Abstract
Cell proliferation is an important determinant of plant form, but little is known about how developmental programs control cell division. Here, we describe the role of microRNA miR396 in the coordination of cell proliferation in Arabidopsis leaves. In leaf primordia, miR396 is expressed at low levels that steadily increase during organ development. We found that miR396 antagonizes the expression pattern of its targets, the GROWTH-REGULATING FACTOR (GRF) transcription factors. miR396 accumulates preferentially in the distal part of young developing leaves, restricting the expression of GRF2 to the proximal part of the organ. This, in turn, coincides with the activity of the cell proliferation marker CYCLINB1;1. We show that miR396 attenuates cell proliferation in developing leaves, through the repression of GRF activity and a decrease in the expression of cell cycle genes. We observed that the balance between miR396 and the GRFs controls the final number of cells in leaves. Furthermore, overexpression of miR396 in a mutant lacking GRF-INTERACTING FACTOR 1 severely compromises the shoot meristem. We found that miR396 is expressed at low levels throughout the meristem, overlapping with the expression of its target, GRF2. In addition, we show that miR396 can regulate cell proliferation and the size of the meristem. Arabidopsis plants with an increased activity of the transcription factor TCP4, which reduces cell proliferation in leaves, have higher miR396 and lower GRF levels. These results implicate miR396 as a significant module in the regulation of cell proliferation in plants.
Keywords: microRNAs, miR396, GRFs, Arabidopsis, Cell proliferation, Leaf, TCP4
INTRODUCTION
In contrast to animals, plants continue to produce new organs throughout their life cycle. The above-ground organs are derived from the shoot apical meristem (SAM), which includes a pool of stem cells residing at the growing tip of the plant. Proliferating SAM cells produce an excess of daughter cells that are either incorporated into the developing leaf primordia at the SAM periphery or become part of the shoot. The core machinery controlling the progression of the cell cycle in plants, as well as in other eukaryotes, relies on the activity of cyclin-dependent kinases (Inze and De Veylder, 2006). Many aspects of cell cycle regulation are highly conserved among eukaryotes. It is, however, the integration of the basic cell cycle mechanisms with the developmental program that generates the enormous phenotypic variation among multicellular organisms, a process that is much less understood (Inze and De Veylder, 2006).
In contrast to the indeterminate SAM in Arabidopsis thaliana, leaves are determinate organs that have a defined morphology. Leaf development involves the concerted action of various hormone signalling pathways and transcription factor networks. Some of the major transcriptional regulators involved in the control of cell proliferation in leaves include AINTEGUMENTA (Mizukami and Fischer, 2000), PEAPOD (White, 2006), JAGGED (Dinneny et al., 2004; Ohno et al., 2004), BLADE ON PETIOLE (Ha et al., 2003), TCPs (Nath et al., 2003) and GROWTH-REGULATING FACTORs (GRFs) (Kim et al., 2003).
To obtain their characteristic final size and shape, growth of the developing leaf needs to be tightly coordinated first through cell proliferation and then by cell expansion (Piazza et al., 2005; Tsukaya, 2006). Initially, cell proliferation is observed throughout the developing leaf (Donnelly et al., 1999). Then, the cell cycle stops at the tip of the leaf and a mitotic arrest front moves towards the base of the organ (Donnelly et al., 1999). Once cells cease to divide, they begin to enlarge and cell growth becomes the driving force regulating organ size (Piazza et al., 2005; Tsukaya, 2006).
Currently, little is known about the molecular mechanisms that coordinate cell proliferation throughout a developing leaf. A known regulator is the TCP gene CINCINNATA (CIN), which controls the progression of the mitotic arrest front in snapdragon (Nath et al., 2003). Mutations such as cin (Nath et al., 2003) and triple knock-outs of its Arabidopsis homologues tcp2/4/10 (Schommer et al., 2008) cause changes in leaf morphogenesis and uneven organ curvature due to excess cell proliferation at the leaf margins. Interestingly, five Arabidopsis TCPs (TCP2, 3, 4, 10 and 24), as well as CIN, have a target site for microRNA (miRNA) miR319 (Palatnik et al., 2003). Overexpression of miR319 causes the degradation of these TCPs and the generation of crinkled leaves similar to those observed in tcp loss-of-function mutants (Palatnik et al., 2003). Mutations in the target site of the TCPs that diminish the interaction with the miRNA affect leaf morphology in Arabidopsis (Palatnik et al., 2003; Palatnik et al., 2007) and leaf complexity in tomato (Ori et al., 2007), and are lethal in extreme cases (Palatnik et al., 2003).
The GRF family of transcription factors comprises nine members in Arabidopsis (Kim et al., 2003). Seven of them have a target site for miR396 (Jones-Rhoades and Bartel, 2004). Mutations indifferent GRFs or overexpression of miR396 have been shown to reduce cell number in Arabidopsis leaves (Horiguchi et al., 2005; Kim et al., 2003; Kim and Kende, 2004; Liu et al., 2009). The GRFs work together with GRF-INTERACTING FACTORs (GIFs), a small gene family encoding proteins with homology to the human SYT transcriptional co-activator (Horiguchi et al., 2005; Kim and Kende, 2004). Inactivation of GIF1 (Kim and Kende, 2004), also known as ANGUSTIFOLIA 3 (AN3) (Horiguchi et al., 2005), produces narrower leaves as a result of a reduction in cell proliferation.
Here, we describe the role of miR396 in the regulation of the GRFs. We show that miR396 accumulates during leaf development and limits the domain of the proliferative tissue. The balance between miR396 and the GRFs quantitatively regulates cell number in leaves. Furthermore, we show that miR319-regulated TCP4 induces miR396 and represses GRF activity. We propose that miR396 can operate as a link between the developmental programs of the cell and the more basic cell cycle mechanisms to fine-tune cell proliferation.
MATERIALS AND METHODS
Plant material
Arabidopsis ecotype Col-0 was used for all experiments. Plants were grown in long photoperiods (16 hour light/8 hour dark) or in short photoperiods (8 hour light/16 hour dark) at 23°C.
Transgenes
See Table S1 in the supplementary material for a list of binary plasmids used in this study. The miRNA target motif in GRF2 was altered, introducing synonymous mutations in a cloned GRF2 wild-type genomic fragment, using the QuikChange site-directed mutagenesis kit (Stratagene).
Expression analysis
First, 0.5-1.0 μg of total RNA was treated with RQ1 RNase-free DNase (Promega). Then, first-strand cDNA synthesis was carried out using SuperScript III reverse transcriptase (Invitrogen). PCR reactions were performed in a Mastercycler ep realplex thermal cycler (Eppendorf) using SYBR Green I (Roche) to monitor double-stranded (ds)DNA synthesis. Quantitative (q)PCR of each gene was carried out for at least three biological replicates, with technical duplicates for each biological replicate. The relative transcript level was determined for each sample, normalized using PROTEIN PHOSPHATASE 2A cDNA level (Czechowski et al., 2005). Primer sequences are given in Table S2 in the supplementary material.
Small RNA analysis
RNA was extracted using TRIzol reagent (Invitrogen). Total RNA was resolved on 17% polyacrylamide gels under denaturing conditions (7 M urea). Blots were hybridized using either radioactively labelled or digoxigenin end-labelled locked nucleic acid (LNA) oligonucleotide probes designed against miR396 (Exiqon, Denmark).
