Abstract
To cover the receptive field completely and non-redundantly, neurons of certain functional groups arrange tiling of their dendrites. In Drosophila class IV dendrite arborization (da) neurons, the NDR family kinase Tricornered (Trc) is required for homotypic repulsion of dendrites that facilitates dendritic tiling. We here report that Sin1, Rictor, and target of rapamycin (TOR), components of the TOR complex 2 (TORC2), are required for dendritic tiling of class IV da neurons. Similar to trc mutants, dendrites of sin1 and rictor mutants show inappropriate overlap of the dendritic fields. TORC2 components physically and genetically interact with Trc, consistent with a shared role in regulating dendritic tiling. Moreover, TORC2 is essential for Trc phosphorylation on a residue that is critical for Trc activity in vivo and in vitro. Remarkably, neuronal expression of a dominant active form of Trc rescues the tiling defects in sin1 and rictor mutants. These findings suggest that TORC2 likely acts together with the Trc signalling pathway to regulate the dendritic tiling of class IV da neurons, and thus uncover the first neuronal function of TORC2 in vivo.
Keywords: dendritic fields, sensory neuron, tiling, TORC2, Tricornered kinase
Introduction
The target of rapamycin (TOR) is an evolutionarily conserved Ser/Thr protein kinase that functions in two distinct multiprotein complexes referred as TOR complex 1 (TORC1) and complex 2 (TORC2). TORC1 is composed of TOR, Raptor, and LST8 (also know as GβL), whereas TORC2 contains TOR, Rictor, LST8, and Sin1 (Sarbassov et al, 2005a; Wullschleger et al, 2006; Bhaskar and Hay, 2007). TORC1 regulates cell growth by phosphorylating ribosomal S6 kinase (S6K) and eukaryote initiation factor 4E-binding protein (4E-BP) in a rapamycin-sensitive manner. The function of TORC2 is less well-defined than that of TORC1, but some studies suggest that TORC2 is involved in actin cytoskeleton reorganization (Jacinto et al, 2004; Sarbassov et al, 2004). Both TORC1 and TORC2 are evolutionarily conserved in the functions and the compositions. Indeed, recent studies in both mammalian and Drosophila cell cultures have indicated that TORC2 can directly phosphorylate the serine residue (Ser473 and Ser505 in humans and Drosophila Akt, respectively) in the hydrophobic motif of Akt/PKB (Hresko and Mueckler, 2005; Sarbassov et al, 2005b; Jacinto et al, 2006).
In addition to the growth control in proliferating cells, TOR has critical functions in non-proliferating cells. In particular, recent genetic and pharmacological studies have shown that mammalian TOR (mTOR) is involved in various processes in the nervous system, including cell size control (Kwon et al, 2003), local protein synthesis in dendrites (Takei et al, 2004; Raab-Graham et al, 2006), synaptic plasticity (Tang et al, 2002; Cammalleri et al, 2003; Hou and Klann, 2004), and dendrite arborization (da) (Jaworski et al, 2005). These mTOR functions in neurons are believed to be mediated by TORC1 because rapamycin, a potential TORC1-specific inhibitor, can mimic the neuronal defects induced by mTOR ablation. In contrast, much less is known regarding the function of TORC2 in neurons, although Sin1 and Rictor are enriched in the developing neurons (Makino et al, 2006; Shiota et al, 2006).
Neurons in the same functional class are often organized in characteristic spatial patterns throughout the nervous system (Wassle and Boycott, 1991; Jan and Jan, 2003; Parrish et al, 2007). In many sensory circuits, a complete and non-redundant representation of sensory information is attained by a tiling arrangement of the dendrites, such that the dendritic arbors of the same cell type show little or no overlap. For example, the mammalian retina contains more than 20 distinct functional classes of retinal ganglion cells (RGCs), and the dendritic fields of some RGCs of the same subclass typically cover the retina with little overlap between neighbouring neurons, whereas RGCs of different subtypes have extensively overlapping arbors (Wassle and Boycott, 1991; Rockhill et al, 2002). Tiling of dendritic fields has also been observed in the sensory neurons of the leech Hirudu medicinalis, Manduca, Drosophila, and Caenorhabditis elegans (Gan and Macagno, 1995; Grueber et al, 2001, 2003; Gallegos and Bargmann, 2004), suggesting that tiling is a general mechanism that organizes the dendritic fields.
The Drosophila peripheral nervous system contains identifiable neurons with cell-type-specific dendritic morphologies, including da neurons (Bodmer and Jan, 1987). Dendrites of class IV da neurons tile the body wall in a cell-type-specific manner (Grueber et al, 2003; Parrish et al, 2007). Time-lapse analysis has indicated that terminal dendrites of these class IV neurons often stop growing or turn when they encounter dendrites of the same type (Grueber et al, 2003; Sugimura et al, 2003; Emoto et al, 2004). In addition, laser ablation of class IV neurons causes an invasion of the vacated dendritic territories by neighbouring class IV neurons (Grueber et al, 2003; Sugimura et al, 2003). Conversely, duplication of class IV neurons results in a partitioning of the receptive field. These observations indicate that dendritic tiling in class IV neurons arises from homotypic repulsive interactions between dendrites of neighbouring cells. This tiling mechanism functions in class IV neurons to avoid crossing of homotypic branches in the same neurons (iso-neuronal tiling) as well as between neighbouring neurons (hetero-neuronal tiling). Recent studies indicate that in addition to the tiling mechanism in class IV neurons, the self-avoidance mechanism functions in all da neurons to ensure proper spacing of dendritic branches (Gao, 2007).
From the results of a genetic screen for genes that regulate dendritic tiling, the NDR family kinase Tricornered (Trc) and its activator Furry (Fry) were identified as important components of the intracellular signalling cascade that regulates homotypic repulsion in class IV da neurons (Emoto et al, 2004). Dendrites of trc and fry mutants fail to avoid homologous dendritic branches, resulting in a significant overlap of dendritic fields. The Trc kinase signalling is required for the homotypic repulsion between neighbouring dendrites (hetero-neuronal tiling) and also between dendritic branches within single neurons (iso-neuronal tiling) (Emoto et al, 2004; Gao, 2007; Parrish et al, 2007). The C. elegans Trc (Sax-1) and Fry (Sax-2) homologues have also been found to serve a similar function in mechano-sensory neurons (Gallegos and Bargmann, 2004), indicating an evolutionarily conserved function for the Trc signalling in dendritic tiling. Hence, a more detailed understanding of Trc signalling may provide new insights into dendritic tiling. The NDR family of kinases including Trc is activated by the phosphorylation of a conserved serine in the kinase domain (Ser292 in Trc) and a conserved threonine within the hydrophobic motif (Thr449 in Trc). Recent genetic and biochemical studies have indicated that the Ste20 family of MST kinases can contribute to phosphorylation of this conserved threonine (Mah et al, 2001; Stegert et al, 2005; Emoto et al, 2006; Seiler et al, 2006), whereas the serine residue appears to be phosphorylated by NDR kinases themselves. In Drosophila, for example, the Ste20 kinase Hippo (Hpo) directly phosphorylates Trc on Thr449 in vivo and in vitro (Emoto et al, 2006), yet the regulatory mechanism for Trc activation in neurons still remains elusive.
In this study, we report that TORC2 genes function cell-autonomously to regulate the dendritic tiling of Drosophila class IV da neurons. Mutations in the TORC2 genes cause significant defects in dendritic tiling of class IV da neurons, which are similar to those observed in trc and fry mutants. TORC2 mutations genetically interact with trc mutations to affect dendritic tiling, and both Trc and its human homologue NDR1 can form a complex with TORC2 in neurons and cultured cells. Furthermore, we provide genetic and biochemical evidence that TORC2 is required for Trc activation both in vitro and in vivo. These findings establish TORC2 as a critical regulator of dendritic tiling in Drosophila sensory neurons through the Trc signalling pathway.
