Abstract
The mechanisms underlying the generation of neural cell diversity are the subject of intense investigation, which has highlighted the involvement of different signalling molecules including Shh, BMP and Wnt. By contrast, relatively little is known about FGF in this process. In this report we identify an FGF-receptor-dependent pathway in zebrafish hindbrain neural progenitors that give rise to somatic motoneurons, oligodendrocyte progenitors and differentiating astroglia. Using a combination of chemical and genetic approaches to conditionally inactivate FGF-receptor signalling, we investigate the role of this pathway. We show that FGF-receptor signalling is not essential for the survival or maintenance of hindbrain neural progenitors but controls their fate by coordinately regulating key transcription factors. First, by cooperating with Shh, FGF-receptor signalling controls the expression of olig2, a patterning gene essential for the specification of somatic motoneurons and oligodendrocytes. Second, FGF-receptor signalling controls the development of both oligodendrocyte progenitors and astroglia through the regulation of sox9, a gliogenic transcription factor the function of which we show to be conserved in the zebrafish hindbrain. Overall, for the first time in vivo, our results reveal a mechanism of FGF in the control of neural cell diversity.
Keywords: FGF, Sox9, Olig2, Radial glia, Neural progenitor, Astroglia, Astrocyte, Oligodendrocyte, Somatic motoneuron, Gliogenesis, Neurogenesis, Patterning, Positional identity, Zebrafish
INTRODUCTION
A detailed description of the molecular and cellular events controlling the generation of the vast diversity of neuronal and glial cell types that constitute the adult vertebrate central nervous system (CNS) is fundamental to our understanding of the normal functioning of the CNS and to developing treatments for neurological disorders. It is generally accepted that during embryonic development neuroepithelial cells initially and later radial glial cells are neural progenitors that give rise to all neural cell types (Gotz et al., 2002; Doetsch, 2003). The characterisation of the spatial and temporal cues that control the maintenance of these cells, their fate restriction and differentiation into neurons, oligodendrocytes and astrocytes is the subject of intense investigation.
A number of studies on the spinal cord dorsoventral (DV) axis have established a framework for understanding the molecular mechanisms controlling neural cell diversity. Secreted signalling molecules including sonic hedgehog (Shh), produced by the notochord and floor plate, and Wnt and bone morphogenetic proteins (BMPs), produced by the roof plate, act as morphogens to establish domains of expression of transcription factors (Briscoe and Ericson, 1999; Lee and Jessell, 1999). These positional identity factors control the specification of both neuronal and glial subtypes. The basic helix-loop-helix (bHLH) transcription factor Olig2 is one such factor that is activated ventrally by Shh in ventricular zone precursors and underlies the sequential specification of somatic motoneurons (SMNs) and oligodendrocytes (Rowitch et al., 2002).
Intimately linked to the positional identity factors are transcription factors that control generic neurogenesis and gliogenesis programmes. Neurogenesis is mediated by bHLH factors, vertebrate homologues of the Drosophila proteins Atonal and Achaete-scute, including Neurogenin and Ascl, respectively (Bertrand et al., 2002). These factors are downstream targets of the positional identity genes and promote cell cycle exit and pan-neuronal properties. As development proceeds, gliogenesis replaces neurogenesis and a number of factors are known to participate in this transition (Guillemot, 2007b). One important player is the high mobility group (HMG) factor Sox9, which is necessary for the downregulation of neurogenesis and the specification of both oligodendrocytes and astrocytes in the mouse spinal cord (Stolt et al., 2003). Following the production of oligodendrocyte progenitors (OLPs), Sox9 acts in combination with Sox10 to control their survival and migration (Finzsch et al., 2008). In addition, recent studies have shown that gliogenesis in the vertebrate retina is also under the control of Sox9 (Poche et al., 2008; Yokoi et al., 2009). The central role of Sox9 in gliogenesis raises the question as to the signals controlling this factor. Although a number of studies point to important roles of the cytokine, BMP, Notch and FGF signalling pathways in gliogenesis (Guillemot, 2007a; Miller and Gauthier, 2007), how Sox9 is regulated has not yet been determined.
The FGF signalling pathway controls multiple aspects of nervous system development, including neural progenitor survival, proliferation, maintenance and differentiation, as well as tissue patterning and compartmentalisation (Mason, 2007). Studies of neural progenitor cultures pointed to a role of FGF in both neurogenesis and gliogenesis (Qian et al., 1997). In addition, in vitro and in vivo gain-of-function studies suggest that FGF promotes oligogenesis in the dorsal neural tube, in a Shh-independent manner (Chandran et al., 2003; Kessaris et al., 2004; Fogarty et al., 2005; Vallstedt et al., 2005; Naruse et al., 2006), and astrogenesis, by interacting with the cytokine signalling pathway (Morrow et al., 2001). Despite these advances, in vivo approaches are necessary to elucidate how FGF coordinates neural patterning, and neurogenic and gliogenic factors. Although in vivo loss-of-function studies suggest a possible role of FGF in neurogenesis and astrogenesis (Raballo et al., 2000; Reuss et al., 2003; Shin et al., 2004; Smith et al., 2006), these studies remain short of addressing this important question.
We have been investigating the mechanisms controlling the development of SMNs, OLPs and astroglia in the zebrafish hindbrain. The construction of the hindbrain follows neural plate induction and involves the transient subdivision of this region into reiterated units called rhombomeres (Schneider-Maunoury et al., 1998). Hindbrain segmentation underlies the pattern of neuronal (Lumsden, 2004) and glial development (Spassky et al., 1998; Perez Villegas et al., 1999). In this report we investigate an FGF-receptor signal in rhombomere centre radial glial cells that are progenitors of SMNs, OLPs and differentiating astroglia. We show that FGF-receptor signalling controls the generation of these cells by coordinately regulating the expression of olig2 and sox9. From these results we propose a model that provides important insights into the role and mechanism of action of FGF-receptor signalling in the generation of neural cell diversity.
MATERIALS AND METHODS
Fish lines and genotyping
The wild-type zebrafish strains TU and TL were routinely used throughout this study. The transgenic lines Tg(hsp70l:dnfgfr1-gfp) (Lee et al., 2005; ZIRC), Tg(uas:k) (Hatta et al., 2006; ZIRC) and Tg(olig2:gfp) (Shin et al., 2003) and mutant alleles sox9atw37 (Yan et al., 2002), sox9bb971 (Yan et al., 2005) and smob641 (Varga et al., 2001; ZIRC) were described previously. sox9atw37 heterozygous and homozygous and sox9bb971 homozygous embryos were identified by PCR. For sox9atw37, 5′-TCGACCCCTACCTGAAGATG-3′ and 5′-TTCGATGGACATTTCTGACG-3′ were used in PCR and 5′-AGGTGCTGAAGGGTTACGAC-3′ for PCR product sequencing. For sox9bb971, 5′-AGAAGTTCCCGGTGTGTATCC-3′ and 5′-GAAGCCTCATGAGTGTCAGATG-3′ were used in PCR. smob641 mutant embryos were identified by labelling for nkx2.2a. Embryos were obtained from natural spawns and staged as previously described (Kimmel et al., 1995).
Cloning of erm upstream sequences and generation of transgenic lines
To construct the Tg(ermp:gv) transgenic line, the sequence of ENSDARG00000044511 (Ensembl Gene ID) from 5 kb upstream to the ATG start site of translation was amplified by PCR (Phusion, Finnzymes) from the BAC DKEYp-78G2 (imaGenes; HUKGB738G0278D) using the following primers: 5′-GATCCTCGAGGACATCACCGTCTGATGTGCAG-3′ and 5′-GATCGGATCCGTCGCTTGCTTCTCACTGCG-3′. The product was digested with XhoI/BamHI and cloned into the Tol2 vector (Kawakami et al., 2004) after fusing with the gal4-vp16 coding sequences (Koster and Fraser, 2001). Transgenic lines were generated as described (Kawakami et al., 2004).
Heat shock of Tg(hsp70l:dnfgfr1-gfp) line and treatment with SU5402
Embryos from a Tg(hsp70l:dnfgfr1-gfp) hemizygous outcross were heat shocked at 20 hours post-fertilisation (hpf) by incubation at 37°C for 15 min in 5 ml embryo medium (E3) and allowed to develop at 28.5°C to 36 hpf and fixed or heat shocked at 38°C for 30 min and allowed to develop to 48 hpf. Wild-type embryos in E3 medium were incubated with DMSO (control) or 20 μM SU5402 (Calbiochem) dissolved in DMSO at 10 mM during the stages indicated in the figures. In all cases experiments were performed twice on at least 15 control and test embryos.
