Abstract
Human mediator of DNA damage checkpoint 1 (hMDC1) is an essential component of the cellular response to DNA double strand breaks. Recently, hMDC1 has been shown to associate with a subunit of the anaphase-promoting complex/cyclosome (APC/C) (Coster, G., Hayouka, Z., Argaman, L., Strauss, C., Friedler, A., Brandeis, M., and Goldberg, M. (2007) J. Biol. Chem. 282, 32053–32064), a key regulator of mitosis, suggesting a possible role for hMDC1 in controlling normal cell cycle progression. Here, we extend this work to show that hMDC1 regulates normal metaphase-to-anaphase transition through its ability to bind directly to the APC/C and modulate its E3 ubiquitin ligase activity. In support of a role for hMDC1 in controlling mitotic progression, depletion of hMDC1 by small interfering RNA results in a metaphase arrest that appears to be independent of both BubR1-dependent signaling pathways and ATM/ATR activation. Mitotic cells lacking hMDC1 exhibit markedly reduced levels of APC/C activity characterized by reduced levels of Cdc20, and a failure of Cdc20 to bind the APC/C and CREB-binding protein. We suggest therefore that hMDC1 functionally regulates the normal metaphase-to-anaphase transition by modulating the Cdc20-dependent activation of the APC/C.
Introduction
Mitotic progression is regulated by the APC/C,4 an E3 ubiquitin ligase that controls the ubiquitin-dependent destruction of mitotic cyclins and other substrates in a coordinated manner. The APC/C regulates the metaphase-to-anaphase transition principally through promoting Separase activation by mediating the destruction of its inhibitor, Securin, at metaphase (1). APC/C activity and its specificity toward individual substrates are controlled successively during mitosis by the timely binding to one of two closely related activator proteins, Cdc20 and Cdh1. Cdc20-APC/C controls the metaphase-to-anaphase transition, whereas Cdh1-APC/C controls mitotic exit and progression though G1 (1). More recently, it has been shown that the ability of the APC/C to promote efficient substrate ubiquitylation also requires the presence of CBP, which probably functions through its capacity to act as an E4 ubiquitin ligase (2). To ensure the fidelity of chromosome segregation at anaphase, the activity of Cdc20-APC/C is tightly regulated by proteins that function in the spindle assembly checkpoint, which monitors for the presence of unattached kinetochores. When the spindle checkpoint is activated, Mad2 and BubR1 binding to Cdc20 inhibits Cdc20-APC/C activity and consequently the metaphase-to-anaphase transition (3). The intricacies of mitotic regulation and checkpoint activation are, however, complicated by observations that DNA damage-responsive proteins, such as BRCA1 and Chk1, appear also to function during normal mitosis. Indeed, both BRCA1 and Chk1 reside at centrosomes, with loss of either BRCA1 or Chk1 resulting in premature centrosome separation (4–6), chromosome misalignment during metaphase, chromosome lagging during anaphase, and kinetochore defects within the regions of misaligned lagging chromosomes (7, 8). Interestingly, both BRCA1 and Chk1 regulate the mitotic exit DNA damage checkpoint, which serves to terminate mitosis in the face of DNA damage before cytokinesis (9). These data indicate the potential for cross-talk between the DNA damage response and cell cycle regulation.
Although there is evidence to suggest an involvement of various regulators of the DNA damage response in controlling mitosis and the mitotic checkpoints, relatively little is known about the mechanisms of how these proteins contribute to this process. Recently, another DNA damage-responsive protein, MDC1, was shown to interact with the APC/C subunit APC3 via its BRCT domain in a phosphorylation-dependent manner, although the functional consequence of this interaction was not determined (10). Here, we expand on those findings and define mechanistically a role for hMDC1 in regulating normal mitotic progression. In addition to confirming previous observations that hMDC1 binds to components of the APC/C, we demonstrate that it also binds directly to Cdc20 and regulates APC/C E3 ubiquitin ligase activity by facilitating Cdc20 recruitment to the APC/C. Concordant with these observations, we have determined that loss of hMDC1 results in a mitotic arrest at metaphase, which is characterized by reduced cellular APC/C activity that is independent of the DNA damage and spindle assembly checkpoint pathways.
EXPERIMENTAL PROCEDURES
Cell Lines and Drug Treatment
HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, glutamine, and penicillin/streptomycin. Lymphoblastoid cell lines were routinely maintained in RPMI 1640 medium supplemented with 10% fetal calf serum, glutamine, and penicillin/streptomycin. HeLa cells, where stated, were treated with 100 ng/ml nocodazole (Sigma) for 12–18 h or 8 mm caffeine (Merck) for 12–18 h at 37 °C.
Immunoblot Analysis
Cell extracts were prepared as described previously (11). The generation of antibodies to MDC1 and ATM has been previously described (11). The anti-phospho-H2AX (serine 139) and phospho-ATM (serine 1981) antibodies were obtained from Upstate Biotechnology. The anti-Cdc20, APC3, APC6, cyclin B1, CBP, BubR1, and phosphohistone H3 (serine 10) antibodies were purchased from Santa Cruz Biotechnology. Generation of antibodies to APC7 has been described previously (2). The antibodies to SMC1, phospho-SMC1 (serine 966), Plk1, and hRad21 were obtained from Bethyl, and the anti-histone H3 antibody was purchased from Cell Signaling. Antibodies to Nbs1 and phospho-Nbs1 serine 343 were obtained from Novus. The anti-Mad2 antibody was purchased from Covance.
