Abstract
The CCAAT box is a DNA element present in the majority of human promoters, bound by the trimeric NF-Y, composed of NF-YA, NF-YB, and NF-YC subunits. We describe and characterize novel isoforms of one of the two histone-like subunits, NF-YC. The locus generates a minimum of four splicing products, mainly located within the Q-rich activation domain. The abundance of each isoform is cell-dependent; only one major NF-YC isoform is present in a given cell type. The 37- and 50-kDa isoforms are mutually exclusive, and preferential pairings with NF-YA isoforms possess different transcriptional activities, with specific combinations being more active on selected promoters. The transcriptional regulation of the NF-YC locus is also complex, and mRNAs arise from the two promoters P1 and P2. Transient transfections, chromatin immunoprecipitations, and reverse transcription-PCRs indicate that P1 has a robust housekeeping activity; P2 possesses a lower basal activity, but it is induced in response to DNA damage in a p53-dependent way. Alternative promoter usage directly affects NF-YC splicing, with the 50-kDa transcript being excluded from P2. Specific functional inactivation of the 37-kDa isoform affects the basal levels of G1/S blocking and pro-apoptotic genes but not G2/M promoters. In summary, our data highlight an unexpected degree of complexity and regulation of the NF-YC gene, demonstrating the existence of a discrete cohort of NF-Y trimer subtypes resulting from the functional diversification of Q-rich transactivating subunits and a specific role of the 37-kDa isoform in suppression of the DNA damage-response under growing conditions.
Introduction
Promoters and enhancers that activate RNA polymerase II transcription are composed of a combinatorial puzzle of short DNA elements recognized by sequence-specific regulators. Among these elements, the CCAAT box is known to be one of the most frequent. This has been illustrated by several bioinformatic studies of large sets of eukaryotic promoters (1–7) and more recently by ChIP3 on chip experiments on different platforms (8–10). In particular, pathways such as cell cycle and endoplasmic reticulum stress response were found to be enriched for CCAAT-containing promoters (2, 11, 12).
Several types of evidence indicate that the histone-like NF-Y, also termed CBF and HAP2/3/5 in yeast, is the CCAAT regulator (13, 14). NF-Y is a ubiquitous heteromeric transcription factor (TF) composed of three subunits, NF-YA, NF-YB, and NF-YC, all necessary for DNA binding (15). NF-YB and NF-YC contain a conserved histone-fold motif (HFM) similar to H2B and H2A (16). The HFM-dependent NF-YB/NF-YC dimerization offers docking sites for NF-YA association. Only the resulting trimer contacts DNA with sequence-specific interactions, mainly via NF-YA as well as nonsequence-specific contacts, through the basic residues in the L1-L2 loops of NF-YB and NF-YC (17). NF-YA and NF-YC also contain a Q-rich domain, with a high density of Gln and hydrophobic residues, essential for NF-Y transactivation functions (18–20). NF-Y binding to the CCAAT box has long been considered an almost exclusive promoter event, with NF-Y acting as a crucial promoter organizer involved in the recruitment of polymerase II and of neighboring TFs (21–23). Intriguingly, recent location analysis experiments (ChIP on chip) performed both on CpG islands and chromosome 20, 21, and 22 tiling arrays revealed that the in vivo landscape of NF-Y binding is far more complex than expected. (i) Most NF-Y-binding sites are far from canonical promoter regions. (ii) NF-Y is associated with areas containing negative histone marks, such as H4K20me3 and H3K27me3. Indeed NF-Y acts as a bi-functional TF, directly activating or repressing transcription depending on the cellular and/or chromatin contexts (8, 24).
Increasing genome-wide studies support the idea that most mammalian genes generate alternative transcripts as a source of protein functional diversification. In this context, the use of alternative promoters (AP) has recently emerged as a hallmark of most human genes (25, 26). Most of them also undergo alternative splicing (AS) with exon skipping being the most frequent AS type, generating at least three different transcript variants per locus (27–29). The combinatorial usage of alternative promoter and AS further increases the potential complexity arising from a single gene, and interestingly, the alternative promoter was shown to directly affect AS genome-wide (30).
We have previously reported the presence of two major NF-YA isoforms, “long” and “short,” with further micro-heterogeneity present in a splicing acceptor site (31); the short isoform originates from AS and lacks a 28-amino acid encoding exon within the NF-YA Q-rich domain. Interestingly, the relative abundance of the two isoforms varies in different cell types, and a switch in their relative levels was recently noticed upon differentiation of mouse embryonic stem cells (6). In particular, the levels of short NF-YA are high in embryonic stem cells and decrease during differentiation of embryonal bodies. The same variant is also specifically required for stem maintenance in the hematopoietic system (32), supporting the idea of a functional specification of single NF-YA isoforms. Whether this functional specification also affects other NF-Y subunits remains to be established.
We originally cloned the human NF-YC cDNA from a HeLa library, as a 1480-bp sequence coding for a 335-amino acid polypeptide with a predicted mass of 37 kDa (33). A highly homologous cDNA had been previously cloned from a rat brain cDNA library (34). Interestingly, Northern blot experiments revealed the tissue-specific presence of different NF-YC mRNAs, both in human and mouse, suggesting that the cloned cDNA might only represent the most abundant NF-YC isoform present in the starting library (33, 35). The presence of a shorter NF-YC isoform, lacking 62 amino acids within helices αN and α3 of the HFM, has been reported by Chen and co-workers (36). This isoform, named NF-YCb, is mainly cytoplasmic and, because of the partial lack of the HFM, is not expected to be part of a functional NF-Y trimer. NF-YCb indeed directly targets Smad2 and Smad3, negatively regulating transforming growth factor-β signaling, rather than affecting NF-Y-mediated transcription (36). By combining bioinformatic and molecular biology techniques, we now detail the complexity of the human NF-YC locus.
