Abstract
In plants, high capacity tonoplast cation/H+ antiport is mediated in part by a family of cation exchanger (CAX) transporters. Functional association between CAX1 and CAX3 has previously been shown. In this study we further examine the interactions between CAX protein domains through the use of nonfunctional halves of CAX transporters. We demonstrate that a protein coding for an N-terminal half of an activated variant of CAX1 (sCAX1) can associate with the C-terminal half of either CAX1 or CAX3 to form a functional transporter that may exhibit unique transport properties. Using yeast split ubiquitin, in planta bimolecular fluorescence complementation, and gel shift experiments, we demonstrate a physical interaction among the half proteins. Moreover, the half-proteins both independently localized to the same yeast endomembrane. Co-expressing variants of N- and C-terminal halves of CAX1 and CAX3 in yeast suggested that the N-terminal region mediates Ca2+ transport, whereas the C-terminal half defines salt tolerance phenotypes. Furthermore, in yeast assays, auto-inhibited CAX1 could be differentially activated by CAX split proteins. The N-terminal half of CAX1 when co-expressed with CAX1 activated Ca2+ transport, whereas co-expressing C-terminal halves of CAX variants with CAX1 conferred salt tolerance but no apparent Ca2+ transport. These findings demonstrate plasticity through hetero-CAX complex formation as well as a novel means to engineer CAX transport.
Introduction
Ca2+ is both a signaling molecule and a component in providing the plant cell its structural strength (1–3). Regulated Ca2+ fluctuations are involved in plant growth and responses to environmental stimuli. Ca2+ transporters play an important role in regulating these biological processes, and the ability to manipulate these transporters is an important component in strategies designed to manipulate plant productivity.
Ca2+ can accumulate into various organelles including the vacuole, whereas the cytosolic Ca2+ concentrations are maintained in the nanomolar range (3, 4). This gradient is established across the tonoplast in part by high capacity Ca2+/H+ exchange (5, 6). Plant Ca2+/H+ exchangers were cloned by the ability of N-terminal truncated versions of the proteins, lacking the regulatory domain (7), to function in Saccharomyces cerevisiae mutants defective in vacuolar Ca2+ transport (8–10). The term cation exchanger (CAX)3 is now used to identify CAX1 and CAX2, as well as four other CAX transporters in the Arabidopsis genome (10). Interestingly, CAX3, which is most similar to CAX1 (77% identical at the amino acid level), is a weak vacuolar Ca2+ transporter when expressed in yeast cells (11, 12). The yeast suppression assays have been a robust tool to dissect the function and regulation of CAX transporters (10).
Oligomerization represents a regulatory mechanism for many transporters (13). An interaction between the Arabidopsis Ca2+/H+ exchangers CAX1 and CAX3 has been shown (14, 15). Furthermore, CAX transporters can form homomeric complexes, as has been shown for Arabidopsis CAX1 and mung bean VCAX1 (9, 15). Using both plant and yeast assays, CAX1 and CAX3 combine to form functional transporters with novel transport properties (15). We postulate that like oligomerization of other transporters, CAX1 and CAX3 interactions allow dynamic flexibility for membrane transport, even though the mechanism by which the CAX1-CAX3 complex confers Li+ and Na+ tolerance remains unclear.
It is thus important to generate additional information about the nature of the CAX interactions. Many transporter proteins are made up of two or more homologous domains, possibly as a result of ancient duplication events. For example, plant sucrose transporter members of the major facilitator superfamily possess two homologous, interacting domains (16). Furthermore, both homo- and heteromeric sucrose half proteins can functionally interact (16, 17). CAX transporters similarly appear to consist of two weakly homologous modules (18) that are likely to interact. The analysis of these split CAX modules may allow us to further understand the mechanisms of protein-protein interaction between the CAX transporter isoforms. The potential to generate chimeric CAX proteins via the manipulation of split CAX halves will also allow us to perform structure-function analyses and may afford a novel means to regulate CAX transporters.
In this study the yeast split ubiquitin system, in planta bimolecular fluorescence complementation (BiFC), and gel shift assays were used to demonstrate that two separately expressed halves of CAX1 can interact. Furthermore, a heteromeric interaction between the first half of sCAX1 and the C-terminal part of CAX3 provides insights into the structure of oligomeric CAXs. The interaction is specific, because unrelated membrane proteins do not interact. Yeast suppression studies together with kinetic measurements show that the halves reconstitute a functional transporter and indicate that they are independently targeted to yeast endomembranes. Collectively, these findings provide insights into the interactions among CAX transporters as well as a strategy to manipulate transport function through genetic engineering.
MATERIALS AND METHODS
Plasmid DNA Construction and Yeast Strains
Half Protein Construction
The truncated forms of CAX1 (sCAX1) and CAX3 (sCAX3) were split into N- and C-terminal halves at the 236th and 238th amino acids, respectively. The N-terminal half of sCAX1 (N-sCAX1, amino acids 37–272) was amplified using primers N-sCAX1F and N-sCAX1R (supplemental Table S1). The C-terminal half of CAX1 (C-CAX1, from amino acids 273–463) was amplified using primers C-CAX1F and C-CAX1R. The sCAX1H338N mutant has been previously described (19) (supplemental Table S2) and was used as a template for PCR to create the C-CAX1H338N clones in the same manner as C-CAX1 was constructed (19). The N-terminal half of sCAX3 (N-sCAX3, amino acids 37–274) was amplified using primers N-sCAX3F and N-sCAX3R. The C-terminal half of CAX3 (C-CAX3, amino acids 275–459) was amplified using primers C-CAX3F and C-CAX3R. These N-terminal and C-terminal PCR fragments were first cloned into the pGEM T-Easy vector (Promega). After sequence confirmation, the constructs were subcloned into the SfiI A and B restriction sites of two yeast expression vectors pHGpd (His selectable marker) and pUGpd (Ura selectable marker), respectively (20). Thus, N-terminal halves or C-terminal halves of sCAX1 and sCAX3 were expressed in plasmids containing either −His or −Ura selectable markers.
Epitope-tagged CAX Half Proteins
The HA and GFP epitope-tagged CAX1 and CAX3 half proteins were constructed using similar approaches. The primers N-sCAX1HAF and N-sCAX1HAR were used to amplify N-sCAX1, which was then cloned into the BamHI and NotI sites of the HA cassette in pUGpd (20) to generate triple HA epitope-tagged N-sCAX1. The primers C-CAX3GFPF and C-CAX3GFPR were used to amplify C-CAX3, which was then subcloned into the SpeI and NotI sites of a GFP fusion expression cassette in pHGpd (20) to generate the GFP-tagged C-CAX3. The C-CAX1 fusion with c-Myc at the C terminus (C-CAX1-c-Myc) was constructed by ligating C-CAX1 (amplified using C-CAX1F and C-CAX1R primers) into a cassette containing five tandem copies of the c-Myc epitope (EQKLISEEDL) (21). The primers C-CAX3RFPF and C-CAX3RFPR were used to amplify C-CAX3, which was then subcloned into pUGpd-RFP to generate the C-CAX3-RFP fusion.