Whole-mount in situ hybridisation, immunohistochemistry and TUNEL analysis
Probe synthesis and in situ hybridisation procedures have been described previously (Thisse and Thisse, 1998) (for probes, see Table S1 in the supplementary material). In situ colour reactions were performed using NBT/BCIP, INT/BCIP or Fast Red as substrate (Roche). Immunohistochemistry on whole mount or cryosections was performed as described previously (Ellingsen et al., 2005) using the following antibodies: rabbit polyclonal α-GFP (1:500, Torrey Pines Biolabs); mouse monoclonal (Mab) α-GS (1:1000, BD Biosciences); Mab α-HuC/D (16A11) (1:500, Molecular Probes); Mab α-Isl1/2 (39.4D5) (1:100, DSHB); Mab Zn8 (α-ALCAM) (1:500, DSHB); Mab Zrf1 (α-GFAP) (1:5000, ZIRC); Alexa fluor 488-, 594- and Cy5-conjugated secondary antibodies (1:800, Invitrogen and Jackson ImmunoResearch). Fluorescence images were acquired using a DM5000B with spinning disk and TCS SP2AOBS confocal (Leica). In co-expression studies, all confocal images were generated from single confocal sections using 40× or 63× objectives. In situ apoptosis detection (TUNEL) was performed as described previously (Kozlowski et al., 2005) using the Fluorescein In Situ Cell Death Detection Kit (Roche).
RESULTS
SMNs, OLPs and astroglia in the zebrafish hindbrain
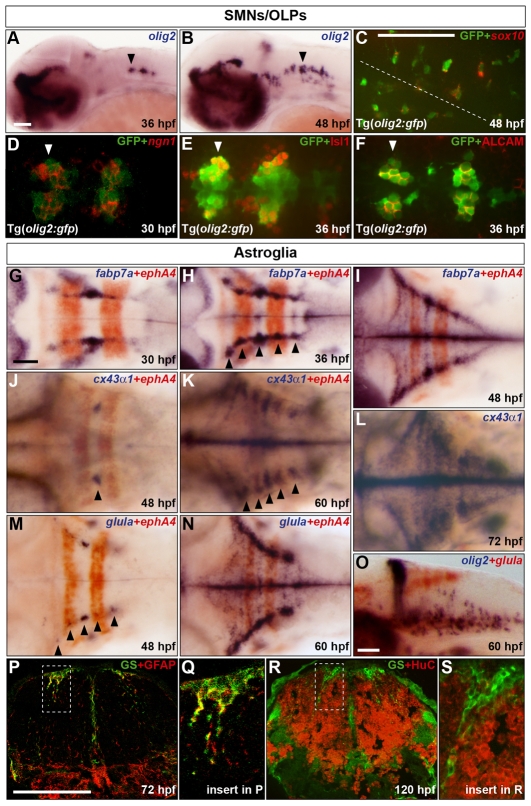
We began this study by characterising the development of SMNs, OLPs and astroglia in the zebrafish hindbrain. Studies in amniotes have established an essential role of the bHLH transcription factor olig2 in the specification of SMNs and oligodendrocytes (Rowitch et al., 2002). In the zebrafish hindbrain, olig2 was first expressed around 30 hpf in the ventromedial ventricular zone of rhombomeres five and six (r5 and r6, data not shown). At 36 hpf, expression was detected in patches in r5-r7 and by 48 hpf olig2 was expressed in r2-r7 and the midbrain/r1 region (Fig. 1A,B). The progeny of these hindbrain olig2-expressing cells was investigated using a bacterial artificial chromosome (BAC) transgenic line Tg(olig2:gfp) that recapitulates the olig2 expression profile (Shin et al., 2003). Double labelling for GFP and the oligodendrocyte lineage-specific transcription factor gene sox10 at 48 hpf revealed sox10 expression in a subset of GFP-positive cells (Fig. 1C). Because olig2 specifies SMNs in combination with neurogenins (Zhou et al., 2001; Zhou and Anderson, 2002), we searched for proneural genes that are expressed in the olig2 lineage. Double labelling at 30 hpf identified ngn1 (neurog1) expression in a subset of olig2:GFP-positive cells (Fig. 1D), suggesting the expression of olig2 in neuronal precursors. In the zebrafish hindbrain, Isl1 is detected in both branchiomotor neurons and SMNs of the abducens nuclei, and this latter population is also positive for ALCAM (DM-GRASP) (Chandrasekhar et al., 1997). Double immunolabelling for olig2:GFP and Isl1 at 36 hpf indicated that only a small proportion of Isl1-positive cells expressed GFP and these were restricted to r5 and r6 (Fig. 1E and data not shown). In addition, double immunolabelling for olig2:GFP and ALCAM at 36 hpf indicated that the majority, if not all, of the ALCAM-positive cells were olig2:GFP-positive (Fig. 1F). Taken together, these results reveal that olig2-expressing precursors in the ventromedial hindbrain give rise to both SMNs of the abducens nuclei and OLPs. Recently, a similar analysis of the olig2 lineage in the zebrafish hindbrain (Zannino and Appel, 2009) resulted in the same conclusions.
Fig. 1.
Characterisation of the development of SMNs, OLPs and astroglia in the zebrafish hindbrain. (A,B) Embryos were analysed for olig2 RNA expression at 36 (A) and 48 (B) hpf. (C-F) Co-expression of GFP protein (green) in Tg(olig2:gfp) line and sox10 (C) or ngn1 (D) RNA and Isl1 (E) or ALCAM (F) protein (red) at 48 (C), 30 (D) and 36 (E,F) hpf. In C the position of the midline is indicated by a dashed line. (D-F) Ventromedial r5-7. Arrowheads indicate r5 (A,B,D-F). (G-O) Wild-type embryos were analysed for RNA expression of fabp7a at 30 (G), 36 (H) and 48 (I) hpf, cx43α1 at 48 (J), 60 (K) and 72 (L) hpf and glula at 48 (M) and 60 (N,O) hpf. Embryos were double labelled for ephA4 (red) RNA to mark r3 and r5 (G-K,M,N) or olig2 (blue) (O). Arrowheads indicate dorsolateral r4 (J) and r2-6 (H,K,M). Note that from about 24 hpf, the hindbrain ventricular zone has a characteristic T-shape, where ventral is located ventromedially and dorsal is located dorsolaterally. Embryos are shown with anterior to the left in lateral (A,B,O) or dorsal (C-N) view. (P-S) Transverse sections of the hindbrain at the level of the otic vesicle were analysed for GS (green) and GFAP (red) at 72 hpf (P) or HuC at 120 hpf (R). The framed areas in P and R are shown at higher magnification in Q and S, respectively. Scale bar: 50 μm.
gfap, the canonical astroglial marker, is widely expressed in the ventricular zone (radial glia) of the embryonic zebrafish CNS and in adults is expressed in the majority of cells with radial glial character and in astrocytes (Tomizawa et al., 2000; Marcus and Easter, 1995; Kawai et al., 2001). Despite the identification of other astroglial markers in teleosts, little is known of their embryonic expression patterns. To address this, we investigated three previously cloned zebrafish genes, fabp7a (brain fatty acid binding protein) (Liu et al., 2004), cx43α1 (connexin 43) (Chatterjee et al., 2005) and glula (glutamine synthetase, GS) (Thisse et al., 2001), which are orthologues of mammalian genes expressed in the astroglial lineage. In mammals, Fabp7 (Blbp) (Feng and Heintz, 1995) and connexin 43 (Gja1 — Mouse Genome Informatics) (Dermietzel et al., 1991; Nadarajah et al., 1997) are predominantly expressed in radial glia and astrocytes, and Glul (Glns) (Pringle et al., 2003) is predominantly expressed in astrocyte precursors and astrocytes. Analysis of the expression profile of fabp7a in the zebrafish hindbrain from 30-48 hpf revealed a progressive amplification within a dorsolateral band of expression, initially in r4 and later in r2-r6 (Fig. 1G-I). Double labelling with the r3/r5 marker, ephA4 (epha4 — Zebrafish Information Network) (Xu et al., 1995), revealed that these patches of fabp7a expression were located in the centre of rhombomeres. Transverse hindbrain sections revealed that fabp7a is expressed throughout the depth of the ventricular zone and at higher levels in the centre compared with boundary regions (see Fig. S1A,B in the supplementary material). Neither cx43α1 nor glula were significantly expressed in the CNS before 48 hpf, at which stage hindbrain expression was first detected in dorsolateral patches in the centre of each rhombomere, initially in r4 and later in r2-r6 (Fig. 1J-N and data not shown). At 72 hpf both fabp7a and glula were expressed throughout the hindbrain ventricular zone with higher levels in the dorsolateral regions (see Fig. S1C,D in the supplementary material). Immunostaining of hindbrain sections at 72-120 hpf identified GS-positive cells in the ventricular zone, particularly in the dorsolateral region, that co-expressed the radial glial marker GFAP but not the neuronal marker HuC (Fig. 1P-S and data not shown). In addition, double RNA in situ labelling revealed that the OLP marker olig2 was not expressed within the dorsal glula expression domain (Fig. 1O). Taken together, these data suggest that radial glia located in the dorsolateral rhombomere centres differentiate within the astroglial lineage. Hereafter these cells are referred to as GS-positive astroglia.