EXPERIMENTAL PROCEDURES
Bioinformatic Analyses
The University of California Santa Cruz Genome Browser was used for initial analyses of human and mouse NF-YC loci. Sequence alignments were performed with MUSCLE, splice site prediction with ASPIC, and TFs consensus search with MOTIF.
Cell Cultures and Treatments
HaCat, HCT116, HCT116/E6, HepG2, and U2OS cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C in a humidified 5% CO2 atmosphere. DU145, PC3, B-Jab, HeLa S3, HT1080, Nalm-6, T98G, U937, and K562 cell lines were maintained in RPMI 1640 medium under the same conditions. HCT116/E6 (kindly provided by Dr. B. Vogelstein, The Johns Hopkins University, Baltimore) were grown in the presence of 0.5 mg/ml G418 (Sigma) to maintain the selection for the E6 construct. Doxorubicin (Sigma) was added to exponentially growing HCT116 and HCT116/E6 cells to a final concentration of 1 μm and incubated for various time points. When HCT116/E6 cells were used, G418 selection was removed just before the addiction of doxorubicin.
Cloning and Transfections
NF-YC P1 and P2 regions were amplified from HCT116 genomic DNA and cloned in the pGL3-Basic vector (Promega). PCR was used to introduce flanking KpnI and HindIII sites for the cloned P1 region (1716 bp) and flanking BglII and HindIII sites for the cloned P2 region (819 bp).
The C-terminal portion of the NF-YC isoforms was amplified from HCT116 cDNA, with primers mapping within the coding exon 4 and the 3′UTR exon 10. PCR products were subcloned, ClaI/BamHI, in the previously described pSG5-based Δ4NF-YC eukaryotic expression vector (20). Final constructs were sequence-verified. All PCR primers used for cloning experiments are reported in supplemental Table S1. Transfection experiments were performed in triplicate. For each point, 150,000 HCT116 cells were seeded in 24-well plates and transfected after 24 h with a transfection kit (GeneSpin). 100 ng of pCMV β-galactosidase and 500 ng of luciferase constructs were used for each transfection. Cells were harvested 36 h post-transfection, and β-galactosidase and luciferase assays were performed according to standard procedures. Luciferase reporters used in this paper had been described previously (41, 48).
Chromatin Immunoprecipitation
Chromatin immunoprecipitations were performed essentially as described previously (10). Briefly, exponentially growing HCT116 cells were washed with phosphate-buffered saline and incubated for 10 min with 1% formaldehyde. The cross-linked material was broken with a Dounce pestle, and chromatin-containing pellets were sonicated to get fragments of an average length of 500 bp. 5 × 106 cell eq were immunoprecipitated for 4 h at 4 °C with 5 μg of anti-FLAG (Sigma), anti-NF-YB, anti-H3 (Abcam), anti-H3K4me3 (Abcam), anti-H3K79me2, anti-H3K9–14ac (Upstate), anti-p53 (Ab7, Calbiochem), and anti-p53 antibodies (DO1, GeneSpin). Semi-quantitative PCRs were performed in the linear range of each amplification product; ChIP-PCR primers used for these experiments are listed in supplemental Table S1.
RNA and RT-PCRs
Total RNA was extracted from 5 × 106 exponentially growing cells, by using RNeasy® mini kit (Qiagen) according to the manufacturer's instructions. 2 μg of RNA were retrotranscribed with SuperScriptTM reverse transcriptase (Invitrogen) and random primers. Semi-quantitative PCRs were performed in the linear range of each amplification product, after normalizing all the cDNAs for ACTAB content. RT-PCR primers used for these experiments are listed in supplemental Table S1.
Cell Extracts and Western Blot Analysis
Nuclear extracts were prepared from each cell line by collecting 3 × 107 cells in ice-cold hypotonic Buffer A (10 mm Hepes, pH 7.9, 10 mm KCl, 0,1 mm EDTA, 0,1 mm EGTA, 1 mm dithiothreitol, protease inhibitors mixture (Sigma), 1 mm phenylmethylsulfonyl fluoride) and incubating on ice for 30 min. Nonidet P-40 was added to a final concentration of 0.625%; cells were vortexed and centrifuged at 13,000 rpm for 5 min. Pelletted nuclei were resuspended in ice-cold Buffer C (20 mm Hepes, pH 7.9, 1 m NaCl, 0,1 mm EDTA, 0,1 mm EGTA, 1 mm dithiothreitol, protease inhibitor mixture, 1 mm phenylmethylsulfonyl fluoride) and rocked at 4 °C for 30 min. Nuclear extracts were then dialyzed against the same buffer, to 150 mm final NaCl. 15 μg of each nuclear extract was loaded on a 12% SDS-polyacrylamide gel, transferred onto a nitrocellulose membrane, and immunoblotted with the following antibodies: anti-NF-YA (Santa Cruz Biotechnology), anti-NF-YB, anti-NF-YC (GeneSpin), and anti-laminin B (Sigma). The protein-antibody complexes were detected using horseradish peroxidase-conjugated secondary antibodies and the protein detection system (GeneSpin).
shRNA Infections
pLKO.1 nontarget shRNA (SHC002) and shRNA-NF-YC targeting exons 8–9 (CCACCAATGGCTCAACAGATTA) were designed by Sigma. For lentivirus production, the shRNA vector plasmid (20 μg) and second generation packaging plasmids (5 μg of pMD2-VSVG and 15 μg of pCMVΔR8.91) were cotransfected into HEK293T cells. Lentivirus-containing supernatant was collected 24 h after transfection, centrifuged at 3000 rpm to remove cell debris for 5 min, 0.45-μm-filtered, and frozen at −80 °C until use. HCT116 cells were infected by spinoculation (1800 rpm for 45 min at 30 °C) at a multiplicity of infection of ∼2 (titer determined by transduction of HEK293T cells). 48 h post-infection cells were harvested, and RNA was extracted.