Alternatively, miR396 levels were determined by stem-loop RT-qPCR, as described previously (Chen et al., 2005). The sequences of the oligonucleotides used were: retrotranscription stem-loop oligo, 5′GTCTCCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGGAGACMAGTTC3′; PCR forward primer, 5′GGCGGTTCCACAGCTTTCTT3′; and PCR reverse primer, 5′TGGTGCAGGGTCCGAGGTATT3′.
Microarray analyses
Total RNA was extracted from the aerial part of seedlings grown on plates for 10 days using the RNeasy plant mini kit (QIAGEN). Microarray analyses using the Affymetrix ATH1 platform were performed on two biological replicates as described (Schmid et al., 2005). Differentially expressed genes were identified using per-gene variance, calculated using logit-T (Lemon et al., 2003). The corresponding fold change of the transcripts was obtained by expression estimates using gcRMA (www.bioconductor.org), a modification of the robust multi-array analysis (RMA) algorithm (Irizarry et al., 2003). The expression of gene groups was assessed by gene set enrichment analysis using GSEA-P 2.0 (Subramanian et al., 2007; Subramanian et al., 2005).
Microscopic observations
Tissue was fixed in FAA and embedded in paraffin. Sections 10 μm thick were stained with Toluidine Blue.
To obtain paradermal views of palisade cells, leaves were fixed with FAA and cleared with chloral hydrate solution as described (Horiguchi et al., 2005). Palisade leaf cells were observed using differential interference contrast (DIC) microscopy.
In situ hybridization
DIG-labelled sense and antisense probes were synthesized by transcription with T7 or SP6 RNA polymerase with the DIG RNA labelling kit (SP6/T7) (Roche) using cloned cDNAs of GRF2 and HISTONE H4 as templates. For the miR396 probe, LNA oligonucleotides (Exiqon) were end labelled with the DIG oligonucleotide 3′-end labelling kit (Roche). Shoot apices from 15-day-old plants grown in short photoperiods were dissected and fixed in FAA. Paraffin-embedded material was sectioned to 8 μm thickness. Hybridization and detection were performed as previously described (Palatnik et al., 2003).
GUS assays
To visualize the activity of the reporters, transgenic plants were subjected to GUS staining, according to Donnelly et al. (Donnelly et al., 1999). Stained tissue was paraffin embedded, sectioned and mounted in Canada balsam.
Accession numbers
A list of relevant AGI locus identifiers is provided in Table S2 in the supplementary material. The accession number for the microarray experiments is GSE11250.
RESULTS
miR396 accumulates during leaf development
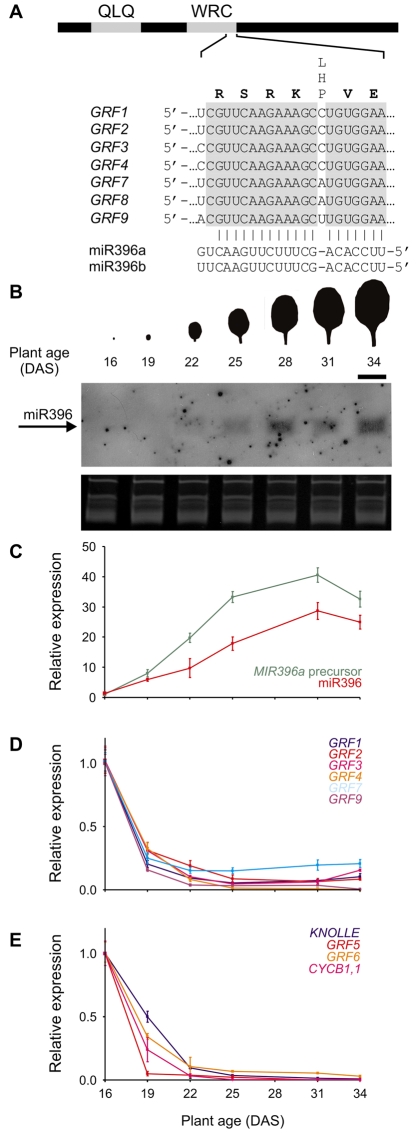
Seven of the nine Arabidopsis GRFs, GRF1-4 and GRF7-9, have a sequence that is partially complementary to that of miR396 (Fig. 1A). The miR396 target sequence is located at the end of a region that encodes a conserved WRC domain (Fig. 1A). The GRF alignments to miR396 share a bulge at position 7 (counting from the 5′ end of the miRNA), whose identity varies among the target genes (Fig. 1A).
Fig. 1.
Accumulation of miR396 during leaf development. (A) Scheme representing the GRF genes. The interaction of the seven miRNA-regulated GRFs from Arabidopsis with miR396 is shown. QLQ and WRC indicate the conserved domains that define the GRF family (Kim et al., 2003). (B) Pattern of miR396 accumulation during development of leaf 5. Samples were collected every 3 days starting 16 DAS. miR396 accumulation was analyzed by small RNA blots. Scale bar: 1 cm. (C) Accumulation of MIR396a precursor and mature miR396 during development of leaf 5. Both precursor and mature miRNA were detected by RT-qPCR. The expression levels were normalized to the earliest time point. The data shown are mean ± s.e.m. of three biological replicates. (D) Expression of miR396-regulated GRFs during leaf development. The GRFs were detected by RT-qPCR. Determination procedures were carried out as in C. (E) Expression of GRF5-6, GIF1, KNOLLE and CYCB1;1 during leaf development. Determination procedures were carried out as in C.
To characterize the functional role of miR396, we followed the expression pattern of the miRNA and its targets during leaf development. We collected the fifth rosette leaf at three-day intervals, starting from the day that it first became visible (~1 mm) to the naked eye, which was 16 days after sowing (DAS). We performed small RNA blots on these samples and found that miR396 was expressed at low levels in young leaves and that it steadily accumulated during the development of the organ (Fig. 1B).
To accurately quantify the expression levels of the miRNA, we turned to an RT-qPCR approach (Chen et al., 2005). We found that miR396 levels increased up to 30-fold during leaf development (Fig. 1C). We also measured the level of the MIR396a precursor and found that it was also upregulated during organogenesis, in good correlation with the level of the mature miRNA (Fig. 1C).
Next, we determined the level of the miR396-regulated GRFs by RT-qPCR. We observed that these transcription factors were expressed during the early stages of leaf development (Fig. 1D). Then, we observed a decrease in the level of miR396-regulated GRFs during leaf development that was complementary to the increase in the miRNA level (Fig. 1B-D).
We then analyzed the expression of GRF5 and GRF6, which are not regulated by miR396. We found that these GRFs had a similar expression pattern to the miR396-regulated transcription factors during organ growth (Fig. 1E). To correlate the expression pattern of the GRFs with the developmental stage of the leaf, we determined the levels of CYCLINB1;1 (CYCB1;1) and KNOLLE, which are known to be expressed during mitosis (Lukowitz et al., 1996; Menges et al., 2005). We observed that the expression of these cell cycle markers decreased in a similar way to the GRFs during early leaf development (Fig. 1E).
