Abstract
Motivation: Prochlorococcus possesses the smallest genome of all sequenced photoautotrophs. Although the number of regulatory proteins in the genome is very small, the relative number of small regulatory RNAs is comparable with that of other bacteria. The compact genome size of Prochlorococcus offers an ideal system to search for targets of small RNAs (sRNAs) and to refine existing target prediction algorithms.
Results: Target predictions for the cyanobacterial sRNA Yfr1 were carried out with INTARNA in Prochlorococcus MED4. The ultraconserved Yfr1 sequence motif was defined as the putative interaction seed. To study the impact of Yfr1 on its predicted mRNA targets, a reporter system based on green fluorescent protein (GFP) was applied. We show that Yfr1 inhibits the translation of two predicted targets. We used mutation analysis to confirm that Yfr1 directly regulates its targets by an antisense interaction sequestering the ribosome binding site, and to assess the importance of interaction site accessibility.
Contact: backofen@informatik.uni-freiburg.de; claudia.steglich@biologie.uni-freiburg.de
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Bacterial small RNAs (sRNAs) are regulatory RNAs that often act as post-transcriptional regulators by base pairing to trans-encoded target mRNAs. The sRNA–mRNA interaction can result in translational repression and/or mRNA degradation, as well as translational activation, mostly in response to changing environmental conditions (Waters and Storz, 2009). The few sRNA–mRNA interactions experimentally characterized so far have been particularly studied in the two model organisms Escherichia coli (E.coli) and Salmonella typhimurium LT2 (Salmonella) (Gottesman, 2005; Vogel, 2009).
However, sRNA regulators are not restricted to model bacteria, but occur ubiquitously in bacteria. In this study, we investigated the ecologically important cyanobacterium Prochlorococcus. This photoautotrophically dwelling organism often accounts for up to 50% of the organic biomass in the oligotrophic areas of the open oceans, and is thus a crucial component of the food web (Goericke and Welschmeyer, 1993; Vaulot et al., 1995). A recent systematic survey of sRNAs in Prochlorococcus MED4 revealed a large number of potential regulatory RNAs comparable with those found in other bacteria (Steglich et al., 2008). This finding was very surprising, as Prochlorococcus has experienced an evolutionary streamlining of its genome, leading to very compact genomes between 1.64 and 2.68 Mb, which notably results in a small number of regulatory proteins (Kettler et al., 2007). The identification of sRNA targets in Prochlorococcus constitutes a big challenge, since common experimental approaches such as knockouts of these sRNAs cannot be applied. Instead, the only possible approach is a combination of in silico target prediction, followed by in vivo experimental validation (in a heterologous expression system).
An interesting sRNA candidate to study is Yfr1, which is an abundant RNA with ubiquitous appearance in all lineages of cyanobacteria except for two Prochlorococcus strains (Voss et al., 2007). Recent studies have shown that Yfr1 is constitutively expressed and accumulates up to 18 000 copies per cell in Synechococcus elangatus PCC6301 (Nakamura et al., 2007). The high copy numbers of Yfr1 raise the question of whether this RNA acts as a trans-encoded sRNA through base pairing with its targets, or whether it modulates protein activity. An example of such modulation activity is the 6S RNA, which downregulates mRNA transcription by mimicking an open promoter complex (Wassarman, 2007). However, a prominent feature of Yfr1 is the ultraconserved 11 nt long sequence motif located in an unpaired sequence stretch flanked by two stem–loops (Fig. 1A). Similar to Yfr1, the two Salmonella sRNAs GcvB and RybB show a conserved single-stranded region. In both the GcvB and RybB sRNAs, these regions are involved in the binding of multiple targets, which results in reduced translation of the targets (Vogel, 2009). To verify whether Yfr1 analogously regulates trans-encoded mRNAs via base pairing, we predicted putative interaction partners of Yfr1 in the cyanobacterium Prochlorococcus MED4 and experimentally validated these candidates by a reporter system based on green fluorescent protein (GFP).