RESULTS
Identification of New NF-YC Splicing Isoforms
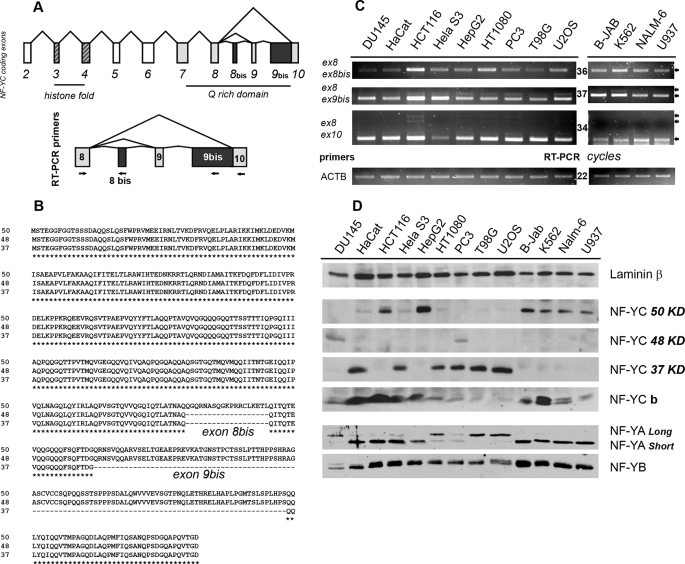
Two types of evidence prompted us to look for new NF-YC isoforms. The first was the reported presence of NF-YCb (36). The second was that performing routine Western blots with our highly specific anti-NF-YC antibody, we often observed bands in addition to the canonical 37-kDa band. Initially, we were reluctant to consider them anything more than antibody cross-reactions, until we realized that in some cell lines the bands were manifestly dominant. The human NF-YC locus maps to chromosome 1, p34.2, and the corresponding open reading frame is composed of 10 annotated exons. The 1st exon contains the 5′UTR, whereas the remaining exons are mostly coding. The ATG is at the beginning of exon 2, and exon 10 contains the Stop codon and a long 3′UTR. The genomic structure of the NF-YC gene is modular in terms of functional domains, with exons 3 and 4 coding the HFM and the region spanning exons 7–10 of the Q-rich transactivation domain.
To identify possible NF-YC splicing isoforms, we aligned 13 human NF-YC coding mRNAs present in the GenBankTM data base and compared them with the exonic structure of the annotated NF-YC mRNA (supplemental Fig. 1). We decided to pursue our analyses only on sequences that contained both the first and the last coding exons of the annotated NF-YC, and that missed the entire annotated exons or contained extra coding sequences, possibly indicative of alternative exons skipped in the originally reported NF-YC cDNAs. With this approach, we identified three nonredundant NF-YC splicing isoforms as follows: the canonical 1005-bp CDS, producing the 37-kDa protein, and two new coding sequences of 1317 and 1374 bp potentially generating polypeptides of 48- and 50-kDa, respectively. These new transcripts contain an extra 312-bp exon downstream of exon 9 (hereafter named exon 9bis), and the longer mRNA also contains an additional exon (57 bp) downstream to exon 8 (hereafter named exon 8bis) (Fig. 1, A and B). The occurrence of spliced messengers containing exons 8bis and 9bis was confirmed by splice site detection and full-length transcript modeling using ASPIC (37) (data not shown).
FIGURE 1.
Identification of new NF-YC splicing isoforms. A, upper panel, genomic structure of the NF-YC coding sequence. Exons coding for the HFM domain are striped; the ones coding for the Q-rich domain are gray, and the newly identified exon 8bis and 9bis are black. Lower panel, primers used for RT-PCR experiments are mapped within the NF-YC C terminus. B, sequence alignments of the 37-, 48-, and 50-kDa polypeptides generated by the in silico identified NF-YC mRNAs. C, semi-quantitative RT-PCRs were performed with the indicated NF-YC primer pairs, after normalizing the 13 human cDNA for β-actin content (lower panel). Cell lines are indicated on the top. Ex8-Ex10 PCR product corresponds to the 37-kDa isoform, Ex8-Ex9bis to the 48-kDa isoform, and Ex8-Ex8bis to the 50-kDa NF-YC. RT-PCRs were performed in the linear range of amplification; number of cycles for each product is indicated in the middle. D, Western blots analysis performed with anti NF-YA, NF-YB, and NF-YC antibodies, on nuclear extract derived from the same 13 cell lines used for RT-PCRs. β-Laminin was used as an internal control for nuclear extracts loading (upper panel).
To assess for the presence of different NF-YC splicing mRNAs in vivo, we performed RT-PCR experiments with RNAs from 13 human cell lines, using a common forward primer mapping within exon 8, shared by all isoforms, and isoform-specific reverse primers mapping within exons 8bis, 9bis, and 10 (Fig. 1A). β-Actin was used as internal control. As shown in Fig. 1C, the three NF-YC mRNAs are expressed in all the cell lines, with cell type-specific differences in the relative levels; the shorter mRNA (8–10 pairs), coding for the 37-kDa NF-YC, is more abundant, being detectable 2–3 PCR cycles earlier in all the cell lines tested. Finally, pushing the PCRs further yielded several additional products with exon 8–10 primers (supplemental Fig. 2), possibly suggesting the presence of other NF-YC isoforms missed during the initial bioinformatic “filtering.” RT-PCR experiments performed with exon 2–5 primers surrounding the HFM region also confirmed the presence of two splicing isoforms within this region, as previously reported, confirming the reliability of our approach (data not shown) (36).