Dynamic pattern of miR396 expression
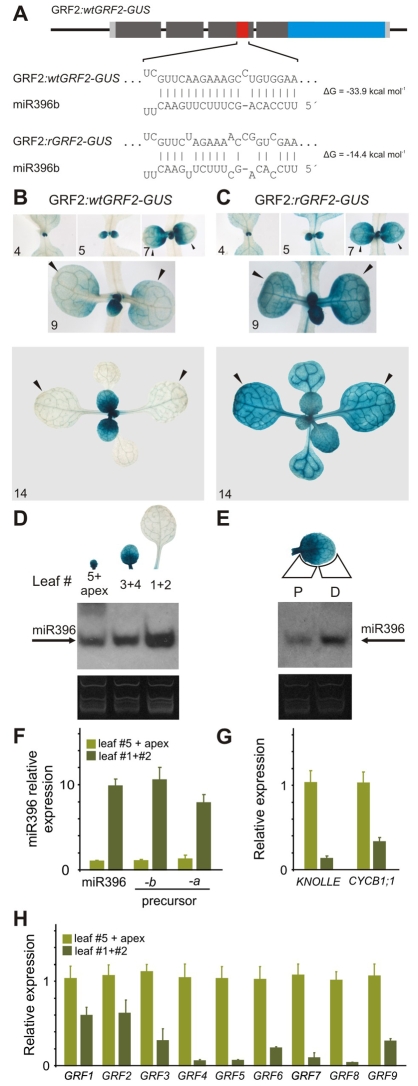
Our previous results showed that miR396 and the GRFs have contrasting expression patterns during leaf development. To study the expression of miR396 in more detail and uncover its function in the regulation of the GRFs, we prepared two reporters. First, we inserted the β-glucuronidase (GUS) gene in frame after exon 4, downstream of the miR396 target site, in the GRF2 genomic region including 2 kb upstream of the 5′-UTR (Fig. 2A). This wild-type reporter (GRF2:wtGRF2-GUS) should monitor GRF2 transcriptional and post-transcriptional regulation.
Fig. 2.
Dynamic pattern of miR396 expression. (A) Scheme representing two reporters with different sensitivity to miR396: GRF2:wtGRF2-GUS, which is sensitive to the miRNA, and GRF2:rGRF2-GUS, which is insensitive. (B) GUS staining of GRF2:wtGRF2-GUS plants. Arrowheads point to leaves 1 and 2. Numerals indicate plant age expressed as DAS. (C) GUS staining of GRF2:rGRF2-GUS plants. Arrowheads point to leaves 1 and 2. Numerals indicate plant age expressed as DAS. (D) Accumulation of mature miR396 in different leaves of a rosette. The GRF2:wtGRF2-GUS pattern of the same leaves is indicated at the top of each lane. (E) Expression of miR396 in the proximal (P) and distal (D) parts of leaf 3 (taken from same plants as for panel D). (F-H) Quantification of miR396, cell proliferation markers and GRFs in proliferative (leaf 5 plus apex) and non-proliferative (leaves 1 and 2) organs by RT-qPCR. The expression levels were normalized to the sample: leaf 5 plus apex. The data shown are mean ± s.e.m. of three biological replicates. (F) Accumulation of MIR396a and MIR396b precursors and mature miR396. (G) KNOLLE and CYCB1;1. (H) GRF1-9.
During organ growth, GRF2:wtGRF2-GUS was strongly expressed in young leaves (Fig. 2B). At later stages of development, reporter activity was detected mainly at the base of the leaf, with a gradient that tapered off towards the distal part of the leaf (Fig. 2B). Finally, we did not observe activity of the wild-type GRF2 reporter in fully expanded leaves. This dynamic pattern of expression of the reporter was repeated with the generation of each new leaf (Fig. 2B).
To reveal the activity of miR396 in leaves and its effect on the expression pattern of GRF2, we prepared a second reporter, with synonymous mutations in the miR396 target site that disrupt the interaction with the miRNA (Fig. 2A). Compared to the wild-type reporter, the expression domain of GRF2:rGRF2-GUS was extended in the leaves of 14 out of 20 T1 lines (Fig. 2C), indicating that the GRF2 promoter is apparently uniformly active throughout young leaves. As the reporters differ only in their sensitivity to the miRNA, the concomitant difference in their expression pattern should reflect the activity of miR396 in vivo. Therefore, these results indicated that miR396 accumulates during leaf development (Fig. 2B,C); in good agreement with our previous results (Fig. 1).
At a given developmental time, each individual rosette leaf showed a particular expression level of the wild-type GRF2 reporter (Fig. 2B). We determined the levels of miR396 in each leaf and found that the miRNA was expressed at the highest level in the oldest leaf, in good correlation with the activity of the reporters (Fig. 2D). RT-qPCR experiments with these samples showed that miR396 levels vary up to tenfold between the apices (containing the smallest leaves) and fully expanded leaves (Fig. 2F). The levels of the MIR396a and MIR396b precursors were also increased tenfold, suggesting that both miRNA genes have a redundant pattern of expression (Fig. 2F). The GRFs, together with cell proliferation markers such as CYCB1;1 and KNOLLE, were expressed in young proliferative tissues, whereas their expression decreased in older tissues, contrasting with the expression pattern of miR396 (Fig. 2G,H).
During the initial stages of leaf development, the wild-type GRF2 reporter displayed a high and homogeneous level of expression, whereas in mature leaves the reporter was turned off. Interestingly, at intermediate developmental stages, the wild-type GRF2 reporter showed a proximo-distal gradient of expression; this is in contrast to the miRNA-resistant GRF2 reporter, which was expressed throughout the leaf (Fig. 2B,C). To test whether miR396 was differentially expressed at the tip and the base of the leaf, we dissected a developing leaf and determined the level of miR396 in the proximal and distal parts. Small RNA blots confirmed that miR396 accumulated to higher levels in the distal part of the leaf (Fig. 2E).
Taken together, these results indicate a direct relationship between miR396 levels and the developmental age of the organ, with high levels in mature organs. Furthermore, miR396 activity varied along the proximo-distal axis of developing leaves, with higher expression levels in the distal part. This suggests that miR396 could also be a marker along the longitudinal axis of a single organ.
Effects of altered miR396 levels on GRF expression and leaf development
To gain insights into the biological role of miR396, we prepared transgenic plants with increased levels of the miRNA by expressing MIR396b under the control of the viral 35S promoter. Most of the transgenic plants (63 out of 81 T1 plants) had smaller rosettes (Fig. 3A,B). This is in good agreement with previous observations showing that GRF mutants (Horiguchi et al., 2005; Kim et al., 2003) or high miR396 levels reduce leaf size (Liu et al., 2009).
Fig. 3.