Presence of an FGF-receptor signal in progenitors of hindbrain SMNs, OLPs and GS-positive astroglia
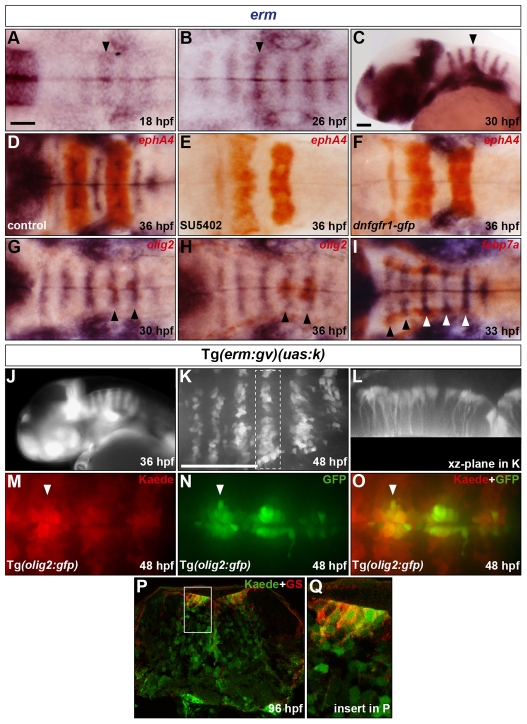
During development, FGF-receptor signalling can be revealed by analysing the expression profile of general targets of the FGF pathway, including the Ets transcription factors erm and pea3 (Raible and Brand, 2001; Roehl and Nusslein-Volhard, 2001). We analysed the profile of erm following hindbrain segmentation. At 18 hpf, erm expression was detected throughout r3-6, except at rhombomere boundaries; progressively these expression domains narrowed, and by 30 hpf were restricted to six transverse stripes in r2-r7 (Fig. 2A-C). Double labelling with ephA4 revealed that erm expression was restricted to the centre of rhombomeres and remained so until at least 48 hpf (Fig. 2D and data not shown).
Fig. 2.
Presence of an FGF-receptor signal in progenitors of hindbrain SMNs, OLPs and GS-positive astroglia. (A-C) Evolution of erm RNA expression in the hindbrain from 18 to 30 hpf. Arrowheads indicate the centre of r4. (D-F) Embryos were analysed at 36 hpf for erm (blue) and ephA4 (red) RNA expression. Wild-type embryos were either treated with DMSO (D) or with SU5402 at 20 hpf (E) and Tg(hsp70l:dnfgfr1-gfp) embryos were heat shocked at 20 hpf (F). (G-I) Embryos were analysed for erm (blue) and olig2 (G,H) or fabp7a (I) (red) RNA expression at 30 (G), 36 (H) and 33 (I) hpf. Arrowheads indicate the centre of r5 and r6 (G,H) or r2-r6 (I). (J) Kaede fluorescence profile in a Tg(erm:gv)(uas:k) embryo at 36 hpf. (K,L) Kaede of a 46 hpf Tg(erm:gv)(uas:k) embryo was completely photoconverted and the embryo was allowed to develop to 48 hpf and was analysed for de novo Kaede fluorescence. Images show half the hindbrain in dorsal (K) and xz-plane of r5 (L). (M-O) Kaede of a 48 hpf Tg(erm:gv)(uas:k)(olig2:gfp) embryo was completely photoconverted and analysed for Kaede (M,O; red) and GFP (N,O; green) fluorescence. The arrowhead indicates the centre of r5. (P,Q) Transverse hindbrain section at the level of the otic vesicle of a Tg(erm:gv)(uas:k) embryo at 96 hpf analysed for Kaede fluorescence (green) and immunolabelling for GS (red). The framed area in P is shown at higher magnification in Q. Embryos are oriented with anterior to the left, in dorsal (A,B,D-I,K,M-O) or lateral (C,J) view. Scale bars: 50 μm.
To confirm that the erm expression pattern reflects FGF-receptor signalling, we generally and specifically blocked FGF-receptor signalling in a temporally controlled manner using two approaches. First, the chemical inhibitor SU5402 was shown previously to effectively inhibit the kinase activity of all FGF-receptor subtypes but not EGF or PDGF receptors (Mohammadi et al., 1997). Second, heat shock induction of the Tg(hsp70l:dnfgfr1-gfp) line was shown to block FGF-receptor signalling by the dimerisation of a dominant-negative FGFR1 with all FGF-receptor subtypes (Lee et al., 2005). At 20 hpf, wild-type embryos were treated with SU5402 or Tg(hsp70l:dnfgfr1-gfp) hemizygous embryos were heat shocked and allowed to develop to 36 hpf. Analysis of the expression of ephA4 showed no significant effect on hindbrain segmentation, whereas the FGF target gene erm was completely downregulated in the hindbrain with both methods of FGF-signal inhibition compared with control embryos (Fig. 2D-F).
We then investigated whether the FGF-receptor signal in rhombomere centres is active in radial glial cells. For this, transgenic lines were generated carrying an approximately 5 kb region upstream of the erm translation start site, fused to a gal4-vp16 construct Tg(erm:gv) (Koster and Fraser, 2001). F1s were crossed to a transgenic line carrying UAS sequences fused to the photoconvertible fluorescent protein Kaede gene Tg(uas:k) (Hatta et al., 2006). Embryos from two of these crosses were analysed in detail and found to recapitulate the expression of the endogenous erm gene throughout embryonic development with the exception of a prolongation of the signal due to the stability of Kaede protein (Fig. 2J and data not shown). Treatment with SU5402 from 20 to 36 hpf downregulated Kaede mRNA expression in both of these lines, revealing a requirement for FGF-receptor signalling (data not shown). Using confocal microscopy we analysed Tg(erm:gv)(uas:k) hindbrains 2 hours after complete photoconversion of the Kaede protein at 46 hpf and found that the de novo Kaede-positive cells were restricted to the rhombomere centre ventricular zone and extended fibres from their apical location towards the basal surface (Fig. 2K,L). Taken together, these results reveal that following hindbrain segmentation, an FGF-receptor signal becomes restricted to a subset of radial glia occupying a domain in the centre of r2-r7.
Because radial glia are known neural progenitors in mammals (Gotz et al., 2002; Doetsch, 2003), we investigated whether FGF-receptor signalling-radial glia are progenitors for hindbrain SMNs, OLPs and GS-positive astroglia. First, in double RNA in situ experiments, we found that at 30 and 36 hpf, erm expression in rhombomere centres is in register with the ventromedial olig2 expression domain in r5 and r6 (Fig. 2G,H) and intersects with the fabp7a expression domain dorsolaterally in r2-r6 (Fig. 2I). We then undertook lineage-tracing studies using the Tg(erm:gv)(uas:k) line. Tg(erm:gv)(uas:k) was crossed with Tg(olig2:gfp) and Kaede was completely photoconverted in Tg(erm:gv)(uas:k)(olig2:gfp) embryos allowing us to distinguish the Kaede from the GFP signal. At 48 hpf we detected numerous Kaede-positive cells in ventral r5-r7 that were also GFP-positive (Fig. 2M-O). In addition, analysis of hindbrain sections of Tg(erm:gv)(uas:k) embryos at 72 hpf revealed a co-expression of Kaede and GS throughout the ventricular zone, which was prominent in the dorsolateral region (Fig. 2P,Q). Taken together, these results suggest that FGF-receptor signalling is active in precursors of SMNs, OLPs and GS-positive astroglia and raise the question as to the role of FGF-receptor signalling in the genesis of these cell types.