We next asked whether the transcriptional complexity of the human NF-YC locus was maintained at the protein level. We prepared nuclear extracts from the same human cell lines and performed Western blots with antibodies specific for the three NF-Y subunits; an anti-laminin B antibody was used as internal control for nuclear extracts loading. As shown in Fig. 1D, at least three NF-YC isoforms are present, with molecular weights consistent with predictions; bands of 48 and 50 kDa were observed, in addition to the expected 37 kDa of NF-YC. The previously reported NF-YCb isoform is also present. Upon longer exposures, additional intermediate bands also emerged, supporting our previous RT-PCR findings (supplemental Fig. 3). Interestingly, the levels of the different NF-YCs are highly variable among cell lines; HaCat, HeLa S3, HT1080, PC3, T98G, and U2OS express the canonical 37-kDa NF-YC; HCT116, HepG2, B-JAB, K562, NALM-6 and U937 are essentially devoid of the latter but significantly express the 50-kDa protein. Indeed, much to our surprise, the 37- and 50-kDa isoforms are almost mutually exclusive. Only two cell lines, DU145 and PC3, express significant levels of the 48-kDa isoform. Overall, only one major NF-YC isoform is present in a given cell type, a finding that does not match the more ubiquitous presence of NF-YC transcripts, implying post-transcriptional mechanisms affecting NF-YC mRNAs stability or translation.
Finally, we noticed striking pair wise preferences between NF-YC and NF-YA isoforms: cells harboring the 50-kDa NF-YC preferentially express the short NF-YA isoform, whereas the long NF-YA is enriched in cells containing the 37-kDa NF-YC (Fig. 1D). The levels of the other HFM subunit, NF-YB, are almost constant. Taken together, these data demonstrate the existence of a discrete cohort of NF-Y trimer subtypes, whose presence is dependent on the cellular context.
Multiple Promoters at the Human NF-YC Locus
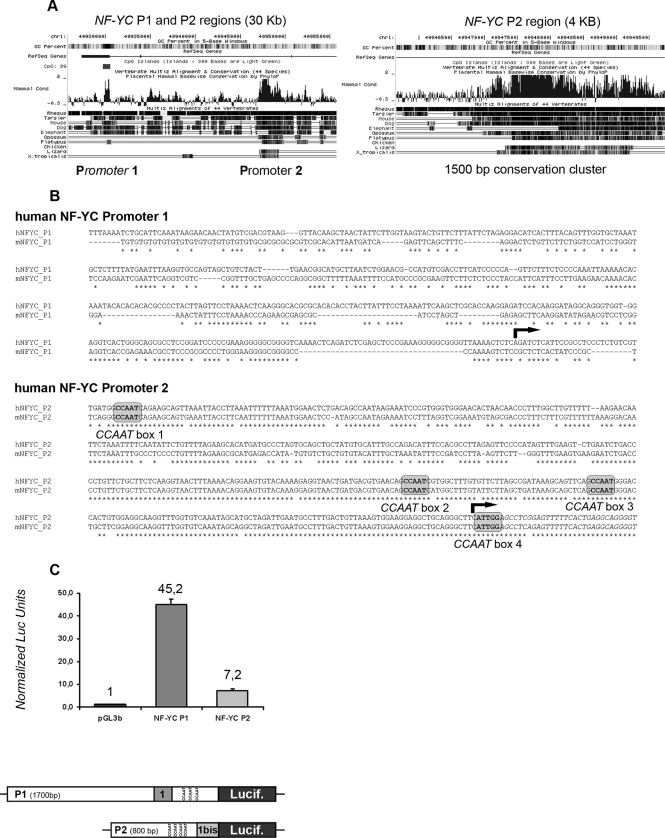
Recent genome-wide analyses suggest that genes undergoing alternative splicing often contain multiple promoters, with alternative promoters directly affecting the resulting splicing pattern (30). This might be the case of the human NF-YC locus. By sequence inspection, we identified a hot spot of vertebrate sequence conservation within the first intron of the human NF-YC gene (Fig. 2A). A human mRNA arising from this region is present within the GenBankTM sequence data base (AK000346). When compared with the annotated NF-YC, this mRNA contains a mutually exclusive 5′UTR 1st exon, hereafter named exon Ibis. It is therefore possible that the NF-YC locus contains an alternative promoter (P2) downstream to the annotated promoter (P1); we aligned human and mouse P2 regions in a window of ±500 bp surrounding exon Ibis and found four canonical CCAAT boxes in the core P2 region and a TATA box embedded in the exon Ibis sequence. The P1 region is TATA-less and possesses overall lower sequence conservation (Fig. 2B). However, three well conserved CCAAT boxes are present within the first intron, downstream to the annotated P1 transcription start site (supplemental Fig. 4). To verify whether the P1 and P2 regions possess promoter activities, we cloned them into the pGL3 basic/luciferase vector. As shown in Fig. 2C, when the two reporter constructs were transiently transfected in HCT116 cells, both regions had significant activity over the pGL3basic empty vector, with P1 being clearly more active than P2 in triplicate experiments. These data clearly indicate that the NF-YC locus contains two promoters as follows: P1 in the canonical position and P2 within the first intron downstream to P1.
FIGURE 2.