Immunoprecipitation
Lymphoblastoid cell lines or HeLa cells were lysed for 30 min in NETN lysis buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm EDTA, 2 mm MgCl2, 1% Nonidet P-40) supplemented with protease inhibitors (Roche Applied Science) and Benzonase (Novagen). The clarified extract was precleared with the appropriate IgG (Dako) and protein A or G beads (GE Healthcare) for 1 h at 4 °C. Immunoprecipitating antibody (5 μg) was added with protein A or G beads to the precleared supernatant and incubated for 3 h at 4 °C. The immunocomplexes were washed four times in NETN lysis buffer (containing 0.5% Nonidet P-40), boiled in SDS sample buffer, and loaded on an SDS-polyacrylamide gel. Proteins were analyzed by immunoblotting using standard methods and detected as described above.
siRNA and Transfections
The siRNA duplexes were 21 bp with a 2-base deoxynucleotide overhang (Dharmacon Research). The sequences of MDC1 siRNA oligonucleotides used were ACAGUUGUCCCCACAGCCCdTdT, ACAACATGCAGAGATTGAAdTdT, and GUUAUGGAGUGCUACUAAAdTdT. The control, Cdc20, and APC3 siRNAs used were CGUACGCGGAAUACUUCGAdTdT, CCUUGUGGAUUGGAGUUCUdTdT, and GGAAATAGCCGAGAGGTAAdTdT, respectively. The BubR1 siRNA used was a Dharmacon on target plus SMARTpool. Cells were transfected with siRNA duplexes by using Oligofectamine (Invitrogen), following the manufacturer's instructions. Cells were routinely harvested 72 h after siRNA transfection. HeLa cells were transfected with a siRNA-resistant full-length or delta PST repeat GFP-hMDC1 expression construct using Lipofectamine 2000 according to the manufacturer's instructions.
Time Lapse Microscopy
Cells were followed by time lapse microscopy maintained at 37 °C in CO2-independent medium. Phase contrast images were acquired at 200-s intervals over a period of 12 h using a Nikon Eclipse E600 microscope and analyzed using ImageJ software (National Institutes of Health).
Immunofluorescence
Cells seeded on to poly-l-lysine-coated coverslips were fixed and permeabilized in 100% methanol cooled to −20 °C for 10–15 min. Coverslips were incubated for 1 h at room temperature with the primary antibody diluted in 2% fetal calf serum, washed three times in phosphate-buffered saline, and then incubated for an additional hour with the secondary antibody. Coverslips were washed again three times in phosphate-buffered saline and then sealed onto slides with Vectashield containing DAPI (Vectorlabs). Primary antibodies used are described above. The anti-α-tubulin and anti-Mad2 antibodies were purchased from Sigma and Covance, respectively. The anti-CENPE antibody was obtained from Abcam. All secondary antibodies were either Alexa Fluor 488- or 596-coupled and purchased from Invitrogen. Cells were analyzed by confocal microscopy.
In Vitro APC/C E3 Ubiquitin Ligase Assays, Flow Cytometry, and GST Pulldowns
APC/C ubiquitin ligase assays, flow cytometry, and GST pulldown assays were carried out as described previously (2).
RESULTS
hMDC1 Directly Binds APC/C Subunits and Is Associated with E3 Ubiquitin Ligase Activity
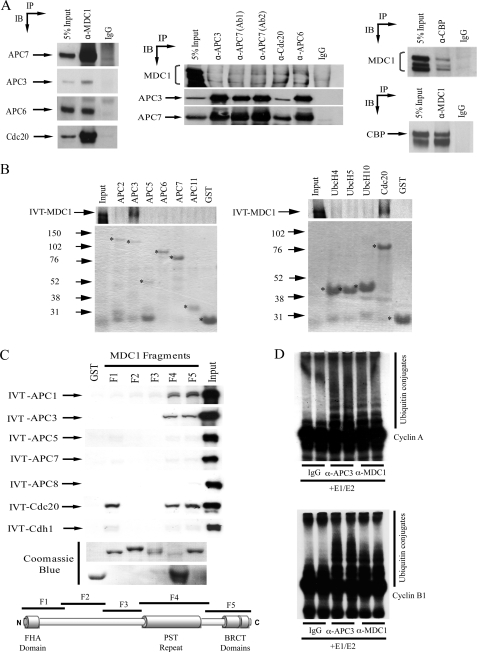
Consistent with the recent study by Coster et al. (10), coimmunoprecipitation studies confirmed that endogenous hMDC1 associates with the APC/C component APC3 in vivo (Fig. 1A). Our coimmunoprecipitation studies also revealed that hMDC1 associates with other components of the APC/C including, APC6 and APC7, and the APC/C activators, Cdc20 and CBP, suggesting that hMDC1 binds to an intact APC/C holoenzyme (Fig. 1A). Furthermore, in vitro binding revealed that hMDC1 specifically binds APC1, APC3, Cdc20, and CBP (Fig. 1, B and C, and data not shown). Indeed, l-α-[35S]methionine-labeled APC1, APC3, and Cdc20 all bound to GST-MDC1 fragments encompassing the proline-, serine-, threonine-rich (PST) repeat region and the C-terminal BRCT domains, whereas Cdc20 additionally bound the N-terminal FHA domain containing fragment (Fig. 1C). The possible involvement of both the BRCT and FHA domains of MDC1 in mediating the interaction with APC/C subunits and Cdc20 suggests that this binding may be modulated by phosphorylation/dephosphorylation cycles. Interestingly, Cdc20 contains a number of potential SQ/TQ motifs and CK2 phosphorylation sites that could facilitate interaction with the BRCT and FHA domains of MDC1, respectively.
FIGURE 1.
MDC1 binds to components of the APC/C. A, hMDC1, CBP, and components of the APC/C were immunoprecipitated (IP) from whole cell extract, separated by SDS-PAGE, and Western blotted (IB) for coprecipitating proteins. B, hMDC1 binds recombinant APC1, APC3, CBP, and Cdc20. GST fusion proteins of APC2, APC3, APC5, APC6, APC7, APC11, UbcH4, UbcH5, UbcH10, and Cdc20 were purified and mixed with [35S]methionine-labeled in vitro translated (IVT) MDC1. GST-bound proteins were purified with glutathione-Sepharose beads and separated by SDS-PAGE, and binding was assessed by autoradiography. The Coomassie Blue-stained gel indicates the level of the GST fusion proteins (marked with an asterisk) added to each reaction. Molecular masses (in kDa) are indicated to the left of the Coomassie gels. C, five GST fusion proteins (F1–5) spanning the whole hMDC1 open reading frame were mixed with [35S]methionine-labeled in vitro translated (IVT) APC1, APC3, APC5, APC7, APC8, Cdc20, or Cdh1. GST-bound proteins were purified with glutathione-Sepharose beads and separated by SDS-PAGE, and binding was assessed by autoradiography. The Coomassie Blue-stained gel indicates the level of GST protein added to each reaction. The hMDC1 protein is represented diagrammatically, and the regions encompassed by the five GST fusion proteins are indicated. D, hMDC1 is associated with ubiquitin ligase activity. hMDC1 and APC3 were immunoprecipitated from whole cell extract and mixed with recombinant E1 and E2 ubiquitin-activating proteins, recombinant ubiquitin, and 35S-labeled cyclin B1 or cyclin A at 37 °C. Ubiquitylation reactions were separated by SDS-PAGE and then visualized by autoradiography.