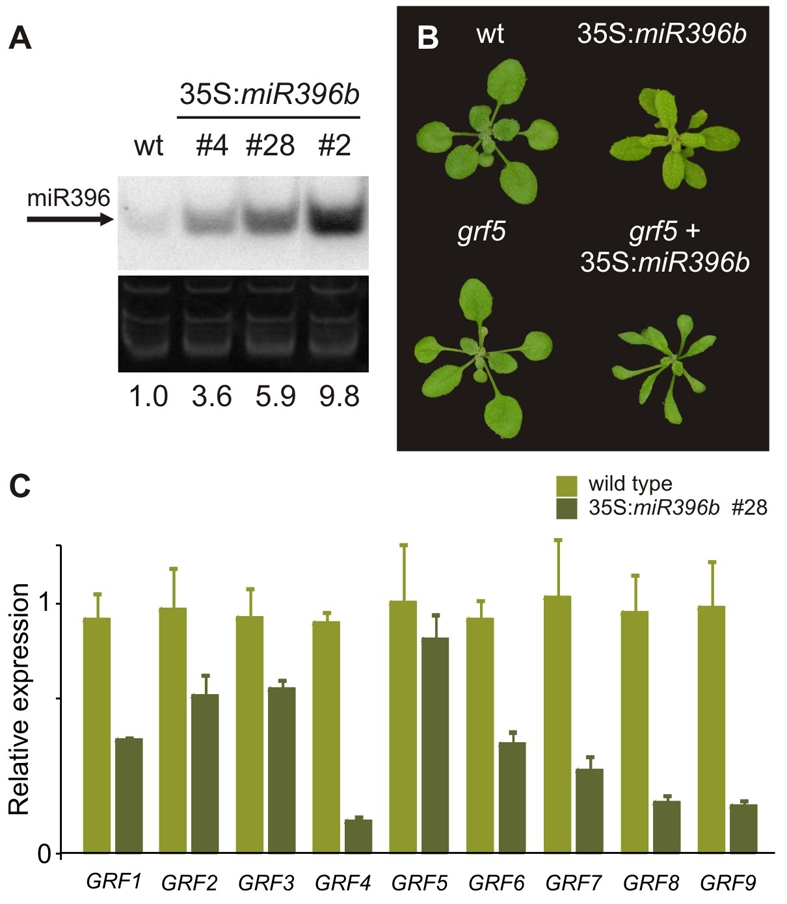
Effects of miR396 on GRF expression and leaf development. (A) Expression of miR396 in 10-day-old rosettes of wild-type plants and miR396b overexpressers. Numerals indicate the miR396 content of transgenic plants relative to that of wild-type plants. (B) Effect of miR396 overexpression on wild-type plants and grf5 mutants. (C) Expression levels of GRFs in 10-day-old rosettes of wild-type and 35S:miR396b #28 plants. The GRF levels were determined by RT-qPCR and normalized to wild-type plants. The data shown are mean ± s.e.m. of five biological replicates.
We then measured the levels of the GRFs in 35S:miR396b stable transgenics by RT-qPCR. We observed that the high levels of miR396 caused a reduction in the expression of the miRNA-regulated transcription factors; however, the magnitude of the decrease in their mRNA levels varied among the different GRFs (Fig. 3C). Similar results have been obtained from the overexpression of other miRNAs (Schwab et al., 2005).
GRF5 and GRF6, which do not have a miR396 target site, responded in different ways to miR396 overexpression. Whereas GRF5 was largely unaffected, GRF6 expression was decreased (Fig. 3C). These results suggested a feed-forward effect of the miR396-regulated GRFs on the levels of GRF6 in 35S:miR396b plants or that the expression of GRF6 is affected by the developmental changes caused by miR396.
Previous results have indicated that mutations in GRF5 produced moderately smaller leaves with fewer cells (Horiguchi et al., 2005). Primary transgenics harbouring an artificial miRNA against GRF5 and GRF6 also revealed a moderate effect on leaf development (V. Crosa and J.F.P., unpublished). Our results indicated that the miR396-regulated GRFs and GRF5-6 were expressed in a similar way during leaf development (Fig. 1), which might suggest that they have redundant functions. Therefore, we overexpressed miR396 in the context of a grf5 insertional mutant (for a list of mutants used in this work, see Table S2 in the supplementary material). Transgenic plants with this large decrease in GRF activity were severely affected, and their leaves were smaller and narrower, reflecting synergistic effects between the 35S:miR396b transgene and grf5 (Fig. 3B).
Genome-wide response to miR396
To identify molecular targets of the miR396 network, we compared global expression profiles in the aerial parts of 10-day-old wild-type and 35S:miR396b #28 seedlings using Affymetrix microarrays. We chose a relatively early stage of a moderate miR396 overexpresser to minimize the consequences of the different final anatomy caused by high miRNA levels. As expected, the expression levels of the miR396-regulated GRFs were lower in 35S:miR396b seedlings (see Table S3 in the supplementary material). Using a per-gene variance of P<0.05 calculated using the logit-T algorithm (Lemon et al., 2003), we found only 12 genes that were downregulated in 35S:miR396b plants (see Table S4 in the supplementary material).
Interestingly, KNOLLE was one of the genes downregulated in 35S:miR396b. KNOLLE expression is coordinated with 79 other Arabidopsis genes present on the ATH1 array that have peaks of expression when cells enter mitosis (Menges et al., 2005), probably through mitosis-specific activator elements in their promoters (Haga et al., 2007; Menges et al., 2005). Therefore, we further analyzed the expression of all mitosis-specific genes using Gene Set Enrichment Analysis, a test designed to detect changes in the expression of coordinated groups of genes (Subramanian et al., 2005). We found that the mitosis-specific genes were significantly downregulated as a group in 35S:miR396b compared to wild-type plants (nominal P<0.0001, false discovery rate<0.0001, see Tables S5 and S6 in the supplementary material).
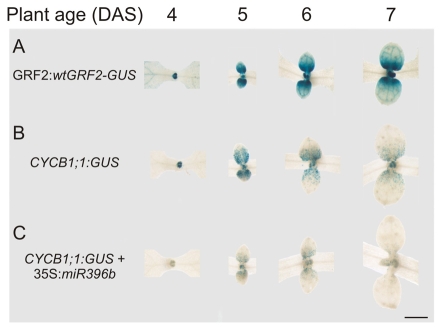
To validate the microarray results experimentally, we analyzed a cyclin reporter (Donnelly et al., 1999). The expression patterns of the CYCB1;1 and the wild-type GRF2-GUS reporters were very similar (Fig. 4A,B). In good agreement with this, a closer look at the gradient of wild-type GRF2-GUS revealed that the reporter activity started to fade away before cell expansion begins (see Fig. S1 in the supplementary material).
Fig. 4.
Control of cell proliferation by miR396. (A) GRF2:wtGRF2-GUS expression 4-7 DAS. (B) CYCB1;1:GUS reporter expression in plants the same age as in A. (C) CYCB1;1:GUS reporter expression in 35S:miR396b #28 transgenic plants. Scale bar: 1 mm.
We then analyzed CYCB1;1 in plants overexpressing miR396 and observed that high levels of the miRNA greatly reduced the number of CYCB1;1 positive cells in developing leaves (Fig. 4C), confirming the results of the microarray analysis. Furthermore, cycling cells were closer to the base of developing leaves in 35S:miR396b than in the wild type, suggesting that the mitotic arrest front occurs earlier in transgenic plants with high miRNA levels (Fig. 4B,C).
The balance between miR396 and the GRFs quantitatively regulates leaf cell number
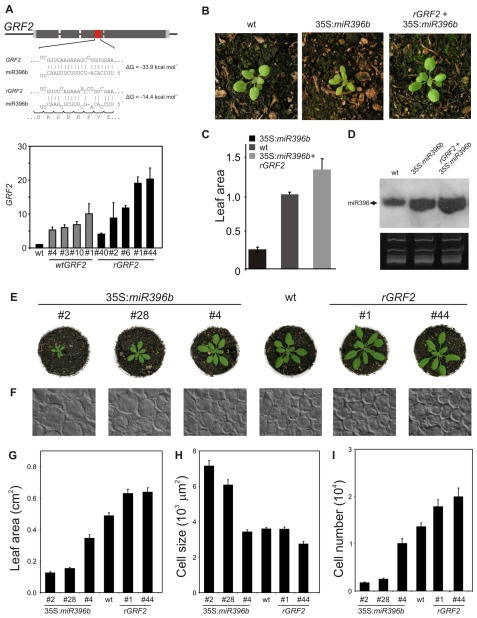
To determine the requirement for miR396 to restrict GRF activity during leaf development, we introduced two GRF2 genomic fragments into plants, one with the wild-type GRF2 coding sequence and one in which the miRNA-targeting motif was altered through synonymous mutations that should prevent miR396 targeting (rGRF2) (Fig. 5A). We analyzed several independent insertion lines for each construct at seedling stage, when the levels of miR396 are low, and found that additional copies of wild-type or miRNA-resistant GRF2 transgenes raised the GRF2 mRNA level several fold (Fig. 5A). However, the rGRF2 version led to higher levels of mRNA accumulation compared to the wild-type transgene, up to 20-fold (Fig. 5A).