Characterization of human NF-YC promoters. A, University of California Santa Cruz genomic view of human NF-YC promoters region (30-kb window, left plot). A 4-kb zoom of the P2 region is also shown (right plot). B, sequence alignments of human and mouse P1 and P2 regions, 500 bp upstream of the transcription start site (arrow). P2 CCAAT boxes are shaded gray. C, basal activity of P1 and P2 regions in HCT116 cells. 500 ng of each reporter construct were cotransfected with 100 ng of pCMV β-galactosidase, and normalized luciferase (Lucif) units were calculated 36 h after transfection. The structure of NF-YC P1 and P2 reporter constructs is also depicted (bottom panel).
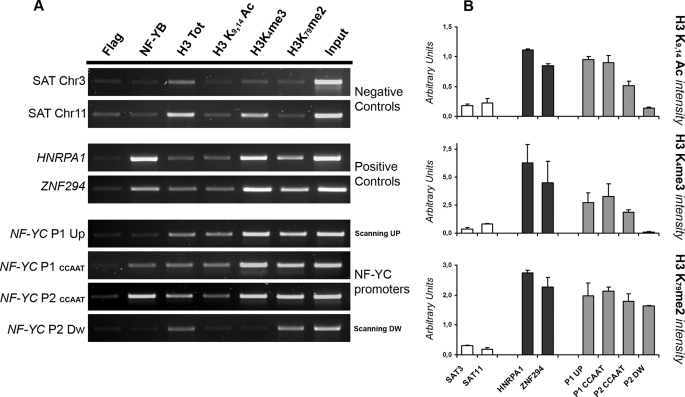
To further characterize P1 and P2 in vivo, we performed chromatin immunoprecipitation experiments in HCT116 cells, with antibodies against histone post-translational modifications that are hallmarks of active chromatin environments, as well as with NF-Y antibody, because of the presence of conserved CCAAT sequences. Fig. 3A shows that NF-Y is bound to both P1 and P2 in vivo, with P2 showing the highest NF-Y enrichment. Two bona fide NF-Y active target genes, HNRPA1 and ZNF29, served as positive controls (8) and heterochromatic satellite sequences as negative ones. Moreover, both promoters are enriched for H3K4me3 and H3K9–14ac, and H3K79me2; the formers mark the promoter region of active genes, whereas H3K79me2 is usually scattered along a transcribed area (38–40). ChIP PCR data were quantified and normalized versus the total unmodified H3. Histone mark plots indicate that P1 possesses a more robust euchromatic environment, suggesting a higher promoter activity under the conditions tested (Fig. 3B). Further promoter scanning upstream of P1 and downstream of P2 confirmed the specificity of our ChIPs because of the following: (i) no NF-Y binding is detected far from core promoters (Fig. 3A, NF-YC P1 upper and NF-YC P2 lower panels), and (ii) only H3K79me2 is present downstream of the P2 region, whereas H3K4me3 and H3K9–14 Ac are absent from the body of the gene (Fig. 3A, NF-YC P2 lower panel).
FIGURE 3.
Analysis of NF-YC promoters chromatin environment. A, ChIP analysis and semi-quantitative PCR were performed for the indicated promoters (on the left of each panel) in HCT116 cells, with anti-NF-YB, anti-H3, anti H3K9, K14Ac, anti-H3K4me3, and anti-H3K79me2 antibodies. The anti-FLAG antibody was used as a negative control. ChIP PCRs were performed in the linear range for each amplification products. B, data obtained with the semi-quantitative PCRs were quantified using the ImageJ software. Active histone marks intensity was scored against the level of total unmodified H3 and plotted as arbitrary units. Error bars refer to two independent PCR analyses.
P2 Promoter Is Regulated and Affects Alternative Splicing
The RT-PCR and Western blots described in Fig. 1, C and D, demonstrate a diversification of NF-YC isoforms and of the resulting NF-Y trimers depending on the cellular context. We thus wondered whether the activities of the two NF-YC promoters is also modulated in a cell type-dependent way and if promoter choice can directly affect alternative splicing.
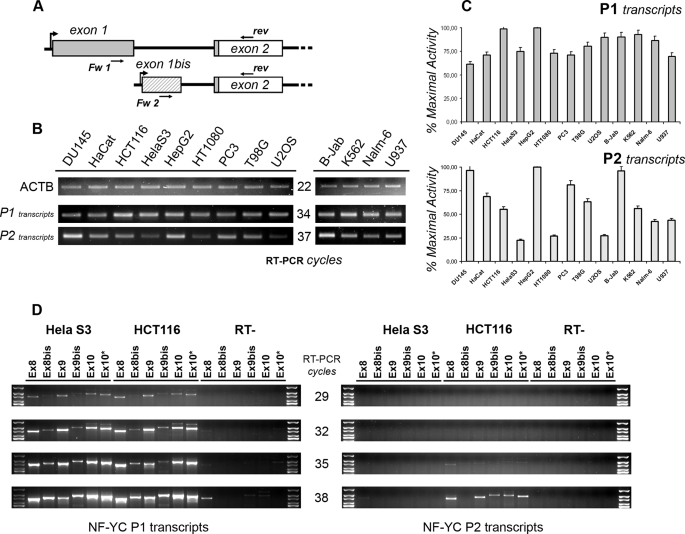
To address the first question, we performed RT-PCRs with RNAs from the 13 human cell lines described above, using two different forward primers specific for P1 (exon 1) or P2 (exon 1bis) and a common reverse primer mapping within exon 2, thus measuring promoters activities in terms of mRNA transcripts specifically arising from one of them (Fig. 4A). The P1 transcripts are generally more abundant, being detectable 3–4 PCR cycles before transcripts arising from P2. With few exceptions, the P1 transcripts levels are similar in abundance, whereas P2 transcripts show variations among the different cell lines (Fig. 4B). Normalization of the relative abundance of each transcript versus the level of the cell line possessing the maximal promoter activity (HepG2 cells for both P1 and P2) confirmed that the variation of P2 transcripts is greater, suggesting that P2 promoter might be a target of specific regulatory events (Fig. 4C).
