Abstract
ADP-glucose pyrophosphorylase controls starch synthesis in plants and is an interesting case to study the evolution and differentiation of roles in heteromeric enzymes. It includes two homologous subunits, small (S) and large (L), that originated from a common photosynthetic eukaryotic ancestor. In present day organisms, these subunits became complementary after loss of certain roles in a process described as subfunctionalization. For instance, the potato tuber enzyme has a noncatalytic L subunit that complements an S subunit with suboptimal allosteric properties. To understand the evolution of catalysis and regulation in this family, we artificially synthesized both subunit genes from the unicellular alga Ostreococcus tauri. This is among the most ancient species in the green lineage that diverged from the ancestor of all green plants and algae. After heterologous gene expression, we purified and characterized the proteins. The O. tauri enzyme was not redox-regulated, suggesting that redox regulation of ADP-glucose pyrophosphorylases appeared later in evolution. The S subunit had a typical low apparent affinity for the activator 3-phosphoglycerate, but it was atypically defective in the catalytic efficiency (Vmax/Km) for the substrate Glc-1-P. The L subunit needed the S subunit for soluble expression. In the presence of a mutated S subunit (to avoid interference), the L subunit had a high apparent affinity for 3-phosphoglycerate and substrates suggesting a leading role in catalysis. Therefore, the subfunctionalization of the O. tauri enzyme was different from previously described cases. To the best of our knowledge, this is the first biochemical description of a system with alternative subfunctionalization paths.
Introduction
Starch synthesis in photosynthetic eukaryotes such as higher plants and unicellular algae is controlled by a heterotetrameric ADP-glucose pyrophosphorylase (ADP-Glc PPase,4 EC 2.7.7.27). This enzyme catalyzes the reaction of ATP and Glc-1-P to form ADP-glucose (ADP-Glc) and PPi, and it is primarily activated by 3-phosphoglycerate (3-PGA) and inhibited by Pi. In photosynthetic eukaryotes the enzyme includes two distinct S and L homologous subunits (S2L2 or α2β2), but in photosynthetic bacteria the enzyme is a homotetramer (α4). There has been debate regarding the role of the different subunits of ADP-Glc PPase. The S subunit homotetramer from potato (Solanum tuberosum) tuber (StuS), Arabidopsis thaliana (APS1), and barley (Hordeum vulgare) endosperm have a catalytic function with defective regulatory properties (1–3). It is not clear if there is a set of universal roles for the L subunit in plants. The L subunit from potato tuber (StuL) is catalytically deficient and plays more of a regulatory role by modifying the apparent affinity of the S subunit toward allosteric regulators (1, 4). On the other hand, the Arabidopsis APL1 and APL2 isoforms have both catalytic and regulatory functions, whereas the APL3 and APL4 isoforms behave like StuL (2, 5). The maize (Zea mays) endosperm L subunit has been postulated to have a role in catalysis due to effects on apparent affinity of substrates (6), but this may be explained by allosteric effects. Even when the L subunit is not catalytic, it binds substrate and activator molecules. This may contribute to the synergistic effect seen when the L subunit influences the properties of the S subunit and vice versa (7–9).
Different subunit roles have involved gene duplication, divergence, and functional evolution from a common ancestor. In plants and green algae, the presence of two types of subunits expanded the sequence space to explore new roles. Recently, it was shown that L subunit isoforms from Arabidopsis had different properties based on the tissue expression (2, 10). On the other hand, the presence of two types of subunits may have constrained each subunit to adopt complementary roles. For instance, S and L subunits need to interact to form tetramers in vivo (11). Because fewer S isoforms (e.g. only one in Arabidopsis) are available to combine with many L isoforms in different tissues, there is extra pressure for S subunits to remain catalytic in cases where the L subunits are not. The evolutionary path for these ADP-Glc PPase subunits to acquire or lose different roles is not clear despite recent success in determining the ancient catalytic role of StuL. This L subunit from potato tuber is defective in catalysis, but 10–20% of its catalytic activity was resurrected by mutating only two key residues (4). Phylogenetic analysis indicated that it evolved from a catalytic ancestor common to all flowering plants. However, it is uncertain how catalytic roles of the subunits diverged in early photosynthetic eukaryotes.
The lineage of the unicellular alga Ostreococcus tauri is in a unique position to understand the functional evolution of the ADP-Glc PPase subunits. This organism belongs to the Prasinophyceae class (12), potentially the most ancient species in the green lineage that diverged from the ancestor of all green plants and algae (13–15). O. tauri has a single starch granule and the full set of genes present in higher plants for polysaccharide metabolism (16). In addition, it is known that the Prasinophyceae use the ADP-Glc pathway to synthesize starch (17).
To study the evolution of structure and function of subunits in the ADP-Glc PPase family, we artificially synthesized O. tauri OtaS and OtaL subunits because of their unique evolutionary position. We gained insightful information about the properties of the common ancestor of all ADP-Glc PPases from photosynthetic eukaryotes and how regulation of starch synthesis evolved. We found that under certain conditions OtaL, more than OtaS, may play a leading role in catalysis. As a result, we propose that the retention of catalytic properties is not necessarily identical in all branches of the phylogenetic tree. Differences in roles of O. tauri subunits also indicate that alternative divergent functional paths may have occurred in the evolution of ADP-Glc PPases.
EXPERIMENTAL PROCEDURES
Materials
α-d-[U-14C]Glucose 1-phosphate (Glc-1-P) was purchased from GE Healthcare. Glc-1-P, ATP, ADP-glucose (ADP-Glc), 3-phosphoglycerate (3-PGA), NADP, glucose-6-phosphate dehydrogenase from Leuconostoc mesenteroides, phosphoglucomutase from rabbit muscle, and inorganic pyrophosphatase were purchased from Sigma. Phusion DNA polymerase and restriction enzymes were purchased from New England Biolabs (Ipswich, MA). StrataClone™ Blunt PCR cloning kit was purchased from Stratagene (La Jolla, CA). All other reagents were purchased at the highest quality available. Escherichia coli glycogen synthase for the ADP-Glc synthesis assay was expressed and purified as described (18).
DNA Methods
Oligonucleotides for gene synthesis and site-directed mutagenesis were synthesized by Integrated DNA Technologies (IDT, San Diego). Automated DNA sequencing was performed by University of Chicago Cancer Research Center.
Gene Synthesis
The OtaS and OtaL subunit genes were artificially synthesized with optimized codon usage (19), following a PCR-based method with some modifications (20) (supplemental Fig. 1). The genes were divided into three pieces of ∼500 bp to facilitate synthesis (supplemental Fig. 2). Oligonucleotides of 50 bp with a 25-bp overlap corresponding to top and bottom strand sequences of each ∼500-bp piece were purchased (supplemental Table 1). These oligonucleotides (40 pmol each) were combined in a single 50-μl PCR with Phusion DNA polymerase, following the manufacturer's instructions. After an initial denaturation of 30 s at 98 °C, we performed 30 cycles of 98 °C for 5 s, 50 °C for 20 s, and 72 °C for 1 min, followed by a 5-min extension at 72 °C. The PCR products were purified after agarose gel electrophoresis and inserted into the pSC-B vector using the StrataClone™ Blunt PCR cloning kit. Colonies were screened and sent to be sequenced. When unwanted mutations were present, they were eliminated linking correct fragments by PCR, and the screening and sequencing process was repeated. Whole genes were constructed using subsequent Phusion PCRs where all ∼500-bp pieces for each gene were used as template DNA in a single reaction and subcloned into pSC-B. The final products were confirmed by DNA sequencing. OtaS and OtaL genes were then subcloned into the compatible pMAB5 and pMAB6 expression vectors (21), respectively, using NdeI and SacI restriction sites. The vector pMAB5 is a derivative of the pMON17335 and pACYC177 with a tac promoter and a phage T7 gene10 leader translation enhancer (22). Also, pMAB6 is a derivative of pMON17336 and pBR327 with a PrecA and a gene10 leader translation enhancer expression cassette (22). These vectors do not express proteins with affinity tags.