FGF-receptor signalling controls the generation of SMNs and OLPs by acting upstream of olig2
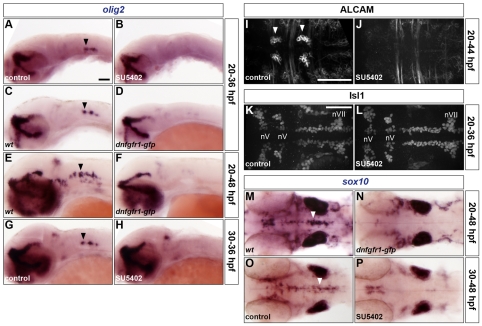
Using the conditions outlined above, we tested the effect of blocking FGF-receptor signalling on olig2 expression and the development of both SMNs and OLPs. Treatment of wild-type embryos with SU5402 or heat shock of Tg(hsp70l:dnfgfr1-gfp) embryos at 20 hpf largely prevented olig2 expression at 36 hpf in the ventromedial hindbrain but did not affect its expression in other tissues, including the forebrain and spinal cord (Fig. 3A-D, data not shown). Similarly, in Tg(hsp70l:dnfgfr1-gfp) embryos heat shocked at 20 hpf and analysed at 48 hpf, olig2 was only very weakly expressed in r2-r7 and in the midbrain/r1 region compared with controls but remained untouched in other tissues (Fig. 3E,F). In addition, treatment of wild-type embryos with SU5402 at 30 hpf, at the time of onset of olig2 expression, downregulated its expression at 36 hpf in the ventral hindbrain (Fig. 3G,H). This loss of olig2 expression suggested that both SMN and oligodendrocyte development would be affected by blocking FGF-receptor signalling. Indeed, treatment of wild-type embryos with SU5402 at 20 hpf prevented the appearance of SMNs of the abducens nuclei at 44 hpf (Fig. 3I,J) but did not affect the Olig2-independent branchiomotor neurons at 36 hpf (Fig. 3K,L). In addition, heat shock of Tg(hsp70l:dnfgfr1-gfp) embryos at 20 hpf or treatment of wild-type embryos with SU5402 at 30 hpf largely eliminated sox10 expression at 48 hpf, consistent with a loss of OLPs (Fig. 3M-P).
Fig. 3.
FGF-receptor signalling controls olig2 expression and the generation of both OLPs and SMNs. (A-P) Embryos were analysed for olig2 RNA expression at 36 (A-D,G,H) and 48 hpf (E,F), ALCAM (I,J) and Isl1 (K,L) protein at 44 and 36 hpf, respectively, and sox10 RNA expression at 48 hpf (M-P). Wild-type embryos were either treated with DMSO (A,G,I,K,O) or treated with SU5402 at 20 hpf (B,J,L) or at 30 hpf (H,P) and wild-type (C,E,M) and Tg(hsp70l:dnfgfr1-gfp) (D,F,N) embryos were heat shocked at 20 hpf. Arrowheads indicate ventromedial r5 (A,C,E,G,M,O) and the abducens nuclei in r5 and r6 (I). nV and nVII; branchiomotor nuclei of the V and VII nerves, respectively. All embryos are shown with anterior to the left in lateral (A-H) or dorsal (I-P) view. Scale bars: 50 μm.
Because FGF maintains neural progenitors in the CNS (Mason, 2007), we asked whether the loss of olig2 expression, SMNs and OLPs following a block in FGF-receptor signalling was a result of cell death or differentiation of rhombomere centre progenitors. To test this, embryos were analysed for apoptosis using the TUNEL assay (Gavrieli et al., 1992). Compared to controls, no increase in hindbrain apoptosis was detected in wild-type embryos treated with SU5402 at 20 or 30 hpf and analysed at several stages (see Fig. S2A-H in the supplementary material) or Tg(hsp70l:dnfgfr1-gfp) embryos heat shocked at 20 hpf and analysed at 36 hpf (see Fig. S2I,J in the supplementary material). To investigate whether rhombomere centre progenitors were still present following a block in FGF-receptor signalling, we exploited the stability of the Kaede protein in the Tg(erm:gv)(uas:k) line. Tg(erm:gv)(uas:k) embryos were allowed to express Kaede in rhombomere centres and then treated with SU5402 at 30 hpf. Similar to control embryos, numerous Kaede-positive cells were still present in the ventricular zone at 48 hpf (see Fig. S3 in the supplementary material). These cells did not express the neuronal marker HuC, suggesting that they maintained their progenitor characteristics. Importantly, under identical conditions of SU5402 treatment, olig2 and sox10 expression were eliminated (Fig. 3 and data not shown). Taken together, these data indicate that FGF-receptor signalling in rhombomere centre progenitors controls olig2 expression and consequently the generation of both SMNs and oligodendrocytes.
We then investigated how FGF-receptor signalling in rhombomere centre progenitors is integrated with known mechanisms controlling olig2 expression. Previous studies in the mouse showed that Pax6 and possibly Nkx2-2 are necessary for Olig2 expression in the hindbrain (Mizuguchi et al., 2001; Vallstedt et al., 2005). We found that, compared to control embryos, blocking FGF-receptor signalling with SU5402 did not affect the expression of the DV patterning genes pax6a (see Fig. S4A,B in the supplementary material), nkx2.2a (see Fig. S4C,D in the supplementary material) or pax3 (see Fig. S4E,F in the supplementary material), suggesting that FGF-receptor signalling acts in parallel to DV patterning genes to control olig2 expression.
Because previous studies identified an essential role of Shh in ventral CNS patterning (Briscoe and Ericson, 1999), we analysed hindbrain olig2 expression in the Shh signalling mutant smoothened (smo) (Varga et al., 2001). Compared with wild-type and heterozygous embryos, in smo mutants, olig2 (see Fig. S5A,B in the supplementary material) and sox10 (see Fig. S5C,D in the supplementary material) expression were eliminated, whereas, FGF-receptor signalling, based on erm expression, was only mildly affected (see Fig. S5E,F in the supplementary material). Taken together, these results suggest that FGF-receptor signalling cooperates with the DV patterning signal Shh to control olig2 expression.
FGF-receptor signalling controls the generation of GS-positive astroglia
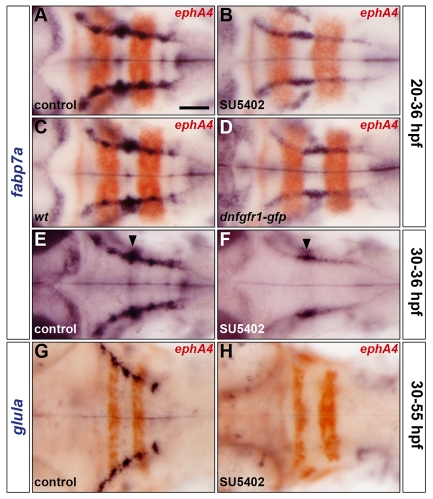
We then tested the effect of blocking FGF-receptor signalling on the development of astroglia by analysing the expression of fabp7a and glula. Treatment with SU5402 or heat shock of the Tg(hsp70l:dnfgfr1-gfp) line at 20 hpf prevented the upregulation of fabp7a dorsolaterally in rhombomere centres at 36 hpf compared with control embryos (Fig. 4A-D). Treatment with SU5402 at 30 hpf similarly affected fabp7a expression at 36 hpf (Fig. 4E,F), suggesting that FGF-receptor signalling is required at the time of upregulation of fabp7a expression. Consistent with the effect on fabp7a, treatment with SU5402 at 30 hpf prevented the expression of glula at 55 hpf compared with control embryos (Fig. 4G,H). As described above, blocking FGF-receptor signalling during this period neither promoted apoptosis nor the depletion of rhombomere centre progenitors (see Figs S2 and S3 in the supplementary material). In addition, the pattern of hindbrain GFAP was not altered following a block in FGF-receptor signalling (see Fig. S6 in the supplementary material), indicating that radial glia are not generally affected. Taken together, these results suggest that FGF-receptor signalling in rhombomere centres controls the generation of GS-positive astroglia.
Fig. 4.
FGF-receptor signalling controls astroglial development. (A-H) Embryos were analysed for fabp7a (A-F) and glula (G,H) RNA expression at 36 hpf and 55 hpf, respectively. Wild-type embryos were either treated with DMSO (A,E,G) or treated with SU5402 at 20 hpf (B) or at 30 hpf (F,H) and wild-type (C) and Tg(hsp70l:dnfgfr1-gfp) (D) embryos were heat shocked at 20 hpf. Embryos were double labelled for ephA4 (red: A-D,G,H) RNA to mark r3 and r5. Arrowheads indicate the centre of r4 (E,F). All embryos are shown in dorsal view with anterior to the left. Scale bar: 50 μm.