FIGURE 4.
NF-YC P2 is highly regulated and affects alternative splicing. A, scheme showing the structure of P1- (exon 1, gray) and P2 (exon 1bis, striped)-specific 5′UTR. The ATG is present at the beginning of exon 2, and the CDS is depicted in white. Primers used for “promoter” RT-PCR are also mapped. B, semi-quantitative RT-PCRs were performed to assess for P1 and P2 activity after normalizing the 13 human cDNAs for ACTAB content (upper panel). RT-PCRs were performed in the linear range of amplification; the number of cycles for each product is indicated in the middle. C, semi-quantitative RT-PCRs were quantified using the ImageJ software. The relative abundance of each transcript type was normalized versus the level of HepG2 cells. Error bars refer to two PCR cycles considered for the analysis. D, semi-quantitative RT-PCRs were performed with P1- (left plot) and P2 (right plot)-specific primers and a pool of reverse primers mapping within NF-YC Q-rich domain. RT-PCR cycles are indicated in the middle. Reverse primers identify the following NF-YC subtypes: Ex8 = total NF-YC; Ex 8bis = 50-kDa NF-YC; Ex9 = 37-kDa NF-YC; Ex9bis = 48-kDa NF-YC; Ex10 = 37 kDa + longer NF-YCs.
Interestingly, we noticed that HeLa S3 cells possess the lowest P2 activity, and the NF-YC 37-kDa mRNA is under-represented in this cell line (Fig. 1C), possibly linking P2 with a specific transcript. To verify whether the choice of one of the two promoters is per se sufficient to affect NF-YC splicing events, we performed RT-PCR experiments using the same forward primers described above, P1 (exon 1) or P2 (exon 1bis), and a set of reverse primers mapping within the different exons coding for the C-terminal part of NF-YC isoforms (Exon 8, 8bis, 9, 9bis, and 2 different primers mapping within exon 10). As shown in Fig. 4D (left panel), the mRNA patterns arising from P1 is essentially identical between HCT116 and HeLa cells; all three transcript types are present, with the mRNAs generating the 37-kDa protein being detectable already at 29 cycles. The picture emerging for P2 transcription is very different (Fig. 4D, right panel); the activity of P2 is clearly reduced in both cell lines when compared with P1, with no transcript being detectable before 35 PCR cycles. Transcripts specifically arising from P2 are present in HCT116 cells (38 RT-PCR cycles and more), whereas the same promoter is almost inactive in HeLa S3 cells. Moreover, the transcript corresponding to the 50-kDa NF-YC is specifically excluded from P2 transcription, indicating that P2 acts as an isoform-specific promoter, at least in this cell line (Fig. 4D, Ex 8bis reverse primer, compare left (P1) and right (P2) panels). In summary, we conclude that the P2 is highly regulated, and the P2 choice directly affects NF-YC splicing.
NF-YC Isoforms Are Functionally Different
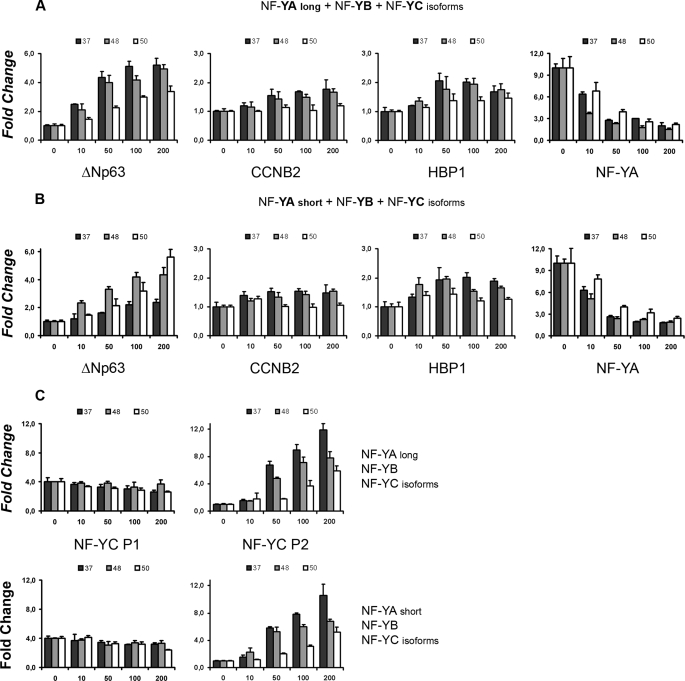
The newly identified 48- and 50-kDa NF-YC isoforms contain one or two additional exons within the NF-YC C-terminal Q-rich domain, which is important for NF-Y transactivation. Exon 8bis and Exon 9bis are not Q-rich, with the latter coding for a stretch of 104 amino acids rich in Ser/Thr (25%). We thus wondered whether NF-YC isoforms behave differently in terms of transcriptional activity. To answer this question, we cloned them in eukaryotic expression vectors and performed luciferase transactivation assays with three bona fide NF-Y target promoters as follows: ΔNp63, CCNB2, and HBP1. Each NF-YC isoforms was co-transfected in HCT116 cells together with NF-YA, either long or short, and NF-YB. Interestingly, the 37-kDa isoform is clearly more potent in transactivating the ΔNp63 promoter combined with long NF-YA, whereas the 50-kDa NF-YC teams up better with the short NF-YA isoform (Fig. 5, A and B, left panel), mirroring the preferential pairing assessed by Western blots. The 48-kDa subunit is always intermediate between the two configurations. Transactivation of the CCNB2 and HBP1 is minimal, as reported previously (41). This is true for all the possible NF-Y timers (Fig. 5, A and B, middle panels). We next assessed NF-YC repressive functions on the NF-YA promoter. As shown in Fig. 6, A and B (right panels), this promoter is strongly repressed by all the trimeric combinations, and only minor differences are present among different NF-YCs. Finally, because the NF-YC P1 and P2 regions contain CCAAT boxes, we performed transfection experiments with these constructs as well; none of the NF-Y combinations activate P1 (less than 1.5-fold reduction at the highest NF-Y dose, most probably due to squelching effects), whereas the 37-kDa NF-YC is a powerful P2 transactivator (about 12 times), with the 50-kDa isoform being about 50% less active, in combination with both NF-YA long and short. We conclude that the new NF-YC isoforms possess different transcriptional activities, and specific NF-YA/NF-YC configurations are required for optimal activation or repression of selected promoters.