Results
Sin1 and Rictor are required cell-autonomously to control dendritic tiling
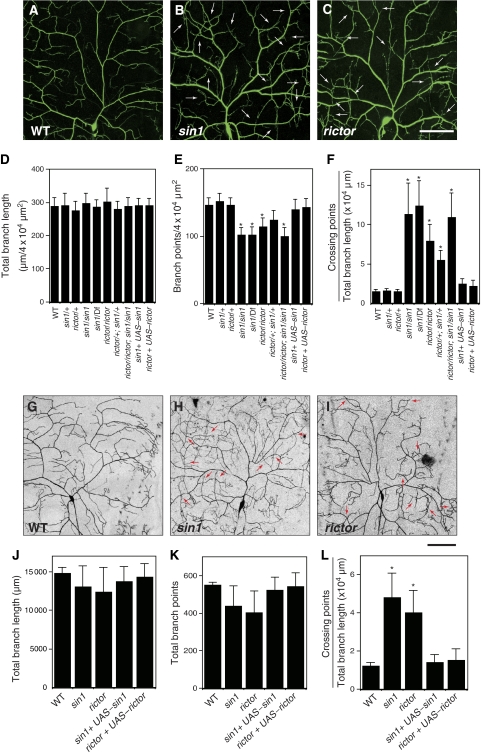
To isolate the genes required for dendritic tiling of class IV neurons, we carried out a genetic screen using the pickpocket-EGFP (ppk-EGFP) reporter, which specifically labels class IV da neurons (Grueber et al, 2003). From ∼300 mutant lines carrying PiggyBac transposon (PBc) insertions on the second chromosome (Thibault et al, 2004), we isolated one PBc insertion line with a robust dendritic tiling defect in class IV neurons (Figure 1A and B). This PBc is inserted into the single coding exon of sin1 (Hietakangas and Cohen, 2007), and is therefore likely to eliminate the Sin1 activity (hereafter this PBac insertion line is referred as sin1PBac). Homozygosity of sin1PBac or trans-heterozygous combinations of sin1PBac and a chromosomal deficiency (Df) that uncovers sin1 showed identical dendritic tiling defects (Figure 1F). In contrast, a heterozygosity of sin1PBac or hemizygosity of sin1 caused no such defects, indicating that the tiling defects we observed in sin1PBac result from the loss of sin1 functions. Quantification of the crossing points between dendritic branches indicated that ∼10% of dendritic branches crossed one another in both sin1PBac homozygotes (11.8±2.8%, n=25) and sin1PBac/Df heterozygotes (12.6±2.1%, n=25), compared with ∼1% of crossing in wild-type (WT) dendrites (1.2±0.2%, n=15) (Figure 1F). The excessive overlap of mutant dendrites is unlikely to result from abnormal stratification of terminal branches, as the terminal branches were sandwiched between the epidermis and muscles, which were typically ∼1 μm apart in both mutant and WT larvae. In addition to the dendritic tiling defect, the total number of dendrite branches in sin1 mutants was reduced to ∼80% of WT (146.0±11.4; sin1PBac/sin1PBac, 101.5±10.7; and sin1PBac/Df, 102.3±12.2/4 × 104 μm2) (Figure 1A, B, D, and E). Thus, in addition to the dendritic tiling, Sin1 may have a function in dendritic branching of class IV da neurons.
Figure 1.
sin1 and rictor function cell-autonomously in regulation of dendritic tiling in class IV neurons. (A–C) Live images of ddaC dendrites visualized by the pickpocket-EGFP (ppk-EGFP) reporter in wild-type (WT) (A), sin1PBac homozygote (B), rictorΔ2 homozygote (C). Anterior is left and dorsal is up. Arrows indicate crossing points of dendritic branches. Scale bar=50 μm. (D–F) Quantification of the total branch length (D), the branch number (E), and the crossing points (F) of WT and mutant ddaC dendrites. Error bars indicate mean±s.d. (WT, n=15; others, n=25), *P<0.01 (Student's t-test). Note that larvae heterozygous for sin1PBac over a deletion [Df(2R)BSC11] uncovering the sin1 gene show dendritic tiling defects identical to those of sin1 homozygotes. Genotypes: (A) yw; +/+; ppk-EGFP/ppk-EGFP, (B) yw; sin1PBac/sin1PBac; ppk-EGFP/ppk-EGFP, and (C) yw, rictor Δ2/yw, rictorΔ2; +/+; ppk-EGFP/ppk-EGFP. (G–I) MARCM clones of WT (G), sin1 (H), and rictor (J) are shown. Arrows indicate the crossing points of the dendrites. Scale bar=50 μm. (J–L) Quantification of the branch length (J), the branch points (K), and the crossing points (L) of MARCM clones. (WT, n=5; sin1, n=11; rictor, n=9) Clone genotypes: (G) hsFLP, elav-Gal4, UAS-mCD8-GFP/+; FRT42D, (H) hsFLP, elav-Gal4, UAS-mCD8-GFP/+; FRT42D, sin1PBac, AND (I) FRT19A, rictorΔ2; UAS-Gal4[109(2)80], UAS-mCD8GFP/hsFLP. Error bars indicate mean±s.d., *P<0.01 (Student's t-test).
Sin1 is implicated in various signalling processes through its formation of a complex with different partners, including stress-activating protein kinase (Wilkinson et al, 1999; Schroder et al, 2005), Ras small GTPase (Lee et al, 1999), and the components of TORC2 Rictor and TOR (Jacinto et al, 2006; Yang et al, 2008). To determine whether Sin1 functions together with any of these known interactors to control dendritic tiling, we examined dendrite phenotypes in mutants for the potential Sin-binding partners and found a prominent tiling defect of class IV dendrites in mutants for rictor, which encodes a component unique to TORC2 (Figure 1C). The phenotypes observed in rictor dendrites were quantitatively similar to those observed in sin1 mutants: the number of dendritic crossings was significantly higher (8.1±1.7%, n=25) than that in WT, whereas the terminal branch number was decreased to ∼80% (114.2±13.4/4 × 104 μm2) of that in WT (Figure 1D–F). Consistent with the earlier finding that Sin1 and Rictor function together to regulate tiling, trans-heterozygous combinations of sin1 and rictor alleles caused significant dendritic defects that were qualitatively similar to sin1 and rictor null mutants, whereas heterozygosity of sin1 or rictor had no obvious dendritic phenotype on its own (Figure 1D–F). Finally, sin1 rictor double mutants showed dendritic tiling defects that were indistinguishable from the single mutants (Figure 1D–F). Hence, Sin1 and Rictor most probably function in the same signalling pathway to regulate dendritic tiling.
As sin1 appears to be expressed ubiquitously (Supplementary Figure S1), the dendritic phenotypes in sin1 and rictor mutants may reflect a cell-autonomous requirement of these genes in neurons or could be a consequence of non-autonomous functions of these genes in surrounding tissues such as the epidermis and muscles. To distinguish between these two possibilities, we carried out MARCM (mosaic analysis with a repressible cell marker) analysis (Lee and Luo, 1999) to generate single-cell clones that are homozygous for null mutations in sin1 or rictor in a heterozygous background and analysed the effects on dendritic tiling. Similar to the sin1 and rictor homozygous mutants, dorsal class IV neuron MARCM clones of sin1 or rictor mutants showed defects in dendritic tiling, indicating that TORC2 genes are cell-autonomously required for dendritic tiling (Figure 1G–I). In contrast, dendrites of class I MARCM clones were not significantly affected in sin1 or rictor mutants (Supplementary Fugure S2). Class IV neuron-specific expression of sin1 and rictor largely rescued the dendritic phenotypes of sin1 and rictor MARCM clones, respectively (Figure 1J–L), further confirming the cell-autonomous functions of Sin1 and Rictor in class IV da neurons. In general, the tiling defects in sin1 or rictor clones were less severe than those observed in the sin1 or rictor homologous mutant larvae. This could be an effect of protein perdurance in MARCM clones (Lee and Luo, 1999). Alternatively, there may be cell-nonautonomous functions of the TORC2 genes in dendrite development.
Sin1 and Rictor are required for dendritic tiling between neighbouring class IV neurons
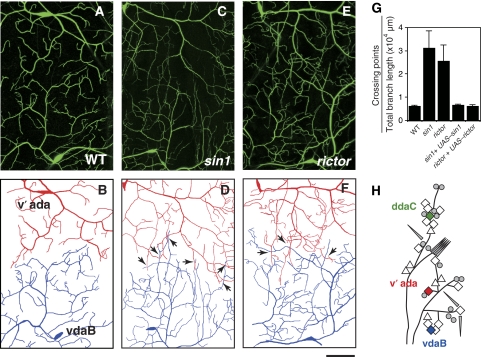
Given the essential roles of Sin1 and Rictor in tiling of terminal branches from the same neuron (iso-neural tiling), we next tested for the requirement in tiling of dendrites from different class IV neurons (hetero-neural tiling). The dendrites of the three class IV neurons found in each hemisegment normally cover the whole epidermis with very little overlap (Grueber et al, 2003; Emoto et al, 2004; Parrish et al, 2007). For example, the adjacent v'ada and vdaB neurons appeared to restrict themselves to their respective dendritic territories and rarely branched into dendritic fields of their neighbours (Figure 2A, B and H). However, in sin1 and rictor null mutants, the v'ada and vdaB dendrites often invaded the neighbouring fields (Figure 2C–F). Furthermore, the hetero-neuronal tiling defects in sin1 and rictor mutants were largely rescued by the neuronal expression of sin1 or rictor, respectively (Figure 2G). These observations suggest that Sin1 and Rictor regulate both iso-neuronal and hetero-neuronal tiling, presumably through the same mechanisms.
Figure 2.
Hetero-neuronal dendritic tiling defect in sin1 and rictor mutants. (A–F) Live images and their traces of adjacent v'ada and vdaB dendrites. In wild-type (WT) larvae (A), the dendrites of the adjacent class IV neurons, v'ada and vdaB, do not overlap; however, class IV dendrites overlap extensively in sin1 (C) and rictor (E) mutants, as evident from the tracing of dendrites derived from v'ada (red) and vdaB (blue) neurons in WT (B), sin1 (D), and rictor (F) larvae. Arrows indicate the crossing points of dendritic branches between the neighbouring neurons. Scale bar=50 μm. Genotypes: (A) yw; +/+; ppk-EGFP/ppk-EGFP, (C) yw; sin1PBac/sin1PBac; ppk-EGFP/ppk-EGFP, and (E) yw, rictorΔ2/yw, rictorΔ2; +/+; ppk-EGFP/ppk-EGFP. (G) Quantification of the crossing points in v'ada and vdaB dendrites of the WT and the mutant third instar larvae. We normalized the crossing number to the total branch length of the ventral area of v'ada dendrites and the dorsal area of vdaB dendrites. Error bars indicate mean±s.d. (WT, n=15, sin1, n=11; rictor, n=9), *P<0.01 (Student's t-test). Rescue genotypes: sin1PBac, UAS-sin1-Flag/sin1PBac, ppkGal4; ppk-EGFP/ppk-EGFP and rictorΔ2/rictorΔ2; +/ppkGal4; UAS-rictor, ppk-EGFP/ppk-EGFP. (H) Schematic representation of an abdominal hemisegment of the Drosophila larval peripheral nervous system (PNS). Dendritic arborization (da) neurons are indicated by diamonds; triangles, other multidendritic neurons; circles, extra sensory neurons; and cylinders, chordotonal organs.