Site-directed Mutagenesis
Site-directed mutagenesis was performed by PCR overlap extension (23) using Phusion DNA polymerase. Plasmids encoding the wild-type S subunit (OtaS) or wild-type L subunit (OtaL) were used as templates unless stated otherwise. Mutations on OtaS were accomplished using the following oligonucleotides: D148A, 5′-TGA GCG GTG CGC ACC TGT AC-3′; K201R, 5′-TTT GCA GAA CGC CCG AAA GGC-3′. To obtain the double mutant OtaSD148A/K201R, the OtaSD148A construct was used as a template. The oligonucleotides used to obtain the homologous mutations in OtaL were as follows: D171A, 5′-TGG CGG GCG CGC ACC TGT AT-3′; K224R, 5′-TTT ACC GAA CGC CCA AAA GGT-3′. Flanking primers for OtaS were as follows: forward, 5′-CAT ATG GCG GGT ACT AAC ACT CCT G-3′; reverse, 5′-GAA TTC GAG CTC TTA GAT CAC AGT GCC-3′. Flanking primers for OtaL were as follows: forward, 5′-CAT ATG TCT GAA CGT CGT ATT GCT C-3′; reverse, 5′-GAG CTC TCA TTA AAT GAT CGT ACC ATC TGG GAT CGT GCA GTT G-3′. All mutations were confirmed by DNA sequencing.
Protein Methods
The bicinchoninic acid reagent from Pierce was used to measure protein concentration during enzyme purification. Bovine serum albumin was used as a standard, and all interfering substances were removed by precipitation (12% trichloroacetic acid and 0.025% sodium deoxycholate and dissolving in 5% SDS, 0.1 n NaOH). Protein concentration after purification was determined by UV absorbance at 280 nm with an extinction coefficient of 0.803 ml cm−1 mg−1 for OtaS and 0.774 ml cm−1 mg−1 for heterotetramers. These extinction coefficients were calculated as described (24). Samples were desalted with Bio-Rad 10 DG chromatography columns. Centricon-30 devices (Amicon Inc., Billerica, MA) were used to concentrate the enzymes. SDS-PAGE was performed using reducing conditions on a 10% BisTris polyacrylamide gel. The BenchMark protein ladder from Invitrogen was used, and the gel was stained with GelCode Blue Stain Reagent from Pierce.
Enzyme Assays
Pyrophosphorolysis Direction
To follow the activity during the purification, the enzyme was assayed with a coupled enzyme spectrophotometric assay. The reaction mixture contained 80 mm HEPPS buffer, pH 8.0, 5 mm MgCl2, 2 mm dithiothreitol, 1 mm 3-PGA, 0.5 mm ADP-Glc, 0.6 mm NADP, 10 mm NaF, 0.01 mm glucose 1,6-bisphosphate, 2 units/ml phosphoglucomutase, 2 units/ml glucose-6-phosphate dehydrogenase, 0.2 mg/ml bovine serum albumin, and enzyme in a total volume of 0.3 ml. The reaction was initiated with 0.1 mm NaPPi, and absorbance at 340 nm was followed for 20 min every 15 s at 37 °C using a BioTek EL808 microplate reader (Winooski, VT). Under these conditions, the path length was 0.75 cm, and an absorbance of 0.0155 was equivalent to 1 nmol of NADPH produced and Glc-1-P synthesized.
Synthesis Direction
The method of Yep et al. (18) was used to measure synthesis of ADP-[14C]Glc from [14C]Glc-1-P and ATP. Unless stated otherwise, the standard reaction mixture contained 50 mm HEPPS buffer, pH 8.0, 10 mm MgCl2, 4 mm dithiothreitol, 1.5 mm [14C]Glc-1-P (100–1000 cpm/nmol) for heterotetramers, 4 mm for OtaS homotetramer, 2.5 mm ATP, 10 mm 3-PGA, 0.75 unit/ml inorganic pyrophosphatase, and 0.2 mg/ml bovine serum albumin plus enzyme in a total volume of 0.2 ml. Reaction mixtures were incubated for 10 min at 37 °C and terminated by heating in a boiling water bath for 1 min. The amount of ADP-[14C]Glc formed during the reaction was measured as described. To obtain saturation curves of a given effector, its concentration was varied, although the other conditions remained constant.
Unit Definition
One unit of enzyme activity is equal to 1 μmol of product, ADP-[14C]glucose, formed per min.
Enzyme Expression and Purification
E. coli AC7OR1-504 cells that lack endogenous ADP-Glc PPase activity were transformed with plasmids for expression (22). The OtaS subunit in plasmid pMAB5 was capable of being expressed alone to form a homotetramer, whereas the OtaL subunit in plasmid pMAB6 expressed alone was insoluble. For co-expression of both subunits, cells were co-transformed with pMAB5 containing wild-type OtaS or derivatives and pMAB6 containing wild-type OtaL or derivatives. Transformed cells were grown in 1 liter of LB medium in a 2.8-liter Fernbach flask at 25 °C, 250 rpm, until the A600 nm reached 1.1–1.3. If the OtaS homotetramer was being expressed, 0.4 mm isopropyl β-d-1-thiogalactopyranoside was used to induce protein expression. Otherwise, the heterotetramers were induced with 5 μg/ml nalidixic acid and 0.4 mm isopropyl β-d-1-thiogalactopyranoside. Protein was expressed for 16 h at 20 °C and harvested by centrifugation at 1000 × g for 5 min at 4 °C in a Sorvall RC5B Plus centrifuge. Cells were resuspended in 5 ml of buffer A (50 mm HEPES, pH 8.0, 5 mm MgCl2, 0.1 mm EDTA, 10% sucrose) per g of cells and sonicated on ice eight times for 30 s with 45-s intervals. Crude extracts were the supernatants obtained after centrifugation at 10,000 × g for 20 min at 4 °C.
Proteins OtaS, OtaSD148A, OtaSD148A/OtaL, OtaS/OtaL, and OtaS/OtaLD171A were purified to homogeneity using the following method. Crude extracts were loaded onto a 10- or 30-ml DEAE-Sepharose Fast Flow weak anion exchange column (Amersham Biosciences) equilibrated with buffer A and eluted with a linear gradient of NaCl (10 column volumes, 0–0.5 m, 1 ml/min flow rate). Fractions that had the greatest enzyme activity were pooled and precipitated with ammonium sulfate at 70% saturation. The protein was resuspended in buffer A, desalted, and loaded onto a 1.7- or 10-ml Source 15Q 4.6/100 PE strong anion exchange column (GE Healthcare). The column was equilibrated with buffer A and eluted with a linear gradient of NaCl (20 column volumes, 0–0.5 m, 1 ml/min flow rate). Fractions with the highest activity were again pooled and precipitated with ammonium sulfate at 70% saturation. Protein was resuspended in buffer H (buffer A plus 1 m (NH4)2SO4) and loaded onto two Resource PHE 1-ml HIC hydrophobic columns in tandem (GE Healthcare). The columns were equilibrated with buffer H, and protein was eluted with a linear gradient of (NH4)2SO4 (50 column volumes, 1 to 0 m, 0.5 ml/min flow rate). Fractions containing the highest activity were pooled, desalted, and concentrated. Chromatograms for each step of the purification of the wild-type enzyme are shown in supplemental Figs. 3–5. Typically, these enzymes were purified close to 30-fold with specific activity doubling after each column. The yield from 1 liter of culture was ∼3 mg. Enzymes OtaSD148A/K201R/OtaL, OtaSD148A/OtaLK224R, OtaSD148A/OtaLR53K/K63T, and OtaSD148A/OtaLD171A were purified using the DEAE-Sepharose column and desalted before kinetic analysis. These were more than 80% pure based on densitometry after SDS-PAGE (data not shown).
Molecular Weight Determination
The molecular weight of the OtaS/OtaL protein was determined using a Superdex 200 10/300 GL column (GE Healthcare). Protein standards included thyroglobulin (669 kDa) (GE Healthcare), apoferritin (443 kDa) (MP Biomedicals), alcohol dehydrogenase (150 kDa) (United States Biochemical Corp.), glyceraldehyde-6-phosphate dehydrogenase (104 kDa) (Sigma), and bovine serum albumin (68 kDa) (Sigma). The column void volume was determined using a blue dextran loading solution (Promega). The purified E. coli ADP-Glc PPase homotetramer was also used as a standard with a molecular mass of 194 kDa.
Densitometry
Gels were visualized on a Canon 4400F scanner, and the software UN-Scan IT™ version 5.1 was used for gel densitometry to determine the subunit stoichiometry for the OtaS/OtaL wild-type protein on a 10% BisTris SDS-PAGE.