In keeping with the in vitro binding data and consistent with the notion that hMDC1 binds the APC/C holoenzyme, anti-hMDC1 antibodies immunoprecipitate functional APC/C ubiquitin ligase activity capable of promoting the polyubiquitylation of both cyclin A and cyclin B1 in vitro (Fig. 1D).
Loss of hMDC1 Results in a Mitotic Block at Metaphase
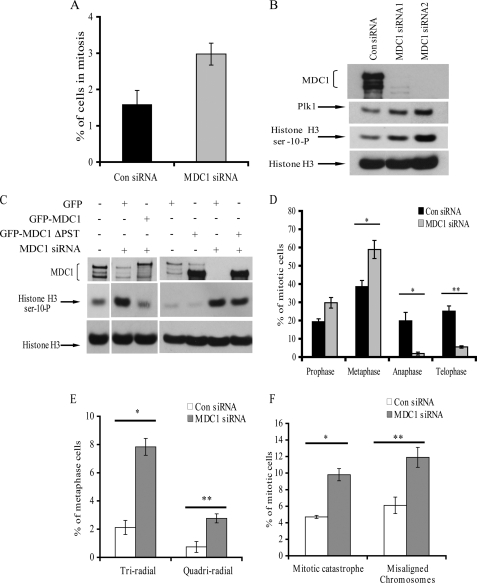
To assess the functional significance of the hMDC1 interaction with the APC/C, HeLa cells were depleted of hMDC1 using siRNA. Initially, FACS analysis was carried out on asynchronous cells treated with either control or MDC1 siRNA using an anti-phosphohistone H3 serine 10 antibody as a marker of mitotic cells. Interestingly, cells depleted of MDC1 exhibited a 2-fold increase in the percentage of mitotic cells compared with the control cells (Fig. 2A), suggesting that MDC1 is required for progression through mitosis. The increase in the number of mitotic cells in the absence of hMDC1 was confirmed by Western blotting for phosphohistone H3 serine 10 in cells treated with either control or two different hMDC1 siRNAs (Fig. 2B). In support for a role for hMDC1 in regulating mitosis, expression of a siRNA-resistant full-length GFP-MDC1 construct supported mitotic progression in the absence of endogenous hMDC1, as the observed increase in mitotic cells caused by MDC1 depletion was returned to normal levels following exogenous hMDC1 expression, judged by Western blotting for phosphohistone H3 serine 10 (Fig. 2C). Interestingly, expression of an exogenous GFP-MDC1 construct lacking the PST repeat domain, which we had demonstrated could bind directly to APC1, APC3, and Cdc20 in vitro, did not correct the mitotic accumulation caused by loss of endogenous MDC1 (Fig. 2C). This observation indicates that the loss of binding of MDC1 to components of the APC/C contributes to the observed mitotic phenotype. Given that the APC/C has discrete roles during mitosis, we wanted to establish whether mitotic cells lacking MDC1 were blocked in a specific phase of mitosis. To address this, asynchronous cells were treated with either control or hMDC1-specific siRNA, and the distribution of mitotic cells was analyzed by immunofluorescence. Notably, mitotic cells lacking hMDC1 exhibited a pronounced arrest in metaphase with a concomitant decrease in the proportion of cells in anaphase and telophase (Fig. 2D). These observations indicate a potential role of hMDC1 in regulating the metaphase-to-anaphase transition during the normal mitotic cycle. Interestingly, in addition to the block in mitotic progression, hMDC1-depleted cells also exhibited an increased number of aberrant mitoses, including multiradial metaphases, lagging chromosomes, and mitotic catastrophes (Fig. 2, E and F).
FIGURE 2.
Loss of hMDC1 causes a block in the metaphase-to-anaphase transition. A, depleting cells of hMDC1 increases the mitotic index. Cells were treated with either control or MDC1-specific siRNA, fixed, and subjected to FACS analysis using phosphohistone H3 serine 10 as a marker of mitotic cells. B, cells were treated with either control (Con) or one of two different MDC1-specific siRNAs. Extracts were subjected to SDS-PAGE, and phosphohistone H3 serine 10 was used as a marker of mitotic cells. Unphosphorylated histone H3 was used as a loading control. C, cells were treated with either control or MDC1-specific siRNA and transfected with the GFP vector alone, full-length GFP-MDC1, or GFP-MDC1 lacking the PST repeat domain. Extracts were subjected to SDS-PAGE, and phosphohistone H3 serine 10 was used as a marker of mitotic cells. Unphosphorylated histone H3 was used as a loading control. D, the distribution of mitotic stages in cells treated with control or hMDC1 siRNA was determined by microscopy. Different mitotic stages were differentiated by DAPI and α-tubulin staining. A minimum of 150 mitoses was counted for each siRNA. Three independent experiments were counted. *, p < 0.05; **, p < 0.005. E, loss of hMDC1 results in multiple mitotic abnormalities. Microscopy was used to score multiradial metaphases in cells treated with control or hMDC1-specific siRNA. Cells were stained with DAPI to visualize the DNA and with an anti-α-tubulin antibody to visualize the spindle. A minimum of 150 metaphases were counted for each siRNA. Three independent experiments were counted and are represented in the histogram. *, p < 0.00001; **, p < 0.004. F, mitotic catastrophes and lagging chromosomes were again determined, again using microscopy of cells treated with either control or hMDC1-targeted siRNA stained with both DAPI and an anti-α-tubulin antibody. A minimum of 150 mitoses were scored for each siRNA. Three independent experiments were counted and are represented in the histogram. *, p < 0.001; **, p < 0.01.