TOR controls dendritic arborization and tiling through distinct complexes
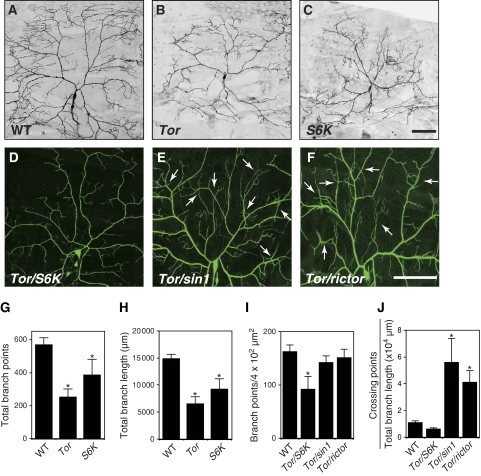
Sin1 and Rictor form a complex together with the TOR kinase referred as the TORC2 (Sarbassov et al, 2005a; Wullschleger et al, 2006; Bhaskar and Hay, 2007). We thus next examined Tor mutant MARCM clones for defects in dendritic tiling and found that unlike sin1 and rictor mutant MARCM clones, Tor MARCM clones showed a severe and highly penetrant simplification of dendritic arbors, with significant reductions in the number and length of dendritic branches, and hence in the overall size of the receptive field (Figure 3B, G and H). In addition to TORC2, TOR is also found in the functionally distinct TORC1, which is composed of TOR, Raptor, and LST8 (Sarbassov et al, 2005a; Wullschleger et al, 2006) and has recently been reported to regulate the elaboration of dendritic arbors by phosphorylating ribosomal S6K and 4E-BP in cultured hippocampal neurons (Jaworski et al, 2005). We thus next examined S6K null mutant MARCM clones and observed dendritic defects similar to Tor MARCM clones (Figure 3C, G, and H). Furthermore, Tor and S6K trans-heterozygotes showed simplified dendrites qualitatively similar to Tor and S6K mutant MARCM clones (Figure 3D, I, and J). Thus, as observed in cultured neurons, the TORC1-S6K signalling seems to have a critical function in dendrite growth and branching in post-mitotic class IV neurons. In contrast to the Tor/S6K trans-heterozygotes, a significant tiling defect was observed in larvae trans-heterozygous for mutations in Tor and either sin1 or rictor (Figure 3E, F, I and J), supporting the model in which TORC2 composed of TOR, Sin1, and Rictor together regulates the dendritic tiling of class IV neurons. Collectively, our data indicate that TORC1 and TORC2 have distinct functions in the dendrite developments of class IV neurons: TORC1 for dendritic growth and branching, and TORC2 for dendritic tiling.
Figure 3.
TORC1 and TORC2 regulate dendritic growth/branching and tiling, respectively. (A–C) Tor and S6K MARCM clones are defective in both dendritic arborization and branching. MARCM clones of (A) wild-type (WT), (B) TorΔP, and (C) S6Kl−1 are shown. Bar represents 50 μm. Clone genotypes: (A) hsFLP, elavGal4, UAS-mCD8-GFP/+; FRT40A, (B) hsFLP, elavGal4, UAS-mCD8-GFP/+; FRT40A, TorΔP; (C) hsFLP, elavGal4, UAS-mCD8-GFP/+; +/+; FRT82B, S6Kl−1. (D–F) Live images of ddaC dendrites visualized by the ppk-EGFP reporter in third instar larvae trans-heterozygous for TorΔP and S6Kl−1 (D), for TorΔP and sin1PBac (E), and for TorΔP and rictorΔ2 (F). Arrows in (E) and (F) indicate the crossing points of the dendritic branches. Genotypes: (D) TorΔP/+; S6Kl−1, ppk-EGFP/ppk-EGFP, (E) TorΔP/sin1PBac; ppk-EGFP/ppk-EGFP, and (F) rictorΔ2/+; TorΔP/+; ppk-EGFP/ppk-EGFP. (G, H) Quantification of the total branch points (G) and the branch length (H) of MARCM clones. Error bars indicate the mean±s.d. (WT, n=6; Tor, n=11; S6K, n=5), *P<0.01 relative to WT controls (Student's t-test). (I, J) Quantification of the branch points (I) and the crossing points (J) per μm2 (4 × 104) of the dendritic branches at the third instar larval stage in ddaC neurons. Error bars indicate the mean±s.d. (n=15), *P<0.01 (Student's t-test).
TORC2 interacts with the Trc kinase signalling pathway to control dendritic tiling
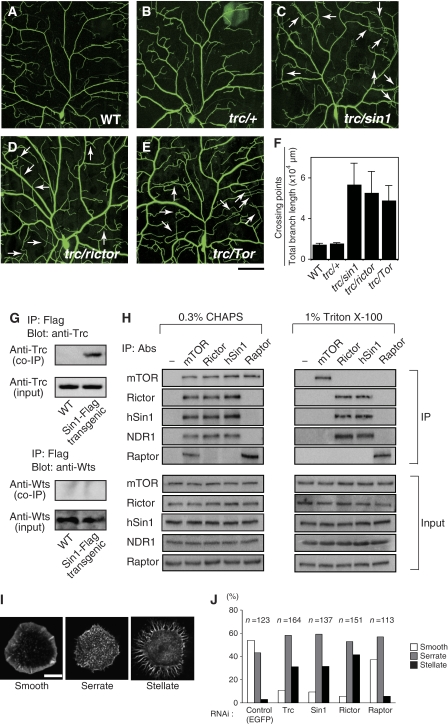
Previous studies have shown that the NDR family kinase Trc/Sax-1 and its activator Fry/Sax-2 control both the iso-neuronal and hetero-neuronal dendritic tiling of sensory neurons in Drosophila and C. elegans (Emoto et al, 2004, 2006; Gallegos and Bargmann, 2004). To examine whether TORC2 genes and trc might function in the same genetic pathway to regulate dendritic tiling, we examined genetic interactions between trc and the TORC2 genes. As mentioned above, heterozygosity for null mutations in the TORC2 genes sin1, rictor, or Tor caused no significant defects in dendritic arborization including tiling (Figure 1D–F). Similarly, heterozygosity for null alleles of trc caused no discernable defects in dendrite development (Figure 4B; Emoto et al, 2004). However, trans-heterozygous combinations of mutations in trc together with sin1 caused a significant tiling defect that was comparable to trans-heterozygous combinations of TORC2 mutants (Figure 4C and F). Similarly, trans-heterozygous combinations of trc together with rictor or Tor caused similar tiling defects (Figure 4D–F). Thus, Trc and the TORC2 genes genetically interact to regulate dendritic tiling.
Figure 4.
TORC2 genetically and physically interacts with Trc. (A–F) Live images of third instar class IV neuron visualized using the pickpocket-EGFP reporter in (A) wild-type (WT), (B) trc/+, (C) trc/sin1 trans-heterozygous, (D) trc/rictor trans-heterozygous, (E) and trc/Tor trans-heterozygous larvae. Anterior is left and dorsal is up. Bar=50 μm. Genotypes: (A) yw; +/+; ppk-EGFP/ppk-EGFP, (B) yw; +/+; trc1, ppk-EGFP/ppk-EGFP, (C) yw; sin1PBac/+; trc1, ppk-EGFP/ppk-EGFP, (D) yw, rictorΔ2/+; +/+; trc1, ppk-EGFP/ppk-EGFP, and (E) yw; TorΔP/+; trc1, ppk-EGFP/ppk-EGFP. (F) Quantification of the crossing points in ddaC of WT and trans-heterozygotes. Error bars indicate the mean±s.d. (n=15) (G) Trc can form a complex with Sin1 in Drosophila neurons. Trc and Wts were co-immunoprecipitated with neuronally expressed Sin1-Flag from transgenic fly embryos as indicated by western blot analysis using anti-Trc and anti-Wts antibodies, respectively. (H) Association of endogenous TORC2 and NDR1 in human HeLa cells. The cells were lysed in buffer containing either 0.3% CHAPS or 1% Triton X-100 as indicated. −, immunoprecipitation control (no primary antibody was used). Co-immunoprecipitation of the TORC components was detected by specific antibodies as described. (I) Morphologies of phalloidin-labeled S2 cells on concanavalin (Con) A-coated coverslips were classified into three groups (stellate, serrate, and smooth). Cells were treated with dsRNA against indicated genes for 7 days and then plated on Con A and then stained with rhodamine-phalloidin to visualize filamentous actin. (J) Quantification of cell shape on knockdown of the indicated genes. RNAi knockdown of Trc- or TORC2-specific components (Sin1and Rictor) causes a significant increase in the number of stellate cells. Note that the cell morphology was not significantly affected by genetic ablation of Raptor, a TORC1-specific component.