Calculation of Kinetic Constants
We performed kinetic assays using saturated concentrations or optimal conditions for all reaction mixture components, while varying the concentration of the effector being studied. After we plotted the data as specific activity (units/mg) (v) versus effector concentration in millimolars ([S]), we fit the parameters of a modified Hill equation, v = Vmax [S]n/(S0.5n + [S]n), using the Levenberg-Marquardt nonlinear least squares algorithm in Origin® 7.5 (25). The concentration of substrate necessary to obtain 50% of the maximum activity (Vmax) is S0.5. We used the equation v = v0 + (Vmax − v0) [A]n/(A0.5n + [A]n) to fit the activation curves, in which the activity with no activator [A] present is v0. A0.5 is the concentration of activator necessary to obtain 50% of the maximal activation (Vmax − v0). We fit the inhibition curves with the equation v = v0 + (Vinh − v0) [I]n/(I0.5n + [I] n), in which the activity with no inhibitor present is v0, and the activity at saturated concentration of inhibitor [I] is Vinh. I0.5 is the concentration of inhibitor necessary to obtain 50% of the maximal inhibition (v0 − Vinh). To calculate catalytic efficiency (ce = Vmax/Km) with accuracy in cases in which complete saturation was not reached (OtaSD148A/OtaLK224R), we used a modified Michaelis-Menten equation. We fit the data with the equation v = ce Km [S]/(Km + [S]) because ce = Vmax/Km. This is more accurate because Vmax/Km is the slope of the curve at very low concentrations of substrate ([S] ≪ Km) and that parameter is well determined even with curves that have not reached saturation. Standard deviations were provided by the fitting software. Kinetic experiments were performed at least twice with similar results.
Structure Prediction and Homology Modeling
Homology models of the O. tauri large subunit (OtaL) and potato tuber large subunit (StuL) proteins were constructed using Modeler 8 version 2. The crystal structure of the potato tuber small subunit homotetramer (Protein Data Bank code 1YP4 chain B) was used as a template, and models were evaluated as described previously (4). To visualize the models we used Swiss-PdbViewer (26) and POV-ray.
Phylogenetic Analysis
Sequences for phylogenetic analysis were downloaded from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) (supplemental Table 2), and ClustalW was used initially to align sequences (27). The alignments were later manually refined. A phylogenetic tree was constructed using the maximum likelihood method, TREE-PUZZLE 5.2 (28). Conditions were as follows: analysis with tree reconstruction; quartet puzzling with 1000 puzzling steps; approximate quartet likelihood; parameter estimates exact (slow); model of substitution with VT (29) estimated from data set; model of rate heterogeneity with γ distributed rates estimated from a data set; and eight γ rate categories. The outgroup sequence was from the cyanobacterium Nostoc sp.
RESULTS
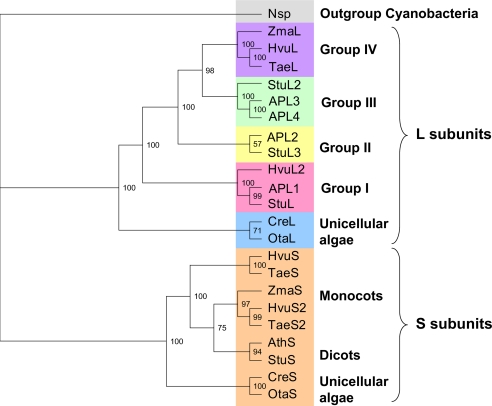
Duplication of an ancestral photosynthetic eukaryote gene gave rise to the ADP-Glc PPase S and L subunits present today in plants and green algae (30). In the phylogenetic tree of these genes the L subunits are further divided into five different branches (Fig. 1), including group I to IV and unicellular algae (4). Previously, L subunits from group III (StuL, APL3, and APL4) were shown to have a regulatory function with a defective catalytic role (2, 4, 5, 31, 32). On the other hand, isoforms APL1 and APL2 (groups I and II) were catalytic with a critical role in regulation (2, 5). The unicellular algae branches of the tree for S and L subunits have not been characterized (Fig. 1), and OtaS and OtaL belong to these two branches.
FIGURE 1.
Phylogenetic tree of representative ADP-Glc PPase proteins from photosynthetic eukaryotes. This tree was constructed using maximum likelihood methods where the cyanobacteria Nostoc sp. was used as the outgroup (Nsp., gray). The L subunits were separated into five main groups as follows: group IV (purple), group III (green), group II (yellow), group I (pink), and unicellular algae (blue). Orange is used to show S subunits from monocots, dicots, and unicellular algae. Accession numbers and abbreviations are in supplemental Table 2. Confidence values for each branch calculated by the TREE-PUZZLE program are included in the tree.
Homology Modeling
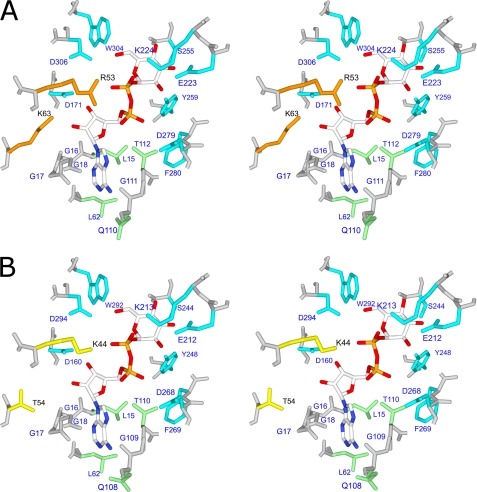
To examine the structure of OtaL, we built a homology model (Fig. 2) and compared the position of the active site residues found in ADP-Glc PPases from other organisms. We found that the model contains all residues in the active site that were previously found to be critical. Residues with a Glc-1-P binding role in the E. coli enzyme (Glu-194, Lys-195, Ser-212, Tyr-216, Asp-239, Phe-240, Trp-274, and Asp-276) (33, 34) and in the StuS subunit (Lys-198 and Asp-252, also studied in E. coli) (32, 35) were present in the corresponding positions (Fig. 2). In OtaL these homologous residues are Glu-223, Lys-224, Ser-255, Tyr-259, Asp-279, Phe-280, Trp-304, and Asp-306. A key catalytic residue found in both E. coli and the potato tuber enzyme (Asp-142 and Asp-145, respectively (31, 36)) was also present in the model of OtaL (Asp-148) (Fig. 2). Interestingly, two residues that were necessary to resurrect the activity of the noncatalytic StuL subunit (4) were also present in the OtaL model (Arg-53 and Lys-63). Conversely, the native StuL subunit has Lys and Thr, respectively, which explains its deficiency in catalysis (4). Based on the model structure and the presence of previously studied residues, there was no indication that OtaL had a defective active site. To test this hypothesis, we artificially synthesized the gene for heterologous expression and further characterization.
FIGURE 2.
Stereodiagram of OtaL and StuL ADP-Glc PPase proteins with ADP-glucose. OtaL is shown in A with key residues Arg-53 and Lys-63 in orange, and StuL is shown in B with Lys-44 and Thr-54 in yellow. Glc-1-P-binding residues are shown in blue, and other residues within 4 Å of ADP-glucose are shown in green.
Expression and Structure of O. tauri ADP-Glc PPase Subunits
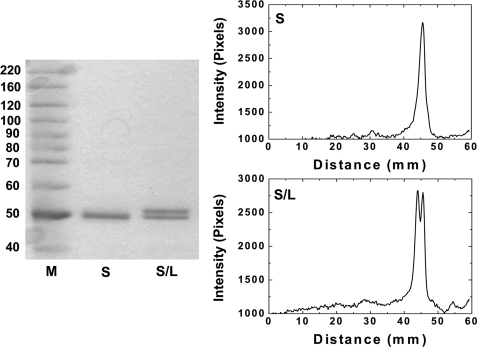
The recombinant O. tauri ADP-Glc PPase subunits were overexpressed in E. coli and purified to homogeneity. The OtaL protein could not be expressed in a soluble form, possibly because it needs the S subunit for folding and solubility as demonstrated in other organisms (4, 11, 37). When OtaL was expressed alone, it went to the insoluble fraction after ultrasonic disruption of the cells. On SDS-PAGE, the OtaS protein expressed alone ran as a single band of ∼50 kDa after purification, whereas the co-expression of OtaS and OtaL yielded a protein with two distinct bands. The upper band was ∼52 kDa (OtaL) and the lower band was ∼50 kDa (OtaS). The stoichiometry of the co-expressed OtaS/OtaL protein was determined by densitometry (Fig. 3). The ratio of peak areas of the two subunits (OtaS/OtaL) was 1.03, indicating that the stoichiometry in the quaternary structure of the enzyme is 1:1. Using size exclusion chromatography, the OtaS/OtaL protein was determined to have a molecular mass of 222 kDa, which is very close to the calculated molecular mass (204 kDa) of a heterotetramer. In addition, the OtaS/OtaL enzyme and the purified homotetrameric E. coli ADP-Glc PPase (194 kDa) have indistinguishable retention volumes. Based on the 1:1 stoichiometry and the molecular size of the native protein, we concluded that the quaternary structure is α2β2 (S2L2). This is in good agreement with the stoichiometry and quaternary structure of the potato tuber enzyme determined by N-terminal sequencing and sucrose density gradient centrifugation (31, 38).