Yeast Strains and Split Ubiquitin Vectors
The Ca2+-sensitive S. cerevisiae strain K667 (MATa cnb1:: LEU1 pmc1::TRP1 vcx1Δ ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1) was used for most yeast assays (22). The N-terminal ubiquitin half (Nub) and C-terminal ubiquitin half (Cub) vectors, KAT1-Cub and Nub-KAT1 constructs, and yeast strains THY.AP4 (MATa leu2-3,112 ura3-52 trp1-289 lexA::HIS3 lexA::ADE2 lexA::lacZ) and THY.AP5 (MATα URA3 leu2-3,112 trp1-289 his3-Δ1 ade2Δ::loxP) used for the mating-based split ubiquitin system were obtained from Dr. Wolf B. Frommer (Stanford University) (23). The CAX1, CAX3, sCAX1, sCAX1H338N, and sCAX3 expression vectors have been described previously (supplemental Table S2).
Construction of CAX Split Ubiqutin Plasmids
CAX1, CAX3, and their N-terminal and C-terminal halves were cloned into split ubiquitin plasmids using in vivo recombination cloning (24). The full-length CAX1 and CAX3 constructs have been described previously (15). For the construction of Nub X and NubWT fusions, the split ubiquitin vectors Nub X and NubWT were cleaved with EcoRI/SmaI and mixed with the purified PCR products for CAX1, CAX3, N-CAX1, and C-CAX3 to transform THY.AP5 cells and transformants were selected on SC medium lacking Trp and Ura. For Cub fusions, the vector metYCgate was cleaved with PstI/HindIII and mixed with the same PCR products to transform yeast strain THY.AP4, and transformants were selected on SC medium lacking Leu. After the initial interaction assay, plasmids were extracted from the yeast strains and amplified in Escherichia coli. The interactions were then verified by repeating the assays using the purified plasmids. After the interaction was confirmed, the insert plasmids were sequenced.
Construction of CAX BiFC Plasmids
To generate CAX BiFC plasmids, full-length CAX1 and CAX3 proteins, and split half N-sCAX1, C-CAX1, and C-CAX3 proteins were fused to N- or C-terminal halves of yellow fluorescent protein (YFP). CAX constructs were amplified using specific primers (supplemental Table S1) and then subcloned into the SacI and BamHI sites of pSY735 or pSY736 (25) for N-terminal fusion with the C-terminal half of YFP (CYFP) or the N-terminal half of YFP (NYFP), respectively. CAX3 constructs were amplified from a CAX3 plasmid, which was engineered to remove the BamHI site at nucleotide position 1103, by site-directed mutagenesis (26) using primers CAX3BamHIF and CAX3BamHIR. The negative actin control construct (ACT8-NYFP) was generated by subcloning the ACT8F and ACT8R PCR product into the XbaI and KpnI sites of pSPYNE (27).
Yeast Growth Conditions
Suppression Assay
S. cerevisiae strain K667 was transformed with His and Ura selectable expression constructs as described previously (15). Transformed cells were tested for their capability to suppress the K667 Ca2+-sensitive phenotype (8). Growth assays were conducted using YPD medium supplemented with or without CaCl2 (50, 100, 150, or 200 mm), LiCl (30, 50, 60, or 100 mm), or NaCl (400, 500, or 600 mm).
Inductively Coupled Plasma Mass Spectroscopy Analysis
Yeast culture conditions and sample processing were slightly modified from a previous study (28). For growth in Li+ and Ca2+ supplemented medium, K667 cells expressing the various constructs were inoculated in 5 ml of YPD + 1/100 volume of 100 × mineral supplement stock (28), supplemented with 10 mm CaCl2, 100 mm NaCl, or 500 mm LiCl (final concentrations). The cultures were grown at 30 °C to stationary phase, and 2.5 ml of each culture was collected by vacuum filtration using isopore membrane filters (1.2-μm pore size) (Fisher Scientific). The cells were washed three times with 1 ml of 1 μm EDTA disodium salt solution, pH 8.0, by vacuum filtration followed by three washes with 1 ml of distilled, deionized H2O. The filters were dried in a 70 °C oven for 48 h before inductively coupled plasma mass spectroscopy analysis was performed as previously described (29). The data from five repeats were calculated with the following formula: (Ioncax − Ionvector)/Ionvector × 100, and expressed as the means ± S.E. (n = 5).
Ca2+ Transport Assay
Ca2+/H+ exchange activity into vacuole-enriched membrane vesicles was measured using 45Ca2+, performed exactly as described previously (15).
Gel Mobility Shift Assay for Split Half Protein Interaction
Microsomal fractions from yeast cells expressing CAX variants were prepared using the glass bead method (30). Briefly, glass bead buffer (25 mm Hepes-KOH, pH 7.5, 10% sucrose, 3 mm EGTA, 2 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 10 mm benzamidine, and 5 μg/ml leupeptin) was added to the cells with an equal volume of glass beads (Sigma), and the mixture was vortexed three times for 1 min at maximum speed with intervals of 10 min on ice. Triton X-100 was added to the lysate to a final concentration of 0.5%. The lysate was centrifuged at 5,000 × g for 5 min, and the supernatant was saved. After solubilization in buffers containing 1% Triton X-100 (15), equal amounts of native protein (about 15 μg) from samples were resolved on native 12% PAGE with SDS-PAGE running buffer followed by transferring membranes and immunoblotting as described above. The proteins were detected with anti-HA, anti-c-Myc, and anti-GFP antibodies (Covance) used at 1:2000.
Split Ubiquitin Assay
The split ubiquitin system used in this assay was a mating-based split ubiqutin system developed previously (24, 31). Approximately 40 clones of each THY.AP5 and THY.AP4 transformation with various Nub and Cub constructs were mixed and incubated in appropriate selective SC with and without G418. Cultures of the lag phase were used for mating as described previously (23). After mating THY.AP4 and THY.AP5 strains on YPD for 8 h of incubation at 28 °C, diploid cells were selected by replica plating on SC without Trp, Leu, and Ura for 2–3 days. Diploid cells were used for analysis on SC medium supplemented with 150 mm Met for 3 days. CAX Nub and Cub constructs were used for retransformation, and the interaction assay was repeated using the confirmed constructs.
For the filter assay, diploid cells were grown for 3 days on sterilized filter sets placed on SC medium supplemented with His, Ade, and 150 mm Met. β-Galactosidase activity assays were conducted according to the standard X-gal filter assay protocol described previously (24). Color changes in the positive interactions could be detected within 3 h.