Given the genetic interactions between Trc and TORC2 components in dendritic tiling control, we next tested whether Trc could physically associate with TORC2 proteins. We expressed an epitope-tagged version of Sin1 (Sin1-Flag) in larval neurons using a nervous-system-specific Gal4 driver and found that Trc could be co-immunoprecipitated with Sin1-Flag (Figure 4G). This co-immunoprecipitation appeared to be specific, because Warts, another NDR kinase present in neurons, did not co-immunoprecipitate with Sin1-Flag (Figure 4G). These results suggest that Trc might associate with TORC2 in the Drosophila nervous system.
To further examine the physical interaction between TORC2 and Trc, we evaluated whether endogenous TOR complexes can be immunoprecipitated with Trc (Figure 4H). As no reliable antibodies are available for Drosophila TORC2 components, we carried out co-immunoprecipitations using HeLa cell extracts and antibodies specific for human TORC2 components and a human Trc homologue NDR1 (Hergovich et al, 2006). We found that the mTOR protein isolated with a specific antibody interacted with NDR1 as well as with hSin1, Rictor, and a TORC1-specific component Raptor (Figure 4H). In contrast, the protein complexes isolated with hSin1 or Rictor antibodies contained mTOR and NDR1 but not Raptor, while those isolated with the Raptor antibody contained mTOR but not NDR1, hSin1, nor Rictor (Figure 4H). These results indicate that NDR1 interacts, at least in part, with TORC2 but not with TORC1. As reported previously, both TORC1 and TORC2 were stable in 0.3% CHAPS buffer but were disrupted by 1% Triton X-100 (Figure 4H). Interestingly, although the interaction between mTOR and NDR1 was disrupted by Triton X-100, the interactions between NDR1 and hSin1 or Rictor were stable under these conditions (Figure 4H), suggesting that NDR1 associates with hSin1 and/or Rictor, rather than mTOR.
Previous studies suggest that the avoidance behaviour of class IV dendrites requires dynamic remodelling of the cytoskeletons (Grueber et al, 2003; Sugimura et al, 2003; Emoto et al, 2004; Parrish et al, 2007). To examine the possible function of TORC2 and Trc in the cytoskeletal organization, we used an established assay for monitoring actin network reorganization in cultured Drosophila S2 cells (Rogers et al, 2003). When placed on glass coverslips coated with the lectin concanavalin A, S2 cells reorganize their actin network to build a lamellipodium (Figure 4I, smooth). RNA interference (RNAi) knockdown of Sin1 or Rictor resulted in a significant increase in cells with aberrant organizations of their actin filaments (Figure 4I and J). These cells can be classified into three categories: cells with normal lamellae, cells that spread but showed an abnormal serrated edge, and cells that spread but showing a stellate morphology. Although the stellate morphology was observed in <5% of the control cells (3.0%, n=123) or Raptor RNAi-treated cells (4.2%, n=113), more than 30% of the Sin1 (31.3%, n=137)- or Rictor (41.4%, n=150)-treated RNAi cells exhibited a stellate shape (Jacinto et al, 2004; Sarbassov et al, 2004; Yang et al, 2008). It is interesting that, Trc knockdown cells also showed severe cytoskeletal defects (stellate cells: 31.2%, n=164), which were found to be similar to those observed in Sin1 and Rictor knockdown cells (Figure 4I and J). These observations suggest that TORC2 and Trc may regulate actin cytoskeletal organizations via the same signalling pathway.
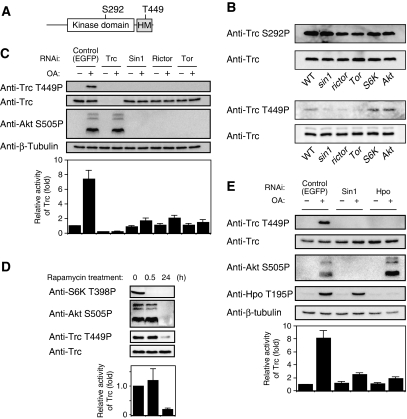
TORC2 is essential for the Trc kinase activity
Trc has conserved phosphorylation sites at Ser292 and Thr449 (Figure 5A), and phosphorylation of both of these residues is essential for maximal activation of the NDR family kinases (Millward et al, 1999; Mah et al, 2001; Tamaskovic et al, 2003; Emoto et al, 2004, 2006; Hergovich et al, 2006). In particular, phosphorylation on the threonine residue in the C-terminal hydrophobic motif (Thr449 in Trc) is tightly correlated with the NDR kinase activity in mammalian cultured cells (Tamaskovic et al, 2003; Hergovich et al, 2006). To test whether TORC2 might have a function in Trc activation, we generated rabbit polyclonal antibodies directed against phospho-epitopes of Ser292 and Thr449 (Supplementary Figure S3). Using these reagents, we examined phosphorylation states of Trc in WT and mutant embryos and found that phosphorylation on Thr449 was significantly reduced in sin1, rictor, and Tor mutant embryos compared with WT (Figure 5B). This reduction likely reflects a specific requirement of TORC2 for Trc phosphorylation as Thr449 phosphorylation was not obviously reduced in either the S6K or Akt mutants (Figure 5B). In contrast to Thr449 phosphorylation, phosphorylation on Ser292 was not significantly altered in the sin1, rictor, or Tor mutants (Figure 5B). These results suggest that TORC2 has a potential function in Trc activation in vivo.
Figure 5.
TORC2 is essential for the maximal activation of Trc. (A) Schematic representation of the Trc domain structure and phosphorylation sites. HM indicates the hydrophobic motif, which is highly conserved in the NDR kinase family. (B) Trc phosphorylation of Thr449 but not Ser292 was reduced in sin1, rictor, and Tor mutant embryos. Embryos (stage 16/17) homozygous for sin1PBac, rictorΔ2, TorΔP, S6Kl−1, and Akt1 were selected by their lack of a GFP-expressing balancer chromosome, and then homogenized in sample buffer, boiled, and analyzed by immunoblotting with the indicated antibodies. (C) Knockdown of Sin1, Rictor, or Tor inhibits Trc activation in Drosophila S2 cells. Different dsRNAs used in the experiments are indicated. ‘Control' denotes control dsRNA targeting EGFP. S2 cells were treated with 100 nM okadaic acid (OA+) or with solvent alone (OA−) for 30 min before harvesting where indicated. Cell lysates were analyzed by immunoblotting for the phosphorylation level of Akt (Ser505 for TORC2 activity) and Trc (Thr449). The bottom panel indicates Trc kinase activity relative to the control (‘control ‘ without OA treatment). The kinase activity was determined using the NDR kinase substrate peptides. Error bars indicate the mean±s.d. (n=3). (D) Prolonged treatment of cells with rapamycin inhibits Trc activation. S2 cells were treated with 100 nM rapamycin for the indicated times. Cell lysates were then analyzed by immunoblotting for phosphorylated S6K (Thr398 for TORC1 activity), Akt (Ser505 for TORC2 activity), and Trc (Thr449). The bottom panel indicates Trc kinase activity relative to the control (0 h treatment). (E) TORC2 and Hpo regulate Trc activity through different pathways. S2 cells were incubated with dsRNA against the indicated genes for 7 days and then treated with 100 nM okadaic acid (OA+) or with solvent alone (OA−) for 30 min before harvesting. Cell lysates were then analyzed by immunoblotting for the phosphorylated Akt (Ser505 for TORC2 activity), Hpo (Thr195 for Hpo activity), and Trc (Thr449). Bottom panel indicates Trc kinase activity relative to the control (‘control' without OA treatment). Error bars indicate the mean±s.d. (n=3).
To further examine whether TORC2 is essential for Trc activity, we next carried out RNAi experiments in cultured Drosophila S2 cells. Under basal conditions, Trc phosphorylation on Thr449 was too low to be detected by our phospho-specific antibodies, likely due to the low level of basal Trc phosphorylation on Thr449 (Millward et al, 1999; Tamaskovic et al, 2003). We therefore examined these RNAi effects under okadaic acid (OA) treatment conditions, which stimulate the basal Trc phosphorylation, and thus render the RNAi inhibitory effects more visible (Figure 5C). Consistently, the Trc kinase activity was elevated by about seven-fold by OA treatment (Figure 5C). Similarly, Trc phosphorylation on Thr449 was significantly increased after OA stimulation, confirming a close correlation between the Trc kinase activity and Thr449 phosphorylation. Knockdown of the TORC2 components Sin1, Rictor, or Tor with double-stranded RNAs (dsRNAs) largely eliminated both OA-induced Trc activation and phosphorylation on Thr449, although the total amount of Trc protein was not significantly affected (Figure 5C). This is consistent with the results of our analysis of Trc phosphorylation in TORC2 mutant embryos (Figure 5B). Conversely, the knockdown of Trc did not affect Akt phosphorylation on Ser505, a direct target of TORC2 (Figure 5C), indicating that Trc is not required for the TORC2 activity. It therefore appears that TORC2 likely functions upstream of the Trc signalling pathway.