hMDC1 Regulates APC/C E3 Ubiquitin Ligase Activity in Vitro and in Vivo
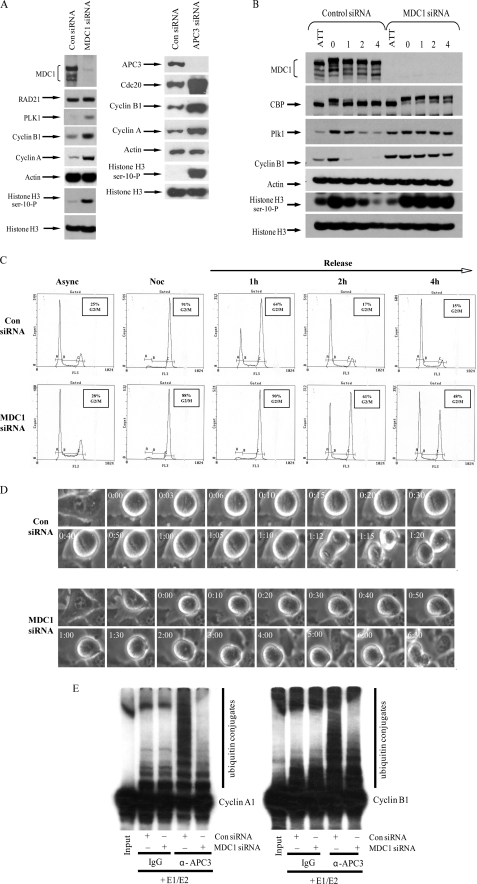
Given the pivotal role of the APC/C ubiquitin ligase activity in regulating mitotic progression, including the metaphase-to-anaphase transition (1) and the metaphase arrest seen in cells lacking hMDC1, we reasoned that the binding of hMDC1 to components of the APC/C might regulate its ubiquitin ligase activity. To assess the functional significance of the hMDC1 interaction with the APC/C, HeLa cells were depleted of hMDC1 or APC3 using siRNA, and the levels of APC/C substrates were determined by Western blot analysis. In support of a role for hMDC1 in regulating APC/C activity, cells lacking hMDC1 or APC3 exhibited elevated levels of the APC/C substrates, cyclin A, cyclin B1, and PLK1 compared with the control siRNA-treated cells (Fig. 3A), indicating that the ability of the APC/C to function as a ubiquitin ligase may be compromised in cells that have lost hMDC1.
FIGURE 3.
Loss of hMDC1 reduces APC/C E3 ubiquitin ligase activity. A, depleting cells of hMDC1 increases the mitotic index. The mitotic index of cells treated with control (Con), hMDC1, or APC3-specific siRNA was monitored by Western blotting using antibodies to cyclin B1, cyclin A, PLK1, and phosphohistone H3 serine 10. Levels of histone H3, Rad21, and actin were used as loading controls. B, in vivo APC/C activity was determined by releasing mitotic cells arrested with nocodazole into medium without nocodazole and monitoring progression (1–4 h) through mitosis by Western blot using anti-cyclin B1, PLK1, and phosphohistone H3 serine antibodies. ATT, indicates attached cells left behind following mitotic shake-off. C, cells treated as in B were stained with propidium iodide and subjected to FACS analysis to monitor cell cycle progression. D, time lapse microscopy of cells treated with either control or MDC1-specific siRNA progressing through mitosis is shown. A representative time (hh:mm) is indicated for each stage of mitosis shown. A minimum of 10 cells were used to calculate the average time for mitotic progression. E, APC/C was immunoprecipitated from cells treated with either control or hMDC1 siRNA, and its E3 ubiquitin ligase activity was determined in vitro by mixing the immunopurified APC/C with recombinant E1 and E2 proteins, recombinant ubiquitin, and 35S-labeled cyclin B1 or cyclin A at 37 °C. Ubiquitylation reactions were separated by SDS-PAGE and then visualized by autoradiography.
To substantiate this further, we treated cells with either control or hMDC1 siRNA and subsequently arrested these cells in mitosis, with nocodazole. Mitotic “shake-off” cells were then released back into the cell cycle by withdrawal of nocodazole treatment, and their progress through mitosis was monitored. Significantly, mitotic cells collected after hMDC1 knock-down took considerably longer to exit mitosis after their release from the nocodazole block, as determined by FACS analysis and by the amounts of cyclin B1, PLK1, and phosphohistone H3 serine 10 than did mitotic cells collected from cells treated with control siRNA (Fig. 3, B and C). To rule out the possibility that the mitotic delay in the MDC1 knock-down cells was caused by prior activation of the spindle assembly checkpoint, we carried out time lapse microscopy in cells treated with either control or MDC1-specific siRNA. Consistent with the nocodazole release experiment, the MDC1-depleted cells took ∼7 times longer to transit through mitosis than the control siRNA-treated cells, with the average time for mitotic progression being 50 min (±17 min) and 5 h 48 min (±2 h 10 min) for control or MDC1 siRNA-treated cells, respectively (Fig. 3D). Interestingly, many of the MDC1 siRNA-treated cells failed to complete mitosis and underwent mitotic catastrophe (data not shown). These data suggest that APC/C function in vivo is impaired after hMDC1 knock-down. To determine whether ablation of hMDC1 gene expression specifically affected APC/C function, we assayed APC/C ubiquitin ligase activity from cells following treatment with control or hMDC1 siRNA. Significantly, APC/C ubiquitin ligase activity directed toward both cyclin A and cyclin B1 was markedly reduced, relative to controls, following hMDC1 knock-down (Fig. 3E).