Co-localization of N-sCAX1-GFP and C-CAX3-RFP
N-sCAX1-GFP and C-CAX3-RFP were co-transformed into K667 cells and selected on SC−His and −Ura media. N-sCAX1-GFP and C-CAX3-RFP localization was visualized using a confocal microscope (Olympus Fluoview, FV500; Olympus Optical, Tokyo, Japan). An argon laser was used for excitation at 488 nm for GFP imaging with emission at 525 nm or excitation at 543 nm for RFP imaging with emission at 605 nm. Co-fractionation of N-sCAX1-GFP and C-CAX3-RFP on vacuolar membrane-enriched fractions was confirmed with Western blotting. Microsomal fractions from N-sCAX1-GFP and C-CAX3-RFP co-expressing K667 yeast cells were subjected to discontinuous sucrose gradient fractionation, and Western blotting was used to detect the vacuolar membrane and endoplasmic reticulum markers alkaline phosphatase and BiP, respectively, N-sCAX1-GFP and C-CAX3-RFP, on these fractions. N-sCAX1-GFP and C-CAX3-RFP were detected with GFP and RFP antibodies, respectively.
Expression and Visualization of CAX BiFC
BiFC plasmid DNA was biolistically transferred into onion (Allium cepa) slices. DNA (3 μg) for each construct was precipitated onto 1-μm gold microparticles (Bio-Rad) as previously described (32). Sections of onion tissue were placed on 1% agar plates and bombarded using a PDS-1000 He system gene gun (Bio-Rad) under a vacuum (0.925 bar) and a helium pressure of 1350 p.s.i. Following bombardment, tissue samples were incubated for 16 h in the dark at 22 °C. The epidermis was then removed for analysis. Imaging was performed on a Leica TCS SP5 AOBS upright confocal microscope. Excitation was with an argon laser at 514 nm and spectral detection between 525 and 580 nm for YFP and 458 nm with spectral detection between 465 and 600 nm for cyan fluorescent protein (CFP).
RESULTS
CAX Half Proteins Functionally Assemble in Yeast
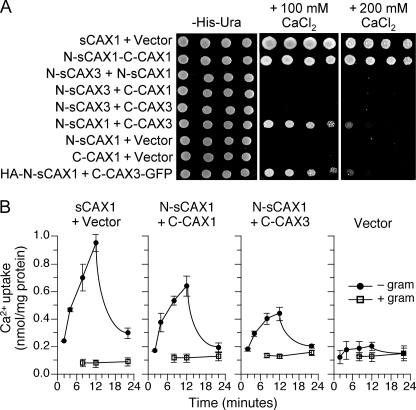
CAXs can be divided into two weakly homologous halves at the short negatively charged loop between transmembranes 6 and 7, previously termed the “acidic motif” (18) (supplemental Fig. S1). To investigate the interaction between these CAX1 and CAX3 split proteins, we utilized K667 yeast expression assays (19, 33). Co-expression of N-sCAX1 and C-CAX1 suppressed the Ca2+ sensitivity phenotype (Fig. 1A), thus effectively reconstituting the N-terminal truncated CAX1 (sCAX1). Yeast co-expressing N-sCAX1 and C-CAX3 could similarly grow well on high Ca2+ medium. All other combinations, such as N-sCAX1+N-sCAX3, N-sCAX3+N-sCAX1, C-CAX3+C-CAX1, N-sCAX3+C-CAX3, and N-sCAX3+C-CAX1 were unable to suppress the Ca2+-sensitive phenotype, and expression of the N- or C-halves of the transporters alone were nonfunctional (Fig. 1A and data not shown). These observations suggested that N-sCAX1 could form an active Ca2+ transporter with either C-CAX1 or C-CAX3.
FIGURE 1.
Functionality of co-expressed CAX half proteins in yeast. A, suppression of Ca2+ sensitivity in K667 yeast through co-expression of various combinations of sCAX1 N-terminal half protein (N-sCAX1), sCAX3 N-terminal half protein (N-sCAX3), CAX1 C-terminal half protein (C-CAX1), and CAX3 C-terminal half protein (C-CAX3), plus epitope-tagged HA-N-sCAX1+C-CAX3-GFP. Saturated liquid cultures of K667 yeast cells were diluted in stepwise 5-fold dilutions and spotted onto selection medium and YPD medium containing either 100 mm or 200 mm CaCl2. Yeast cells expressing sCAX1 and empty vector were used as controls. The plates were incubated and photographed after 2 days at 30 °C. B, time course of 45Ca2+ uptake into endomembrane vesicles prepared from K667 yeast co-expressing sCAX1+empty vector, N-sCAX1+C-CAX1, N-sCAX1+C-CAX3, and empty vector alone. Solid circle, pH-dependent 45Ca2+ uptake; open square, uptake in the presence of the protonophore gramicidin. The Ca2+ ionophore A23187 (5 μm) was added at 12 min. The reduction in Ca2+ uptake following A23187 addition demonstrated that accumulation into the vesicles occurred. The data represent the means ±S.E. of three independent experiments.
To confirm Ca2+ transport activity, the H+-gradient dependent 45Ca2+ uptake assay was utilized (Fig. 1B). Membrane vesicles from yeast expressing N-sCAX1+C-CAX3 showed significantly increased Ca2+/H+ exchange activity compared with vector control (Fig. 1B) and sCAX3-expressing cells (data not shown), but the activity was lower than from cells expressing sCAX1 and slightly lower than from cells expressing N-sCAX1+C-CAX3. This suggests that N-sCAX1 and C-CAX3 half proteins can together form an active Ca2+ transporter.
Half Protein Complexes Have Distinct Transport Properties
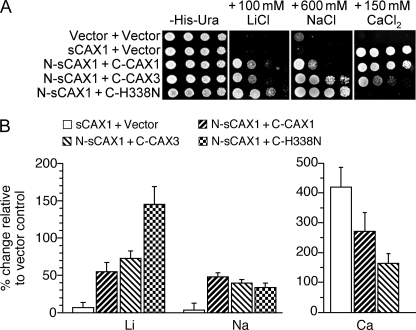
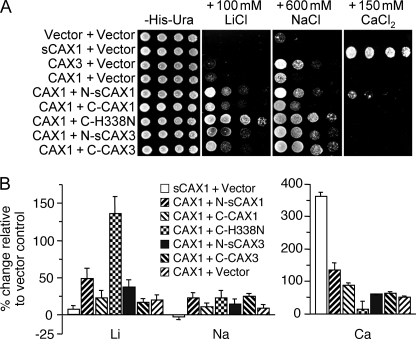
To further examine the transport properties of the CAX half protein complexes, we tested the growth sensitivity of K667 yeast cells expressing these constructs on Li+-, Na+-, and Ca2+-containing media (Fig. 2). sCAX1-expressing yeast are tolerant to Ca2+ in the media but very sensitive to Na+ and Li+, whereas CAX3 cells are slightly tolerant to Na+ and Li+, but sensitive to Ca+ (15). Yeast K667 cells co-expressing N-sCAX1+C-CAX1 were Ca2+-, Li+-, and Na+-tolerant (Fig. 2A). In contrast, both N-sCAX3+C-CAX3- and N-sCAX1+C-CAX3-expressing yeast were Ca2+-sensitive but tolerant to Na+ (Fig. 2; data not shown). Whole yeast cell metal accumulation was measured by inductively coupled plasma mass spectroscopy to infer the transport properties of the half protein complexes. In high Ca2+ and high Li+ growth conditions, the N-sCAX1+C-CAX1 and N-sCAX1+C-CAX3 cells significantly accumulated Ca2+, Li+, and Na+ compared with vector control (Fig. 2B). N-sCAX1+C-CAX1 accumulated more Ca2+ than the N-sCAX1+C-CAX3 cells, whereas the N-sCAX1+C-CAX3 cells accumulated more Li+ (Fig. 2B).