We next attempted to confirm the function of TORC2 in the Trc signalling pathway using a pharmacological approach. Sarbassov et al (2006) have shown that although a short rapamycin treatment (0.5–1 h) specifically disrupts TORC1, a prolonged rapamycin treatment (48–72 h) inhibits both TORC1 and TORC2 functions in mammalian cultured cells. We thus treated S2 cells with 100 nM rapamycin for different time periods and examined the effects of this on Akt phosphorylation of Ser505 and S6K phosphorylation on Thr398, well-known phosphorylation sites for TORC2 and TORC1, respectively. Although a 30-min treatment of S2 cells eliminated S6K phosphorylation but not Akt phosphorylation, a 48-h treatment caused a strong inhibition of both S6K phosphorylation and Akt phosphorylation (Figure 5D), indicating that a short rapamycin treatment causes specific disruption of TORC1, whereas a prolonged rapamycin treatment inhibits both TORC1 and TORC2 functions in S2 cells. This is similar to what has been observed in mammalian cultured cells. Under the same rapamycin treatment conditions, neither the Trc activity nor Trc phosphorylation on Thr449 was affected after a 30-min treatment, but both were suppressed by a 48-h treatment (Figure 5D). These results strongly suggest that TORC2, but not TORC1, is required for Trc activation in S2 cells.
Finally, we examined whether TORC2 regulates Trc activity through the Ste20 family kinase Hpo, as Hpo activates Trc by phosphorylating Thr449, thereby controlling the dendritic tiling of class IV neurons (Emoto et al, 2006). On the basis of the results of previous reports (Praskova et al, 2004), we used Hpo autophosphorylation on Thr195 as a marker of Hpo activation. Under OA treatment conditions, the knockdown of Hpo with dsRNAs largely inhibited both Trc activity and Trc phosphorylation on Thr449, consistent with a previous report that Hpo is predominantly responsible for Thr449 phosphorylation (Emoto et al, 2006). However, in Hpo knockdown cells, Akt phosphorylation on Ser505 was not significantly reduced (Figure 5E). Similarly, Sin1 knockdown did not affect Hpo activation, although Trc activity and phosphorylation were inhibited (Figure 5E). These results suggest that both TORC2 and Hpo are required for full activation of the Trc kinase, and that TORC2 and Hpo appear to control Trc kinase activity through independent pathways.
A dominant active form of Trc rescues the dendritic tiling defects in sin1 and rictor neurons
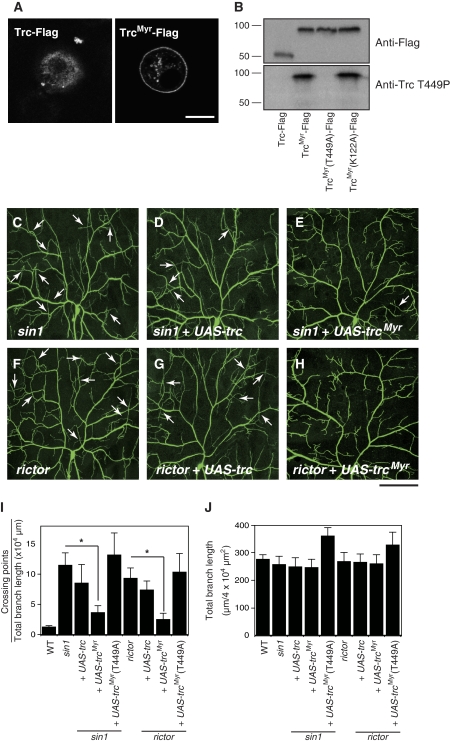
We reasoned that if TORC2 acts upstream of the Trc signalling pathway in class IV neurons to control dendritic tiling, overexpression of Trc might rescue some aspects of sin1 and rictor mutant phenotypes. We first overexpressed WT Trc using a class IV neuron-specific Gal4 driver and found a slight reduction in the tiling phenotypes of sin1 and rictor mutants (Figure 6D and G). As full activation of Trc/NDR kinases requires activator proteins such as MOBs (designated as Mats in Drosophila) and Fry (Emoto et al, 2004; Hergovich et al, 2005; Hirabayashi et al, 2008), overexpression of Trc alone might be insufficient to activate the Trc signalling in neurons.
Figure 6.
Neuronal expression of a constitutive active form of Trc (TrcMyr) rescues the dendritic tiling defects of the sin1 and rictor mutants. (A) S2 cells transfected with wild-type Trc (Trc-Flag), a membrane-targeted Trc (TrcMyr-Flag), or a membrane-targeted Trc with a mutation at the Thr449 site (TrcMyr(T449A)-Flag) were stained with anti-Flag antibodies. Images were taken under a confocal microscope. Scale bar=10 μm. (B) Lysates of S2 cells expressing a Flag-tagged wild-type (WT) (Trc-Flag), a membrane-targeted Trc (TrcMyr-Flag), a membrane-targeted Trc with a mutation at the Thr449 site (TrcMyr(T449A)-Flag), or a membrane-targeted Trc with a kinase-dead mutation at the Lys122 site (TrcMyr (K122A)-Flag) were analyzed by blotting using anti-Flag (top panel) and anti-Thr449P (bottom panel) antibodies. (C–H) In sin1 and rictor mutant larvae carrying a single copy of UAS-trcMyr under the control of ppk-Gal4 driver, tiling defects in ddaC dendrites were largely rescued. Scale bar=50 μm. Genotypes: (C) yw; sin1PBac/sin1PBac; ppk-EGFP/ppk-EGFP, (D) yw; UAS-trc, sin1PBac/sin1PBac; ppk-Gal4, ppk-EGFP/ppk-EGFP, (E) yw; sin1PBac, UAS-trcMyr/sin1PBac; ppk-Gal4, ppk-EGFP/ppk-EGFP, (F) yw, rictorΔ2/yw, rictorΔ2; +/+; ppk-EGFP/ppk-EGFP, (G) yw, rictorΔ2/yw, rictorΔ2; UAS-trc/+; ppk-Gal4, ppk-EGFP/ppk-EGFP, and (H) yw, rictorΔ2/yw, rictorΔ2; UAS-trcMyr/+; ppk-Gal4, ppk-EGFP/ppk-EGFP. (I, J) Quantification of the dendritic crossing points (I) and the total branch length (J) in the rescue experiments. −, sin1 or rictor mutants carrying no transgene. Error bars indicate the mean±s.d. (n=15), *P<0.01 (Student's t-test).
Previous studies indicate that the membrane targeting of human NDR1 leads to its constitutive activation (Hergovich et al, 2005). Accordingly, we generated a membrane-anchored version of Trc by fusing a myristylation signal from Drosophila Src1 to the N-terminus of Trc (TrcMyr) and expressed it in cultured Drosophila S2 cells. Although WT Trc was predominantly localized in the cytosol, the TrcMyr was largely observed at the plasma membrane (Figure 6A), confirming that the myristylation signal facilitates the membrane targeting of Trc. A similar membrane localization was observed with the phosphorylation point mutant TrcMyr T449A and the kinase-dead mutant TrcMyr T122A (Figure 6A). Thus, neither phosphorylation on Thr449 nor kinase activity is necessary for the membrane localization of TrcMyr. Remarkably, TrcMyr became phosphorylated on Thr449 even in the absence of OA treatment (Figure 6B), consistent with a previous report that the myristylated NDR kinases act as constitutively active forms in cultured cells (Hergovich et al, 2005). We further found that a single copy of trcMyr rescues the tiling defects in trc mutants (Supplementary Figure S4), indicating that TrcMyr retains WT activity.
We next introduced a single copy of the transgene into a sin1 or rictor null background. Strikingly, the expression of TrcMyr in class IV neurons substantially rescued both iso-neuronal and hetero-neuronal tiling defects in sin1 and rictor dendrites (Figure 6E, H and I, Supplementary Figure S5). The total branch length of class IV neurons was not significantly affected by TrcMyr, indicating that the rescue of tiling phenotype is not secondary to the growth defects of dendritic branches (Figure 6J). Furthermore, the ability of TrcMyr to rescue the tiling defects of sin1 and rictor mutants was dependent on the phosphorylation on Thr449, as the TrcMyr T449A transgene in which Thr449 was replaced with alanine was unable to rescue these mutants (Figure 6I). These data are consistent with the model in which TORC2 regulates dendritic tiling at least in part by signalling through the Trc signalling pathway.
Discussion
TORC2 regulates dendritic tiling in da neurons
In this study, we have shown that TORC2, composed of Tor, Sin1, and Rictor, is essential for the dendrite tiling of Drosophila sensory neurons. Dendrites of class IV neurons rely on homotypic repulsion to ensure that dendrites do not cross over into other boundary, and thus resulting in a complete and non-redundant coverage of the body wall. However, sin1 mutant class IV dendrites showed significant tiling phenotypes, presumably due to defects in both iso-neuronal and hetero-neuronal repulsion (Figures 1B and 2C, D). Similarly, mutations in rictor, which encodes a component of TORC2 but not TORC1, caused similar defects in dendritic tiling (Figures 1C and 2E, F). In addition, sin1 strongly interacts with rictor in the regulation of dendritic tiling (Figure 1D–F), suggesting that Sin1 and Rictor act together as TORC2 in the dendritic tiling control. The tiling defects can be substantially ameliorated by the expression of sin1 or rictor in mutant neurons (Figure 1L), indicating that TORC2 largely functions cell-autonomously to regulate dendritic tiling.