FGF-receptor signalling controls gliogenesis via sox9
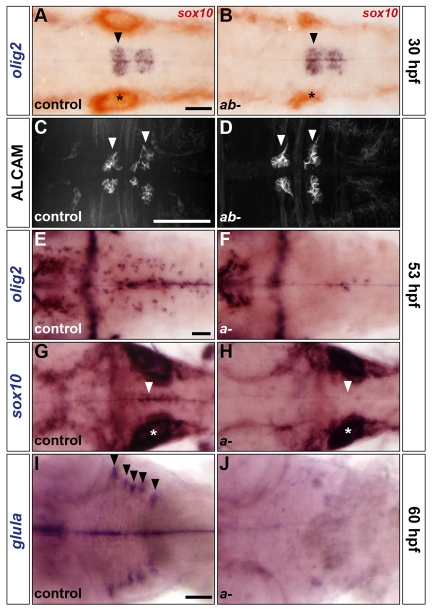
In the mouse, the HMG transcription factor Sox9 specifies both oligodendrocytes and astrocytes (Stolt et al., 2003). In zebrafish, duplicate sox9 genes have been identified (Chiang et al., 2001); however, their role in CNS gliogenesis has not been established. To investigate this problem, we analysed embryos from incrosses of single and double heterozygotes carrying loss-of-function alleles of sox9a and sox9b (Yan et al., 2002; Yan et al., 2005). We first examined olig2 expression at 30 hpf and ALCAM at 53 hpf and found no significant difference between wild type and simple or double mutants (Fig. 5A-D and data not shown). This indicates that olig2 expression and the development of SMNs are independent of Sox9. Previous studies have shown that the levels of apoptosis are higher in the CNS of sox9b but not sox9a mutants (Yan et al., 2005). To confirm this finding, we analysed sox9a mutants at 30 and 48 hpf and found no significant difference in the level of apoptosis compared to controls (see Fig. S7 in the supplementary material). We further confirmed the integrity of the sox9a mutant hindbrain by analysing the expression of a number of markers including erm, GFAP and deltaA, which were found to be unaffected (data not shown). Consequently, we restricted our analyses of glial marker expression to sox9a mutants. Analysis of OLPs, olig2 and sox10 (Fig. 5E-H), and astroglial, glula (Fig. 5I,J), marker expression at 53 and 60 hpf, respectively, revealed a strong reduction in sox9a mutants compared with controls. The persisting low levels of glial gene expression in the sox9a mutants are likely to be a result of the presence of the sox9b gene. Taken together, these results support a conserved role of zebrafish sox9a in gliogenesis.
Fig. 5.
sox9 genes control the development of OLPs and astroglia but not SMNs. (A-J) Analysis of olig2 RNA (A,B,E,F), ALCAM protein (C,D), sox10 RNA (G,H) and glula RNA (I,J) expression in control (A,C,E,G,I), sox9atw37/tw37; sox9bb971/b971 double mutants (B,D) and sox9atw37/tw37 simple mutants (F,H,J) at 30 (A,B), 53 (C-H) and 60 (I,J) hpf. Embryos in A,B were double labelled for sox10 RNA. Control embryos are either homozygous wild type or heterozygous. Arrowheads indicate ventromedial r5 (A,B,G,H), abducens nuclei in r5 and r6 (C,D) and dorsolateral r2-r6 (I). Asterisks, otic vesicle. All embryos are shown in dorsal view with anterior to the left. Scale bars: 50 μm.
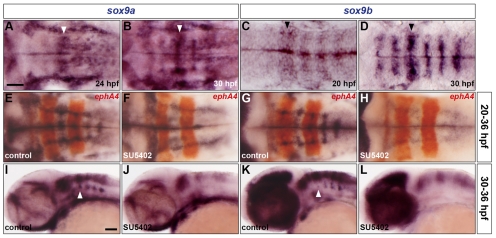
We then investigated whether FGF-receptor signalling in the zebrafish hindbrain acts upstream of the sox9 genes. We first analysed the expression profile of sox9a and sox9b and found that, although sox9a was delayed relative to sox9b, both genes were expressed in rhombomere centres; by 36 hpf this expression was prominent in the ventromedial and dorsolateral regions corresponding to sites of oligogenesis and astrogenesis, respectively (Fig. 6A-E,G,I,K). We then tested the role of FGF-receptor signalling in the expression of both sox9 genes. Compared with control embryos, SU5402 treatment beginning at either 20 or 30 hpf prevented the expression of sox9a and sox9b in rhombomere centres at 36 hpf, most notably in the ventromedial and dorsolateral patches, but did not significantly affect their expression elsewhere in the hindbrain (Fig. 6E-L). Taken together, these results indicate that FGF-receptor signalling controls hindbrain gliogenesis, in part by regulating sox9 gene expression.
Fig. 6.
sox9 genes are expressed in rhombomere centres under control of FGF-receptor signalling. (A-D) Expression of sox9a (A,B) and sox9b (C,D) RNA at 24 hpf (A), 30 hpf (B,D) and 20 hpf (C). Arrowheads indicate r4. (E-L) Wild-type embryos were either treated with DMSO (E,G,I,K) or treated with SU5402 at 20 hpf (F,H) and at 30 hpf (J,L) and analysed for sox9a (E,F,I,J) and sox9b (G,H,K,L) RNA expression at 36 hpf. Embryos were double labelled for ephA4 (red: E-H) RNA to mark r3 and r5. Arrowheads indicate ventromedial r4 (I,K). All embryos are shown with anterior to the left in dorsal (A-H) or lateral (I-L) view. Scale bars: 50 μm.
DISCUSSION
Due to pleiotropy and ligand and receptor redundancy, much of our prior knowledge concerning the role of FGF in the generation of neural cell diversity in the vertebrate CNS was based on in vitro studies. In this report we have established an important role of FGF-receptor signalling in the generation of neural cell diversity in the zebrafish hindbrain. We identified an FGF-receptor signal in rhombomere centre radial glia that are a source of precursors for late neural cell subtypes including SMNs, OLPs and differentiating astroglia. We then showed that FGF-receptor signalling neither affects progenitor survival nor is essential for the maintenance of these progenitors; rather we found that FGF-receptor signalling controls the expression of key genes that underlie neural subtype specification. Below we discuss the significance of these findings and present a model for the generation of neural cell diversity in the zebrafish hindbrain.
FGF-receptor signalling in the regulation of olig2 expression
Among amniotes, Olig2 is an essential positional identity factor specifying both SMNs and oligodendrocytes (Rowitch et al., 2002) and we showed that olig2-expressing cells in the zebrafish hindbrain indeed give rise to SMNs of the abducens nuclei and OLPs. Importantly, these experiments showed that FGF-receptor signalling in hindbrain progenitors controls olig2 expression and consequently the generation of both SMNs and OLPs, thereby providing the first evidence for a role of FGF in this process. Previous studies in mouse and chick revealed that Olig2 expression in the ventral spinal cord and hindbrain is established by the combinatorial action of patterning genes including Pax6 (Mizuguchi et al., 2001), Nkx6.1, Nkx6.2 (Cai et al., 2005) and possibly Nkx2.2 (Nkx2.2a — Zebrafish Information Network) (Vallstedt et al., 2005), which are regulated by the DV patterning signal Shh (Briscoe and Ericson, 1999). In agreement with this conclusion, Shh signalling was also required for olig2 expression in the zebrafish hindbrain. Blocking FGF-receptor signalling did not affect the expression of pax3, pax6 or nkx2.2. Moreover, these and other DV patterning genes, such as nkx6.1, are expressed throughout the rostrocaudal axis of the hindbrain (Cheesman et al., 2004) suggesting that FGF-receptor signalling does not contribute to DV patterning events. Based on these data, we propose that Shh signalling establishes a progenitor domain that is competent to express olig2 wherein FGF-receptor signalling permits or promotes olig2 gene transcription. Interestingly, dorsal BMP signalling prevents the generation of OLPs (Mekki-Dauriac et al., 2002; Vallstedt et al., 2005) by downregulating olig2 expression, a process that is antagonised by FGF (Bilican et al., 2008). Further studies will be required to establish whether FGF-receptor signalling controls olig2 expression in the zebrafish hindbrain by antagonising BMP signalling or via another transcriptional regulatory pathway.
In addition to our proposed role of FGF-receptor signalling in the control of olig2 in the ventral hindbrain, in vitro studies on the mouse and chick revealed that FGF promotes Olig2 expression and the generation of OLPs in dorsal neural tube progenitors in a Shh-independent manner (Chandran et al., 2003; Kessaris et al., 2004; Fogarty et al., 2005; Vallstedt et al., 2005; Naruse et al., 2006). This suggests that FGF-receptor signalling may play a more general role in controlling olig2 expression in different progenitor domains along the DV axis of the neural tube.