FIGURE 2.
Diverse functions of co-expressed CAX half proteins in yeast. A, suppression of Li+, Na+, and Ca2+ sensitivity of K667 yeast cells expressing sCAX1 or co-expressing sCAX1 N-terminal half protein (N-sCAX1) with CAX1 C-terminal half protein (C-CAX1), CAX3 C-terminal half protein (C-CAX3), or CAX1H338N C-terminal half protein (C-H338N). Saturated liquid cultures of K667 yeast were diluted in stepwise 5-fold dilutions and spotted onto selection medium and YPD medium containing 150 mm CaCl2, 100 mm LiCl, or 600 mm NaCl. Yeast cells expressing empty vector were used as a negative control. The plates were incubated and photographed after 2 days at 30 °C. B, Li+, Na+ and Ca2+ ion content analysis in K667 yeast cells co-expressing sCAX1, N-sCAX1+C-CAX1, N-sCAX1+C-CAX3, and N-sCAX1+C-H338N grown to stationary phase in YPD medium supplemented with 500 μm LiCl (for the Li+ and Na+ content measurements) or YPD medium supplemented with 10 mm CaCl2 (for the Ca2+ content measurements). The data from five replicate experiments were calculated with a formula: (Ioncax − Ionvector)/Ionvector × 100, and expressed as the means ± S.E. (n = 5). Ion content varied depending on the growth medium used, but levels of Ca2+ ion content differed among the lines only when the medium was supplemented with CaCl2, whereas Li+ and Na+ content differed among the lines only when the medium was supplemented with LiCl2 and NaCl (data not shown).
The Li+ and Na+ transport activity by the N-sCAX1+C-CAX1 complex was further inferred by measuring the direct competition of 45Ca2+ uptake by these ions (data not shown). The N-sCAX1+C-CAX1-expressing cells had a similar ion competition profile to sCAX1-expressing cells, including significant Li+ and Na+ inhibition of 45Ca2+ uptake; however, unlike for sCAX1, 45Ca2+ uptake was not significantly inhibited by Cd2+.
To further assess the role of specific CAX domains in substrate specificity, we used a previously characterized CAX1 variant. We have shown that expressing a sCAX1 variant containing a single amino acid mutation H338N, termed sCAX1H338N, decreases yeast Ca2+ uptake but increases Cd2+ uptake (19). sCAX1H338N in yeast is also unable to provide Na+ or Li+ tolerance (19) (data not shown). Here we co-expressed N-sCAX1 with the C terminus of sCAX1H338N (C-H338N) in yeast. The N-sCAX1+C-H338N yeast grew very well on high Na+ and Li+ media but was very sensitive to Ca2+ (Fig. 2A). Significant accumulation of Li+ and poor accumulation of Ca2+ was observed in N-sCAX1+C-H338N cells growing in Li+-containing medium (Fig. 2B and data not shown).
Co-localization of CAX Split Half Proteins in Yeast Cells
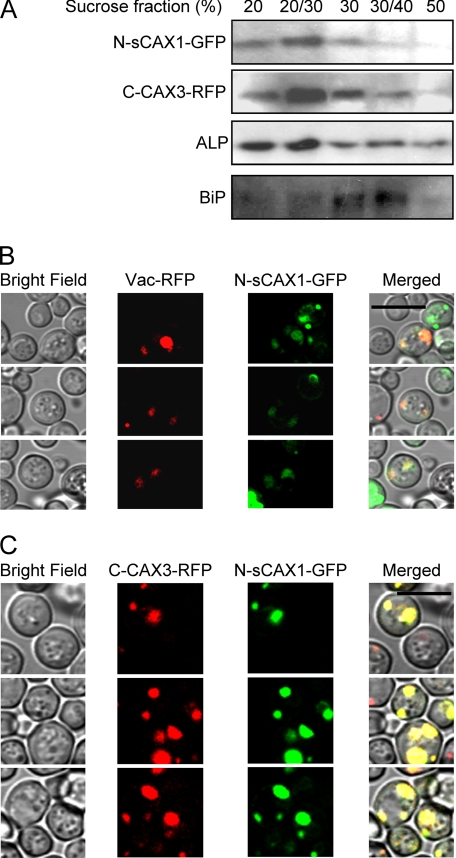
To confirm that the CAX half proteins were correctly folded and co-localized, N-sCAX1-GFP and C-CAX3-RFP were expressed in yeast cells. The tagged half proteins remained functional in K667 yeast with respect to providing Ca2+ tolerance (supplemental Fig. S2). Vacuolar membrane localization of CAX1 and CAX3 have been well documented in both yeast and plant cells (12, 33). When N-sCAX1-GFP and C-CAX3-RFP were co-expressed in yeast, both half proteins were co-fractionated and detected in vacuolar membrane-enriched fractions as indicated by the tonoplast marker alkaline phosphatase (Fig. 3A). Furthermore, both N-sCAX1 and C-CAX3 were expressed to equivalent levels. Similarly, when a predominantly tonoplast-localized Vac-RFP and N-sCAX1-GFP were co-expressed, both fusions predominantly co-localized to similar endomembranes (Fig. 3B). We postulate that the multi-compartmental localization of Vac-RFP prohibits a perfect overlap with N-CAX1-GFP (34). High copy co-expression of N-sCAX1-GFP and C-CAX3-RFP in yeast cells also showed overlap of the fluorescent signal predominantly at intracellular vesicles similar to where Vac-RFP was localized (Fig. 3C). Together, these observations suggest that both CAX half proteins can co-localize independently at the same endomembrane.
FIGURE 3.