Fig. 5.
Regulation of leaf development by the balance between miR396 and the GRFs. (A) Diagram of rGRF2 indicating the synonymous mutations that alter the interaction with miR396. The graph below shows expression levels of GRF2 in seedlings with extra copies of a wild-type (GRF2) or miRNA-resistant (rGRF2) form of the transcription factor. The data shown are mean ± s.e.m. of three biological replicates. (B-D) Complementation of the miR396 overexpression phenotype by rGRF2. (B) Rosette phenotype of wild-type, 35S:miR396b and rGRF2 plants overexpressing miR396. (C) Leaf area determined for fully expanded first leaves of wild-type, 35S:miR396b and rGRF2+35S:miR396b plants. Values expressed relative to wild-type plants. (D) miR396 expression in wild-type, 35S:miR396b and rGRF2 plants overexpressing miR396. (E) Rosette phenotype of 18-day-old plants with different GRF levels. 35S:miR396b plants have reduced GRF levels, whereas rGRF2 plants have increased GRF levels. (F) Paradermal view of palisade cells in the subepidermal layer of leaf 1 of 20-day-old plants. (G-I) Leaf area (G), cell size (H) and palisade cell number (I) per leaf in wild-type, 35S:miR396b and rGRF2 transgenic plants. The data shown are the mean ± s.e.m. of eight leaves or 50 cells.
The reduction in leaf size caused by miR396 overexpression was suppressed by the introduction of rGRF2, demonstrating that leaf development was disturbed in 35S:miR396b plants because of a decrease in GRF activity (Fig. 5B-D, see Fig. S2 in the supplementary material). Plants expressing rGRF2 in a wild-type background had larger leaves with more cells than control plants (Fig. 5E-I). Transgenic lines expressing high levels of wild-type GRF2 also had a moderate increase in cell number (see Fig. S2 in the supplementary material), which is in good agreement with a quantitative interaction between miR396 and the GRFs. However, we noticed that cells in rGRF2 leaves could also be reduced in their size (Fig. 5H), an effect that we have not observed in plants expressing the wild-type version of the transcription factor.
35S:miR396b plants with the highest levels of miR396 (Fig. 3A) had fewer palisade cells in the subepidermal layer (Fig. 5E-I). In these cases, the final number of cells in leaf 1 was estimated to be less than 13% that of the wild type (Fig. 5I). Concomitant with this reduction in leaf cell number was an increase in cell size: cells from 35S:miR396b plants were twice as large as wild-type cells (Fig. 5H). Determination of leaf area, cell size and palisade cell number in the transgenic plants indicated a quantitative effect of miR396 levels on the leaf parameters (Fig. 5E-I).
Cell size, however, was only altered in plants with at least a fivefold increase in miR396, which partially compensated for the reduction in cell number (Fig. 5). Compensatory changes during leaf development are well known and have been observed in an3 mutants or strong overexpressers of the cyclin-dependent kinase inhibitor KIP-RELATED PROTEIN2 (De Veylder et al., 2001; Ferjani et al., 2007; Fujikura et al., 2009; Horiguchi et al., 2006; Tsukaya, 2005). Such changes have also been shown to be activated once the cell number deviates beyond a certain threshold. Taken together, our results suggest that the balance between miR396 and the GRFs quantitatively regulates cell proliferation in leaves.
Regulation of cell proliferation by miR396 in the SAM
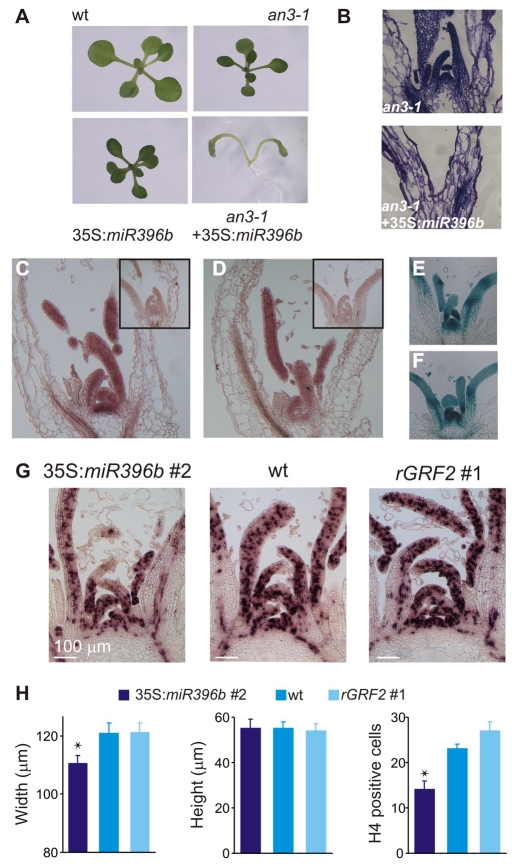
In addition to their post-transcriptional regulation by miR396, the GRFs have been shown to interact with GIF proteins, which have been proposed to act as transcriptional co-activators (Horiguchi et al., 2005; Kim and Kende, 2004). The an3-1 mutant, which has a mutation in GIF1, has smaller leaves and petals (Horiguchi et al., 2005; Kim and Kende, 2004). To further decrease the GRF activity, we overexpressed miR396 in an3-1 and observed that half of the 35S:miR396b an3-1 plants (48 out of 90 independent T1s) lacked a functional SAM (Fig. 6A). Sections revealed the absence of a meristematic dome and developing leaf primordia (Fig. 6B). These results indicated synergistic effects between miR396 overexpression, which post-transcriptionally regulates the GRFs, and mutations in the co-activator GIF1, and also suggested functions of miR396 in SAM development and maintenance.
Fig. 6.
Control of meristem function by miR396. (A) Effect of miR396 overexpression on an3-1 mutants. (B) Medial-longitudinal sections of 15-day-old seedlings stained with toluidine blue. Note the lack of a meristem dome in 35S:miR396b an3-1 plants. (C) In situ hybridization of vegetative shoot apices with an miR396 probe in the wild type. The inset shows an in situ hybridization to a section of hyl1-1 mutant, which has reduced miRNA levels. (D) In situ hybridization of vegetative shoot apices with an antisense probe against GRF2 mRNA. An in situ hybridization with a sense probe is shown in the inset. (E-F) Expression pattern in sections of shoot apices of GRF2:wtGRF2-GUS (E) and GRF2:rGRF2-GUS (F) reporters. (G) HISTONE H4 expression in shoot apices of 30-day-old plants grown in short photoperiods, detected by in situ hybridization. (H) Morphology and cell proliferation of the SAM. Width and height were estimated from medial-longitudinal sections of the SAMs of 30-day-old plants grown in short photoperiods. HISTONE H4 positive cells were counted from sections such as those of panel G. Numbers are mean ± s.e.m. (n=9). Asterisks indicate significant differences between transgenic and wild-type plants, as determined by ANOVA (P<0.05).