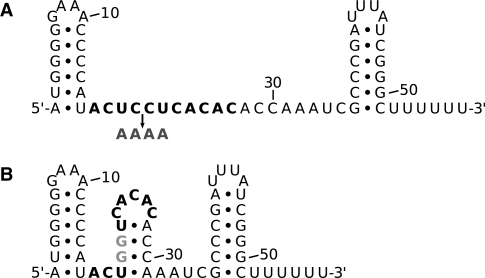
Fig. 1.
(A) Secondary structure of Prochlorococcus MED4 Yfr1, as predicted by RNAfold (Hofacker et al., 1994). The ultraconserved region is set in bold. The arrow indicates the introduced mutation M2 (dark grey). (B) Secondary structure resulting from mutation M1 (substituted positions highlighted in light grey).
2 METHODS
2.1 Computational prediction of Yfr1 targets
For the target prediction, a 400 nt subsequence including 250 nt upstream and 150 nt downstream of the start codon was extracted for all annotated genes of the Prochlorococcus MED4 genome [GenBank accession number BX548174 (Rocap et al., 2003) using the updated annotation by Kettler et al. (2007)]. In total, we obtained 1964 sequences covering the full 5′ untranslated region (5′ UTR) (if not >250 nt) and the beginning of the coding sequence of each gene to search for interactions with Yfr1.
Putative interactions with Yfr1 were predicted with IntaRNA based on hybridization energy and accessibility of the interaction sites (Busch et al., 2008). The IntaRNA approach also incorporates interaction seeds, i.e. short regions of (nearly) perfect sequence complementarity. Accessibility is defined as the energy required to unfold the region of interaction in each molecule. In the calculation of these unfolding energies, we assumed global folding of Yfr1. In contrast, the mRNA does not fold globally due to helicase activity of the ribosome (Takyar et al., 2005). Hence, the mRNA subsequence was locally folded in a 200 nt window with a maximal base pair distance of 100 nt. For each gene, the optimal interaction and up to five suboptimal interactions were computed.
In Prochlorococcus MED4, the ultraconserved motif 5′-ACUCCUCACAC−3′ covers positions 17–27 of Yfr1 RNA (Fig. 1A). This motif was predicted to be single-stranded in the consensus secondary structure of Yfr1 orthologs from 31 cyanobacteria (Voss et al., 2007). In order to search for interactions with this motif as seed region, we extended the IntaRNA program by adding optional constraints that allow to fix the seed position to a given interval of the sRNA sequence. For the target search, we defined an interaction seed of eight paired bases and at most one unpaired base within the aforementioned conserved Yfr1 motif (IntaRNA parameters -p 8 -u 1 -f 17,27). To investigate the influence of interaction seeds, the target prediction was additionally conducted without requiring a seed region (IntaRNA parameter -p 2 for at least 2 bp).
We also tested a modified energy score that weights the accessibility against the hybridization energy with factor α:
where Ehybrid denotes the hybridization energy of the interaction and EDx denotes the energy required to make the interaction site accessible in sequence x. The original IntaRNA scoring does not weight the unfolding energy of the interaction sites, i.e. α=1.
In addition to the IntaRNA energy score, the location of the interaction in the mRNA is used as a further criterion to evaluate the quality of prediction. The majority of characterized trans-encoded sRNAs downregulate their targets by base pairing to the 5′ UTR in the vicinity of the ribosome binding site (RBS) (reviewed in Aiba, 2007). Therefore, the predicted target candidates were filtered for interactions that involve the mRNA region from −39 to +19 relative to the start codon, which is the maximal region covered by ribosomes (Hüttenhofer and Noller, 1994).
The Yfr1-target interactions predicted with fixed seed and full accessibility scoring are provided in Supplementary Material 1. Target candidates resulting from each parameter setting are listed in Supplementary Table 1.