FIGURE 5.
NF-YC isoforms are functionally different. A, increasing amounts (0–200 ng) of long NF-YA, NF-YB, and NF-YC isoforms were cotransfected in HCT116 cells along with ΔNp63, CCNB2, HBP1, and NF-YA luciferase reporters. β-Galactosidase-normalized luciferase units were scored 36 h post-transfection, and fold changes were calculated versus the basal activity of each promoter. B, same as in A, but the short NF-YA isoform was used. C, CCAAT-containing NF-YC P1 and NF-YC P2 reporter constructs were used in transfection experiments with the same layout described in A: long NF-YA, NF-YB, and NF-YC isoforms (upper plots); short NF-YA, NF-YB, and NF-YC isoforms (lower plots).
FIGURE 6.
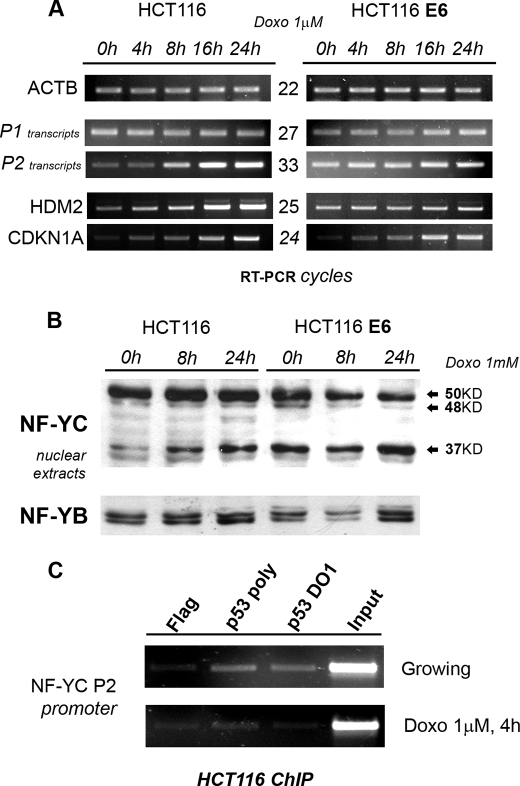
NF-YC P2 and 37-kDa NF-YC are induced by DNA damage. A, HCT116 and HCT116/E6 were treated with doxorubicin (Doxo) (1 μm) for the indicated time points, and semi-quantitative RT-PCR analyses were performed for the indicated genes, after normalizing samples for β-actin content (upper panel). RT-PCRs were performed in the linear range of amplification; number of cycles for each product is indicated in the middle. B, Western blot analysis was performed with anti-NF-YC antibody on nuclear extracts from HCT116 and HCT116/E6 treated with 1 μm doxorubicin for the indicated time points. NF-YB was used as an internal control for nuclear extracts loading (lower panel). C, ChIP analysis and semi-quantitative PCRs were performed for NF-YC P2 promoter in HCT116 cells, before and 4 h after 1 μm doxorubicin, with anti-p53 Ab7 (polyclonal) and anti-p53 DO1 (monoclonal) antibodies. The anti-FLAG antibody was used as a negative control. ChIP PCRs were performed in the linear range for each amplification product.
P2 Is Activated and NF-YC 37-kDa Is Induced upon DNA Damage
NF-Y-mediated transcription is involved in cell cycle control and response to checkpoint activation (10, 41); we thus assessed P1 and P2 activities in response to DNA damage by treating HCT116 and the p53-inactivated HCT116/E6 cells with doxorubicin for different time points. As shown in Fig. 6A, RT-PCR shows that P2 transcripts are strongly induced following doxorubicin treatment, with kinetics that essentially mimics that of HDM2 and CDKN1A, two bona fide p53 target genes. P2 activation is clearly p53-dependent, because high basal levels, and no induction, are observed in HCT116/E6 cells. On the other hand, P1 activity is unaffected following DNA damage. We next asked if P2 activation per se is sufficient to affect, globally or specifically, NF-YC isoform levels. Western blot experiments in the two cell lines indicate a strong induction of the 37-kDa isoform, whereas the 48- and 50-kDa isoforms are essentially unaffected. The 37-kDa NF-YC accumulation is also p53-dependent, as protein levels of this isoform are high in basal conditions and unaffected in HCT116/E6 cells (Fig. 6B, compare left and right parts). The P2 promoter region contains multiple CCAAT boxes that could recruit p53 through NF-Y; to address this possibility, we performed ChIPs in HCT116 cells, with anti-p53 antibodies and a control FLAG antibody, before and after DNA damage. As shown in Fig. 6C, p53 is indeed bound to the P2 region in growing conditions, whereas binding is lost following drug treatment. Because P2 basal activity and 37-kDa isoform levels are higher in HCT116/E6 (Fig. 6B, compare 1st and 4th lanes), these results suggest that the NF-YC P2 promoter is under negative regulation by p53, most probably through direct binding to conserved promoter elements, including the CCAAT boxes.