FIGURE 3.
Gel densitometry of OtaS and OtaS/OtaL proteins. The SDS-PAGE shown was used to determine the stoichiometry of the OtaS:OtaL subunits. M corresponds to the molecular weight protein marker; S refers to the OtaS homomeric protein; and S/L is the OtaS/OtaL heteromeric protein. The purified protein loaded in each lane was 1 μg. The electrophoresis and the gel staining proceeded as described under “Experimental Procedures.”
Kinetic Analysis
The ability to express OtaS alone allowed us to compare its kinetic characteristics with the OtaS/OtaL heterotetrameric protein, showing the effect of the presence of the L subunit.
Glc-1-P Saturation Curves
The OtaS homotetramer has a uniquely high S0.5 for Glc-1-P (3.1 mm), which corresponds to an apparent affinity of about 2 orders of magnitude lower than all native ADP-Glc PPases studied so far. Typical homotetrameric and heterotetrameric ADP-Glc PPases from plants and bacteria have a high apparent affinity for Glc-1-P with an S0.5 range 0.015 to 0.092 mm (Table 1). Unfortunately, we do not know the apparent affinity for Glc-1-P of the Chlamydomonas reinhardtii homotetramer because only the native heterotetrameric form was purified from the source (39). However, both C. reinhardtii and recombinant OtaS/OtaL heterotetramers have a high affinity for Glc-1-P (0.03 and 0.092 mm, respectively) (Table 1). The S0.5 of the OtaS homotetramer is 33-fold higher than that of the OtaS/OtaL heterotetramer. Both OtaS and OtaS/OtaL displayed hyperbolic curves (the Hill coefficients “n” were very close to 1; Table 2). For this reason, S0.5 was equivalent to Km, which allowed us to calculate the catalytic efficiency (Vmax/Km). OtaS and OtaS/OtaL did not have a significantly different Vmax (Fig. 4), and the catalytic efficiency of OtaS (3.3 units mg−1 mm−1) was 33-fold lower than OtaS/OtaL (122 units mg−1 mm−1).
TABLE 1.
Kinetic parameters of various ADP-Glc PPase enzymes for substrates in the synthetic direction
The S0.5 values for ATP, Mg2+, and Glc-1-P for representative bacterial, unicellular algae, and plant homotetrameric and heterotetrameric proteins studied to date are shown. Stu indicates S. tuberosum; Ota indicates O. tauri; Eco indicates E. coli; Syn indicates Synochocystis; Ana indicates Anabaena, and Cre indicates C. reinhardtii. APS1 and APL1–4 are from A. thaliana.
| S0.5 |
|||
|---|---|---|---|
| ATP | Mg2+ | Glc1P | |
| mm | |||
| APS1a | 0.402 | b | 0.076 |
| StuSc,d | 0.20 | 2.2 | 0.029 |
| OtaS | 0.42 | 3.6 | 3.1 |
| Ecoe | 0.30 | 1.4 | 0.015 |
| Synf | 0.80 | 3.8 | 0.05 |
| Anaf | 0.46 | 1.6 | 0.08 |
| Cre S + Lg | 0.08 | 2.2 | 0.03 |
| Ota S + L | 0.37 | 4.2 | 0.092 |
| APS1 + APL1a | 0.067 | NDh | 0.019 |
| APS1 + APL2a | 0.575 | ND | 0.085 |
| APS1 + APL3a | 0.094 | ND | 0.052 |
| APS1 + APL4a | 0.118 | ND | 0.060 |
| Stu S + Li | 0.076 | 2.2 | 0.057 |
TABLE 2.
Kinetic parameters of O. tauri ADP-Glc PPase enzymes for substrates
| Enzyme | S0.5 ATP | n | S0.5 Mg2+ | n | S0.5 Glc-1-P | n |
|---|---|---|---|---|---|---|
| mm | mm | mm | ||||
| OtaS | 0.415 ± 0.052 | 1.58 | 3.65 ± 0.21 | 2.76 | 3.08 ± 0.28 | 0.89 |
| OtaS/OtaL | 0.373 ± 0.036 | 1.93 | 4.23 ± 0.18 | 3.14 | 0.092 ± 0.014 | 1.07 |
| OtaSD148A/OtaL | 0.145 ± 0.011 | 1.54 | 1.56 ± 0.07 | 3.40 | 0.047 ± 0.003 | 1.60 |
| OtaS/OtaLD171A | 0.491 ± 0.046 | 1.53 | 4.08 ± 0.28 | 2.37 | 0.044 ± 0.005 | 1.50 |
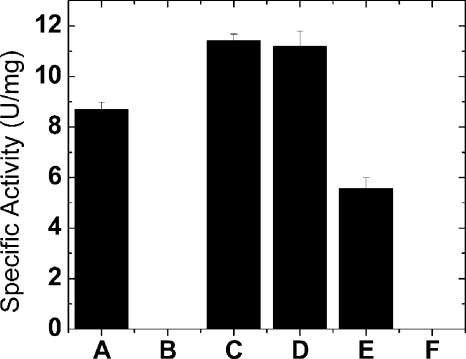
FIGURE 4.
Specific activity of various O. tauri ADP-Glc PPase forms. Proteins shown are OtaS (A), OtaSD148A (B), OtaSD148A/OtaL (C), OtaS/OtaL (D), OtaS/OtaLD171A (E) and OtaSD148A/OtaLD171A (F). In the histogram, B and F had negligible activity (0.0056 ± 0.0001 and 0.00014 ± 0.00001 unit/mg, respectively). The concentration of substrates and effectors were saturating as described under “Experimental Procedures.”
ATP Saturation Curves
The affinity for ATP of OtaS is very similar to the other recombinant homotetrameric plant S subunits characterized in the literature (Arabidopsis and potato tuber; Table 1). The S subunit from barley endosperm was expressed, but no ATP saturation curve has been reported (3). Moreover, the apparent affinity of OtaS is similar to representative bacterial homotetrameric forms (Anabaena, Synechocystis, and E. coli; Table 1), indicating that the structure of the ATP binding pocket is not significantly different.
The apparent affinity for ATP of the OtaS/OtaL protein is slightly lower than other unicellular algae and plant heterotetrameric ADP-Glc PPases, and it most closely resembles the affinity observed for other homotetrameric proteins (Table 1). This indicates that the presence of OtaL in the heterotetramer did not alter the apparent affinity for ATP. In potato tuber and Arabidopsis, the combination of any L with the S subunit yielded heterotetramers with a lower S0.5 for ATP than the S subunit alone. The only exception is APL2 from Arabidopsis (Table 1). In the case of O. tauri, both OtaS and OtaS/OtaL exhibited similar apparent affinities (S0.5) for ATP (0.415 and 0.373 mm, respectively), differing only in the maximum velocity achieved (Table 2). Conversely, the S0.5 values of ATP for the heterotetramers were 6-, 4-, 3-, and 3-fold lower than their respective homotetramers for the combinations of APL1, APL3, APL4, and StuL with APS1 and the StuS subunit, respectively (Table 1).
Mg2+ Saturation Curves
The S0.5 of Mg2+ is of the same order of magnitude for all the proteins previously analyzed. Comparing the unicellular algae heterotetramers, however, the S0.5 for Mg2+ is nearly two times greater (4.2 mm) than that of C. reinhardtii (2.2 mm). There was no difference in kinetic parameters for the StuS homotetramer or heterotetrameric proteins (2.2 mm) (Table 1).