Localization of CAX half proteins in yeast cells. Yeast cells expressing GFP-tagged sCAX1 N-terminal half protein (N-sCAX1-GFP) and/or RFP-tagged CAX3 C-terminal half protein (C-CAX3-RFP) were grown overnight in selection medium then diluted with water and observed using sucrose fractionation and confocal microscopy. A, discontinuous sucrose gradient fractionation of microsomes from yeast cells co-expressing N-sCAX1-GFP and C-CAX3-RFP. N-sCAX1-GFP and C-CAX3-RFP were co-fractionated in vacuolar membrane-enriched fractions (20% and 20/30% sucrose interfaces). Alkaline phosphatase (ALP) was used as a vacuolar membrane marker, and BiP was used as an endoplasmic reticulum marker. N-sCAX1-GFP was detected using a GFP antibody and C-CAX3-RFP was detected using an RFP antibody. B and C, co-localization of N-sCAX1-GFP with a prevacuole marker (Vac-RFP) (B) and co-localization of N-sCAX1-GFP and C-CAX3-RFP (C). The panels from left to right are: yeast cells in bright field, RFP image (Vac-RFP or C-CAX3-RFP), GFP image (N-sCAX1-GFP), and merged image. Bar, 10 μm.
Physical Interaction between CAX1 and CAX3 in Yeast
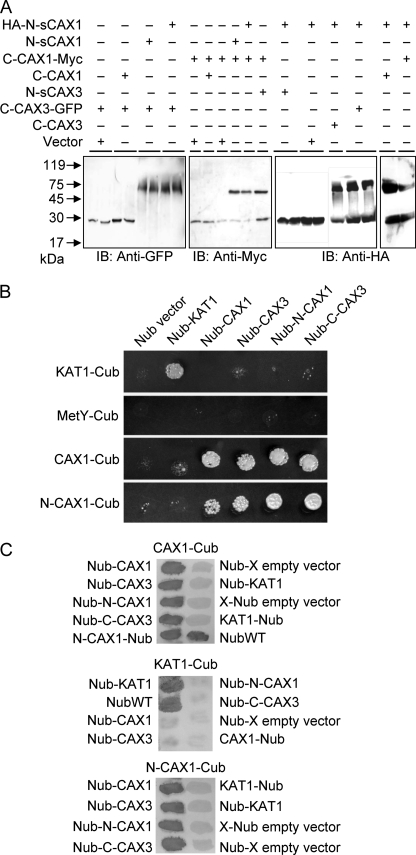
Although the previous assays show a functional interaction among the split transporters, they do not establish a physical interaction. To test this, we conducted native protein gel shift assays using functional GFP-, c-Myc-, or HA-tagged versions of the proteins (supplemental Fig. S2). After solubilizing microsomal proteins from yeast expressing the tagged CAX half proteins, the proteins were resolved by denaturing SDS-PAGE. Using antibodies against the epitope tags, a band consistent with the tagged proteins and a higher molecular weight complex were observed (Fig. 4A). Higher molecular weight bands, indicating half protein interaction, were observed for samples co-expressing N-sCAX1+C-CAX1 and N-sCAX1+C-CAX3. Half protein complexes could not be observed in samples from yeast co-expressing either single half protein with empty vector or N-sCAX1+N-sCAX3, N-sCAX3+N-sCAX1, or C-CAX3+C-CAX1. These findings suggest a physical interaction between half proteins only where the N-CAX half interacts with the C-CAX half.
FIGURE 4.
Yeast protein interaction assays. A, physical interaction in yeast of N- and C-terminal halves of CAX1 and CAX3 by gel mobility shift assay. The proteins were isolated and solubilized from yeast cells co-expressing HA-tagged sCAX1 N-terminal half (HA-N-sCAX1), sCAX1 C-terminal half (C-CAX1-Myc), or CAX3 C-terminal-GFP fusion (C-CAX3-GFP) with tagged or nontagged sCAX1 N-terminal half (HA-N-sCAX1 or N-sCAX1), CAX1 C-terminal half (C-CAX1-Myc, C-CAX1), CAX3 C-terminal half (C-CAX3 or C-CAX3-GFP), sCAX3 N-terminal half (N-sCAX3), or empty vectors. Following 12% SDS-PAGE and transfer to polyvinylidene difluoride membranes, single tagged split half proteins or protein complexes were detected with immunoblotting (IB) using c-Myc, HA, and GFP antibodies. B, split ubiquitin interaction growth assay. Interaction between CAX1 and CAX3 was determined by growth assay on synthetic minimal medium lacking all six amino acids but supplemented with 150 μm Met for 3 days. The empty Cub and Nub vectors were used as a negative control, and the K+ channel KAT1 was used as a positive control. CAX1-Cub, N-CAX1-Cub, KAT1-Cub, or empty Cub vector (MetY-Cub) were bait for interaction in combination with empty Nub vector, Nub-KAT1, Nub-CAX1, Nub-CAX3, Nub-N-CAX1, and Nub-C-CAX3, respectively. All of the experiments were repeated at least three times with similar results obtained in each replicate. C, X-gal filter assay. Yeast filters set on synthetic minimal medium lacking Ade and His but supplemented with 150 μm Met were grown for 3 days then incubated for 30 min in X-gal solution. All of the experiments were repeated at least three times with similar results obtained in each replicate. The interaction assays are shown with CAX1-Cub (top panel), KAT1-Cub (middle panel), or N-CAX1-Cub (bottom panel) as bait construct and tested in combination with the various Nub-containing prey constructs.
We also tested whether the half CAX1 and CAX3 proteins can physically interact using an optimized yeast split ubiquitin system, which allows detection of interactions between membrane proteins in vivo (23). In addition, we examined whether the half proteins could associate with full-length CAX proteins. Protein fusions to the Nub and to the Cub were constructed (supplemental Fig. S1, C–F), and protein interaction was determined by selective cell growth on Met or LacZ reporter gene expression. As a positive control, we demonstrated the interaction between AtKAT1 subunits (Fig. 4B). As negative controls, none of the CAX constructs interacted with the membrane protein AtKAT1. The N-terminal half of CAX1 including the regulatory domain (N-CAX1) interacted with C-CAX3, full-length CAX1, and CAX3 and with another copy of N-CAX1 (Fig. 4B). The interaction among the proteins was also strong enough to be measured by β-galactosidase activity because of LacZ expression (Fig. 4C). Although N-CAX1+N-CAX1 interacted in the split ubiquitin assay, we did not detect interaction between N-sCAX1+N-sCAX1 (data not shown), suggesting that the N-terminal 36-amino acid regulatory region may mediate N-CAX1 interaction.