FGF-receptor signalling in astroglia
We have investigated the expression pattern of zebrafish orthologues of brain fatty acid binding protein (fabp7a) (Liu et al., 2004), connexin 43 (cx43α1) (Chatterjee et al., 2005) and glutamine synthetase (glula) (Thisse et al., 2001), genes known to be expressed in the astroglial lineage in mammals (Dermietzel et al., 1991; Feng and Heintz, 1995; Nadarajah et al., 1997; Pringle et al., 2003). Whereas radial glia are present throughout the zebrafish hindbrain ventricular zone from early stages (Marcus and Easter, 1995), fabp7a expression was restricted to the dorsolateral hindbrain and subsequently both cx43α1 and glula were expressed within the fabp7a expression domain coincident with the rhombomere centres. Because these genes were not co-expressed with either neuronal or OLP markers, these data indicate that hindbrain radial glia, located in the dorsolateral rhombomere centres, differentiate within the astroglial lineage. The de novo expression of glula and cx43α1 in radial glia is likely to represent the early stages of differentiation of astroglia present in the adult teleost brain, including cells with radial glial character, such as ependymoglia, perivascular glia and astrocytes (Kalman, 1998). Our studies on the larval hindbrain revealed that, whereas the majority of GS-positive cells are located in the ventricular zone, a small number of cells were found in the mantle region that was consistent with their identity as astrocytes (data not shown). Nevertheless, this result implies that, at least until larval stages, the majority of hindbrain astroglia do not migrate. Similarly, previous studies in the adult teleost brain showed that GFAP-positive cells are mostly radial glia and there are relatively few true astrocytes (Kalman, 1998). Radial astroglia may act as a substrate for neuronal migration and neural connectivity during development and regeneration in the adult (Singer et al., 1979; Levitt and Rakic, 1980; Marcus and Easter, 1995). However, the predominance of radial astroglia in adult teleosts has led to the suggestion that these cells may also serve functions similar to astrocytes in higher vertebrates (Wicht et al., 1994; Kawai et al., 2001).
Although Fgf10 promotes the differentiation of neuroepithelial cells into radial glia (Sahara and O'Leary, 2009), at subsequent stages of astroglial development, in vivo analyses have identified a role of FGF-receptor signalling only in the migration of astrocytes (Smith et al., 2006). That FGFs may act at other stages of astroglial development has been suggested by in vitro studies investigating the role of FGF in the specification of astrocytes (Qian et al., 1997; Morrow et al., 2001).
Our results showed that FGF-receptor signalling controls the generation of GS-positive astroglia in the zebrafish hindbrain, thus revealing a previously unknown level of FGF regulation of the astroglial lineage in vivo. Because GS-positive astroglia appear in the dorsal hindbrain long after the FGF-receptor signal is active throughout the rhombomere centre, additional cues are likely to be required. Notch, BMP and cytokine signalling pathways are candidates because numerous in vitro studies have revealed their potential as positive regulators of astrogenesis (Miller and Gauthier, 2007). Interestingly, both the Notch target genes her6 and her9, zebrafish orthologues of Hes1, and the cytokine receptor cntfr, are expressed in the dorsolateral hindbrain throughout the period of astroglial differentiation (Thisse et al., 2001; data not shown), suggesting that the Notch and cytokine pathways are likely contributors.
FGF-receptor signalling and Sox9 in gliogenesis
Studies in the mouse spinal cord have shown that Sox9 controls the specification of both oligodendrocytes and astrocytes and subsequently, in combination with Sox10, promotes the further development of OLPs (Stolt et al., 2003; Finzsch et al., 2008). We have shown that in the zebrafish hindbrain, sox9a is necessary for the generation of both OLPs and GS-positive astroglia, supporting a phylogenetically conserved role. Because sox9atw37 behaves as a null allele (Yan et al., 2002), residual expression of glial markers in these mutants is probably due to the sox9b co-orthologue, which has an overlapping expression profile.
The importance of Sox9 in gliogenesis has raised the question as to what signals control this factor and our experiments showed that FGF-receptor signalling promotes the expression of both sox9a and sox9b in rhombomere centre progenitors throughout the period of gliogenesis. To further investigate the FGF/Sox9 connection we attempted to rescue the defect in gliogenesis, following a block in FGF-receptor signalling, by ectopic expression of Sox9. We found no evidence for rescue in these experiments (data not shown) suggesting that FGF-receptor signalling controls factors, in addition to Sox9, are essential for gliogenesis.
Although FGF-receptor signalling was necessary for olig2 expression in hindbrain progenitors and the generation of SMNs, eliminating both sox9 genes had no effect on either the early expression of olig2 or the development of SMNs. This suggests that FGF-receptor signalling controls olig2 expression independently of Sox9.
Model for FGF-receptor signalling in the generation of neural cell diversity
Experiments reported here showed that during development of the zebrafish hindbrain, an FGF-receptor signal is progressively restricted to neural progenitors in the centres of r2-r7. This pattern suggests it is intimately linked to hindbrain segmentation events. Although the source and species of FGF are unknown, the restricted FGF signal may be due to the restricted expression of FGF-receptors. Indeed, we have found that fgfr2 is expressed in rhombomere centres (data not shown). Because the FGF-receptor signalling territory is circumscribed by a neurogenic zone, we speculate that a factor(s) within the pathway promoting neurogenesis downregulates the FGF signal. Because previous studies suggested that the rhombomere boundaries control the segmental pattern of neurogenesis (Riley et al., 2004; Amoyel et al., 2005), these boundaries may be the source of signals that restrict the FGF signal to rhombomere centres.
In summary, our studies reveal that in the zebrafish hindbrain, FGF-receptor signalling specifies SMNs and OLPs by promoting olig2 expression, and supports gliogenesis, by promoting sox9 expression (Fig. 7). Ventrally, FGF-receptor signalling cooperates with the Shh pathway to control olig2 expression. Olig2 promotes the generation of SMNs by acting in combination with proneural factors, such as Ngn1. Subsequently, FGF-receptor signalling, by controlling sox9 expression, promotes oligogenesis in the Olig2 domain. Dorsally, FGF-receptor signalling acting via Sox9 cooperates with signalling pathways such as Notch, BMP and cytokine to promote astroglial development. This in vivo model explains previous in vitro studies using CNS progenitors that revealed a role of FGF in promoting gliogenesis and that the particular glial subtype specified by FGF was context dependent (Qian et al., 1997; Morrow et al., 2001). Because FGF ligands and receptors are widely expressed in the developing CNS, this model is likely to be of general importance in the generation of neural cell diversity throughout the CNS.
Fig. 7.
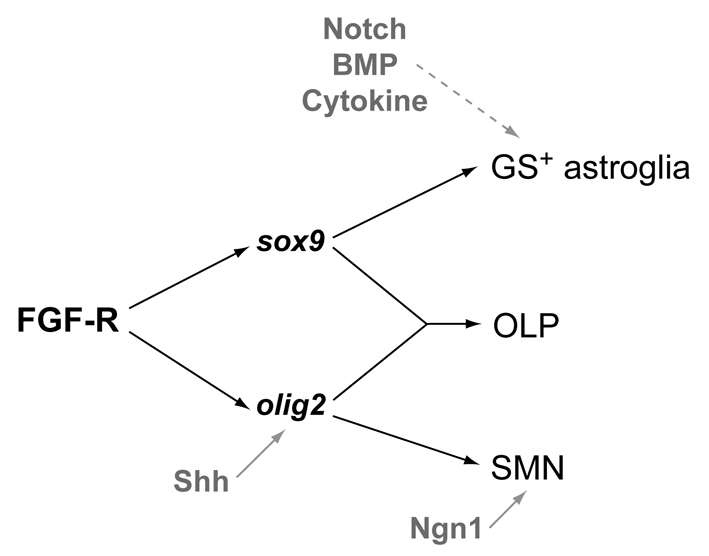
Model for FGF-receptor signalling in the generation of hindbrain neural cell diversity. FGF-receptor signalling in the hindbrain specifies both SMNs and OLPs by promoting olig2 expression, and gliogenesis, by promoting sox9 expression. In ventral neural progenitors FGF-receptor signalling cooperates with the Shh pathway to control olig2 expression. Olig2 promotes the generation of SMNs by acting in combination with proneural factors, such as Ngn1. Subsequently, FGF-receptor signalling controls gliogenesis by promoting sox9 expression in neural progenitors throughout the DV axis. In Olig2 expressing progenitors, this results in oligogenesis, and in dorsal progenitors, in the genesis of GS-positive astroglia. Other signals including Notch, BMP and cytokine, are likely to participate in the latter.