Activity in Absence of 3-PGA
Another unique feature of the OtaS homotetramer is its relatively high activity in the absence of activator (1.96 units/mg; Table 3). It seems to be in a slightly preactivated state, in which the activity is 16.6% of the maximum at saturating concentrations of the activator 3-PGA. The other S subunit homotetramers studied from potato tuber, Arabidopsis, and barley endosperm had negligible or undetectable activity in the absence of activator. In all those cases, the activity was at least 500-fold lower than the maximum at saturated concentrations of 3-PGA (1–3). On the other hand, OtaS has a lower fold activation (6-fold) than the OtaS/OtaL enzyme (33-fold; Table 3) due to its slightly preactivated state in absence of 3-PGA.
TABLE 3.
Kinetic parameters of O. tauri ADP-Glc PPase enzymes for activator and inhibitor
Enzymes were purified to homogeneity as described under “Experimental Procedures.” The A0.5 and I0.5 values for 3-PGA and Pi, respectively, are shown for O. tauri enzymes purified to homogeneity.
| A0.5 3-PGA | Fold activation | I0.5 Pia | Fold decrease | |
|---|---|---|---|---|
| mm | mm | |||
| OtaS | 1.99 ± 0.29 | 6 | 2.34 ± 0.32 | 3 |
| OtaS/OtaL | 0.544 ± 0.074 | 33 | 0.64 ± 0.09 | 3 |
| OtaSD148A/OtaL | 0.0080 ± 0.0015 | 3 | 3.05 ± 0.87 | 48 |
| OtaS/OtaLD171A | 0.059 ± 0.010 | 8 | 0.137 ± 0.017 | 3 |
a Concentration of 3-PGA was at their respective A0.5 values.
Activation by 3-PGA
OtaS and OtaS/OtaL ADP-Glc PPases are activated by 3-PGA and inhibited by Pi as are other ADP-Glc PPases from oxygenic photosynthetic organisms, including cyanobacteria, unicellular algae, and plants (30). OtaS/OtaL has a higher apparent affinity for the activator than OtaS (A0.5 of 0.544 and 1.99 mm, respectively) (Table 3). The specific activities of ADP-Glc synthesis at optimal concentrations of 3-PGA and substrates for OtaS/OtaL and OtaS were 11.2 and 8.7 units/mg, respectively (Fig. 4). This activity was lower but on the same order of magnitude as those reported for the corresponding potato tuber recombinant enzymes (30 and 32 units/mg) (1). Similarly, the recombinant heterotetramer maize endosperm was 26.7 units/mg (40).
Inhibition by Pi
OtaS/OtaL has a higher apparent affinity for Pi than OtaS. The I0.5 measured at the A0.5 concentration of 3-PGA was 0.64 and 2.34 mm for OtaS/OtaL and OtaS, respectively (Table 3). Interestingly, the maximum inhibition by Pi of both of these forms was only about 3-fold. In other ADP-Glc PPases from plants the Pi inhibition is more dramatic (30).
Catalytic Role of OtaL
The OtaL properties were studied by co-expressing it with OtaS because it could not be expressed alone. To observe potential catalysis coming from OtaL without interference, we used a catalytically defective OtaS for the co-expression. A similar strategy was previously used to study the roles of subunits in Arabidopsis and potato tuber ADP-Glc PPases (4, 5). In OtaS, we chose to replace Asp-148 with alanine to obtain a catalytically defective S subunit. This residue was proven to be critical for catalysis, and it is homologous to Asp-145 and Asp-142 in the ADP-Glc PPases from potato tuber and E. coli, respectively (31, 36).
OtaSD148A was expressed in the homotetrameric form to confirm that there was no significant catalytic activity coming from this subunit. We observed that this mutation decreased the activity nearly 1500-fold (Fig. 4), in good agreement with its predicted critical role. When OtaL was co-expressed with OtaSD148A, the activity was 2000-fold higher than OtaSD148A alone, strongly suggesting a direct catalytic role for OtaL (Fig. 4). The S0.5 values of ATP, Mg2+, and Glc-1-P for the OtaSD148A/OtaL protein were 0.145, 1.56, and 0.047 mm, respectively (Table 2). These kinetic parameters were similar to the ones from OtaS/OtaL and other described ADP-Glc PPases (30), indicating that OtaL interacts normally with substrates.
To further show that catalysis in OtaSD148A/OtaL was coming from the L subunit, we decided to probe the apparent affinity for Glc-1-P in the putative active site. In potato tuber and E. coli ADP-Glc PPases, there is a conserved lysine responsible for high affinity binding of Glc-1-P (Lys-195 and Lys-198, respectively). This residue forms a salt bridge with the β-phosphate of the product ADP-Glc, which corresponds to the phosphate of the substrate Glc-1-P (41). Even an arginine could not effectively replace this lysine to retain normal binding properties (32, 34). In OtaS and OtaL the homologous residues are Lys-201 and Lys-224, which also interact with ADP-Glc in a similar manner according to our homology model (Fig. 2).
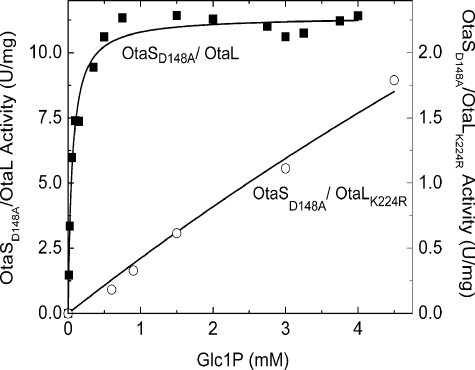
Mutation of this lysine to arginine on OtaL hindered the ability of the tetramer to catalyze the production of ADP-Glc in presence of a catalytically defective OtaSD148A. The Glc-1-P S0.5 of OtaSD148A/OtaL was 0.047 mm, whereas OtaSD148A/OtaLK224R did not even reach saturation at 10 mm. The Glc-1-P S0.5 of OtaSD148A/OtaLK224R shifted more than 200-fold when compared with the Glc-1-P S0.5 of OtaSD148A/OtaL (Fig. 5). Because saturation was not reached, S0.5 or Km values could not be measured for OtaSD148A/OtaLK224R. Instead, we calculated the catalytic efficiency from the slope at lower concentrations of Glc-1-P as described under “Experimental Procedures” (Fig. 5). The catalytic efficiency of OtaSD148A/OtaLK224R (0.41 unit mg−1 mm−1) was 590-fold lower than OtaSD148A/OtaL (243 units mg−1 mm−1). This result indicates that Lys-224 is important for the interaction with Glc-1-P during catalysis, which confirms the catalytic role of OtaL. As a control, the corresponding mutation K201R was performed on the already catalytically deficient OtaSD148A, and the apparent affinity for Glc-1-P did not change significantly (data not shown).
FIGURE 5.
Glc-1-P curves for OtaSD148A/OtaL and OtaSD148A/OtaLK224R proteins. The synthetic assay was used to determine the affinity of the L subunit for Glc-1-P when it was in the presence of a catalytically defective S subunit OtaSD148A. Black squares indicate OtaSD148A/OtaL, and open circles indicate OtaSD148A/OtaLK224R. The maximum specific activity was 11.41 and 1.79 units/mg, respectively, for each respective curve.
Properties of OtaS in the Heterotetramer
We investigated the properties of the S subunit in the presence of a catalytically deficient L subunit by mutating a catalytic aspartic acid to alanine (OtaS/OtaLD171A). This Asp-171 is homologous to the Asp-148 residue in OtaS as mentioned above (Fig. 2). In OtaS/OtaLD171A, catalysis occurs only at the OtaS active site. As a confirmation, a double mutant OtaSD148A/OtaLD171A rendered an enzyme that was >30,000-fold less active than OtaS/OtaLD171A (Fig. 4). A comparison of the kinetic parameters from OtaS/OtaLD171A with those of the OtaS homotetramer indicated there is a distinct effect of OtaL on the efficiency to bind Glc-1-P by OtaS. The S0.5 for Glc-1-P of OtaS/OtaLD171A was 0.044 mm, 2 orders of magnitude lower than OtaS alone. Other kinetic parameters did not change significantly after the co-expression of OtaLD171A with OtaS. The S0.5 values for ATP and Mg2+ of OtaS/OtaLD171A were only 20 and 10% higher than OtaS (Table 2). In addition, the presence of the L subunit seems to increase the affinity for regulators (3-PGA and Pi) when co-expressed with the S subunit. OtaS/OtaLD171A had a higher affinity for 3-PGA (0.059 mm) than OtaS by 2 orders of magnitude (Table 3). The I0.5 of Pi at the A0.5 concentration of 3-PGA was 0.137 mm, which is 1 order of magnitude lower than OtaS (Table 3).