Physical Interaction between CAX1 and CAX3 in Plant Cells
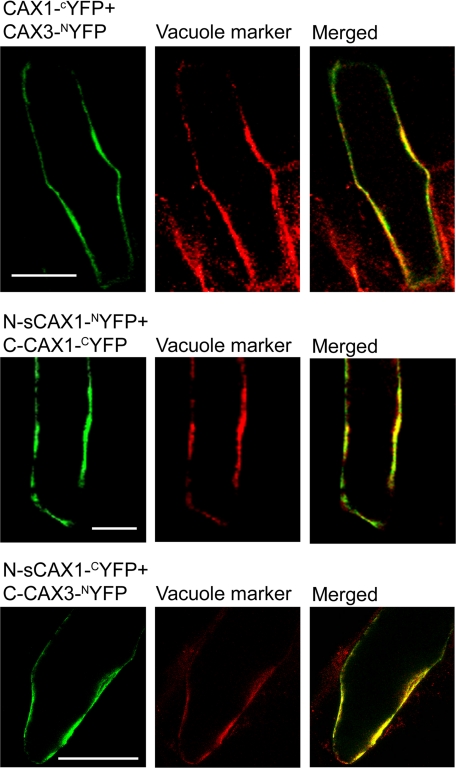
To confirm that the interaction between the half CAX proteins is relevant to plant cells, direct interaction was examined using the BiFC approach by using protein fusions to the N-terminal YFP half (NYFP) and to the C-terminal YFP half (CYFP) and detecting the reconstitution of YFP fluorescence, which indicates interaction between the tested proteins (25, 27). Protein fusions were transiently expressed in onion epidermal cells. We previously confirmed the in planta interaction of full-length CAX1 and CAX3 (15). As a positive control, we demonstrated the reconstitution of YFP fluorescence and thus interaction between CAX1 and CAX3, which co-localized with the CFP-vacuolar membrane marker (Fig. 5). N-sCAX1 interacted with C-CAX1 and with C-CAX3. None of the CYFP-tagged CAX constructs were able to reconstitute YFP fluorescence when co-expressed with actin-NYFP (data not shown). As an additional negative control, CAX2-NYFP or CAX2-CYFP was co-expressed with CYFP- or NYFP-tagged CAX1 and CAX3 constructs. None of the CAX1 and CAX3 constructs interacted with CAX2 (data not shown).
FIGURE 5.
Interaction of CAX half proteins at the tonoplast in plant cells. Onion epidermal cells transiently co-expressing CAX constructs fused to the N-terminal half of YFP (NYFP) and the C-terminal half of YFP (CYFP) (left panels) with a CFP-tagged vacuolar marker (middle panels), and the merged image (right panels). Top row panels, co-expression of CAX1-CYFP and CAX3-NYFP; middle row panels, co-expression of sCAX1 N-terminal half protein (N-sCAX1)-NYFP and CAX1 C-terminal half protein (C-CAX1)-CYFP; bottom row panels, co-expression of N-sCAX1-CYFP and CAX3 C-terminal half protein (C-CAX3)-NYFP. Reconstituted YFP fluorescence signal is shown as false color green, CFP fluorescence signal is shown as false color red, and overlapping YFP/CFP fluorescence signal in the merged image is yellow. Bar, 100 μm.
Cross-activation of CAX Transporters by Half Proteins
When expressed in yeast, full-length CAX1 is autoinhibited and is unable to efficiently transport Ca2+ (7, 35), whereas interaction with soluble activator proteins (36) or with CAX3 (15) activates CAX1. We were interested to see whether the split CAX halves are able to co-assemble with the autoinhibited CAX1 and activate transport. Co-expressing N-sCAX1 with the full-length autoinhibited CAX1 in yeast activated CAX Ca2+ transport as measured by increased Ca2+ tolerance (Fig. 6A) and increased total Ca2+ accumulation (Fig. 6B). The magnitude of this activation was similar to yeast cells co-expressing CAX1+CAX3 (14, 15). Furthermore, co-expression of N-sCAX1 or C-CAX1 with full-length CAX1 lead to increased tolerance and accumulation of Na+ and Li+ compared with sCAX1-, CAX1-, or CAX3-expressing cells (Fig. 6). We again analyzed the influence of the CAX1 C-H338N variant. When C-H338N was co-expressed with full-length CAX1, the yeast cells were less tolerant to Ca2+ and accumulated less Ca2+ than sCAX1-expressing cells; however, the CAX1+C-H338N cells were more tolerant to Na+ and Li+ (Fig. 6A) and significantly accumulated more Li+ (Fig. 6B). Interestingly, neither N-sCAX3 or C-CAX3 expression could cause cells expressing CAX1 to become Ca2+ tolerant, but they did improve salt tolerance.
FIGURE 6.
Functionality of CAX1 and CAX3 when co-expressed with CAX half proteins. A, suppression of Li+, Na+, and Ca2+ sensitivity of K667 yeast cells expressing sCAX1, CAX1, or CAX3, or co-expressing CAX1 with sCAX1 N-terminal half protein (N-sCAX1), CAX1 C-terminal half protein (C-CAX1), sCAX3 N-terminal half protein (N-sCAX3), CAX3 C-terminal half protein (C-CAX3), or CAX1H338N C-terminal half protein (C-H338N). Saturated liquid cultures of K667 yeast were diluted in stepwise 5-fold dilutions and then spotted onto selection medium or YPD medium containing either 150 mm CaCl2, 100 mm LiCl, or 600 mm NaCl. Yeast cells expressing empty vector were used as a negative control. The plates were incubated and photographed after 2 days at 30 °C. B, Li+, Na+, and Ca2+ ion content analysis in K667 yeast cells co-expressing sCAX1 or CAX1 with vector or CAX1 in combination with N-sCAX1, C-CAX1, N-sCAX3, C-CAX3, or C-H338N grown to stationary phase in YPD medium supplemented with 500 μm LiCl (for the Li+ and Na+ content measurements) or 10 mm CaCl2 (for the Ca2+ content measurements). The data from five repeats were calculated with a formula: (Ioncax − Ionvector)/Ionvector × 100, and expressed as the means ± S.E. (n = 5). As discussed previously, the ion content fluctuated depending on the growth medium used.
DISCUSSION
Many transporter proteins are made up of two or more similar modules that may be products of ancient gene duplication events. Indeed, CAX transporters appear to consist of two weakly homologous modules (18). The N-terminal-truncated CAX1 (sCAX1) was initially identified in a yeast screen through the ability to suppress the Ca2+ sensitivity of yeast strains deficient in vacuolar Ca2+ transport (8). Meanwhile, sCAX3 fails to function in these yeast assays (11). In this present study, we have utilized the ability of the N- and C-terminal halves of sCAX1 and sCAX3 to co-localize, fold properly, and be functional to begin to further understand the mechanisms of substrate specificity, CAX transport regulation, and CAX complex protein interaction.