Metaphase Block in Cells Lacking hMDC1 Is Unlikely to Be Caused by Spindle or DNA Damage Checkpoint Activation
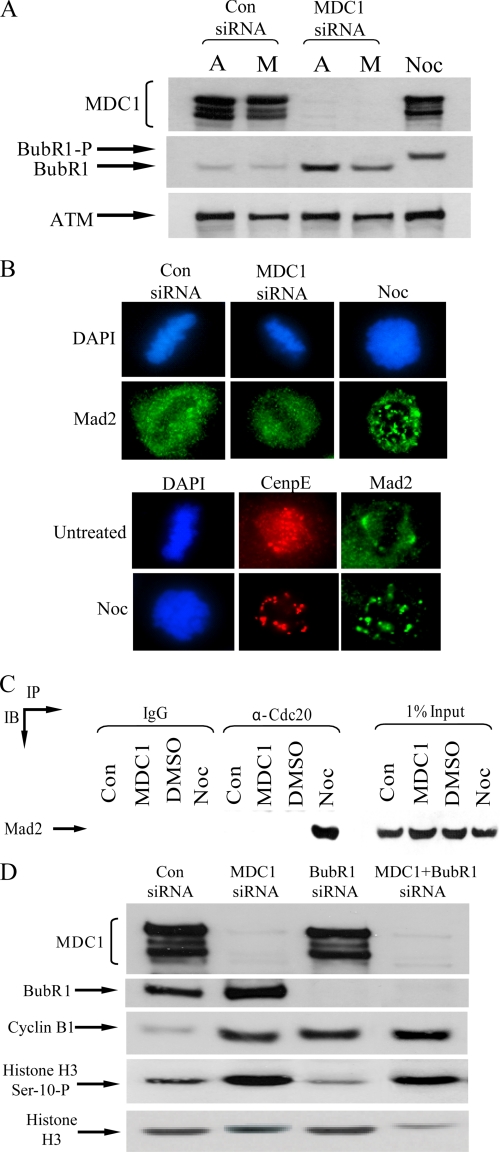
To ascertain whether the reduction in APC/C activity observed in cells lacking hMDC1 arose indirectly through the activation of the spindle checkpoint, we next investigated whether hMDC1 knock-down increased the phosphorylation status of the spindle checkpoint kinase, BubR1. Notably, depletion of hMDC1 by RNA interference did not result, relative to control cells, in the hyperphosphorylation of BubR1 (Fig. 4A). BubR1 was, however, phosphorylated and activated in normal cells treated with nocodazole, as judged by its slower migration following SDS-PAGE (Fig. 4A). These data suggest that the spindle checkpoint is unlikely to be activated when hMDC1 expression is compromised. To substantiate these findings, the localization of the spindle checkpoint protein, Mad2, was determined in mitotic cells that had been treated with nocodazole, control siRNA, or an siRNA targeted against hMDC1. Consistent with spindle checkpoint activation, Mad2 was redistributed into foci at kinetochores following treatment of cells with nocodazole (Fig. 4B). In contrast, Mad2 was not relocalized following treatment of cells with control or hMDC1 siRNA (Fig. 4B). Indeed, Mad2 could only be coimmunoprecipitated with Cdc20 in cells treated with nocodazole but not in cells treated with control or hMDC1-specific siRNA (Fig. 4C). It would be anticipated that if hMDC1 depletion specifically activated the spindle checkpoint the subsequent depletion of a component of the spindle checkpoint would override the mitotic arrest imposed by the loss of hMDC1. To assess this, we treated HeLa cells with control, hMDC1, BubR1, or MDC1 and BubR1 siRNA. Interestingly, knockdown of hMDC1 together with BubR1 failed to alleviate significantly the mitotic block observed in cells lacking hMDC1 as illustrated by the elevated phosphohistone H3 serine 10 and cyclin B1 levels in the double knock-down relative to the controls (Fig. 4D). Taken together, these data suggest that the metaphase arrest arising following hMDC1 knock-down is unlikely to be due to spindle checkpoint activation. However, we cannot rule out the possibility that the classical spindle checkpoint might be activated at very low levels, or an atypical checkpoint is activated. Despite this, in keeping with our interpretation, the elevated levels of cyclin A in MDC1-depleted cells (Fig. 3A) argues against spindle checkpoint activation because disruption of the mitotic spindle does not prevent APC/C-dependent cyclin A degradation (12).
FIGURE 4.
Metaphase arrest caused by hMDC1 knockdown is unlikely to arise as a result of spindle checkpoint activation. A, BubR1 activation is monitored by a decrease in mobility on an SDS-polyacrylamide gel. Cells were treated with control (Con) or hMDC1 siRNA, and mitotic cells (indicated by M) in each case were isolated by shake-off. The remaining attached cells (indicated by A) were also harvested. Mitotic cells treated with nocodazole (Noc) to activate the spindle checkpoint were also isolated by shake-off. Levels of ATM were used as a measure of protein loading. BubR1-P, hyperphosphorylated BubR1. B, cells were treated with control or hMDC1 siRNA or incubated with nocodazole, seeded onto coverslips, fixed, and the subcellular localization of Mad2 was determined by immunofluorescence. Centromeric protein E was used as a marker of kinetochores. C, Cdc20 was immunoprecipitated from cells treated with control siRNA, hMDC1 siRNA, dimethyl sulfoxide (DMSO), or nocodazole, separated by SDS-PAGE, and Western blotting was used to assess whether Mad2 was bound. D, HeLa cells were transfected with control, hMDC1, BubR1, or hMDC1 and BubR1 siRNA. Cells were harvested, and the mitotic population was determined by Western blotting using antibodies to cyclin B1 and phosphohistone H3 serine 10. Histone H3 is used as a loading control. Levels of MDC1 and BubR1 were determined to assess the efficiency of siRNA-mediated knockdown.
