Abstract
Summary: We present Grid Analysis of Time series Expression (GATE), an integrated computational software platform for the analysis and visualization of high-dimensional biomolecular time series. GATE uses a correlation-based clustering algorithm to arrange molecular time series on a two-dimensional hexagonal array and dynamically colors individual hexagons according to the expression level of the molecular component to which they are assigned, to create animated movies of systems-level molecular regulatory dynamics. In order to infer potential regulatory control mechanisms from patterns of correlation, GATE also allows interactive interroga-tion of movies against a wide variety of prior knowledge datasets. GATE movies can be paused and are interactive, allowing users to reconstruct networks and perform functional enrichment analyses. Movies created with GATE can be saved in Flash format and can be inserted directly into PDF manuscript files as interactive figures.
Availability: GATE is available for download and is free for academic use from http://amp.pharm.mssm.edu/maayan-lab/gate.htm
Contact: avi.maayan@mssm.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Advances in experimental molecular biology techniques are allowing us to elucidate the molecular mechanisms cell behavior with ever-increasing detail. Consequently, it is now routine for individual studies to employ a variety of high-throughput techniques—each of which characterizes the molecular state of the cell at different layers of regulation—including microarrays to assess genome-wide mRNA expression; high-throughput chromatin immunoprecipitation techniques to assess protein–DNA interactions and epigenetic status; and mass spectrometry proteomics and phosphoproteomics to assess protein expression profiles. However, despite the success of these emerging experimental techniques, the potential gains from these advances are generally not fully realized since high-throughput techniques often produce considerably more data than our current ability to adequately organize and analyze, resulting in loss of potentially valuable information. This problem is particularly acute when considering high-throughput time series. In order to address this data integration challenge, a number of systematically curated databases which collate data from different experiments have been established. For example, the Molecular Signatures Database provides a set of annotated gene sets representing a large number of gene expression profiles from a variety of organisms for use with Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005). In addition, a variety of regulatory networks, which encode systems-level molecular interactions, have also been reconstructed, with the aim of dissecting coordinated or synergistic regulatory control of cell behavior. Along with these networks, a range of tools to infer functional subnetworks that ‘connect’ molecular species of interest—and thereby uncover pathways or regulatory modules responsible for changes in cellular state—have also been developed (Berger et al., 2007; Ulitsky and Shamir, 2007). However, many interesting cellular phenomena are dynamic and concern the ways in which transitions from one cellular state to another occur and the molecular mechanisms responsible for accompanying changes in cellular phenotype. For this reason, several algorithms have been developed specifically for clustering and interrogating high-dimensional time series (Ma et al., 2006; Segal et al., 2003; Wang et al., 2008). Of particular note is the Gene Expression Dynamics Inspector (GEDI) (Eichler et al., 2003). GEDI makes animated movies of changing gene expression patterns using dynamically colored self-organizing maps. However, although GEDI is a powerful computational resource, it does not allow interrogation of dynamic patterns of expression against databases of prior knowledge, thus making it difficult to identify potential functional mechanisms responsible for observed dynamic gene expression patterns. However, several approaches have been developed specifically for this purpose. For example, the Short Time series Expression Miner (STEM) uses a correlation-based clustering in combination with gene ontology enrichment analysis to identify potential regulatory mechanisms responsible for expression changes (Ernst and Bar-Joseph, 2006). However, STEM results are not dynamically visualized as with GEDI but rather are visualized using line graphs and histograms.