Our findings suggest that N-sCAX1 can functionally associate with C-CAX3 (Figs. 1 and 2), and we have verified a physical interaction between these halves both in yeast and plant cells (Figs. 4 and 5). In the case of N-sCAX1+C-CAX3 our results indicate that: 1) the half protein modules can fold properly; 2) both “half” proteins are localized to the same endomembrane; and 3) there is sufficient interaction to facilitate function.
The ability to split the Ca2+/H+ exchanger into two domains is similar to the situation found with the cardiac Na+/Ca2+ exchanger (37). This transporter can also be divided into two domains that reassemble in the membrane of Xenopus oocytes. The two domains are properly delivered to the membrane, and both domains are required for activity. Like the data presented here, the cardiac exchanger activity was maintained when fusion proteins were made to both half proteins. Likewise, it has been shown that separately expressed halves of the potato sucrose/H+ transporter SUT1 can interact and reconstitute a functional transporter (16). In addition, that same study demonstrated that the N-terminal half of SUT1 can functionally interact with the C-terminal half of SUT2.
This “split protein” approach expands our potential repertoire of functional transporters. For example, we could engineer novel transporters by combining CAXs with unique substrate affinities. We have shown here that expressing a C-terminal CAX1 variant containing a single amino acid mutation, H338A, can modify transport activity of CAX1 and N-sCAX1 (Figs. 2 and 6). Previous transport studies show that sCAX2 has different biochemical properties from sCAX1 (10). We may now be able to generate novel transporters by selecting heteromeric combinations of CAX1/CAX2. Similarly, site-directed mutagenesis has created a series of sCAX1 and sCAX2 variants with different transport properties (21, 33) that could now be combined. Alternatively, we may be able to create dominant negative forms of the half proteins to down-regulate CAX activity. Harnessing this technology should allow further manipulation of the nutritional status of plants.
The cross-activation of CAX1 by CAX half proteins suggests a mechanism that may allow rapid activation. Oligomerization may represent a regulatory mechanism for many transporters. Both plant and animal sugar transporters form regulatory complexes (17, 38). In lipid transport, the small CER5 ABC transporter may form a homo- or heterodimer to facilitate fatty acid transport to the cuticle (39, 40). Oligomerization of S. cerevisiae Nha1p is essential for its Na+/H+ antiporter activity (41). Recently, a cytosolic transactivation domain was identified as being essential for ammonium uptake with AMT transporters (42, 43). A molecular model of AtAMT1;2 provides a mechanism where the C terminus of one monomer directly contacts the neighboring subunit. These alterations in the C-terminal domain may provide conformation coupling between monomers to allow tight regulation of transport and sensing (43, 44). Our results here suggest that activation of CAX1 can be achieved by interactions within either the N- or C-terminal half of the transporter (Fig. 6). Furthermore, these results indicate that the site of interaction of the CAX1-CAX3 complex may be located in both N- and C-terminal domains of the CAX proteins. The lack of interaction between N-sCAX1+N-sCAX1 compared with the positive interaction observed between N-CAX1+N-CAX1 (Fig. 4B) may suggest that the 36-amino acid N-terminal regulatory domain does play a role in protein interaction, as previously seen with other CAX1-activating proteins (36). However, more specific mutation and domain analysis studies will be needed to fully pinpoint all the amino acids required for CAX complex protein interaction.
The split protein studies further implicate specific domains within CAXs defining substrate specificity. Previously, the inability of sCAX3 to transport Ca2+ in yeast was indicated to be due partly to sequence variations in a 9-amino acid region named the Ca2+ domain that permits Ca2+ transport in CAX1 but not CAX3 (33). Here, the interaction between N-sCAX3+C-CAX1 or C-CAX3 was unable to mediate growth on high Ca2+ medium (Fig. 1), presumably because of a lack of the CAX1 Ca2+ transport domain (33). However, an alternative explanation could be that the sCAX3 construct still harbors autoinhibitory sequences.
With regard to the C-terminal half of the CAX proteins, previous studies suggest that this portion of CAX1 may determine substrate specificity for specific cations including Cd2+ (19). For example, yeast cells expressing a sCAX1 variant containing a single point mutation within the C-terminal half of the protein, H338A, have decreased Ca2+ uptake but increased Cd2+ uptake (19). When the C-terminal half of CAX3 (C-CAX3) was co-expressed with N-sCAX1, Na+ tolerance was increased, but Ca2+ tolerance was decreased compared with N-sCAX1+C-CAX1-expressing cells (Fig. 2A), indicating that a region of the CAX3 C-terminal half may be involved in Na+ specificity. When N-sCAX1+C-H338A half proteins were co-expressed, the yeast cells were even more tolerant to Na+ and Li+ but showed increased sensitivity to Ca2+. However, such Na+ and Li+ phenotypes are not be mediated by the original “intact” sCAX1H338A construct (19) (data not shown), indicating that the altered substrate specificity phenotypes in response to split CAX interaction are not solely due to the primary protein sequence, but additional as yet unknown alteration in the folding of the reconstituted transporter may also play a role. In sum, these findings suggest that, in these yeast assays, the N-terminal half of CAXs mediate Ca2+ transport activity, whereas the C-terminal halves of CAXs determine Na+ or Li+ tolerance.
CONCLUSIONS
We have developed a system by which CAX half proteins can be co-expressed to form unique combinations of transporters. These findings allow dynamic flexibility for the regulation of mineral nutrition.
Supplementary Material
This work was supported by funding from the United States Department of Agriculture/Agricultural Research Service under Cooperative Agreement 58-6250-6001, National Science Foundation Grant 90344350, United States Department of Agriculture Cooperative State Research, Education, & Extension Service Grant 2005-34402-17121, a Designing Foods for Health grant (to K. D. H.), and by Biotechnology and Biological Sciences Research Council Grant BB/B502152/1 (to J. K. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1 and S2.
- CAX
- cation exchanger
- BiFC
- bimolecular fluorescence complementation
- HA
- hemagglutinin
- GFP
- green fluorescent protein
- RFP
- red fluorescent protein
- YFP
- yellow fluorescent protein
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
- CFP
- cyan fluorescent protein
- Cub
- C-terminal ubiquitin half
- Nub
- N-terminal ubiquitin half
- IonCAX
- ion content in CAX-expressing yeast
- Ionvector
- ion content in vector-expressing yeast.