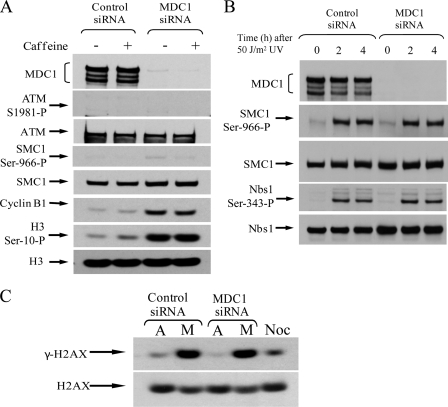
Because hMDC1 functionally regulates DNA damage response pathways, it is conceivable that, following hMDC1 knockdown, ATM- and/or ATR-dependent damage-response checkpoints might be activated that block the transition from metaphase into anaphase. To see whether this was the case, HeLa cells were treated with either control or MDC1 siRNA and then exposed to caffeine to inhibit ATM and ATR function. Notably, neither the loss of ATM nor ATR function caused by caffeine treatment overcame the metaphase block imposed by loss of hMDC1. Indeed, phosphohistone H3 serine 10 and cyclin B1 levels remained high in hMDC1 knock-down cells, where ATM and ATR were inactivated (Fig. 5A). Furthermore, the lack of ATM and SMC1 phosphorylation in cells depleted of MDC1 indicated that the mitotic block did not arise as a consequence of the presence of DNA damage (Fig. 5A), even though MDC1 siRNA-treated cells were capable of phosphorylating known ATM/ATR targets following exposure to DNA damage (Fig. 5B). These data indicate that ATM- and ATR-dependent DNA damage checkpoints are not activated following loss of hMDC1 function and therefore do not account for the mitotic arrest observed in cells lacking hMDC1. Consistent with the notion that ATM- and ATR-dependent checkpoint pathways are not activated following hMDC1 knock-down, γ-H2AX levels, used as a measure of DNA double strand breaks, were not elevated in mitotic cells after treatment with hMDC1 siRNA, relative to mitotic cells treated with control siRNA (Fig. 5C). Collectively, these observations suggest a role for hMDC1 in regulating the onset of anaphase by directly modulating the E3 ubiquitin ligase activity of the APC/C.
FIGURE 5.
Knockdown of hMDC1 does not activate the DNA damage checkpoint. A, HeLa cells were transfected with either control or hMDC1 siRNA. 72 h after transfection, cells were either mock-treated or exposed to 8 mm caffeine for 12 h. Cells were harvested, and the mitotic population was determined by Western blotting using antibodies to cyclin B1 and phosphohistone H3 serine 10. Phospho-ATM and phospho-SMC1 antibodies were used as markers of DNA damage-induced checkpoint activation. ATM, SMC1, and histone H3 levels were used as loading controls. B, loss of MDC1 does not block the ability of cells to respond to DNA damage. Cells were treated with control or MDC1-specific siRNA, irradiated with 50 J/m2 UV, and harvested at the times indicated. Western blotting was performed and phosphospecific antibodies to SMC1 and Nbs1 were used as markers of checkpoint activation. C, mitotic (M) and attached (A) cells were isolated from cells treated with control or MDC1 siRNA and separated by SDS-PAGE. DNA damage was determined by assessing the level of H2AX phosphorylation. Noc, mitotic cells treated with nocodazole.
hMDC1 Regulates Cdc20
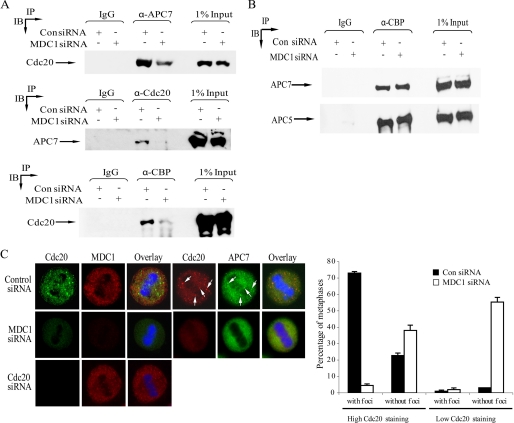
The activity of the APC/C during metaphase is controlled principally by its ability to bind its activator, Cdc20. The metaphase arrest phenotype observed when hMDC1, CBP, and Cdc20 expression is compromised suggested that these proteins function in a common pathway to regulate the APC/C (2, 13). Indeed, like hMDC1, CBP binds APC/C subunits and Cdc20 (Figs. 1A and 6, A and B) directly (2). The capability of hMDC1 to directly bind components of the APC/C, CBP, and Cdc20 in vitro and in vivo (Fig. 1) led us to speculate that hMDC1 might regulate mitotic progression by facilitating the recruitment of Cdc20 to the APC/C and/or CBP. To investigate this possibility, we performed reciprocal coimmunoprecipitation and Western blot analysis to determine whether Cdc20 retained the ability to bind the APC/C and CBP when hMDC1 expression was knocked down. Strikingly, and consistent with our hypothesis, Cdc20 binding to both the APC/C and CBP was compromised significantly in cells lacking hMDC1 compared with cells treated with control siRNA (Fig. 6A). Interestingly, the loss of hMDC1 did not affect the association of CBP with components of the APC/C (Fig. 6B), indicating that although CBP binds hMDC1, it can associate with the APC/C independently of hMDC1.
FIGURE 6.
Loss of hMDC1 compromises the ability of Cdc20 to bind APC/C and CBP. A, APC/C, CBP, or Cdc20 was immunoprecipitated from cells treated with control (Con) or hMDC1-targeted siRNA, separated by SDS-PAGE, and Western blotting was used to assess the binding to Cdc20 and APC7, respectively. B, loss of hMDC1 does not affect the association of CBP with the APC/C. CBP was immunoprecipitated from cells treated with control or hMDC1 siRNA, separated by SDS-PAGE, and Western blotting was used to assess its ability to bind APC/C subunits APC5 and APC7. C, loss of hMDC1 compromises the localization of Cdc20. The localization of Cdc20 and APC7 was determined by confocal microscopy in cells with and without hMDC1. The specificity of Cdc20 staining was verified using Cdc20-specific siRNA. The histogram indicates the number of metaphases exhibiting either high or low Cdc20 staining as well as the presence or absence of Cdc20 foci. An example of high and low Cdc20 staining is given. A minimum of 150 metaphases were counted for each siRNA. Three independent experiments were counted.