In cultured hippocampal neurons, the inhibition of mTOR by RNAi knockdown or rapamycin treatment leads to reductions in the number of dendrite branches and in the complexity of dendritic arbors, thus indicating that mTOR has an important function in regulating dendrite growth (Jaworski et al, 2005). These dendritic defects can be mimicked by the RNAi knockdown of S6K, suggesting that TORC1 may regulate dendrite growth through translational control. Consistent with these findings, we found in our current experiments that TORC1 has a similar important function in regulating dendrite growth in Drosophila, that is, Tor and S6K null mutants were found to be severely defective in dendritic growth and branching in da neurons (Figure 3A–C). In addition, our genetic studies indicated that Tor genetically interacts with S6K in dendritic growth and branching, but shows a strong interaction with sin1 and rictor in dendritic tiling (Figure 3D–F). Thus, TOR likely acts through two distinct complexes to regulate different aspects of dendrite development in class IV da neurons: TORC1 for dendritic growth/branching and TORC2 for dendritic tiling. Given the evolutionarily conserved role of TORC1 in the regulation of dendrite growth/branching, it is feasible therefore that the role of TORC2 in tiling control is also conserved. Indeed, mammalian homologues of Sin1 and Rictor are highly expressed in specific neurons in the brain (Makino et al, 2006; Shiota et al, 2006). Although it has not been established whether a tiling mechanism contributes to dendritic field specification in the central nervous system outside the retina, some neurons are known to exhibit contact-mediated growth inhibition of neurites (Sestan et al, 1999). It will thus be of interest to examine the potential roles of TORC2 in the regulation of dendritic tiling in the vertebrate nervous system. In addition, accumulating evidence now suggests that mTOR is involved in several neuronal diseases, including neurodegeneration (Ravikumar et al, 2004; Khurana et al, 2006), neurofibromatosis (Johannessen et al, 2005), and schizophrenia (Kalkman, 2006). Although the molecular events underlying the onset of mTOR-related diseases remain poorly understood, much of the current knowledge about these pathologies is associated with TORC1 (Jaworski and Sheng, 2006). In the light of our current findings that TORC1 and TORC2 have distinct functions in dendrite development, it is possible that aberrant signalling through TORC2 may account for some aspects of neuronal disorders. It will thus be intriguing to examine the relationship between TORC2 and mTOR-associated neuronal diseases in future studies.
How does TORC2 regulate dendritic tiling in da neurons? One possibility is that TORC2 may modulate the avoidance behaviour of dendritic branches by regulating cytoskeletal rearrangement. In class IV neurons, an unknown repulsion signal induces the avoidance behaviour of dendrites, which presumably requires dynamic remodelling of the cytoskeleton (Grueber et al, 2003; Sugimura et al, 2003; Emoto et al, 2004; Parrish et al, 2007). It can be noted that TORC2 has been implicated in reorganization of the actin cytoskeleton in yeast and mammalian cells (Schmidt et al, 1996; Jacinto et al, 2004; Sarbassov et al, 2004; Yang et al, 2008). Similarly, we found that RNAi knockdown of the TORC2 genes sin1 or rictor in S2 cells results in the aberrant organizations of actin fibres (Figure 4I). Hence, TORC2 can regulate cytoskeletal organization in Drosophila cultured cells as well. Given that Trc is one of the downstream targets of the TORC2 signalling pathway and that NDR kinases has critical functions in cell morphogenesis by modulating the actin cytoskeleton (Hergovich et al, 2006), TORC2 might control the actin cytoskeleton, at least in part, by signalling through Trc in neurons. In support of this model, a Trc knockdown in S2 cells caused defects in actin organization similar to what we observed after TORC2 knockdown (Figure 4I).
Previous reports have suggested that TORC2 phosphorylates PKCα to regulate the cytoskeleton in cultured mammalian cells (Schmidt et al, 1997; Sarbassov et al, 2004). It is unlikely, however, that Trc acts downstream of PKCα in regulation of the actin cytoskeleton as Trc phosphorylation on Thr449 is unaffected by the RNAi ablation of PKC or Akt (Supplementary Figure S6). Thus, TORC2 might use multiple downstream targets to regulate different aspects of actin organization. Further studies will be required to clarify the functional relationship between PKC signalling and Trc signalling with respect to actin organization.
TORC2 functions upstream of the Trc signalling pathway
Although Trc signalling has been implicated in the control of dendritic tiling in sensory neurons (Emoto et al, 2004; Gallegos and Bargmann 2004), little is known about the regulation of Trc activation in neurons. In this study, we provide genetic and biochemical data indicating that Trc functionally interacts with TORC2, but not with TORC1, in class IV neurons and cultured cells (Figure 4). This functional interaction is likely related to the specific association between Trc and the TORC2-specific components Sin1 and/or Rictor (Figure 4G and H). In addition, mutations in TORC2 genes cause a significant reduction in Trc phosphorylation on Thr449 in vivo (Figure 5B). Given that the phosphorylation of this residue is critical for Trc activation, it seems likely that TORC2 regulates the Trc activity in vivo. Reinforcing this notion, Trc phosphorylation and kinase activity were found to be largely suppressed by RNAi ablation of TORC2 components (Sin1, Rictor, or Tor) and by a pharmacological disruption of the TORC2 complex assembly (Figure 5C and D). These data thus indicate that TORC2 probably functions upstream of the Trc signalling. We thus propose a model in which TORC2 regulates dendritic tiling by signalling through the Trc signalling pathway. This contention is based on the following evidence. First, sin1 and rictor mutants show both iso-neuronal and hetero-neuronal dendritic tiling defects similar to those observed in trc mutants (Figures 1 and 2). Second, TORC2 genes including sin1, rictor, and Tor genetically interact with trc in the regulation of dendritic tiling (Figure 4A–F). Third, a constitutively active form of Trc can substantially rescue the dendritic tiling defects in sin1 and rictor mutants (Figure 6).
In addition to the dendritic tiling defects, the total number of dendrite branches in sin1 and rictor mutants was reduced to ∼80% of WT (Figure 1E). This branching defect in TORC2 mutants is inconsistent with that of trc mutants, as trc mutations cause overbranching in class IV dendrites (Emoto et al, 2004). Although it remains unknown why TORC2 and trc mutants show opposite branching phenotypes, this might be due to the reduction in the Akt activity in TORC2 mutants. Previous studies have shown that Akt activity is required for dendrite growth/branching in cultured neurons (Jaworski et al, 2005). Our RNAi experiments in S2 cells indicate that TORC2 is essential for both Trc and Akt activities (Supplementary Figure S6). Therefore, TORC2 mutants might show the combined branching phenotypes of trc and Akt mutations, resulting in a slight reduction in the branch points.
Our present data indicate that TORC2 is essential for Trc phosphorylation on the Thr449 residue that is critical for the maximal activation of Trc (Figure 5B and C). Recent studies have established that members of the Hpo/MST family of kinases directly phosphorylate the Trc/NDR kinases on this conserved threonine residue in the C-terminal hydrophobic motif (Mah et al, 2001; Stegert et al, 2005; Emoto et al, 2006; Seiler et al, 2006). In addition, we found that Hpo and TORC2 are required independently for Trc phosphorylation (Figure 5E). It is thus likely that TORC2 promotes the Hpo-dependent Trc phosphorylation on Thr449 in an indirect manner. The molecular mechanism by which TORC2 regulates Trc phosphorylation is currently unclear. On the basis of the results from recent studies of NDR1, one possible scenario is that TORC2 may have a function in the membrane recruitment of Trc for its activation. Hergovich et al (2005) show that the full activation of human NDR kinases requires the recruitment of NDR kinases to the plasma membrane, which presumably induces their proximity to the upstream kinases Hpo/MST kinases and subsequent phosphorylation of the threonine residue in the hydrophobic motif (Thr449 in Trc). MOBs are proposed to have a function in this recruitment process, although no obvious membrane-binding domain is present in these proteins (Hergovich et al, 2005). Interestingly, Sin1 contains a pleckstrin homology (PH)-like domain at its C-terminus, and this domain has been shown to be essential for the function and the membrane localization of TORC2 in both yeast and mammalian cells (Schroder et al, 2007; Berchtold and Walther, 2009). Similarly, we found that Drosophila Sin1 overexpressed in S2 cells is localized at the plasma membranes via the C-terminal PH-like domain (data not shown). As Trc associates with TORC2, it is possible that Trc might be recruited to the plasma membrane partially through the activity of TORC2. It is also possible that TORC2 may facilitate the membrane targeting of Trc by phosphorylating MOBs, which are suggested to be important for the membrane recruitment of Trc (Wei et al, 2007; Praskova et al, 2008). In either scenario, TORC2 is expected to enhance the membrane recruitment of Trc. Given our current finding that a membrane-targeted form of Trc (TrcMyr) can partially rescue dendritic tiling defects in sin1 and rictor mutants (Figure 6), we propose that TORC2 may regulate dendritic tiling by promoting membrane targeting of Trc. It is likely that the precise regulation of Trc activation within dendrites is crucial to its functions in dendritic tiling as the avoidance behaviour of class IV dendrites is induced only when the branches come within a short distance of each other (Grueber et al, 2003; Sugimura et al, 2003; Emoto et al, 2004). Presumably, therefore, TORC2 cooperates with Hpo to regulate the precise recruitment and activation of Trc signalling in specific spatial domains in dendrites.
In summary, we show in our current analyses that there is a novel neuronal function of TORC2 in the control of dendritic tiling of Drosophila sensory neurons. We also show that TORC2 regulates dendritic tiling through the Trc signalling pathway. Given the widespread function of mTOR in neuronal development and plasticity, as well as potential implications for neurological diseases, it will be important to determine whether TORC2–Trc signalling is a general mechanism underlying dendritic field specification and neural circuit formation in the nervous system.