2.2 Experimental validation of Yfr1 targets
2.2.1 E.coli growth conditions and plasmid constructions
E.coli strain Top10F′ was used for cloning of all target-gfp fusions in plasmid pXG-10 or of Yfr1 gene in plasmid pZE12-luc. All interaction studies were carried out in E.coli strain Top10. E.coli cells were grown in Luria–Bertani broth at 37°C in the presence of 100 μg/ml ampicillin and/or 25 μg/ml chloramphenicol. Plasmids used in this work were obtained from Dr Jörg Vogel (MPI, Berlin). Plasmid constructions of the respective 5′ UTRs and of Yfr1 are described in detail in Urban and Vogel (2007). In brief, full-length 5′ UTRs and the first coding residues of the targets of interest were ligated in pXG-10 plasmid using two complementary oligonucleotides with an Mph1103I restriction site at the 5′ terminus and an NheI restriction site at the 3′ terminus, which were annealed to each other prior to ligation. In the case of the 5′ UTR of PMM0494, a PCR-generated fragment (containing an Mph1103I and an NheI restriction site) was digested and ligated into Mph1103I- and NheI-digested pXG-10 plasmid. The Yfr1 gene was amplified by PCR containing an XbaI restriction site and ligated in pZE12-luc plasmid containing an XbaI restriction site for insertion. Yfr1 mutants (Yfr1 M1: CC at positions 20 and 21 substituted by GG leading to the formation of a stem–loop structure in the normally unpaired region, Yfr1 M2: UCCU at positions 19–22 substituted by AAAA without changing the structure, see Figure 1) were generated by annealing two complementary oligonucleotides containing an XbaI restriction site. The complete list of oligonucleotides used for cloning is provided in Supplementary Table 2.
2.2.2 Analysis of Yfr1-mediated target regulation
We tested potential interactions of Yfr1 sRNA with the 5′ UTRs of the putative targets PMM0050 (argJ, bifunctional ornithine acetyltransferase/N-acetylglutamate synthase), PMM0494 (ppa, putative inorganic pyrophosphatase), PMM0538 (unknown function), PMM1119 (som, outer membrane protein), PMM1121 (som, outer membrane protein) or PMM1697 (type II alternative σ factor). For fluorescence measurement, overnight cultures were grown in 96-well plates (Nunc, Roskilde, Denmark) at 37°C with gentle agitation in an air humidity saturated environment to prevent evaporation. Cells were diluted 1:100, fixed in 1% Histofix (Roth, Karlsruhe, Germany) and kept in darkness until measurements were conducted. Single cell fluorescence was determined by flow cytometry with the flow cytometer LSR II (BD Bioscience, New Jersey, USA). Cell fluorescence was measured with an excitation wavelength of 488nm and the emission was detected at 513/17nm. Target-gfp fusions as well as control plasmids pXG-0 (negative control) and pXG-1 (positive control) were tested in the presence of a nonsense RNA and Yfr1 sRNA, respectively. The mean fluorescence per plasmid combination was calculated from 10 000 events (cells) of six individual clones.
3 RESULTS AND DISCUSSION
3.1 Experimental validation of predicted Yfr1 targets
Table 1 lists the 10 highest scoring candidates of the Yfr1 target prediction. Out of these, we experimentally tested the six monocistronic target candidates with known transcriptional start sites and interaction sites predicted in the 5′ UTR or at the start codon. The predicted interactions for targets with a GFP fluorescence signal above background (indicating measurable expression) are shown in Figure 2. Two of the six tested target candidates are translationally repressed by Yfr1, as shown by a reduced GFP fluorescence signal (Fig. 3). The first clusters of the bar chart in Figure 3 constitute the negative controls (E.coli strain Top10 without plasmid or with plasmid pXG-0 devoid of gfp, respectively) and the positive control (E.coli strain Top10 with plasmid pXG-1 carrying gfp). The remaining clusters represent the 5′ UTR-gfp fusions for the targets of interest. Each gfp fusion plasmid was tested in the presence of a second plasmid containing a nonsense RNA (white bars), Yfr1 sRNA (red bars) and the two mutated Yfr1 sRNAs M1 and M2 (light and dark blue bars) (Fig. 3).
Table 1.