To investigate the relationship among Cdc20, the APC/C, and CBP further, we used immunofluorescent confocal microscopy to examine the cellular localization of Cdc20, the APC/C, and CBP in cells lacking hMDC1. Loss of hMDC1 had no effect on the centrosomal and diffuse cellular speckle localization of CBP during metaphase (data not shown). In normal metaphase cells, Cdc20 displayed a predominantly diffuse cellular staining interspersed by numerous large, distinct foci, the validity of which was verified by Cdc20 siRNA (Fig. 6C). Importantly, the Cdc20 foci colocalized with APC7, indicating that these subnuclear domains may represent a discrete subpopulation of active APC/C (Fig. 6C). Significantly, metaphases lacking hMDC1 exhibited a complete loss of Cdc20 and APC7 foci (Fig. 6C). Interestingly, the levels of Cdc20 staining were reduced in more than half of the metaphases observed in hMDC1 knockdown cells (Fig. 6C). These data suggest that hMDC1 might control APC/C activity during normal mitotic progression by coordinating APC/C-Cdc20 interaction by specifically regulating the cellular localization of Cdc20 and the APC/C as well as maintaining stable levels of mitotic Cdc20.
DISCUSSION
In this study, we demonstrate a novel function of the double strand break repair protein, hMDC1, in regulating the metaphase-to-anaphase transition. Tight control of APC/C activity is essential to ensure equal distribution of genetic material into each daughter cell. However, relatively little is known about what governs the activity of the APC/C throughout mitosis. Modulating the binding of Cdc20 and Cdh1 to the APC/C appears to be a critical mechanism that dictates its activity and substrate specificity. Moreover, the inactivation of Cdc20-bound APC/C by Mad2 and BubR1 forms the underlying mechanism by which the activated spindle checkpoint can halt the mitotic progression at metaphase (3). In addition to confirming the direct binding between MDC1 and APC3 as described by Coster et al. (10), our data demonstrate that hMDC1 binds independently to other components of the APC/C and regulates its activity by affecting the efficacy with which Cdc20 can interact with components of the APC/C and the APC/C-interacting protein, CBP. The absence of an activated classical spindle checkpoint in cells devoid of hMDC1 suggests that the inability of Cdc20 to bind the APC/C is unlikely to be mediated indirectly through the actions of Mad2/BubR1, although we cannot rule out the possibility that cells lacking hMDC1 activate either the spindle checkpoint at a very low level or an as yet uncharacterized component of the spindle checkpoint. Given the function of hMDC1 as a scaffold protein during the cellular response to DNA double strand breaks, it is conceivable that hMDC1 acts in a similar manner during normal mitotic progression. Consistent with this hypothesis is the observed loss of Cdc20 recruitment to APC/C-containing foci in hMDC1 siRNA-transfected cells. The ability of hMDC1 to bind multiple components of the APC/C, CBP, and Cdc20 independently suggests that it might function as a molecular bridge between the APC/C and Cdc20. A reduced ability of Cdc20 to bind the APC/C as well as CBP is likely to account, in part, for both the reduced E3 ubiquitin ligase activity observed in hMDC1-deficient cells and the metaphase-to-anaphase block. Interestingly, a recent report by Aldeman et al. (14) demonstrated that loss of Rad50, a protein known to bind to hMDC1, also resulted in a mitotic block with fewer cells making the transition into anaphase, thus strengthening a role for hMDC1 perhaps in conjunction with the MRN complex in regulating progression through mitosis. In this respect, the addition of recombinant MDC1 failed to increase the E3 ligase activity of the APC/C in vitro, suggesting that additional MDC1-binding proteins not present in the purified MDC1 preparation, such as the MRN complex, may also be required for MDC1 to promote efficient APC/C activation (data not shown).
The reduced levels of metaphase Cdc20 in cells lacking hMDC1 may additionally contribute to the inability of the APC/C to target substrates properly, critical for the progression to anaphase, for degradation. The mechanism underlying the reduced Cdc20 levels remains unclear, although a failure of Cdc20 to localize to APC/C foci may be a factor in its instability.
Again, we cannot rule out the possibility that loss of hMDC1 activates an as yet, unidentified, novel mitotic DNA damage checkpoint that can act in a manner similar to the spindle assembly checkpoint and inhibits APC/C activity by preventing Cdc20 binding. However, the fact that loss of ATM and ATR activity failed to facilitate the progression of hMDC1-deficient mitotic cells into anaphase as well as the absence of detectably increased levels of γ-H2AX in mitotic cells lacking hMDC1 compared with control siRNA-treated mitotic cells suggest that it is unlikely that the metaphase block in these cells stems from the presence of elevated levels of DNA damage.
The elevated percentage of mitotic cells in prophase as well as the increased frequency of other mitotic abnormalities in cells devoid of hMDC1 suggest that it may play additional roles during normal mitotic progression. The increased presence of multiradial metaphases could indicate a role for hMDC1 in centrosome duplication or migration in a manner similar to BRCA1 and Chk1 (4–6), both of which are known to be regulated by MDC1 during the DNA damage response (11). In this respect, a recent observation by Rai et al. would support this notion (15). However, we were unable to detect the presence of hMDC1 at centrosomes using six different anti-MDC1 antibodies and a GFP-tagged MDC1 expression construct (data not shown). The observed increased numbers of lagging chromosomes and mitotic catastrophe in hMDC1-deficient cells again is reminiscent of cells lacking BRCA1 and Chk1, although the severe spindle disruption exhibited in Xenopus laevis extracts depleted of BRCA1/BARD1 was not observed in hMDC1 siRNA-treated cells (7). Overall, the additional mitotic phenotypes caused by compromising hMDC1 expression indicates that hMDC1 may also function together with BRCA1 and Chk1 during normal mitotic progression. Unlike Chk1 and BRCA1, however, MDC1 appears to facilitate the activation of the APC/C and may represent a novel mitotic regulator.
It is interesting to speculate as to the biological consequences of this novel mitotic function of MDC1. Although a knock-out mouse model for MDC1 has been generated, a role for MDC1 in regulating mitotic progression was not addressed (16). Despite the low amino acid conservation between human and mouse MDC1, the growth retardation exhibited by these animals caused by a reduced cellular proliferative capacity and the appearance of giant nondividing cells could be in part accounted for by defective mitotic progression.