Materials and methods
Fly stocks
The following lines were used in this study: sin1PBac (PBac e03756) (Hietakangas and Cohen, 2007), rictorΔ2 (Hietakangas and Cohen, 2007), UAS-rictor (Hietakangas and Cohen, 2007), TorΔP (Knox et al, 2007), S6Kl−1 (Knox et al, 2007), Akt1 (Stocker et al, 2002), trc1 (Emoto et al, 2004), and Df(2R)BSC11 (a deletion line uncovering sin1 gene). To visualize class IV dendrites, we used yw; +/+; ppk-EGFP/ppk-EGFP, yw; sin1PBac/sin1PBac; ppk-EGFP/ppk-EGFP, yw, rictorΔ2; +/+; ppk-EGFP/ppk-EGFP, and yw; +/+; S6Kl−1, ppk-EGFP/S6Kl−1, ppk-EGFP. For pUAS-sin1-Flag transgenic flies, the encoding region of Sin1 cDNA was amplified by PCR and subcloned into the pUAST vector using NotI and Xba sites. The trcMyr constructs were prepared by fusing DNA encoding the first 90 amino acids of Drosophila Src1 to the first codon of trc. Trc mutants were generated using a QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) and confirmed by sequencing. The PBc insertion lines that we screened in this study were obtained from the Bloomington, Exelixis, and Kyoto Stock Centers. For the genetic screening of mutants defective in dendrite development, we introduced all PBc mutant alleles into the ppk-EGFP reporter line and examined the class IV dendrite morphology at the third instar larval stage.
MARCM analysis
MARCM analyses were carried out as described previously, with some modifications (Emoto et al, 2004). Briefly, to generate mosaic clones, yw; sin1PBac, FRT42D/CyO, yw, rictorΔ2, FRT19A/FM7, w; TorΔP, FRT40A/CyO, w; S6Kl−1, FRT82B/TM3 were mated with w, elav-Gal4, UAS-mCD8GFP, hsFLP; FRT42D, tub-Gal4/CyO, w, FRT19A, tub-Gal80, hsFLP/FM7; FRT42D, Gal4[109(2)80], UAS-mCD8GFP/CyO, or w, elav-Gal4, UAS-mCD8GFP, hsFLP; FRT82B, tub-Gal80/TM6B flies. Embryos were kept for 2 h and allowed to grow for 3–5 h at 25°C before being subjected to the following heat-shock regime: 38°C for 45 min, room temperature recovery for 30 min, and finally 38°C for 45 min. The eggs were kept at 25°C and larvae were examined for mutant clones and then dissected, fixed, and stained with anti-mCD8 antibody (Caltag). Dendritic length and branch numbers were quantified using ImageJ (NIH, Bethesda, MD) with a NeuronJ plug-in.
Immunoprecipitation
For immunoprecipitation, HeLa cells were cultured in a 100-mm-diameter dish and then lysed in 200 μl of CHAPS buffer (150 mM NaCl, 50 mM Tris–HCl at pH 7.4, 2 mM EDTA, 1 mM DTT, 20 mM β-glycerophosphate, 0.3% CHAPS, and Complete cocktail (Roche)) or Triton buffer (150 mM NaCl, 50 mM Tris–HCl at pH 7.4, 2 mM EDTA, 1 mM DTT, 20 mM β-glycerophosphate, 1% Triton X-100, and Complete cocktail (Roche)). Extracts were precleaned by Protein G beads (Roche) and then incubated with ∼2 μg of primary antibodies for 2 h, followed by Protein G beads for 1 h. Beads were washed five times in lysis buffer for analyses of associated proteins by SDS–PAGE and western blotting. The following antibodies were obtained commercially: mTOR (Cell Signaling), NDR1 (Santa Cruz Biotechnology), Rictor (Cell Signaling, Abcam), and Raptor (Cell Signaling, Bethyl Laboratories). Anti-hSIn1 antibodies were kindly provided by Dr S Ishii (Makino et al, 2006).
Phospho-specific antibodies
Phospho-Drosophila Akt (Ser505), phospho-Drosophila S6K (Thr398), and phospho-MST1 (Thr195 in Drosophila Hpo) antibodies were purchased from Cell Signaling. The anti-Trc antibody has been reported previously (Emoto et al, 2004). Anti-phospho-Trc antibodies were raised against the synthetic peptides RALAY(pS)TVGT for the Ser292 phosphorylation site and FINY(pT)YKRFE for the Thr449 phosphorylation site. These antibodies were then purified using peptides coupled to Sepharose beads.
RNAi
The primers fused to generate dsRNAs were synthesized mainly as described in previous reports (Sarbassov et al, 2005a, 2005b; Yang et al, 2008) and are listed in Supplementary Table 1. dsRNAs were produced by in vitro transcription using MEGAscript kits (Ambion) according to the manufacturer's instructions. Drosophila S2 cell RNAi experiments were performed as described previously (Rogers et al, 2003). In brief, cells were plated in 24-well plates, with a starting density of 1 × 105 cells per well. Cells were then treated with 15 μM dsRNA every 3 days for 6 days. At the end of the 7-day treatment, cells were recovered for biochemical analysis, or plated on concanavalin A-treated coverslips and allowed to spread for ∼10 h for subsequent phalloidin staining.
Kinase assay
Trc kinase activity was measured as previously described (Tamaskovic et al, 2003). Briefly, S2 cells were lysed in CHAPS buffer (150 mM NaCl, 50 mM Tris–HCl at pH 7.4, 1 mM DTT, 20 mM β-glycerophosphate, 0.3% CHAPS, and Complete cocktail (Roche)) and Trc was immunoprecipitated. In vitro kinase assays were carried out in kinase buffer (20 mM Tris–HCl at pH 7.4, 10 mM MgCl2, 1 mM DTT, 100 μM ATP, 1 μM cAMP-dependent protein kinase inhibitor peptide, 1 mM NDR1 substrate peptide (KKRNRRLSVA), and 20 μCi [γ-32P]ATP) at 25°C for 30 min. The resulting solutions were then spotted onto P81 phosphocellulose paper (Whatman), and then washed five times for 10 min in 1% phosphoric acid and assayed in a liquid scintillation counter.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Table S1
Supplementary Legends
Review Process File
Acknowledgments
We thank Drs Yuh-Nung Jan (University of California, San Francisco), Iswar Hariharan (University of California, Berkeley), Duojia Pan (Johns Hopkins University), Steve Cohen (Temasek Life Sciences Laboratory), Enst Hafen (Universität Zürich), Scott Selleck (University of Minnesota), and the Bloomington, Exelixis, and Kyoto Stock Centers for fly stocks; Drs Shunsuke Ishii (RIKEN) and Duojia Pan for the rabbit anti-human Sin1 polyclonal antibody and the rabbit anti-Drosophila Warts antibodies, respectively; Drs Yash Hiromi (National Institute of Genetics) and Jay Z Parrish (University of California, San Francisco) for critical reading of this paper. This work is supported by Grants-in-Aid for Scientific Research, JSPS, and PRESTO, JST.
Footnotes
The authors declare that they have no conflict of interest.