2 METHODS AND RESULTS
In order to combine systems-level visualization of expression dynamics with interrogation against prior knowledge, we developed an integrated visualization and analysis software package named Grid Analysis of Time Series Expression (GATE; Fig. 1). GATE uses a correlation-based clustering algorithm to arrange molecular time series in a two-dimensional hexagonal grid in a manner similar to that employed by GEDI. However, in order to interrogate more effectively against databases of prior knowledge, GATE assigns genes/proteins (or other molecular components) to hexagons in a strictly one–one manner (rather than clustering them together as GEDI does). Each individual hexagon in the array is then dynamically colored according to the expression level of the molecular component to which it is assigned, allowing the creation of animated movies that illustrate systems level molecular expression dynamics (see supporting text for a detailed description of the algorithm). Additionally, in order to infer potential regulatory control mechanisms from patterns of time series correlations, GATE also allows interactive interrogation of time course movies against a wide variety of background knowledge datasets. In particular, movies can be paused at any time by the user and are interactive, allowing users to highlight areas of interest, examine/save their content and interrogate features of interest for enrichment against a variety of background knowledge datasets including: known protein–protein interactions; known targets of specific transcription factors; known substrates of specific protein kinases (Lachmann and Ma'ayan, 2009); known targets of specific microRNAs; common gene ontologies; common metabolites; common chromosomal locations; common structural domains; common signaling pathways; and common promoter sequences. In fact, GATE allows flexible interrogation of time series data against any dataset in the database of molecular signatures (Subramanian et al., 2005); any other prior knowledge dataset supplied in the GSEA Gene Matrix Transposed (.GMT) flat-file format; or any background network supplied in the Cytoscape-compatible simple interactions format (.SIF). In addition to allowing users to investigate enrichment against background datasets, GATE also allows users to visualize previously reported interactions between molecular components (Berger et al., 2007) directly on the GATE hexagonal array. Subnetworks of interactions can also be exported from GATE as simple interaction files (.SIF) for visualization in other software packages such as Cytoscape (Shannon et al., 2003).
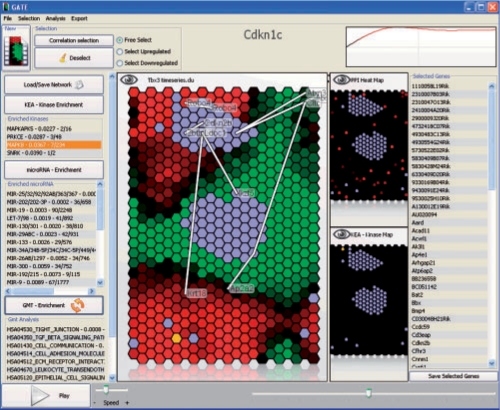
Fig. 1.
Screenshot from the GATE software. Green hexagons represent biomolecular components that decreased in expression compared with the initial state, while red hexagons represent biomolecular components that increased. On the left are the results from enrichment analysis, and on the right is a list of currently selected components. The time-line plot for the currently selected component Cdkn1c is highlighted on the top right.
Additionally, time series expression movies created with GATE can be converted into Flash files and inserted into PDF manuscript files as interactive figures. Users can export movies as Flash ActionScript 3.0 scripts which can then be imported into Adobe Flash ready for conversion into .swf files. Such files can be embedded in PDF manuscript files using the multimedia tool recently added to Adobe Acrobat (Version 9). GATE also provides features to search for specific biomolecular components on the hexagonal array, and to select biomolecular components based on common expression trends interactively sketched with a pen.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Betty Chang, Dung-Fang Lee and Avinash Waghray for useful discussions.
Funding: National Institutes of Health (Grant No. 1P50GM071558-01A27398); start-up fund from Mount Sinai School of Medicine (to A.M.).
Conflict of Interest: none declared.
REFERENCES
- Berger S, et al. Genes2Networks: connecting lists of gene symbols using mammalian protein interactions databases. BMC Bioinformatics. 2007;8:372. doi: 10.1186/1471-2105-8-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler GS, et al. Gene Expression Dynamics Inspector (GEDI): for integrative analysis of expression profiles. Bioinformatics. 2003;19:2321–2322. doi: 10.1093/bioinformatics/btg307. [DOI] [PubMed] [Google Scholar]
- Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann A, Ma'ayan A. KEA: kinase enrichment analysis. Bioinformatics. 2009;25:684–686. doi: 10.1093/bioinformatics/btp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P, et al. A data-driven clustering method for time course gene expression data. Nucleic Acids Res. 2006;34:1261–1269. doi: 10.1093/nar/gkl013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, et al. Discovering molecular pathways from protein interaction and gene expression data. Bioinformatics. 2003;19:i264–i272. doi: 10.1093/bioinformatics/btg1037. [DOI] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shamir R. Identification of functional modules using network topology and high-throughput data. BMC Syst. Biol. 2007;1:8. doi: 10.1186/1752-0509-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, et al. Conditional clustering of temporal expression profiles. BMC Bioinformatics. 2008;9:147. doi: 10.1186/1471-2105-9-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.