Supplementary Material
Acknowledgements
We are grateful to Pierre Drapeau for the use of laboratory space, equipment and reagents and Claudia Maios for technical assistance during the review process of this work. We thank Laure Bally-Cuif and Marion Coolen for critical reading of an earlier version of the manuscript. A. Firas Bouallague, Amanda Rapp and Catherine Wilson for fish care and technical assistance. T. Becker, P. Blader, M. Gajewski, R. N. Kelsh, R. Köster, S. Krauss, H. Roehl, H. C. Seo, D. G. Wilkinson and S. W. Wilson for gifts of plasmids and probes. ZIRC, DSHB and ImaGene for zebrafish lines, antibodies and plasmids. V.E. was supported by a fellowship from the Ministère de l'Enseignement Supérieur et de la Recherche and ARC. This work was supported by Grants from INSERM, MENRT, ANR, AFM, ARC, ARSEP and CNRS in France and the Centre Hospitalier Universitaire (CHU) Sainte-Justine Research Center in Canada. The work in J.H.P. lab was funded by NIH grant 5R01RR020833; the contents of this study are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.038026/-/DC1
References
- Amoyel M., Cheng Y. C., Jiang Y. J., Wilkinson D. G. (2005). Wnt1 regulates neurogenesis and mediates lateral inhibition of boundary cell specification in the zebrafish hindbrain. Development 132, 775-785 [DOI] [PubMed] [Google Scholar]
- Bertrand N., Castro D. S., Guillemot F. (2002). Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3, 517-530 [DOI] [PubMed] [Google Scholar]
- Bilican B., Fiore-Heriche C., Compston A., Allen N. D., Chandran S. (2008). Induction of Olig2 precursors by FGF involves BMP signalling blockade at the Smad level. PLoS ONE 3, e2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J., Ericson J. (1999). The specification of neuronal identity by graded Sonic Hedgehog signalling. Semin. Cell Dev. Biol. 10, 353-362 [DOI] [PubMed] [Google Scholar]
- Cai J., Qi Y., Hu X., Tan M., Liu Z., Zhang J., Li Q., Sander M., Qiu M. (2005). Generation of oligodendrocyte precursor cells from mouse dorsal spinal cord independent of Nkx6 regulation and Shh signaling. Neuron 45, 41-53 [DOI] [PubMed] [Google Scholar]
- Chandran S., Kato H., Gerreli D., Compston A., Svendsen C. N., Allen N. D. (2003). FGF-dependent generation of oligodendrocytes by a hedgehog-independent pathway. Development 130, 6599-6609 [DOI] [PubMed] [Google Scholar]
- Chandrasekhar A., Moens C. B., Warren J. T., Jr, Kimmel C. B., Kuwada J. Y. (1997). Development of branchiomotor neurons in zebrafish. Development 124, 2633-2644 [DOI] [PubMed] [Google Scholar]
- Chatterjee B., Chin A. J., Valdimarsson G., Finis C., Sonntag J. M., Choi B. Y., Tao L., Balasubramanian K., Bell C., Krufka A., et al. (2005). Developmental regulation and expression of the zebrafish connexin43 gene. Dev. Dyn. 233, 890-906 [DOI] [PubMed] [Google Scholar]
- Cheesman S. E., Layden M. J., Von Ohlen T., Doe C. Q., Eisen J. S. (2004). Zebrafish and fly Nkx6 proteins have similar CNS expression patterns and regulate motoneuron formation. Development 131, 5221-5232 [DOI] [PubMed] [Google Scholar]
- Chiang E. F., Pai C. I., Wyatt M., Yan Y. L., Postlethwait J., Chung B. (2001). Two sox9 genes on duplicated zebrafish chromosomes: expression of similar transcription activators in distinct sites. Dev. Biol. 231, 149-163 [DOI] [PubMed] [Google Scholar]
- Dermietzel R., Hertberg E. L., Kessler J. A., Spray D. C. (1991). Gap junctions between cultured astrocytes: immunocytochemical, molecular, and electrophysiological analysis. J. Neurosci. 11, 1421-1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F. (2003). The glial identity of neural stem cells. Nat. Neurosci. 6, 1127-1134 [DOI] [PubMed] [Google Scholar]
- Ellingsen S., Laplante M. A., Konig M., Kikuta H., Furmanek T., Hoivik E. A., Becker T. S. (2005). Large-scale enhancer detection in the zebrafish genome. Development 132, 3799-3811 [DOI] [PubMed] [Google Scholar]
- Feng L., Heintz N. (1995). Differentiating neurons activate transcription of the brain lipid-binding protein gene in radial glia through a novel regulatory element. Development 121, 1719-1730 [DOI] [PubMed] [Google Scholar]
- Finzsch M., Stolt C. C., Lommes P., Wegner M. (2008). Sox9 and Sox10 influence survival and migration of oligodendrocyte precursors in the spinal cord by regulating PDGF receptor alpha expression. Development 135, 637-646 [DOI] [PubMed] [Google Scholar]
- Fogarty M., Richardson W. D., Kessaris N. (2005). A subset of oligodendrocytes generated from radial glia in the dorsal spinal cord. Development 132, 1951-1959 [DOI] [PubMed] [Google Scholar]
- Gavrieli Y., Sherman Y., Ben-Sasson S. A. (1992). Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 119, 493-501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz M., Hartfuss E., Malatesta P. (2002). Radial glial cells as neuronal precursors: a new perspective on the correlation of morphology and lineage restriction in the developing cerebral cortex of mice. Brain Res. Bull. 57, 777-788 [DOI] [PubMed] [Google Scholar]
- Guillemot F. (2007a). Cell fate specification in the mammalian telencephalon. Prog. Neurobiol. 83, 37-52 [DOI] [PubMed] [Google Scholar]
- Guillemot F. (2007b). Spatial and temporal specification of neural fates by transcription factor codes. Development 134, 3771-3780 [DOI] [PubMed] [Google Scholar]
- Hatta K., Tsujii H., Omura T. (2006). Cell tracking using a photoconvertible fluorescent protein. Nat. Protoc. 1, 960-967 [DOI] [PubMed] [Google Scholar]
- Kalman M. (1998). Astroglial architecture of the carp (Cyprinus carpio) brain as revealed by immunohistochemical staining against glial fibrillary acidic protein (GFAP). Anat. Embryol.(Berl.) 198, 409-433 [DOI] [PubMed] [Google Scholar]
- Kawai H., Arata N., Nakayasu H. (2001). Three-dimensional distribution of astrocytes in zebrafish spinal cord. Glia 36, 406-413 [DOI] [PubMed] [Google Scholar]
- Kawakami K., Takeda H., Kawakami N., Kobayashi M., Matsuda N., Mishina M. (2004). A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell 7, 133-144 [DOI] [PubMed] [Google Scholar]
- Kessaris N., Jamen F., Rubin L. L., Richardson W. D. (2004). Cooperation between sonic hedgehog and fibroblast growth factor/MAPK signalling pathways in neocortical precursors. Development 131, 1289-1298 [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253-310 [DOI] [PubMed] [Google Scholar]
- Koster R. W., Fraser S. E. (2001). Tracing transgene expression in living zebrafish embryos. Dev. Biol. 233, 329-346 [DOI] [PubMed] [Google Scholar]
- Kozlowski D. J., Whitfield T. T., Hukriede N. A., Lam W. K., Weinberg E. S. (2005). The zebrafish dog-eared mutation disrupts eya1, a gene required for cell survival and differentiation in the inner ear and lateral line. Dev. Biol. 277, 27-41 [DOI] [PubMed] [Google Scholar]
- Lee K. J., Jessell T. M. (1999). The specification of dorsal cell fates in the vertebrate central nervous system. Annu. Rev. Neurosci. 22, 261-294 [DOI] [PubMed] [Google Scholar]
- Lee Y., Grill S., Sanchez A., Murphy-Ryan M., Poss K. D. (2005). Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development 132, 5173-5183 [DOI] [PubMed] [Google Scholar]
- Levitt P., Rakic P. (1980). Immunoperoxidase localization of glial fibrillary acidic protein in radial glial cells and astrocytes of the developing rhesus monkey brain. J. Comp. Neurol. 193, 815-840 [DOI] [PubMed] [Google Scholar]
- Liu R. Z., Denovan-Wright E. M., Degrave A., Thisse C., Thisse B., Wright J. M. (2004). Differential expression of duplicated genes for brain-type fatty acid-binding proteins (fabp7a and fabp7b) during early development of the CNS in zebrafish (Danio rerio). Gene Expr. Patterns 4, 379-387 [DOI] [PubMed] [Google Scholar]
- Lumsden A. (2004). Segmentation and compartition in the early avian hindbrain. Mech. Dev. 121, 1081-1088 [DOI] [PubMed] [Google Scholar]
- Marcus R. C., Easter S. S., Jr (1995). Expression of glial fibrillary acidic protein and its relation to tract formation in embryonic zebrafish (Danio rerio). J. Comp. Neurol. 359, 365-381 [DOI] [PubMed] [Google Scholar]
- Mason I. (2007). Initiation to end point: the multiple roles of fibroblast growth factors in neural development. Nat. Rev. Neurosci. 8, 583-596 [DOI] [PubMed] [Google Scholar]
- Mekki-Dauriac S., Agius E., Kan P., Cochard P. (2002). Bone morphogenetic proteins negatively control oligodendrocyte precursor specification in the chick spinal cord. Development 129, 5117-5130 [DOI] [PubMed] [Google Scholar]