References
- Berchtold D, Walther TC (2009) TORC2 plasma membrane localization is essential for cell viability and restricted to a distinct domain. Mol Biol Cell 20: 1565–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar PT, Hay N (2007) The two TORCs and Akt. Dev Cell 12: 487–502 [DOI] [PubMed] [Google Scholar]
- Bodmer R, Jan YN (1987) Morphological differentiation of the embryonic peripheral neurons in Drosophila. Rouxs Arch Dev Biol 196: 69–77 [DOI] [PubMed] [Google Scholar]
- Cammalleri M, Lutjens R, Berton F, King AR, Simpson C, Francesconi W, Sanna PP (2003) Time-restricted role for dendritic activation of the mTOR–p70S6K pathway in the induction of late-phase long-term potentiation in the CA1. Proc Natl Acad Sci USA 100: 14368–14373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto K, He Y, Ye B, Grueber WB, Adler PN, Jan LY, Jan YN (2004) Control of dendritic branching and tiling by the Tricornered-Kinase/Furry signaling pathway in Drosophila sensory neurons. Cell 119: 245–256 [DOI] [PubMed] [Google Scholar]
- Emoto K, Parrish JZ, Jan LY, Jan YN (2006) The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature 443: 210–213 [DOI] [PubMed] [Google Scholar]
- Gao FB (2007) Molecular and cellular mechanisms of dendrite morphogenesis. Curr Opin Neurobiol 17: 525–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos ME, Bargmann CI (2004) Mechanosensory neurite termination and tiling depends on SAX-2 and the SAX-1 kinase. Neuron 44: 239–249 [DOI] [PubMed] [Google Scholar]
- Gan WB, Macagno ER (1995) Interactions between segmental homologs and between isoneuronal branches guide the formation of sensory terminal fields. J Neurosci 15: 3243–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber WB, Graubard K, Truman JW (2001) Tiling of the body wall by multidendritic sensory neurons in Manduca sexta. J Comp Neurol 440: 271–283 [DOI] [PubMed] [Google Scholar]
- Grueber WB, Ye B, Moore A, Jan LY, Jan YN (2003) Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr Biol 13: 618–626 [DOI] [PubMed] [Google Scholar]
- Hergovich A, Bichsel SJ, Hemmings BA (2005) Human NDR kinases are rapidly activated by MOB proteins through recruitment to plasma membrane and phosphorylation. Mol Cell Biol 25: 8259–8272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A, Stegert MR, Schmitz D, Hemmings BA (2006) NDR kinases regulate essential cell processes from yeast to humans. Nat Rev Mol Cell Biol 7: 253–264 [DOI] [PubMed] [Google Scholar]
- Hietakangas V, Cohen SM (2007) Re-evaluating AKT regulation: role of TOR complex 2 in tissue growth. Genes Dev 21: 632–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi S, Nakagawa K, Sumita K, Hidaka S, Kawai T, Ikeda M, Kawata A, Ohno K, Hata Y (2008) Threonine 74 of MOB1 is a putative key phosphorylation site by MST2 to form the scaffold to activate nuclear Dbf2-related kinase 1. Oncogene 27: 4281–4292 [DOI] [PubMed] [Google Scholar]
- Hou L, Klann E (2004) Activation of the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin signaling pathway is required for metabotropic glutamate receptor-dependent long-term depression. J Neurosci 24: 6352–6361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hresko RC, Mueckler M (2005) mTOR. RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J Biol Chem 208: 40406–40416 [DOI] [PubMed] [Google Scholar]
- Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huamg Q, Qin J, Su B (2006) SIN1/MIP1 maintains rictor–mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 127: 125–137 [DOI] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN (2004) Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 6: 1122–1128 [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY (2003) The control of dendrite development. Neuron 40: 229–242 [DOI] [PubMed] [Google Scholar]
- Jaworski J, Sheng M (2006) The growing role of mTOR in neuronal development and plasticity. Mol Neurobiol 34: 205–219 [DOI] [PubMed] [Google Scholar]
- Jaworski J, Spangler S, Seeburg DP, Hoogenraad CC, Sheng M (2005) Control of dendritic arborization by the phosphoinositide-3′-kinase–Akt-mammalian target of rapamycin pathway. J Neurosci 25: 11300–11312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen CM, Reczek EE, James MF, Brems H, Legius E, Cichowski K (2005) The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc Natl Acad Sci USA 102: 8573–8578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkman HO (2006) The role of the phosphatidylinositol 3-kinase protein kinase B pathway in schizophrenia. Pharmacol Ther 110: 117–134 [DOI] [PubMed] [Google Scholar]
- Khurana V, Lu Y, Steinhilb ML, Oldham S, Shulman JM, Feany MB (2006) TOR-mediated cell-cycle activation causes neurodegeneration in a Drosophila tauopathy model. Curr Biol 16: 230–241 [DOI] [PubMed] [Google Scholar]
- Knox S, Ge H, Dimitroff BD, Ren Y, Howe KA, Arsham AM, Easterday MC, Neufeld TP, O'Connor MB, Selleck SB (2007) Mechanisms of TSC-medicated control of synapse assembly axon guidance. PLoS ONE 2: e375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CH, Zhu X, Zhang J, Baker SJ (2003) mTor is required for hypertrophy of Pten-deficient neuronal soma in vivo. Proc Natl Acad Sci USA 100: 12923–12928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Parent CA, Insall R, Firtel RA (1999) A novel Ras-interacting protein required for chemotaxis and cyclic adenosine monophosphate signal relay in Dictyostelium. Mol Biol Cell 10: 2829–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L (1999) Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 25: 307–316 [DOI] [PubMed] [Google Scholar]
- Mah AS, Jang J, Deshaies RJ (2001) Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc Natl Acad Sci USA 98: 7325–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino C, Sano Y, Shinagawa T, Miller JBA, Ishii S (2006) Sin1 binds to both ATF-2 and p38 and enhances ATF-2-dependent transcription in an SAPK signaling pathway. Genes Cells 11: 1239–1251 [DOI] [PubMed] [Google Scholar]
- Millward TA, Hess D, Hemmings BA (1999) Ndr protein kinase is regulated by phosphorylation on two conserved sequence motifs. J Biol Chem 274: 33847–33850 [DOI] [PubMed] [Google Scholar]
- Praskova M, Khoklatchev A, Ortiz-Vega S, Avruch J (2004) Regulation of the MST1 kinase by autophosphorylation, by the growth inhibitory proteins, RASSF1 and NORE1, and by Ras. Biochem J 381: 453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praskova M, Xia F, Avruch J (2008) MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr Biol 18: 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish JZ, Emoto K, Kim MD, Jan YN (2007) Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annu Rev Neurosci 30: 399–424 [DOI] [PubMed] [Google Scholar]
- Raab-Graham KF, Haddick PC, Jan YN, Jan LY (2006) Activity- and mTOR-dependent suppression of Kv1-1 channel mRNA translation in dendrites. Science 314: 144–148 [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O'Kane CJ, Rubinsztein DC (2004) Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet 36: 585–595 [DOI] [PubMed] [Google Scholar]
- Rockhill RL, Daly FJ, MacNeil MA, Brown SP, Masland RH (2002) The diversity of ganglion cells in a mammalian retina. J Neurosci 22: 3831–3843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Weidemann U, Stuurman N, Vale RD (2003) Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J Cell Biol 162: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM (2004) Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and Raptor-independent pathway that regulates the cytoskeleton. Curr Biol 14: 1296–1302 [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sabatini DM (2005a) Growing roles for the mTOR pathway. Curr Opin Cell Biol 17: 596–603 [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM (2005b) Phosphorylation and regulation of Akt/PKB by the Rictor–mTOR complex. Science 307: 1098–1101 [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM (2006) Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell 22: 159–168 [DOI] [PubMed] [Google Scholar]
- Schmidt A, Kunz J, Hall MN (1996) TOR2 is required for organization of the actin cytoskeleton in yeast. Proc Natl Acad Sci USA 93: 13780–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Bickle M, Beck T, Hall MN (1997) The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell 88: 531–542 [DOI] [PubMed] [Google Scholar]
- Schroder W, Bushell G, Sculley T (2005) The human stress-activated kinase interacting gene 1 encodes JNK-binding proteins. Cell Signal 17: 761–767 [DOI] [PubMed] [Google Scholar]
- Schroder WA, Buck M, Cloonan N, Hancock JF, Suhrbier A, Sculley T, Bushell G (2007) Human Sin1 contains Ras-binding and pleckstrin homology domains and suppresses Ras signaling. Cell Signal 19: 1279–1289 [DOI] [PubMed] [Google Scholar]
- Seiler AE, Buesen R, Visan A, Spielmann H (2006) The STE20/germinal center kinase POD6 interacts with the NDR kinase COT1 and is involved in polar tip extension in Neurospora crassa. Mol Biol Cell 17: 4080–4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sestan N, Artavanis-Tsakonas S, Rakic P (1999) Contact-dependent inhibition of cortical neurite growth mediated by Notch signaling. Science 286: 741–746 [DOI] [PubMed] [Google Scholar]
- Shiota C, Woo JT, Lindner J, Shelton KD, Magnuson MA (2006) Multiallelic disruption of the rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Dev Cell 11: 583–589 [DOI] [PubMed] [Google Scholar]
- Stegert MR, Hergovich A, Tamaskovic R, Bichsel SJ, Hemmings BA (2005) Regulation of NDR protein kinase by hydrophobic motif phosphorylation mediated by the mammalian Ste20-like kinase MST3. Mol Cell Biol 25: 11019–11029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker H, Andjelkovic M, Oldham S, Laffargue M, Wymann MP, Hemmings BA, Hafen E (2002) Living with lethal PIP3 levels: viability of flies lacking PTEN restored by a PH domain mutation in Akt/PKB. Science 295: 2088–2091 [DOI] [PubMed] [Google Scholar]
- Sugimura K, Yamamoto M, Niwa R, Satoh D, Goto S, Taniguchi M, Hayashi S, Uemura T (2003) Distinct developmental modes and lesion-induced reactions of dendrites of two classes of Drosophila sensory neurons. J Neurosci 23: 3752–3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei N, Inamura N, Kawamura M, Namba H, Hara K, Yonezawa K, Niwa H (2004) Brain-derived neurotrophic factor induces mammalian target of rapamycin-dependent local activation of translation machinery and protein synthesis in neuron dendrites. J Neurosci 24: 9760–9769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaskovic R, Bichsel SJ, Rongniaux H, Stegert MR, Hemmings BA (2003) Mechanism of Ca2+-mediated regulation of NDR protein kinase through autophosphorylation and phosphorylation by an upstream kinase. J Biol Chem 278: 6710–6718 [DOI] [PubMed] [Google Scholar]
- Tang SJ, Reis G, Kang H, Gingras AC, Sonenberg N, Schuman EM (2002) A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus. Proc Natl Acad Sci USA 99: 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, Ryner L, Cheung LM, Chong A, Erickson C, Fisher WW, Greer K, Hartouni SR, Howie E, Jakkula L, Joo D et al. (2004) A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet 36: 283–287 [DOI] [PubMed] [Google Scholar]
- Wassle H, Boycott BB (1991) Functional architecture of the mammalian retina. Physiol Rev 71: 447–480 [DOI] [PubMed] [Google Scholar]
- Wei X, Shimizu T, Lai ZC (2007) Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. EMBO J 26: 1772–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MG, Pino TS, Tournier S, Buck V, Martin H, Christiansen J, Wilkinson DG, Millar JB (1999) Sin1: an evolutionarily conserved component of eukaryotic SAPK pathway. EMBO J 18: 4210–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN (2006) TOR signaling in growth and metabolism. Cell 124: 471–484 [DOI] [PubMed] [Google Scholar]
- Yang Q, Inoki K, Ikenoue T, Guan KL (2008) Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev 20: 2820–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Table S1
Supplementary Legends
Review Process File