2836(92)90681-9. [DOI] [PubMed] [Google Scholar]
- 18.Wold MS, Weinberg DH, Virshup DM, Li JJ, Kelly TJ. J. Biol. Chem. 1989;264:2801–2809. [PubMed] [Google Scholar]
- 19.Brill SJ, Bastin-Shanower S. Mol. Cell. Biol. 1998;18:7225–7234. doi: 10.1128/mcb.18.12.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomes XV, Wold MS. Biochemistry. 1996;35:10558–10568. doi: 10.1021/bi9607517. [DOI] [PubMed] [Google Scholar]
- 21.Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 22.Bochkareva E, Korolev S, Bochkarev A. J. Biol. Chem. 2000;275:27332–27338. doi: 10.1074/jbc.M000620200. [DOI] [PubMed] [Google Scholar]
- 23.Philipova D, Mullen JR, Maniar HS, Lu J, Gu C, Brill SJ. Genes Dev. 1996;10:2222–2233. doi: 10.1101/gad.10.17.2222. [DOI] [PubMed] [Google Scholar]
- 24.Mass G, Nethanel T, Kaufmann G. Mol. Cell. Biol. 1998;18:6399–6407. doi: 10.1128/mcb.18.11.6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bochkareva E, Frappier L, Edwards AM, Bochkarev A. J. Biol. Chem. 1998;273:3932–3936. doi: 10.1074/jbc.273.7.3932. [DOI] [PubMed] [Google Scholar]
- 26.Gomes XV, Wold MS. J. Biol. Chem. 1995;270:4534–4543. doi: 10.1074/jbc.270.9.4534. [DOI] [PubMed] [Google Scholar]
- 27.Henricksen LA, Umbricht CB, Wold MS. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 28.Bochkareva E, Belegu V, Korolev S, Bochkarev A. EMBO J. 2001;20:612–618. doi: 10.1093/emboj/20.3.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 30.Kim C, Wold MS. Biochemistry. 1995;34:2058–2064. doi: 10.1021/bi00006a028. [DOI] [PubMed] [Google Scholar]
- 31.Ferrari ME, Bujalowski W, Lohman TM. J. Mol. Biol. 1994;236:106–123. doi: 10.1006/jmbi.1994.1122. [DOI] [PubMed] [Google Scholar]
- 32.Bochkarev A, Bochkareva E, Frappier L, Edwards AM. EMBO J. 1999;18:4498–4504. doi: 10.1093/emboj/18.16.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobs DM, Lipton AS, Isern NG, Daughdrill GW, Lowry DF, Gomes X, Wold MS. J Biomol NMR. 1999;14:321–331. doi: 10.1023/a:1008373009786. [DOI] [PubMed] [Google Scholar]
- 34.Ikegami T, Kuraoka I, Saijo M, Kodo N, Kyogoku Y, Morikawa K, Tanaka K, Shirakawa M. Nat Struct Biol. 1998;5:701–706. doi: 10.1038/1400. [DOI] [PubMed] [Google Scholar]
- 35.Dornreiter I, Erdile LF, Gilbert IU, von Winkler D, Kelly TJ, Fanning E. EMBO J. 1992;11:769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim DK, Stigger E, Lee SH. J. Biol. Chem. 1996;271:15124–15129. doi: 10.1074/jbc.271.25.15124. [DOI] [PubMed] [Google Scholar]
- 37.Braun KA, Lao Y, He Z, Ingles CJ, Wold MS. Biochemistry. 1997;36:8443–8454. doi: 10.1021/bi970473r. [DOI] [PubMed] [Google Scholar]
- 38.Kim H-S, Brill SJ. Mol. Cell. Biol. 2001;21:3725–3737. doi: 10.1128/MCB.21.11.3725-3737.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walther AP, Gomes XV, Lao Y, Lee CG, Wold MS. Biochemistry. 1999;38:3963–3973. doi: 10.1021/bi982370u. [DOI] [PubMed] [Google Scholar]
- 40.Gomes XV, Henricksen LA, Wold MS. Biochemistry. 1996;35:5586–5595. doi: 10.1021/bi9526995. [DOI] [PubMed] [Google Scholar]
- 41.Din S, Brill S, Fairman MP, Stillman B. Genes Dev. 1990;4:968–977. doi: 10.1101/gad.4.6.968. [DOI] [PubMed] [Google Scholar]
- 42.Brush GS, Morrow DM, Hieter P, Kelly TJ. Proc. Natl. Acad. Sci. USA. 1996;93:15075–15080. doi: 10.1073/pnas.93.26.15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henricksen LA, Wold MS. J. Biol. Chem. 1994;269:24203–24208. [PubMed] [Google Scholar]
- 44.Treuner K, Ramsperger U, Knippers R. J. Mol. Biol. 1996;259:104–112. doi: 10.1006/jmbi.1996.0305. [DOI] [PubMed] [Google Scholar]
- 45.He Z, Wong JM, Maniar HS, Brill SJ, Ingles CJ. J. Biol. Chem. 1996;271:28243–28249. doi: 10.1074/jbc.271.45.28243. [DOI] [PubMed] [Google Scholar]