These results prompted us to analyze the role of miR396 and the GRFs in meristem function. First, we studied the expression of miR396 by in situ hybridization on sections of 15-day-old plants grown in short photoperiods. We detected miR396 throughout the SAM and leaf primordia (Fig. 6C). We also observed the expression of GRF2 in the meristem and young developing leaves (Fig. 6D). In good agreement with this, the GUS staining pattern in sections of wild-type GRF2-GUS was similar to the expression of GRF2 mRNA (Fig. 6D,E). At this stage, we did not see any qualitative difference between the expression patterns of the wild-type and miR396-resistant GRF2 reporters (Fig. 6E,F). Taken together, these results indicated that there is weak expression of miR396 at the SAM and leaf primordia, which overlaps with GRF2 expression.
Next, we investigated whether miR396 overexpression, which we have shown to affect leaf growth by reducing cell proliferation, also compromised the function of the SAM. We performed in situ hybridizations with HISTONE H4, which is expressed during the G1-S phase of the cell cycle in coordination with DNA synthesis (Gaudin et al., 2000; Krizek, 1999). We found that 35S:miR396 plants had fewer proliferating cells in the SAM, paralleled with a reduction in SAM size (Fig. 6G,H). The effects on SAM size indicated that the miR396-targeted GRFs can affect meristem function.
The effect of miR396 on the proliferation of meristematic cells is consistent with the lack of a functional meristem in 35S:miR396b an3-1 plants. Previous studies have shown that some of the progeny of grf multiple mutants have cotyledon fusions (Kim et al., 2003; Kim and Lee, 2006), which often appear to be related to defects in SAM function. Taken together, these data implicate the network formed by miR396, the GRFs and their interacting factors as a relevant regulator of meristem function, at least partially through the control of cell proliferation.
Regulation of the miR396 network by TCP4
The dynamic expression pattern of miR396, which generates a distal-proximal gradient of miRNA activity, and its role in the repression of cell proliferation suggested that miR396 could be part of the mitotic arrest front during leaf development. Interestingly, TCP transcription factors with a target binding site for miR319 have already been implicated in the regulation of the arrest front (Nath et al., 2003). Therefore, we decided to test the interaction between these two miRNA networks.
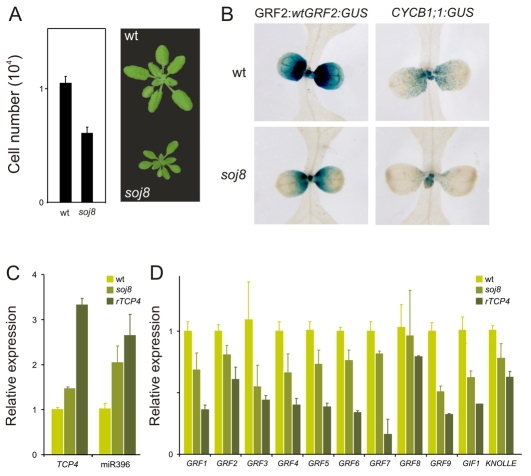
We analyzed two genotypes: soj8, an EMS-generated line that contains a point mutation in the miRNA target site of TCP4 that diminishes the interaction with miR319 (Palatnik et al., 2007); and transgenic plants moderately overexpressing TCP4:rTCP4-GFP (see Fig. S3 in the supplementary material), as higher levels of TCP4 are lethal (Palatnik et al., 2003; Schommer et al., 2008). We observed that the mild increase in TCP4 levels in soj8 (Fig. 7C) caused a reduction in cell number, with a concomitant reduction in leaf size (Fig. 7A). Analysis of the CYCB1;1 reporter in soj8 confirmed a reduction in the number of cycling cells (Fig. 7B).
Fig. 7.
Regulation of miR396 and the GRFs by TCP4. (A) Estimated palisade cell number per leaf in wild-type and soj8 plants. The data shown are the mean ± s.e.m of eight leaves or 50 cells. (B) GRF2:wtGRF2:GUS and CYCB1;1:GUS expression in wild-type and soj8 plants. (C) Accumulation of TCP4 and miR396 in moderate rTCP4 transgenic plants and soj8 mutants. Determinations were carried out by RT-qPCR and normalized to wild-type plants. RNA was prepared from apices of 30-day-old plants grown in short photoperiods, including developing leaves smaller than 3 mm. The data shown are mean ± s.e.m. of three biological replicates. (D) Measurements of GRF1-9, GIF1 and KNOLLE in soj8 and rTCP4 plants. Samples were analyzed as in C.
We then measured the levels of miR396 and found that they were increased two- to three-fold in the apices of soj8 and TCP:rTCP4-GFP (rTCP4) with respect to wild-type plants (Fig. 7C). The increase in miR396 levels was even detected in soj8, which has a mild phenotype and a marginal increase in TCP4 levels of nearly 50% (Fig. 7C). We crossed soj8 to the GRF2:wtGRF2-GUS plants and observed that the reporter was turned off sooner in the mutant than in wild-type plants (Fig. 7B).
Next, we determined the levels of the miRNA-regulated GRFs in soj8 and weak rTCP4 transgenic plants, and found that there was a reduction in the levels of these transcription factors (Fig. 7D). Interestingly, we observed that the levels of GRF5 and GRF6, which are not regulated by miR396, as well as of GIF1, were decreased in soj8 and rTCP4 (Fig. 7D), indicating that TCP4 also regulates GRF activity independently of miR396.
The finding that TCP4 can inhibit the expression of GIF1 as well as the GRFs agreed with the strong effects of the miR319-resistant TCP4 on leaf morphology. We observed a similar phenotype in both rTCP4 and 35S:miR396 an3-1 transgenic plants that were expressing the transgenes at moderate levels (see Fig. S4 in the supplementary material). It has been suggested that the repression of organ boundary genes by the TCPs underlies the lethality of the miR319-resistant TCPs (Koyama et al., 2007). Our results suggest that a major inhibition of the cell cycle machinery could also be involved in the harmful effects caused by rTCP4.
DISCUSSION
miR396 as a marker of leaf development
We followed the expression pattern of miR396 directly using small RNA blots and in situ hybridization in apices, and indirectly through the differential expression of wild-type and miRNA-resistant GRF2-GUS reporters. miR396 was expressed at low levels in the meristem and leaf primordia, and then it steadily accumulated with the development of the leaf. In contrast, the GRFs, which are highly expressed in the SAM and young leaves, decreased during leaf development, in concert with the retreat of cell proliferation.
Temporal antagonistic patterns of expression have been observed for miR156 and miR172 and their targets, the SPL and AP2-like transcription factors, respectively (Chuck et al., 2007; Wu and Poethig, 2006). The heterochronic miR156 and miR172 networks correspondingly regulate juvenile to adult, and vegetative to reproductive phase transitions, which require decisions implicating the whole plant (Aukerman and Sakai, 2003; Chen, 2004; Chuck et al., 2007; Schmid et al., 2005; Wu and Poethig, 2006). Our observations on miR396 indicated that this miRNA is also involved in the coordination of developmental events in plants; however, its role would be restricted to individual organs.