Highest scoring Yfr1 target candidates and their ranks under different IntaRNA parameter settings
| Target | Fixed seed |
No seed |
|||||
|---|---|---|---|---|---|---|---|
| α | 1 | 0.5 | 0 | 1 | 0.5 | 0 | |
| PMM1119 (som) | 1 | 4 | 22 | 1 | 11 | 173 | |
| PMM0494 (ppa) | 2 | 3 | 25 | 2 | 10 | 195 | |
| PMM1121 (som) | 3 | 6 | 32 | 4 | 32 | 184 | |
| PMM1697 | 4 | 7 | 12 | 12 | 19 | 10 | |
| PMED4_09051 | 5 | 1 | 8 | 52 | 2 | 46 | |
| PMM0538 | 6 | 16 | 14 | 26 | 84 | 91 | |
| PMM0130 | 7 | 13 | 72 | 13 | 60 | 238 | |
| PMM1021 | 8 | 22 | 69 | 5 | 51 | 830 | |
| PMM1405 | 9 | 12 | 26 | 37 | 61 | 115 | |
| PMM0050 (argJ) | 10 | 11 | 7 | 40 | 8 | 11 | |
Only interactions at RBS [−39 to +19 relative to start codon, see Hüttenhofer and Noller (1994)] were considered. All ranks are given according to IntaRNA energy score. α is a weighting factor for the accessibility in the energy score.
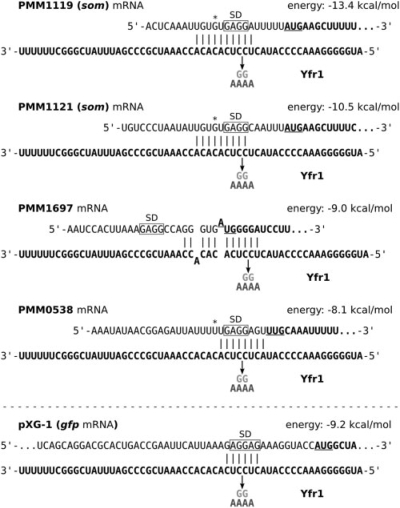
Fig. 2.
Interactions between Yfr1 and target mRNA 5′ UTRs predicted by IntaRNA. Additionally, a putative interaction between Yfr1 and the positive control pXG-1 is presented. The 5′ ends of the mRNAs were experimentally mapped by deep sequencing (C.Steglich, unpublished data). Yfr1 RNA and coding sequences of the mRNAs are set in bold. Start codons are underlined. Shine-Dalgarno (SD) sequences are marked with a box. Asterisks denote start codons that are presumably misannotated in the Prochlorococcus MED4 genome sequence. The arrows indicate mutations M1 (light grey) and M2 (dark grey) introduced in Yfr1.
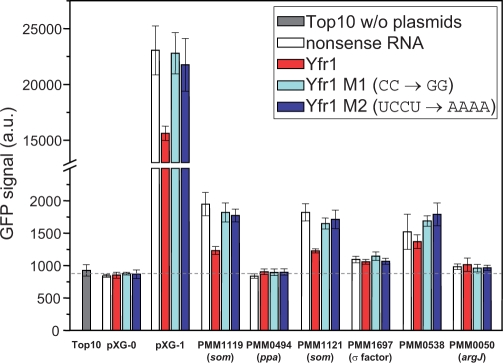
Fig. 3.
Experimental validation of Yfr1 target predictions. The relative decrease in GFP fluorescence as determined by flow cytometry indicates the strength of Yfr1-mediated regulation. The dashed line indicates background fluorescence (i.e. cellular autofluorescence), determined as the mean GFP signal of the negative controls. Fold changes of reduced GFP signal for PMM1119 (3.0-fold), PMM1121 (2.7-fold) and pXG1 (1.5-fold) were calculated after background subtraction from absolute fluorescence values (Urban and Vogel, 2007).
In the presence of the nonsense RNA, no regulation of the 5′ UTR-gfp fusions by an interaction is expected (Fig. 3, white bars), and the fluorescence measured here represents the 5′ UTR-specific translation efficiency. The different GFP fluorescence intensities can be explained by differences in the affinities of the ribosomes for the translation initiation region. The strongest inhibition by Yfr1 was detected for the 5′ UTRs of the two som genes PMM1119 and PMM1121 (3.0- and 2.7-fold reduced GFP signal, red bars in Fig. 3). No change in GFP fluorescence was observed for PMM1697 and PMM0538 5′ UTRs in the presence of Yfr1. For PMM0494 and PMM0050, no fluorescence above the background level (dashed line in Fig. 3) could be detected for any tested plasmid combination.