REFERENCES
- 1.Sanders D., Pelloux J., Brownlee C., Harper J. F. (2002) Plant Cell 14, S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirschi K. D. (2004) Plant Physiol. 136, 2438–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McAinsh M. R., Pittman J. K. (2009) New Phytol. 181, 275–294 [DOI] [PubMed] [Google Scholar]
- 4.Marty F. (1999) Plant Cell 11, 587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schumaker K. S., Sze H. (1986) J. Biol. Chem. 261, 2172–2178 [PubMed] [Google Scholar]
- 6.Sze H., Liang F., Hwang I., Curran A. C., Harper J. F. (2000) Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 433–462 [DOI] [PubMed] [Google Scholar]
- 7.Pittman J. K., Hirschi K. D. (2001) Plant Physiol. 127, 1020–1029 [PMC free article] [PubMed] [Google Scholar]
- 8.Hirschi K. D., Zhen R. G., Cunningham K. W., Rea P. A., Fink G. R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 8782–8786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueoka-Nakanishi H., Tsuchiya T., Sasaki M., Nakanishi Y., Cunningham K. W., Maeshima M. (2000) Eur. J. Biochem. 267, 3090–3098 [DOI] [PubMed] [Google Scholar]
- 10.Shigaki T., Hirschi K. D. (2006) Plant Biol. 8, 419–429 [DOI] [PubMed] [Google Scholar]
- 11.Shigaki T., Hirschi K. (2000) Gene 257, 291–298 [DOI] [PubMed] [Google Scholar]
- 12.Cheng N. H., Pittman J. K., Shigaki T., Lachmansingh J., LeClere S., Lahner B., Salt D. E., Hirschi K. D. (2005) Plant Physiol. 138, 2048–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amtmann A., Blatt M. R. (2009) New Phytol. 181, 35–52 [DOI] [PubMed] [Google Scholar]
- 14.Zhao J., Barkla B. J., Marshall J., Pittman J. K., Hirschi K. D. (2008) Planta 227, 659–669 [DOI] [PubMed] [Google Scholar]
- 15.Zhao J., Shigaki T., Mei H., Guo Y. Q., Cheng N. H., Hirschi K. D. (2009) J. Biol. Chem. 284, 4605–4615 [DOI] [PubMed] [Google Scholar]
- 16.Reinders A., Schulze W., Thaminy S., Stagljar I., Frommer W. B., Ward J. M. (2002) Structure 10, 763–772 [DOI] [PubMed] [Google Scholar]
- 17.Reinders A., Schulze W., Kühn C., Barker L., Schulz A., Ward J. M., Frommer W. B. (2002) Plant Cell 14, 1567–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shigaki T., Rees I., Nakhleh L., Hirschi K. D. (2006) J. Mol. Evol. 63, 815–825 [DOI] [PubMed] [Google Scholar]
- 19.Shigaki T., Barkla B. J., Miranda-Vergara M. C., Zhao J., Pantoja O., Hirschi K. D. (2005) J. Biol. Chem. 280, 30136–30142 [DOI] [PubMed] [Google Scholar]
- 20.Nathan D. F., Vos M. H., Lindquist S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 1409–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shigaki T., Pittman J. K., Hirschi K. D. (2003) J. Biol. Chem. 278, 6610–6617 [DOI] [PubMed] [Google Scholar]
- 22.Cunningham K. W., Fink G. R. (1996) Mol. Cell. Biol. 16, 2226–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ludewig U., Wilken S., Wu B., Jost W., Obrdlik P., El Bakkoury M., Marini A. M., André B., Hamacher T., Boles E., von Wirén N., Frommer W. B. (2003) J. Biol. Chem. 278, 45603–45610 [DOI] [PubMed] [Google Scholar]
- 24.Obrdlik P., El-Bakkoury M., Hamacher T., Cappellaro C., Vilarino C., Fleischer C., Ellerbrok H., Kamuzinzi R., Ledent V., Blaudez D., Sanders D., Revuelta J. L., Boles E., André B., Frommer W. B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12242–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bracha-Drori K., Shichrur K., Katz A., Oliva M., Angelovici R., Yalovsky S., Ohad N. (2004) Plant J. 40, 419–427 [DOI] [PubMed] [Google Scholar]
- 26.Shigaki T., Hirschi K. D. (2001) Anal. Biochem. 298, 118–120 [DOI] [PubMed] [Google Scholar]
- 27.Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004) Plant J. 40, 428–438 [DOI] [PubMed] [Google Scholar]
- 28.Eide D. J., Clark S., Nair T. M., Gehl M., Gribskov M., Guerinot M. L., Harper J. F. (2005) Genome Biol. 6, R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lahner B., Gong J., Mahmoudian M., Smith E. L., Abid K. B., Rogers E. E., Guerinot M. L., Harper J. F., Ward J. M., McIntyre L., Schroeder J. I., Salt D. E. (2003) Nat. Biotechnol. 21, 1215–1221 [DOI] [PubMed] [Google Scholar]
- 30.Liang F., Cunningham K. W., Harper J. F., Sze H. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 8579–8584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandey S., Assmann S. M. (2004) Plant Cell 16, 1616–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hull G., Garrido J. M. G., Parcy F., Menossi M., Martinez-Izquierdo J. A., Gallois P. (1996) Plant Sci. 120, 153–160 [Google Scholar]
- 33.Shigaki T., Cheng N. H., Pittman J. K., Hirschi K. (2001) J. Biol. Chem. 276, 43152–43159 [DOI] [PubMed] [Google Scholar]
- 34.Carter C., Pan S., Zouhar J., Avila E. L., Girke T., Raikhel N. V. (2004) Plant Cell 16, 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pittman J. K., Sreevidya C. S., Shigaki T., Ueoka-Nakanishi H., Hirschi K. D. (2002) Plant Physiol. 130, 1054–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng N. H., Hirschi K. D. (2003) J. Biol. Chem. 278, 6503–6509 [DOI] [PubMed] [Google Scholar]
- 37.Ottolia M., John S., Qiu Z., Philipson K. D. (2001) J. Biol. Chem. 276, 19603–19609 [DOI] [PubMed] [Google Scholar]
- 38.Zottola R. J., Cloherty E. K., Coderre P. E., Hansen A., Hebert D. N., Carruthers A. (1995) Biochemistry 34, 9734–9747 [DOI] [PubMed] [Google Scholar]
- 39.Schulz B., Frommer W. B. (2004) Science 306, 622–625 [DOI] [PubMed] [Google Scholar]
- 40.Pighin J. A., Zheng H., Balakshin L. J., Goodman I. P., Western T. L., Jetter R., Kunst L., Samuels A. L. (2004) Science 306, 702–704 [DOI] [PubMed] [Google Scholar]
- 41.Mitsui K., Yasui H., Nakamura N., Kanazawa H. (2005) Biochim. Biophys. Acta 1720, 125–136 [DOI] [PubMed] [Google Scholar]
- 42.Marini A. M., Springael J. Y., Frommer W. B., André B. (2000) Mol. Microbiol. 35, 378–385 [DOI] [PubMed] [Google Scholar]
- 43.Loqué D., Lalonde S., Looger L. L., von Wirén N., Frommer W. B. (2007) Nature 446, 195–198 [DOI] [PubMed] [Google Scholar]
- 44.Neuhäuser B., Dynowski M., Mayer M., Ludewig U. (2007) Plant Physiol. 143, 1651–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.