Differential Sensitivity to Regulators by OtaS and OtaL
OtaSD148A/OtaL had an apparent affinity for 3-PGA nearly 2 orders of magnitude higher than OtaS/OtaL, with A0.5 values of 0.008 and 0.544 mm, respectively (Table 3). The apparent affinity for Pi, at the A0.5 concentration of 3-PGA, was about 5-fold lower than OtaS/OtaL (Table 3). OtaSD148A/OtaL also had a higher apparent affinity for 3-PGA and a lower apparent affinity for Pi than OtaS/OtaLD171A. In both cases the difference is about 1 order of magnitude (Table 3). This indicates that the catalysis occurring at the L subunit is more sensitive to activation than the catalysis observed in the wild-type heterotetramer, particularly the one observed in the S subunit.
Catalytic Contribution of OtaS and OtaL
After mutation of a catalytic residue in one of the subunits, the remaining specific activity revealed the contribution to catalysis of the other. The OtaSD148A/OtaL specific activity was 2.04-fold higher than OtaS/OtaLD171A (Fig. 4), which indicates that the contribution of the L subunit may be more influential than the S subunit in the heterotetramer. The specific activity of the wild-type heterotetramer is lower than the addition of the specific activities from OtaSD148A/OtaL and OtaS/OtaLD171A, suggesting a slight antagonistic effect between active sites from both subunits with respect to maximum velocity.
DISCUSSION
The two homologous subunits of the O. tauri ADP-Glc PPase complement each other. The S subunit is defective for both 3-PGA activation and Glc-1-P apparent affinity. The L subunit, however, performs catalysis with a higher apparent affinity for these effectors, but it needs the presence of the S subunit to express in a soluble form. The individual properties of the S and L subunits that complement each other can be defined as subfunctions. These are specific subsets of a function of a gene that, when mutated, establish a distinct complementation group (42). In this case, when a property (subfunction) in one of the ADP-Glc PPase subunits becomes suboptimal, the other subunit compensates for it. This process of subfunctionalization is important because it has been described as a transition state for further neofunctionalization. The latter is a process that involves the retention of the ancestral function of one gene while another duplicate acquires new functions (43). The description and characterization of these processes are essential in this system to understand the evolution of roles in the ADP-Glc PPase family.
Functional Similarities and Differences
The scenario of role differentiation in the O. tauri enzyme has similarities and differences with ADP-Glc PPases in other branches of the phylogenetic tree. OtaS shares some similarities with StuS and Arabidopsis APS1. They have good expression by themselves but have defective regulatory properties (low apparent affinity for the activator 3-PGA) (1, 2). OtaL, StuL, and Arabidopsis APL3 and APL4 isoforms confer a high apparent affinity for the activator to the heterotetramer (1, 2) but cannot fold properly by themselves (2, 11, 37). There are two remarkable differences between O. tauri S and L subunits and other ADP-Glc PPases. First, OtaS has a defective catalytic site compared with StuS and APS1. Second, OtaL has a full catalytically competent active site, whereas StuL, APL3, and APL4 have mutated catalytic residues that rendered them unable to perform catalysis (2, 4). Only Arabidopsis isoforms APL1 and APL2 have comparable catalytic properties to OtaL (5).
Alternative Subfunctionalization Path
The differential roles in O. tauri subunits indicate that alternative divergent functional paths occurred in the evolution of ADP-Glc PPases. Groups I and II of the phylogenetic tree (Fig. 1) have not subfunctionalized the catalytic properties of the active site compared with their respective S subunit partners. Arabidopsis L subunits from those two groups (APL1 and APL2) are catalytic and form heteromers with APS1, which is also fully catalytic (5). We do not know the relative catalytic contribution of the L subunits to the heteromers, but we know that it is at least high enough to synthesize starch in vivo in the presence of a mutated S subunit (5). A different scenario has been observed with subunits from group III, in which subfunctionalization of catalytic properties occurred. It has been described previously that this group includes catalytically deficient L subunits, which are complemented by catalytically active S subunits in the respective heterotetramers. Those S subunits kept an active site with all the necessary residues to perform catalysis, but the L subunits from group III did not. This case was described as subfunctionalized catalysis (4). The gene duplication that led to these two types of subunits caused a possible evolutionary unstable overlap of roles. This resulted in a defective catalysis in the L subunits, and a defective regulation in the S subunit. It is not clear why group III subfunctionalized the catalytic properties of the enzyme, whereas groups I and II did not. It is possible that APL1 and APL2 still maintain their ability to bind and catalyze substrates because of homotropic allosteric effects or because just doubling the catalytic capacity is important in certain tissues. Alternatively, we can speculate that the disruption of their active sites has not occurred yet, and because evolution is a dynamic process it may in the future.
Here we observed that the subfunctionalization of O. tauri ADP-Glc PPase was different. The catalytic site of OtaL is fully functional, but catalysis in OtaS is suboptimal because it binds a substrate with an extremely poor apparent affinity (Table 2). In this case, the levels of catalytic efficiency of the subunits are swapped if we compare them to enzymes with L subunits from group III such as potato tuber (Fig. 6). We propose that in organisms from different branches of the phylogenetic tree the retention of catalytic properties in ADP-Glc PPases may not necessarily be identical. In some cases both subunits may be fully catalytic (e.g. Arabidopsis APS1/APL1), but in others one of the subunits may be defective like the L subunit in potato tuber or the S subunit in O. tauri. The subfunctionalization path observed in O. tauri could be present in other branches of the phylogenetic tree. A bioinformatics analysis of other ADP-Glc PPases reveals that the S subunit from Brassica rapa (accession number AAK27684.1) lacks a critical residue. The homologous lysine critical to resurrect the ancestral catalytic activity in StuL (Thr-54 to Lys-54) (4) is replaced by an asparagine in the S subunit from B. rapa. Interestingly, the L subunit cloned from B. rapa (accession number AAK27685.1) belongs to group I and has a lysine in this position. It is possible that at least in certain tissues of B. rapa, the catalysis may occur in the L subunit and not in the S subunit.
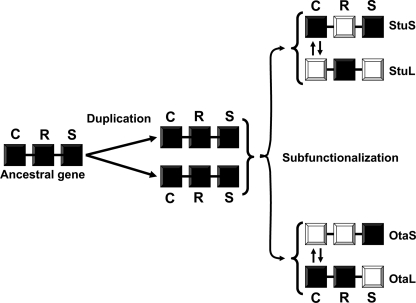
FIGURE 6.
Alternative subfunctionalization paths for O. tauri and S. tuberosum genes. An ancestral eukaryotic ADP-Glc PPase gene with fully functional catalytic (C), regulatory (R), and solubility (S) properties was duplicated to yield at least two copies in the same organism. Copies originally inherited the fully functional properties and are represented by dark squares. These genes followed different paths after speciation of the ancestral eukaryotic organism. Some properties became suboptimal in one of the copies (white squares), whereas the other one complemented it (dark squares). In tissues of some species (e.g. potato tuber), one of the genes corresponding to an ADP-Glc PPase subunit (StuS) became defective in regulation. The other (StuL) compensated for this property but became defective in catalysis and solubility. In other cases, one of the subunits (OtaS) became defective in both catalysis and regulation, although the other subunit (OtaL) compensated for both of those properties. The property that followed a different functional divergence is represented by opposing arrows between the two copies of the genes.
Importance of Arg-53 and Lys-63
To confirm our hypothesis regarding the importance of key Arg-53 and Lys-63 residues for catalysis, we replaced them with Lys-53 and Thr-63 to mimic the primary structure of the inactive L subunit from potato tuber (Fig. 2). The co-expression of an inactive S subunit (OtaSD148A) with the double mutant (OtaLR53K/K63T) resulted in a heterotetramer (OtaSD148A/OtaLR53K/K63T) with a specific activity that was at least 800-fold lower than the wild-type OtaL co-expressed with the same inactive S subunit (OtaSD148A/OtaL) (data not shown). This result emphasizes the importance of these two residues for proper catalysis and suggests that their absence could predict a lack of a catalytic role in newly sequenced L subunits. For instance, all subunits from groups III and IV lack these residues.