Translation inhibition of the two soms was abolished by the introduction of a mutation in the conserved Yfr1 motif exchanging CC by GG (Yfr1 M1, light blue bars in Fig. 3). These two substitutions involve the region predicted to base pair with the RBS of the two som mRNAs. Furthermore, mutation M1 led to a structural change by introducing a stem–loop in the single-stranded region of wild-type Yfr1 (Fig. 1B). Thus, mutation M1 results in both a sequential and structural change at the interaction site. To test whether the destruction of the antisense complementarity alone (without structural change) abolishes regulation by Yfr1, we constructed another Yfr1 mutant. In the Yfr1 mutant M2, nucleotides UCCU were substituted by AAAA without changing the secondary structure of wild-type Yfr1 (Fig. 1A). Again, translation of PMM1119 and PMM1121 was restored (Fig. 3, dark blue bars). These results indicate that Yfr1 inhibits translation of the two som mRNAs by direct base pairing at the RBS. Furthermore, the results strongly indicate that both sequence and structure are important for Yfr1 regulation.
Surprisingly, we also observed a 1.5-fold reduction in GFP fluorescence for the positive control pXG-1 in the presence of Yfr1 and restored translation under the control of Yfr1 M1 and M2. However, the strong RBS in the 5′ UTR of gfp in pXG-1 (Urban and Vogel, 2007) shows a perfect complementarity to part of the conserved Yfr1 motif. Thus, Yfr1 can form a perfect 6 nt duplex with the 5′ UTR (Fig. 2), which can explain the observation of a reduction in translation.
3.2 Influence of seed requirement and accessibility on Yfr1 target prediction
The prediction of sRNA targets with IntaRNA is based on two assumptions: (i) a seed region is required to initiate the interaction [in analogy to the 5′ seed region of miRNAs (Bartel, 2009)] and (ii) the accessibility of the interaction sites is important for target recognition. A previous study on a dataset of 18 different sRNA–mRNA interactions presented evidence that the incorporation of these two requirements improves the prediction quality of IntaRNA (Busch et al., 2008). Here, we investigated the importance of accessibility and of a seed region in a practical application, namely the identification of new targets for the Yfr1 sRNA.
Therefore, we computed lists of putative targets without enforcing a seed region and with enforcing a seed at the conserved Yfr1 motif. When requiring the fixed seed position, we obtained a short list of only 29 target candidates with the experimentally validated Yfr1 targets PMM1119 and PMM1121 ranked at positions 1 and 3, respectively (Table 1). Without the seed requirement, 1418 target candidates were obtained with the two true positives ranked at positions 1 and 4. Even without using a seed constraint, the interactions predicted for the true positives include the conserved single-stranded region of Yfr1. Thus, the combination of complementarity and accessibility alone resulted in interactions with an implicit seed.
In addition to the effect of a seed requirement, we studied the influence of accessibility on the Yfr1 target prediction. In the original IntaRNA scoring, hybridization energy and interaction site accessibilities contribute equally to the energy score. Here, we tested a modified energy score, where the interaction site accessibility of both sequences was weighted by factor α with the values 0, 0.5 and 1. For both seed requirements studied, the true positives PMM1119 and PMM1121 were ranked best with the original scoring (Table 1).
One interesting observation was that in the case of Yfr1, a full weighting of the interaction site accessibility, i.e. α=1, was required for a correct target site prediction. When both the seed region and accessibility were neglected, the two verified Yfr1 targets were not found within the top 150 predictions. When the seed position was fixed to the conserved region but accessibility was not included in the scoring, the validated targets were ranked at positions 22 and 32. However, in this case, predicted interactions involved almost the entire Yfr1 sequence (data not shown). This observation is consistent with the findings of Tjaden et al. (2006) and Busch et al. (2008), who showed that an energy model based solely on hybridization energy tends to maximize the length of hybridization, resulting in a small fraction of correctly predicted base pairs (i.e. low positive predictive value).