The Arabidopsis developmental program directs a basiplastic pattern, whereby leaf maturation begins at the tip and then proceeds towards the base of the organ (Donnelly et al., 1999). Cell division occurs first throughout the primordia and then a mitotic arrest front moves from the tip to the base of the leaf, so that cells in the distal part of the leaf stop cycling and begin to expand, while cells at the base continue to proliferate (Donnelly et al., 1999). Our results showed that the distal part of the leaf accumulates more miR396 and a gradient of miRNA activity proceeds towards the base of the organ. This result was supported by small RNA blots and the observed retreat of the wild-type GRF2-GUS reporter, which then matched the pattern of a CYCB1;1 reporter. These observations prompt us to implicate the repression of GRF expression by miR396 as a component of the mitotic arrest front.
Regulation of the GRFs by miR396
We observed similar spatial patterns of expression for GRF2 mRNA and miR396 in the meristem and leaf primordia, indicating that there is co-expression of the miRNA and its target at this early stage. The situation was different, however, at later stages of leaf development. The wild-type GRF2-GUS reporter was active only in the proximal part of young developing leaves, whereas the rGRF2-GUS reporter was expressed throughout the leaf. This qualitative change in the expression of wild-type GRF2-GUS was paralleled by a large increase in miR396, whose levels change by up to 10-30-fold in leaves with different developmental ages. Interestingly, the decrease in GRF expression occurred before miR396 reached its maximum level, indicating that a partial increase in the miRNA is sufficient to repress the GRFs in vivo; however, we cannot rule out the participation of additional factors that act in concert with miR396.
It has been proposed that miRNAs could have both qualitative effects, leading to complete elimination of their targets, and more subtle quantitative effects (Bartel and Chen, 2004). In plants, these quantitative interactions have been proposed for miR169 (Cartolano et al., 2007) and miR156 (Wang et al., 2008), miR319 (Ori et al., 2007) and miR164 (Baker et al., 2005; Nikovics et al., 2006), and their targets. From a mechanistic point of view, it is tempting to speculate that miR396 has dual functions during leaf development: it might quantitatively regulate GRF expression in the SAM and leaf primordia, while causing a large qualitative effect contributing to the clearance of GRF activity from older organs. This latter functional role in clearing GRF transcripts might explain the continued rise in miR396 levels, even after cell proliferation has ceased. On the other hand, the potential quantitative regulation of GRF activity during early leaf development might play a relevant role in the fine-tuning of cell proliferation, as we have shown that modifications of the balance between miR396 and GRF2 levels have important consequences for the final number of cells in the organ.
Control of cell proliferation by the miR396 network
miR396 was first identified because of its conservation between A. thaliana and rice (Jones-Rhoades and Bartel, 2004). miR396 and GRFs with an miR396 target site are present in many plant species (Axtell and Bartel, 2005; Jones-Rhoades and Bartel, 2004), indicating an ancient origin for the miR396-GRF regulatory network. The function of the GRFs as regulators of cell number in leaves is well established based on the phenotypes of grf (Horiguchi et al., 2005; Kim et al., 2003; Kim and Lee, 2006) and gif (Horiguchi et al., 2005; Kim and Kende, 2004) mutants, and plants with high miR396 levels (Liu et al., 2009).
Here, we have extended these observations and found that the GRFs regulate cell proliferation in the SAM, which at least partially explains the lack of a functional meristem in an3-1 mutants overexpressing miR396 (this study) and in grf multiple knock-outs (Kim et al., 2003; Kim and Lee, 2006). Analysis of the transcriptome of moderate miR396 overexpressers has shown that the downregulation of mitosis-specific genes is one of the main molecular effects of high miR396 levels. However, the GRFs themselves do not change their expression during the cell cycle (Menges et al., 2005) and future work will be required to identify the mechanisms underlying the activity of the GRFs.
Measurements of the GRFs by RT-qPCR indicated that miR396 targets and non-targets are turned off at similar stages of leaf development, and that they act redundantly. Previous studies in which promoters have been fused directly to a GUS reporter have shown that the transcription of the GRF genes can occur in different regions of the leaf (Horiguchi et al., 2005). We observed that the post-transcriptional control of GRF2 by miR396 contributes significantly to its final expression pattern, so it is possible that the miRNA also plays a key role in adjusting the expression of other GRFs.
Regulation of leaf development by TCP4 and miR396
The snapdragon TCP gene CIN has been shown to be expressed in a dynamic pattern during leaf development and to regulate cyclin expression (Nath et al., 2003). CIN-like genes from Arabidopsis, which are regulated by miR319, have also been implicated in the coordination of cell proliferation and differentiation in leaves (Efroni et al., 2008; Koyama et al., 2007; Masuda et al., 2008; Palatnik et al., 2003; Schommer et al., 2008). An increase of TCP4 levels due to mutations that impair the interaction with miR319 produces smaller leaves (Efroni et al., 2008; Palatnik et al., 2003; Schommer et al., 2008).
Here, we observed that plants expressing miR319-resistant forms of TCP4 induced miR396. As the quantitative balance between miR396 and the GRFs regulates cell number in leaves, the increase in miR396 caused by TCP4 might be responsible for at least part of the reduction in cell number in soj8 mutants. We observed, however, that the increase in TCP4 levels also caused a reduction in the GRFs that were not regulated by miR396 and GIF1, indicating an effect at the transcriptional level. Regulatory circuits in which a transcription factor causes both the transcriptional repression of target genes and the induction of an miRNA that in turn post-transcriptionally inhibits the same group of genes are well described in animals, where they are referred to as coherent feed-forward loops (Hornstein and Shomron, 2006).
miR319 overexpressers (Efroni et al., 2008; Ori et al., 2007; Palatnik et al., 2003) and tcp knock-outs (Nath et al., 2003; Schommer et al., 2008) have large changes in leaf morphology, as well as other phenotypic defects, such as a delay in flowering time (Palatnik et al., 2003). This indicates that the TCPs have functions that go beyond leaf development. However, it may be possible that the miR319-regulated TCPs recruit the miR396 network as part of their biological function. We proposed here that the miR396 network could be a link between different developmental inputs or environmental stimuli and the components of the cell cycle machinery.
Supplementary Material
Acknowledgements
We thank Alexis Maizel and J.F.P. laboratory members for discussions, Gabriela Olmedo and Joe Ecker for sharing unpublished data, Virginia Perdomo, Osvaldo Di Sapio and Martha Gattuso for help with cellular observations, Uciel Chorostecki for the microarray analysis, John Celenza for the cyclin reporter and especially Heike Wollmann for help with the in situ hybridization experiments. M.A.M., J.M.D. and C.S. are CONICET fellows, and R.E.R. and J.F.P. are members of CONICET. Initial work was supported by the Max Planck Society (D.W.). The majority of studies were supported by grants to J.F.P. (PICT 22142/04 from ANPCyT, CDA0007/2005-C from HFSPO, and the Howard Hughes Medical Institute). Deposited in PMC for release after 6 months.