Interestingly, however, it has been shown with inherited human syndromes associated with mutations in genes known to regulate mitotic progression and spindle assembly, such as MCPH1, PERICENTRIN, ASPM, CDK5RAP2, and CENPJ, that mitotic defects are usually associated with the development of microcephaly and mental retardation (17). Furthermore, these features are also associated with a number of other human disorders that have mutations in DNA repair and checkpoint genes, for example ATR-Seckel syndrome, Nijmegen breakage syndrome, and radiosensitive severe-combined immunodeficiency (18). In addition, both MCPH1 and Pericentrin have been linked to regulating cellular DNA damage signaling (19–21). Therefore, it is conceivable that defective control of mitotic progression in the absence of MDC1 could lead to or contribute to the development of microcephaly.
Collectively, our data demonstrate an additional biological role for MDC1 during mitosis, where it functions as a key regulator of metaphase-to-anaphase transition. We propose that hMDC1 might integrate signaling pathways to mediate Cdc20 association with the APC/C and hence control APC/C activity.
Acknowledgments
We thank Drs. Hiroyuki Yamano and Andrew Fry for reagents. In addition, we thank Drs. Tanya Paull and Ji-Hoon Lee for supplying recombinant MDC1 and Drs. Elena Odintsova and Fedor Berditchevski for assistance with the time lapse microscopy. Furthermore, we are grateful to Drs. Stephen Jackson, Roger Grand, and Malcolm Taylor for advice, reagents, and critical reading of the manuscript.
This work was supported by Cancer Research-United Kingdom (CR-UK) Career Development Fellowship GSS C17183/A5592 and CR-UK Project Grant AST C10000/A7542.
- APC/C
- anaphase-promoting complex/cyclosome
- ATM
- ataxia-telangiectasia mutated
- ATR
- ATM- and Rad3-related
- CBP
- CREB-binding protein
- CREB
- cAMP-response element-binding protein
- FACS
- fluorescence-activated cell sorter
- GFP
- green fluorescent protein
- GST
- glutathione S-transferase
- h
- human
- H2AX
- histone 2A variant X
- MDC1
- mediator of DNA damage checkpoint 1
- siRNA
- small interfering RNA.
REFERENCES
- 1.Peters J. M. (2006) Nat. Rev. Mol. Cell Biol. 7, 644–656 [DOI] [PubMed] [Google Scholar]
- 2.Turnell A. S., Stewart G. S., Grand R. J., Rookes S. M., Martin A., Yamano H., Elledge S. J., Gallimore P. H. (2005) Nature 438, 690–695 [DOI] [PubMed] [Google Scholar]
- 3.Bharadwaj R., Yu H. (2004) Oncogene 23, 2016–2027 [DOI] [PubMed] [Google Scholar]
- 4.Hsu L. C., White R. L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 12983–12988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krämer A., Mailand N., Lukas C., Syljuåsen R. G., Wilkinson C. J., Nigg E. A., Bartek J., Lukas J. (2004) Nat. Cell Biol. 6, 884–891 [DOI] [PubMed] [Google Scholar]
- 6.Ko M. J., Murata K., Hwang D. S., Parvin J. D. (2006) Oncogene 25, 298–303 [DOI] [PubMed] [Google Scholar]
- 7.Joukov V., Groen A. C., Prokhorova T., Gerson R., White E., Rodriguez A., Walter J. C., Livingston D. M. (2006) Cell 127, 539–552 [DOI] [PubMed] [Google Scholar]
- 8.Tang J., Erikson R. L., Liu X. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11964–11969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang X., Tran T., Zhang L., Hatcher R., Zhang P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1065–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coster G., Hayouka Z., Argaman L., Strauss C., Friedler A., Brandeis M., Goldberg M. (2007) J. Biol. Chem. 282, 32053–32064 [DOI] [PubMed] [Google Scholar]
- 11.Stewart G. S., Wang B., Bignell C. R., Taylor A. M., Elledge S. J. (2003) Nature 421, 961–966 [DOI] [PubMed] [Google Scholar]
- 12.Wolthuis R., Clay-Farrace L., van Zon W., Yekezare M., Koop L., Ogink J., Medema R., Pines J. (2008) Mol. Cell 30, 290–302 [DOI] [PubMed] [Google Scholar]
- 13.Li M., York J. P., Zhang P. (2007) Mol. Cell. Biol. 27, 3481–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adelman C. A., De S., Petrini J. H. (2009) Mol. Cell. Biol. 2, 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rai R., Phadnis A., Haralkar S., Badwe R. A., Dai H., Li K., Lin S. Y. (2008) Cell Cycle 7, 2225–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lou Z., Minter-Dykhouse K., Franco S., Gostissa M., Rivera M. A., Celeste A., Manis J. P., van Deursen J., Nussenzweig A., Paull T. T., Alt F. W., Chen J. (2006) Mol. Cell 21, 187–200 [DOI] [PubMed] [Google Scholar]
- 17.Cox J., Jackson A. P., Bond J., Woods C. G. (2006) Trends Mol. Med. 12, 358–366 [DOI] [PubMed] [Google Scholar]
- 18.O'Driscoll M., Jeggo P. A. (2006) Nat. Rev. Genet. 7, 45–54 [DOI] [PubMed] [Google Scholar]
- 19.Jeffers L. J., Coull B. J., Stack S. J., Morrison C. G. (2008) Oncogene 27, 139–144 [DOI] [PubMed] [Google Scholar]
- 20.Rai R., Dai H., Multani A. S., Li K., Chin K., Gray J., Lahad J. P., Liang J., Mills G. B., Meric-Bernstam F., Lin S. Y. (2006) Cancer Cell 10, 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffith E., Walker S., Martin C. A., Vagnarelli P., Stiff T., Vernay B., Al Sanna N., Saggar A., Hamel B., Earnshaw W. C., Jeggo P. A., Jackson A. P., O'Driscoll M. (2008) Nat. Genet. 40, 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]