Catalytic Contribution of S and L Subunits
We have shown previously that the APL1 and APL2 subunits from Arabidopsis are catalytic (5). Unfortunately, we were unable to determine the individual catalytic contribution of those L subunits in the presence of the S subunit because the enzymes could not be purified to homogeneity. Because the O. tauri heterotetramers were purified to homogeneity, this is the first time we can address this issue. Mutating a catalytic residue in one subunit (OtaSD148A and OtaLD171A, respectively) and assaying the remaining activity in the heterotetramer allowed us to draw two important conclusions. First, when each wild-type subunit was analyzed in the presence of a catalytically defective partner, a considerable specific activity was observed when compared with the wild-type heterotetramer (Fig. 4). This implies that the catalytic sites in one subunit can work without the one from the other subunit being catalytically functional. Second, comparing the OtaSD148A/OtaL and OtaS/OtaLD171A proteins at saturating concentrations of substrates and activator, we concluded that in the heterotetramer the optimal catalytic contribution of the L subunit may be higher than the S subunit (Fig. 4). OtaS alone may be defective in substrate binding (Table 2), but when the active site is fully occupied it performs catalysis almost like OtaL (Fig. 4). A similar case was observed with the activator 3-PGA. The L subunit in the presence of a catalytically defective S subunit had a higher apparent affinity for 3-PGA and lower apparent affinity for Pi than the reverse construct or wild-type heterotetramer (Table 3). This indicates that in the heterotetramer OtaL has a greater tendency to be activated than OtaS at subsaturated concentrations of allosteric effectors. Therefore, under these conditions the active site of OtaL may play a leading role in catalysis.
Interestingly, in Fig. 4 the Vmax values of OtaSD148A/OtaL and OtaS/OtaL are not significantly different even though the former tetramer has two functional catalytic sites (two OtaL subunits) and the latter has four. This suggests that there may be a strong negative cooperativity between the subunits or half-site reactivity. In other words, when the first two active sites are occupied, they cause a change that forbids the binding of the next two. This type of strong interaction between subunits may not be a rare case for ADP-Glc PPases. For instance, in the crystal structure of the homotetrameric potato tuber S subunit, it has been observed that only two subunits bind the substrate ATP (41). This is in good agreement with the fact that the specific activity of the homotetrameric potato tuber S subunit with four functional active sites is not very different from the heterotetrameric potato tuber enzyme with two functional active sites and two noncatalytic sites (32 and 30 units/mg, respectively) (1). A bacterial enzyme such as the one from E. coli binds only two molecules of ATP per tetramer but binds four when the other substrate (Glc-1-P) is present (44).
OtaS-defective Apparent Affinity for Glc-1-P
It is known that the Glc-1-P apparent affinity slightly decreases in the absence of activator for many ADP-Glc PPases (2, 6, 7). For OtaS, we discarded this explanation because a saturating concentration of 3-PGA was used. This is the first time a wild-type ADP-Glc PPase form from either photosynthetic eukaryotes or bacteria displays such poor affinity for Glc-1-P. The structural reason for this is unclear because all the residues characterized as important for Glc-1-P binding in other ADP-Glc PPases are present in OtaS (Fig. 2).
To determine the cause for the OtaS low apparent affinity for Glc-1-P, we constructed chimeric enzymes. We wanted to know if there was a defect in the OtaS Glc-1-P binding domain or the rest of the protein. The high apparent affinity Glc-1-P domain of StuS, composed of residues located in the center of the sequence (33, 41), was switched into OtaS. We also built the opposite construct by replacing the StuS Glc-1-P domain with the OtaS domain. Both chimeric proteins had a low apparent affinity for Glc-1-P, which was comparable with that of OtaS rather than the higher affinity seen by StuS.5 Therefore, the decreased affinity for Glc-1-P may be due to the interaction between domains rather than a specific mutation in the Glc-1-P domain and deserves further investigation.
OtaS has a low apparent affinity for Glc-1-P, but the presence of a catalytically defective L subunit restored its high apparent affinity in the heterotetramer (OtaS/OtaLD171A) (Table 2). There may be several explanations for this effect. First, this could be a homotropic allosteric effect where one subunit is being affected by the binding of a substrate molecule on the other subunit. Second, this could be a synergistic effect where the interaction of the two individual subunits increases the apparent affinity for Glc-1-P. A similar case was actually observed with the activator 3-PGA in the potato tuber enzyme. The mere presence of StuL in the heterotetrameric form improved the apparent affinity for the activator in StuS (1).
Evolution of Redox Regulation
There may be a clear distinction in redox regulation between the unicellular algae and all the other ADP-Glc PPases from photosynthetic tissues. StuS has an inter-subunit disulfide bridge Cys-12–Cys-12 at the N terminus where its reduction activates the heterotetramer in vivo and in vitro (41, 45, 46). Unicellular algae ADP-Glc PPases lack the necessary cysteine residues to form a disulfide bridge responsible for this type of regulation. In fact, the activity of the O. tauri wild-type heterotetramer in the presence and absence of dithiothreitol was not significantly different even when concentrations of 3-PGA were varied (data not shown). Additionally, in the presence and absence of a reducing agent (β-mercaptoethanol) there was no shift in mobility on an SDS-PAGE. This indicated that no dimerization occurred by the formation of disulfide bridges as shown in other redox-regulated ADP-Glc PPases (47, 48). The lack of redox regulation in the O. tauri enzyme strongly suggests that this regulatory mechanism appeared later in the evolution of ADP-Glc PPase from multicellular photosynthetic organisms.
Conclusions
The literature describing subfunctionalization of genetic processes, observed by gene expression, and genomics analysis is vast and has been reviewed (49). However, biochemical examples emphasizing protein-structure relationships are scarce (4, 50, 51). To the best of our knowledge, this is the first biochemical description of a system with alternative subfunctionalization paths (Fig. 6). From the results of this work, it is clear that the evolution of subunit role differentiation has taken different routes in the phylogenetic tree. It seems like there is not one defined role for subunits in the family of ADP-Glc PPases in photosynthetic eukaryotes. Artificial synthesis of predicted ancestors (52) may help elucidate at what point in evolution some of these changes occurred. The characterization of these evolutionary paths is important because inactivation of dimers and further complementation with the partner subunit could be common features. The formation of nonenzyme homologues from enzymes has been observed (53, 54), and sometimes they could be reverted by the mutation of only one or two residues (4, 55). This subfunctionalization process could be among the first steps into the generation of new roles.
Supplementary Material
Acknowledgments
We thank F. Bryan Pickett for stimulating discussions and Kenneth W. Olsen for revision of the manuscript.
This work was supported in part by National Science Foundation Grant MCB 0615982.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5 and Tables 1 and 2.
M. L. Kuhn, D. A. Falaschetti, and M. A. Ballicora, unpublished results.
- ADP-Glc PPase
- ADP-glucose pyrophosphorylase
- OtaS
- O. tauri S subunit
- OtaL
- O. tauri L subunit
- 3-PGA
- 3-phosphoglycerate
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- HEPPS
- 3-[4-(2-hydroxyethyl)-1-piperazinyl]propanesulfonic acid.