Footnotes
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.043067/-/DC1
References
- Aukerman M. J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15, 2730-2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M. J., Bartel D. P. (2005). Antiquity of microRNAs and their targets in land plants. Plant Cell 17, 1658-1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. C., Sieber P., Wellmer F., Meyerowitz E. M. (2005). The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr. Biol. 15, 303-315 [DOI] [PubMed] [Google Scholar]
- Bartel D. P., Chen C. Z. (2004). Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 5, 396-400 [DOI] [PubMed] [Google Scholar]
- Cartolano M., Castillo R., Efremova N., Kuckenberg M., Zethof J., Gerats T., Schwarz-Sommer Z., Vandenbussche M. (2007). A conserved microRNA module exerts homeotic control over Petunia hybrida and Antirrhinum majus floral organ identity. Nat. Genet. 39, 901-905 [DOI] [PubMed] [Google Scholar]
- Chen C., Ridzon D. A., Broomer A. J., Zhou Z., Lee D. H., Nguyen J. T., Barbisin M., Xu N. L., Mahuvakar V. R., Andersen M. R., et al. (2005). Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33, e179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022-2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G., Cigan A. M., Saeteurn K., Hake S. (2007). The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 39, 544-549 [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M. K., Scheible W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T., Beemster G. T., Krols L., Terras F., Landrieu I., van der Schueren E., Maes S., Naudts M., Inze D. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13, 1653-1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny J. R., Yadegari R., Fischer R. L., Yanofsky M. F., Weigel D. (2004). The role of JAGGED in shaping lateral organs. Development 131, 1101-1110 [DOI] [PubMed] [Google Scholar]
- Donnelly P. M., Bonetta D., Tsukaya H., Dengler R. E., Dengler N. G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215, 407-419 [DOI] [PubMed] [Google Scholar]
- Efroni I., Blum E., Goldshmidt A., Eshed Y. (2008). A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20, 2293-2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferjani A., Horiguchi G., Yano S., Tsukaya H. (2007). Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiol. 144, 988-999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura U., Horiguchi G., Ponce M. R., Micol J. L., Tsukaya H. (2009). Coordination of cell proliferation and cell expansion mediated by ribosome-related processes in the leaves of Arabidopsis thaliana. Plant J. 59, 499-508 [DOI] [PubMed] [Google Scholar]
- Gaudin V., Lunness P. A., Fobert P. R., Towers M., Riou-Khamlichi C., Murray J. A., Coen E., Doonan J. H. (2000). The expression of D-cyclin genes defines distinct developmental zones in snapdragon apical meristems and is locally regulated by the Cycloidea gene. Plant Physiol. 122, 1137-1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha C. M., Kim G. T., Kim B. C., Jun J. H., Soh M. S., Ueno Y., Machida Y., Tsukaya H., Nam H. G. (2003). The BLADE-ON-PETIOLE 1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development 130, 161-172 [DOI] [PubMed] [Google Scholar]
- Haga N., Kato K., Murase M., Araki S., Kubo M., Demura T., Suzuki K., Muller I., Voss U., Jurgens G., et al. (2007). R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development 134, 1101-1110 [DOI] [PubMed] [Google Scholar]
- Horiguchi G., Kim G. T., Tsukaya H. (2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43, 68-78 [DOI] [PubMed] [Google Scholar]
- Horiguchi G., Ferjani A., Fujikura U., Tsukaya H. (2006). Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J. Plant Res. 119, 37-42 [DOI] [PubMed] [Google Scholar]
- Hornstein E., Shomron N. (2006). Canalization of development by microRNAs. Nat. Genet. 38Suppl, S20-S24 [DOI] [PubMed] [Google Scholar]
- Inze D., De Veylder L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40, 77-105 [DOI] [PubMed] [Google Scholar]
- Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., Speed T. P. (2003). Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31, e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades M. W., Bartel D. P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14, 787-799 [DOI] [PubMed] [Google Scholar]
- Kim J. H., Kende H. (2004). A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. USA 101, 13374-13379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Lee B. H. (2006). GROWTH-REGULATING FACTOR4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J. Plant Biol. 49, 463-468 [Google Scholar]
- Kim J. H., Choi D., Kende H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36, 94-104 [DOI] [PubMed] [Google Scholar]
- Koyama T., Furutani M., Tasaka M., Ohme-Takagi M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19, 473-484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B. A. (1999). Ectopic expression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Dev. Genet. 25, 224-236 [DOI] [PubMed] [Google Scholar]
- Lemon W. J., Liyanarachchi S., You M. (2003). A high performance test of differential gene expression for oligonucleotide arrays. Genome Biol. 4, R67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Song Y., Chen Z., Yu D. (2009). Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol. Plant 136, 223-236 [DOI] [PubMed] [Google Scholar]
- Lukowitz W., Mayer U., Jurgens G. (1996). Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84, 61-71 [DOI] [PubMed] [Google Scholar]
- Masuda H. P., Cabral L. M., De Veylder L., Tanurdzic M., de Almeida Engler J., Geelen D., Inze D., Martienssen R. A., Ferreira P. C., Hemerly A. S. (2008). ABAP1 is a novel plant Armadillo BTB protein involved in DNA replication and transcription. EMBO J. 27, 2746-2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M., de Jager S. M., Gruissem W., Murray J. A. (2005). Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant. J. 41, 546-566 [DOI] [PubMed] [Google Scholar]
- Mizukami Y., Fischer R. L. (2000). Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 97, 942-947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath U., Crawford B. C., Carpenter R., Coen E. (2003). Genetic control of surface curvature. Science 299, 1404-1407 [DOI] [PubMed] [Google Scholar]
- Nikovics K., Blein T., Peaucelle A., Ishida T., Morin H., Aida M., Laufs P. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18, 2929-2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno C. K., Reddy G. V., Heisler M. G., Meyerowitz E. M. (2004). The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 131, 1111-1122 [DOI] [PubMed] [Google Scholar]
- Ori N., Cohen A. R., Etzioni A., Brand A., Yanai O., Shleizer S., Menda N., Amsellem Z., Efroni I., Pekker I., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39, 787-791 [DOI] [PubMed] [Google Scholar]
- Palatnik J. F., Allen E., Wu X., Schommer C., Schwab R., Carrington J. C., Weigel D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257-263 [DOI] [PubMed] [Google Scholar]
- Palatnik J. F., Wollmann H., Schommer C., Schwab R., Boisbouvier J., Rodriguez R., Warthmann N., Allen E., Dezulian T., Huson D., et al. (2007). Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev. Cell 13, 115-125 [DOI] [PubMed] [Google Scholar]
- Piazza P., Jasinski S., Tsiantis M. (2005). Evolution of leaf developmental mechanisms. New Phytol. 167, 693-710 [DOI] [PubMed] [Google Scholar]
- Schmid M., Davison T. S., Henz S. R., Pape U. J., Demar M., Vingron M., Scholkopf B., Weigel D., Lohmann J. U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501-506 [DOI] [PubMed] [Google Scholar]
- Schommer C., Palatnik J. F., Aggarwal P., Chetelat A., Cubas P., Farmer E. E., Nath U., Weigel D. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6, e230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Palatnik J. F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8, 517-527 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545-15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Kuehn H., Gould J., Tamayo P., Mesirov J. P. (2007). GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 23, 3251-3253 [DOI] [PubMed] [Google Scholar]
- Tsukaya H. (2005). Leaf shape: genetic controls and environmental factors. Int. J. Dev. Biol. 49, 547-555 [DOI] [PubMed] [Google Scholar]
- Tsukaya H. (2006). Mechanism of leaf-shape determination. Annu. Rev. Plant Biol. 57, 477-496 [DOI] [PubMed] [Google Scholar]
- Wang J. W., Schwab R., Czech B., Mica E., Weigel D. (2008). Dual effects of miR156-tTargeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell. 5, 1231-1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White D. W. (2006). PEAPOD regulates lamina size and curvature in Arabidopsis. Proc. Natl. Acad. Sci. USA 103, 13238-13243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Poethig R. S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133, 3539-3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.