- Miller F. D., Gauthier A. S. (2007). Timing is everything: making neurons versus glia in the developing cortex. Neuron 54, 357-369 [DOI] [PubMed] [Google Scholar]
- Mizuguchi R., Sugimori M., Takebayashi H., Kosako H., Nagao M., Yoshida S., Nabeshima Y., Shimamura K., Nakafuku M. (2001). Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron 31, 757-771 [DOI] [PubMed] [Google Scholar]
- Mohammadi M., McMahon G., Sun L., Tang C., Hirth P., Yeh B. K., Hubbard S. R., Schlessinger J. (1997). Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science 276, 955-960 [DOI] [PubMed] [Google Scholar]
- Morrow T., Song M. R., Ghosh A. (2001). Sequential specification of neurons and glia by developmentally regulated extracellular factors. Development 128, 3585-3594 [DOI] [PubMed] [Google Scholar]
- Nadarajah B., Jones A. M., Evans W. H., Parnavelas J. G. (1997). Differential expression of connexins during neocortical development and neuronal circuit formation. J. Neurosci. 17, 3096-3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse M., Nakahira E., Miyata T., Hitoshi S., Ikenaka K., Bansal R. (2006). Induction of oligodendrocyte progenitors in dorsal forebrain by intraventricular microinjection of FGF-2. Dev. Biol. 297, 262-273 [DOI] [PubMed] [Google Scholar]
- Perez Villegas E. M., Olivier C., Spassky N., Poncet C., Cochard P., Zalc B., Thomas J. L., Martinez S. (1999). Early specification of oligodendrocytes in the chick embryonic brain. Dev. Biol. 216, 98-113 [DOI] [PubMed] [Google Scholar]
- Poche R. A., Furuta Y., Chaboissier M. C., Schedl A., Behringer R. R. (2008). Sox9 is expressed in mouse multipotent retinal progenitor cells and functions in Muller glial cell development. J. Comp. Neurol. 510, 237-250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle N. P., Yu W. P., Howell M., Colvin J. S., Ornitz D. M., Richardson W. D. (2003). Fgfr3 expression by astrocytes and their precursors: evidence that astrocytes and oligodendrocytes originate in distinct neuroepithelial domains. Development 130, 93-102 [DOI] [PubMed] [Google Scholar]
- Qian X., Davis A. A., Goderie S. K., Temple S. (1997). FGF2 concentration regulates the generation of neurons and glia from multipotent cortical stem cells. Neuron 18, 81-93 [DOI] [PubMed] [Google Scholar]
- Raballo R., Rhee J., Lyn-Cook R., Leckman J. F., Schwartz M. L., Vaccarino F. M. (2000). Basic fibroblast growth factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. J. Neurosci. 20, 5012-5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raible F., Brand M. (2001). Tight transcriptional control of the ETS domain factors Erm and Pea3 by Fgf signaling during early zebrafish development. Mech. Dev. 107, 105-117 [DOI] [PubMed] [Google Scholar]
- Reuss B., Dono R., Unsicker K. (2003). Functions of fibroblast growth factor (FGF)-2 and FGF-5 in astroglial differentiation and blood-brain barrier permeability: evidence from mouse mutants. J. Neurosci. 23, 6404-6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley B. B., Chiang M. Y., Storch E. M., Heck R., Buckles G. R., Lekven A. C. (2004). Rhombomere boundaries are Wnt signaling centers that regulate metameric patterning in the zebrafish hindbrain. Dev. Dyn. 231, 278-291 [DOI] [PubMed] [Google Scholar]
- Roehl H., Nusslein-Volhard C. (2001). Zebrafish pea3 and erm are general targets of FGF8 signaling. Curr. Biol. 11, 503-507 [DOI] [PubMed] [Google Scholar]
- Rowitch D. H., Lu Q. R., Kessaris N., Richardson W. D. (2002). An ‘oligarchy’ rules neural development. Trends Neurosci. 25, 417-422 [DOI] [PubMed] [Google Scholar]
- Sahara S., O'Leary D. D. (2009). Fgf10 regulates transition period of cortical stem cell differentiation to radial glia controlling generation of neurons and basal progenitors. Neuron 63, 48-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Maunoury S., Gilardi-Hebenstreit P., Charnay P. (1998). How to build a vertebrate hindbrain. Lessons from genetics. C. R. Acad. Sci. III 321, 819-834 [DOI] [PubMed] [Google Scholar]
- Shin D. M., Korada S., Raballo R., Shashikant C. S., Simeone A., Taylor J. R., Vaccarino F. (2004). Loss of glutamatergic pyramidal neurons in frontal and temporal cortex resulting from attenuation of FGFR1 signaling is associated with spontaneous hyperactivity in mice. J. Neurosci. 24, 2247-2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Park H. C., Topczewska J. M., Mawdsley D. J., Appel B. (2003). Neural cell fate analysis in zebrafish using olig2 BAC transgenics. Methods Cell Sci. 25, 7-14 [DOI] [PubMed] [Google Scholar]
- Singer M., Nordlander R. H., Egar M. (1979). Axonal guidance during embryogenesis and regeneration in the spinal cord of the newt: the blueprint hypothesis of neuronal pathway patterning. J. Comp. Neurol. 185, 1-21 [DOI] [PubMed] [Google Scholar]
- Smith K. M., Ohkubo Y., Maragnoli M. E., Rasin M. R., Schwartz M. L., Sestan N., Vaccarino F. M. (2006). Midline radial glia translocation and corpus callosum formation require FGF signaling. Nat. Neurosci. 9, 787-797 [DOI] [PubMed] [Google Scholar]
- Spassky N., Goujet-Zalc C., Parmantier E., Olivier C., Martinez S., Ivanova A., Ikenaka K., Macklin W., Cerruti I., Zalc B., et al. (1998). Multiple restricted origin of oligodendrocytes. J. Neurosci. 18, 8331-8343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt C. C., Lommes P., Sock E., Chaboissier M. C., Schedl A., Wegner M. (2003). The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 17, 1677-1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B., Pflumio S., Fürthauer M., Loppin B., Heyer V., Degrave A., Woehl R., Lux A., Steffan T., Charbonnier X. Q., et al. (2001). Expression of the zebrafish genome during embryogenesis. ZFIN Direct Data Submission (http://zfin.org).
- Thisse C., Thisse B. (1998). High-resolution whole-mount in situ hybridization. Zebrafish Science Monitor, Vol. 5, Issue 1 Eugene: University of Oregon Press; (http://zfin.org/zf_info/monitor/vol5.1/vol5.1.html). [Google Scholar]
- Tomizawa K., Inoue Y., Nakayasu H. (2000). A monoclonal antibody stains radial glia in the adult zebrafish (Danio rerio) CNS. J. Neurocytol. 29, 119-128 [DOI] [PubMed] [Google Scholar]
- Vallstedt A., Klos J. M., Ericson J. (2005). Multiple dorsoventral origins of oligodendrocyte generation in the spinal cord and hindbrain. Neuron 45, 55-67 [DOI] [PubMed] [Google Scholar]
- Varga Z. M., Amores A., Lewis K. E., Yan Y. L., Postlethwait J. H., Eisen J. S., Westerfield M. (2001). Zebrafish smoothened functions in ventral neural tube specification and axon tract formation. Development 128, 3497-3509 [DOI] [PubMed] [Google Scholar]
- Wicht H., Derouiche A., Korf H. W. (1994). An immunocytochemical investigation of glial morphology in the Pacific hagfish: radial and astrocyte-like glia have the same phylogenetic age. J. Neurocytol. 23, 565-576 [DOI] [PubMed] [Google Scholar]
- Xu Q., Alldus G., Holder N., Wilkinson D. G. (1995). Expression of truncated Sek-1 receptor tyrosine kinase disrupts the segmental restriction of gene expression in the Xenopus and zebrafish hindbrain. Development 121, 4005-4016 [DOI] [PubMed] [Google Scholar]
- Yan Y. L., Miller C. T., Nissen R. M., Singer A., Liu D., Kirn A., Draper B., Willoughby J., Morcos P. A., Amsterdam A., et al. (2002). A zebrafish sox9 gene required for cartilage morphogenesis. Development 129, 5065-5079 [DOI] [PubMed] [Google Scholar]
- Yan Y. L., Willoughby J., Liu D., Crump J. G., Wilson C., Miller C. T., Singer A., Kimmel C., Westerfield M., Postlethwait J. H. (2005). A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development 132, 1069-1083 [DOI] [PubMed] [Google Scholar]
- Yokoi H., Yan Y. L., Miller M. R., Bremiller R. A., Catchen J. M., Johnson E. A., Postlethwait J. H. (2009). Expression profiling of zebrafish sox9 mutants reveals that Sox9 is required for retinal differentiation. Dev. Biol. 329, 1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannino D. A., Appel B. (2009). Olig2+ precursors produce abducens motor neurons and oligodendrocytes in the zebrafish hindbrain. J. Neurosci. 29, 2322-2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Anderson D. J. (2002). The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell 109, 61-73 [DOI] [PubMed] [Google Scholar]
- Zhou Q., Choi G., Anderson D. J. (2001). The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron 31, 791-807 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.