REFERENCES
- 1.Ballicora M. A., Fu Y., Nesbitt N. M., Preiss J. (1998) Plant Physiol. 118, 265–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crevillén P., Ballicora M. A., Mérida A., Preiss J., Romero J. M. (2003) J. Biol. Chem. 278, 28508–28515 [DOI] [PubMed] [Google Scholar]
- 3.Doan D. N., Rudi H., Olsen O. A. (1999) Plant Physiol. 121, 965–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballicora M. A., Dubay J. R., Devillers C. H., Preiss J. (2005) J. Biol. Chem. 280, 10189–10195 [DOI] [PubMed] [Google Scholar]
- 5.Ventriglia T., Kuhn M. L., Ruiz M. T., Ribeiro-Pedro M., Valverde F., Ballicora M. A., Preiss J., Romero J. M. (2008) Plant Physiol. 148, 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cross J. M., Clancy M., Shaw J. R., Greene T. W., Schmidt R. R., Okita T. W., Hannah L. C. (2004) Plant Physiol. 135, 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang S. K., Hamada S., Okita T. W. (2006) FEBS Lett. 580, 6741–6748 [DOI] [PubMed] [Google Scholar]
- 8.Hwang S. K., Hamada S., Okita T. W. (2007) Phytochemistry 68, 464–477 [DOI] [PubMed] [Google Scholar]
- 9.Hwang S. K., Salamone P. R., Okita T. W. (2005) FEBS Lett. 579, 983–990 [DOI] [PubMed] [Google Scholar]
- 10.Crevillén P., Ventriglia T., Pinto F., Orea A., Mérida A., Romero J. M. (2005) J. Biol. Chem. 280, 8143–8149 [DOI] [PubMed] [Google Scholar]
- 11.Wang S. M., Lue W. L., Yu T. S., Long J. H., Wang C. N., Eimert K., Chen J. (1998) Plant J. 13, 63–70 [DOI] [PubMed] [Google Scholar]
- 12.Courties C., Perasso R., Chretiennot-Dinet M.-J., Gouy M., Guillou L., Troussellier M. (1998) J. Phycol. 34, 844–849 [Google Scholar]
- 13.Hicks G. R., Hironaka C. M., Dauvillee D., Funke R. P., D'Hulst C., Waffenschmidt S., Ball S. G. (2001) Plant Physiol. 127, 1334–1338 [PMC free article] [PubMed] [Google Scholar]
- 14.Piganeau G., Moreau H. (2007) Gene 406, 184–190 [DOI] [PubMed] [Google Scholar]
- 15.Misumi O., Yoshida Y., Nishida K., Fujiwara T., Sakajiri T., Hirooka S., Nishimura Y., Kuroiwa T. (2008) J. Plant Res. 121, 3–17 [DOI] [PubMed] [Google Scholar]
- 16.Derelle E., Ferraz C., Rombauts S., Rouzé P., Worden A. Z., Robbens S., Partensky F., Degroeve S., Echeynié S., Cooke R., Saeys Y., Wuyts J., Jabbari K., Bowler C., Panaud O., Piégu B., Ball S. G., Ral J. P., Bouget F. Y., Piganeau G., De Baets B., Picard A., Delseny M., Demaille J., Van de Peer Y., Moreau H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11647–11652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ral J. P., Derelle E., Ferraz C., Wattebled F., Farinas B., Corellou F., Buléon A., Slomianny M. C., Delvalle D., d'Hulst C., Rombauts S., Moreau H., Ball S. (2004) Plant Physiol. 136, 3333–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yep A., Bejar C. M., Ballicora M. A., Dubay J. R., Iglesias A. A., Preiss J. (2004) Anal. Biochem. 324, 52–59 [DOI] [PubMed] [Google Scholar]
- 19.Hoover D. M., Lubkowski J. (2002) Nucleic Acids Res. 30, e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong A. S., Yao Q. H., Peng R. H., Li X., Fan H. Q., Cheng Z. M., Li Y. (2004) Nucleic Acids Res. 32, e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bejar C. M., Ballicora M. A., Gómez-Casati D. F., Iglesias A. A., Preiss J. (2004) FEBS Lett. 573, 99–104 [DOI] [PubMed] [Google Scholar]
- 22.Iglesias A. A., Barry G. F., Meyer C., Bloksberg L., Nakata P. A., Greene T., Laughlin M. J., Okita T. W., Kishore G. M., Preiss J. (1993) J. Biol. Chem. 268, 1081–1086 [PubMed] [Google Scholar]
- 23.Sambrook J., Russell D. W. (2001) in Molecular Cloning: A Laboratory Manual (Argentine J. ed) 3rd Ed., pp. 13.36–13.39, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24.Gill S. C., von Hippel P. H. (1989) Anal. Biochem. 182, 319–326 [DOI] [PubMed] [Google Scholar]
- 25.Press W. H., Flannery B. P., Teukolsky S. A., Vetterling W. T. (1988) Numerical Recipes in C: The Art of Scientific Computing, pp. 347–383, Cambridge University Press, New York [Google Scholar]
- 26.Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 27.Jeanmougin F., Thompson J. D., Gouy M., Higgins D. G., Gibson T. J. (1998) Trends Biochem. Sci. 23, 403–405 [DOI] [PubMed] [Google Scholar]
- 28.Schmidt H. A., Strimmer K., Vingron M., von Haeseler A. (2002) Bioinformatics 18, 502–504 [DOI] [PubMed] [Google Scholar]
- 29.Müller T., Vingron M. (2000) J. Comput. Biol. 7, 761–776 [DOI] [PubMed] [Google Scholar]
- 30.Ballicora M. A., Iglesias A. A., Preiss J. (2004) Photosynth. Res. 79, 1–24 [DOI] [PubMed] [Google Scholar]
- 31.Frueauf J. B., Ballicora M. A., Preiss J. (2003) Plant J. 33, 503–511 [DOI] [PubMed] [Google Scholar]
- 32.Fu Y., Ballicora M. A., Preiss J. (1998) Plant Physiol. 117, 989–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bejar C. M., Jin X., Ballicora M. A., Preiss J. (2006) J. Biol. Chem. 281, 40473–40484 [DOI] [PubMed] [Google Scholar]
- 34.Hill M. A., Kaufmann K., Otero J., Preiss J. (1991) J. Biol. Chem. 266, 12455–12460 [PubMed] [Google Scholar]
- 35.Laughlin M. J., Payne J. W., Okita T. W. (1998) Phytochemistry 47, 621–629 [DOI] [PubMed] [Google Scholar]
- 36.Frueauf J. B., Ballicora M. A., Preiss J. (2001) J. Biol. Chem. 276, 46319–46325 [DOI] [PubMed] [Google Scholar]
- 37.Hwang S. K., Nagai Y., Kim D., Okita T. W. (2008) J. Biol. Chem. 283, 6640–6647 [DOI] [PubMed] [Google Scholar]
- 38.Ballicora M. A., Laughlin M. J., Fu Y., Okita T. W., Barry G. F., Preiss J. (1995) Plant Physiol. 109, 245–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iglesias A. A., Charng Y. Y., Ball S., Preiss J. (1994) Plant Physiol. 104, 1287–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boehlein S. K., Sewell A. K., Cross J., Stewart J. D., Hannah L. C. (2005) Plant Physiol. 138, 1552–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin X., Ballicora M. A., Preiss J., Geiger J. H. (2005) EMBO J. 24, 694–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L., Postlethwait J. (1999) Genetics 151, 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rastogi S., Liberles D. A. (2005) BMC Evol. Biol. 5, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haugen T. H., Preiss J. (1979) J. Biol. Chem. 254, 127–136 [PubMed] [Google Scholar]
- 45.Tiessen A., Hendriks J. H., Stitt M., Branscheid A., Gibon Y., Farré E. M., Geigenberger P. (2002) Plant Cell 14, 2191–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ballicora M. A., Frueauf J. B., Fu Y., Schürmann P., Preiss J. (2000) J. Biol. Chem. 275, 1315–1320 [DOI] [PubMed] [Google Scholar]
- 47.Fu Y., Ballicora M. A., Leykam J. F., Preiss J. (1998) J. Biol. Chem. 273, 25045–25052 [DOI] [PubMed] [Google Scholar]
- 48.Hendriks J. H., Kolbe A., Gibon Y., Stitt M., Geigenberger P. (2003) Plant Physiol. 133, 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lynch M. (2006) Mol. Biol. Evol. 23, 450–468 [DOI] [PubMed] [Google Scholar]
- 50.Tocchini-Valentini G. D., Fruscoloni P., Tocchini-Valentini G. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8933–8938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D'Ovidio R., Raiola A., Capodicasa C., Devoto A., Pontiggia D., Roberti S., Galletti R., Conti E., O'Sullivan D., De Lorenzo G. (2004) Plant Physiol. 135, 2424–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thornton J. W. (2004) Nat. Rev. Genet. 5, 366–375 [DOI] [PubMed] [Google Scholar]
- 53.Pils B., Schultz J. (2004) J. Mol. Biol. 340, 399–404 [DOI] [PubMed] [Google Scholar]
- 54.Todd A. E., Orengo C. A., Thornton J. M. (2002) Structure 10, 1435–1451 [DOI] [PubMed] [Google Scholar]
- 55.Truman A. W., Fan Q., Röttgen M., Stegmann E., Leadlay P. F., Spencer J. B. (2008) Chem. Biol. 15, 476–484 [DOI] [PubMed] [Google Scholar]
- 56.Iglesias A. A., Kakefuda G., Preiss J. (1991) Plant Physiol. 97, 1187–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.