4 CONCLUSIONS
In this study, we show that Yfr1 sRNA modulates the translation of two high-scoring predicted targets by an antisense interaction. Both target mRNAs code for outer membrane proteins (Hansel et al., 1998). This class of proteins constitutes a major functional class that is regulated by bacterial sRNAs in E.coli and Salmonella (Waters and Storz, 2009). The result was surprising as, until now, no highly abundant sRNAs have been shown to act via base pair interaction. However, both mRNA targets identified herein are also highly abundant [among the 10 most expressed mRNAs and with long half-lives of about 30 min (C.Steglich, unpublished data)], which may require a high copy number of Yfr1 for efficient regulation. Furthermore, an mRNA with a long half-life can be regulated more efficiently by translational control than by transcriptional control.
Additionally, we assessed the influence of seed regions and interaction site accessibility on the prediction quality of Yfr1 targets. As with the Salmonella sRNAs GcvB and RybB, Yfr1 contains a conserved single-stranded region, which seems to constitute a perfect interaction seed. When requiring this region as seed for the target prediction, the number of putative Yfr1 targets was remarkably smaller without seed requirement (29 versus 1418 candidates), although the two true positives were under the highest ranking candidates in both settings. When neglecting both accessibility and a seed region, the true Yfr1 targets could not be found amongst the top 150 predictions.
In conclusion, the combination of computational and experimental methods, as presented in this study, proved to be an appropriate approach for the identification of sRNA targets in organisms where genetic manipulation constitutes a great challenge.
Funding: German Research Foundation DFG Priority Program SPP1258 Sensory and Regulatory RNAs in Prokaryotes (grant number BA 2168/2-1 to R.B., Ste 1119/2-1 to C.S.]; German Federal Ministry of Education and Research FRISYS - Freiburg Initiative for Systems Biology (grant number 0313921 to R.B.).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A, et al. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics. 2008;24:2849–2856. doi: 10.1093/bioinformatics/btn544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goericke R, Welschmeyer NA. The marine prochlorophyte Prochlorococcus contributes significantly to phytoplankton biomass and primary production in the sargasso sea. Deep Sea Res. Part I Oceanogr. Res. Pap. 1993;40:2283–2294. [Google Scholar]
- Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Hansel A, et al. Cloning and characterization of the genes coding for two porins in the unicellular cyanobacterium Synechococcus PCC 6301. Biochim. Biophys. Acta. 1998;1399:31–39. doi: 10.1016/s0167-4781(98)00090-6. [DOI] [PubMed] [Google Scholar]
- Hofacker IL, et al. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 1994;125:167–188. [Google Scholar]
- Hüttenhofer A, Noller HF. Footprinting mRNA-ribosome complexes with chemical probes. EMBO J. 1994;13:3892–3901. doi: 10.1002/j.1460-2075.1994.tb06700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettler GC, et al. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 2007;3:e231. doi: 10.1371/journal.pgen.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, et al. A cyanobacterial non-coding RNA, Yfr1, is required for growth under multiple stress conditions. Plant Cell Physiol. 2007;48:1309–1318. doi: 10.1093/pcp/pcm098. [DOI] [PubMed] [Google Scholar]
- Rocap G, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- Steglich C, et al. The challenge of regulation in a minimal photoautotroph: non-coding RNAs in Prochlorococcus. PLoS Genet. 2008;4:e1000173. doi: 10.1371/journal.pgen.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takyar S, et al. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- Tjaden B, et al. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 2006;34:2791–2802. doi: 10.1093/nar/gkl356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35:1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaulot D, et al. Growth of Prochlorococcus a Photosynthetic Prokaryote, in the Equatorial Pacific Ocean. Science. 1995;268:1480–1482. doi: 10.1126/science.268.5216.1480. [DOI] [PubMed] [Google Scholar]
- Vogel J. A rough guide to the non-coding RNA world of Salmonella. Mol. Microbiol. 2009;71:1–11. doi: 10.1111/j.1365-2958.2008.06505.x. [DOI] [PubMed] [Google Scholar]
- Voss B, et al. A motif-based search in bacterial genomes identifies the ortholog of the small RNA Yfr1 in all lineages of cyanobacteria. BMC Genomics. 2007;8:375. doi: 10.1186/1471-2164-8-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM. 6S RNA: a small RNA regulator of transcription. Curr. Opin. Microbiol. 2007;10:164–168. doi: 10.1016/j.mib.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.