Functional Inactivation of the 37-kDa Isoforms
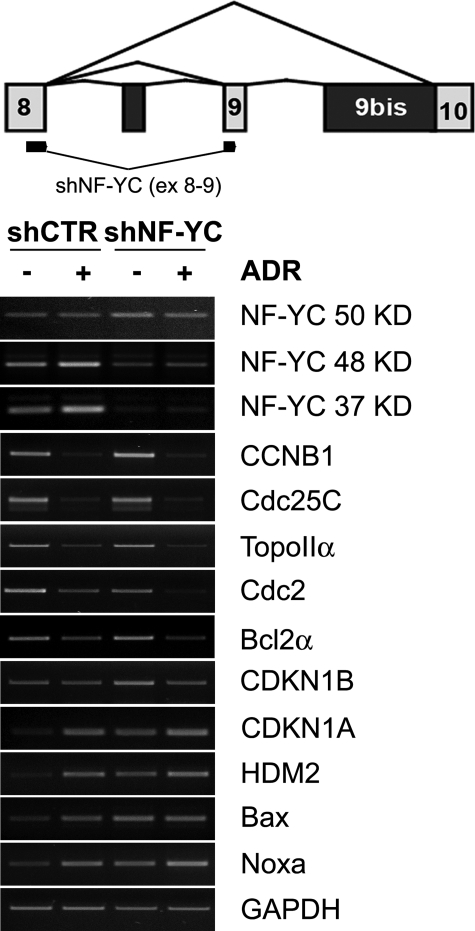
The DNA damage induction of the 37-kDa isoform prompted us to selectively inactivate it with an shRNA that maps across exons 8 and 9. We infected HCT116 cells with lentiviruses expressing a control and an NF-YC-37 shRNA, in the presence or absence of doxorubicin. We performed semi-quantitative RT-PCR analysis of several genes under NF-Y control, 48-h post-infections. Fig. 7 shows that indeed the mRNAs for the 37-kDa isoforms and, to a lesser degree, the 48-kDa isoforms are affected, whereas the mRNA of the 50-kDa isoform, if anything, is slightly increased. In particular, the DNA damage induction of the 37-kDa isoform is completely abolished. The G2/M genes CCNB1, CDC25C, TOPOIIA, and CDC2, all under NF-Y control, are not affected in growing conditions and repressed after DNA damage, with a particularly strong effect on CDC2. The same is also true for the anti-apoptotic BCL2A. On the other hand, elimination of the 37-kDa subunit profoundly affects CDKN1A, HDM2, and the pro-apoptotic BAX and NOXA, substantially increasing the basal levels of these genes, to the point where no further induction is observed upon doxorubicin addition. More modest effects are observed on CDK1B. These results indicate an important functional diversification, with possible counteracting activities, within the 37- and 50-kDa NF-YC isoforms, in terms of regulation of genes that control cell cycle progression and apoptosis.
FIGURE 7.
Functional inactivation of the 37-kDa isoform. The upper panel indicates the position in exon 8 and exon 9 of the shRNA used to functionally inactivate the 37-kDa isoform. HCT116 were infected with lentiviruses producing control and shRNA37, and treated with doxorubicin (1 μm) for 8 h. Semi-quantitative RT-PCR analysis was performed for the indicated genes, after normalizing samples for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (lower panel) and β-actin (data not shown). RT-PCRs were performed in the linear range of amplification.
DISCUSSION
In this study, similarly to what was previously reported for NF-YA (31), we uncover a high and unexpected complexity of the human NF-YC locus both in terms of alternative promoter usage and splicing isoforms. Interestingly, multiple genes encoding NF-Y subunits have been identified in plant genomes (42). Arabidopsis thaliana contains 10 NF-YA, 9 NF-YB, and 10 NF-YC genes. The expression pattern of most subunits is tissue-specific, with the resulting trimers specifically regulating processes as diverse as flowering, embryo maturation, and meristem development (43). Most eukaryotic genomes however, including human and mouse, only possess one gene encoding for each NF-Y subunit; deletion of NF-YA alleles indeed results in early lethality in mouse (44). One way to explain this apparently simple genomic structure could be the presence of alternative splicing events generating NF-Y variants. Recent analyses suggest that the vast majority of multiple exon genes in the human genome are subject to alternative promoter usage and alternative splicing. This has the potential to expand the proteome exponentially, and alternatively spliced exons that encode as few as one or two amino acids can modify protein function. Alternative splicing events have been reported to often be unique to specific cell types, and some evidence supports the idea of brain and testis being the tissues that undergo most alternative promoters and alternative splicing events (29, 45). NF-Y derives transcriptional activities from functional subdomains present within NF-YA and NF-YC. Two major NF-YA splicing isoforms are present and possess differences both in terms of transcriptional potential and developmental function. Microheterogeneity of the splicing acceptor site impacts the capacity to synergize with Sp1 (46), and Grskovic et al. (6) reported a switch in the relative levels of NF-YA isoforms upon differentiation of embryonic stem cells, with the NF-YA short being significantly down-regulated after commitment through differentiation of embryonal bodies. The overexpression of the same isoform, lacking an alternate exon within the NF-YA transactivation “Q-rich” domain, activates multiple genetic pathways involved in the maintenance of the hematopoietic stem cell compartment (32), suggesting that the short NF-YA specifically affects stemness.
NF-YC Splicing Isoforms
In this study, we provide evidence that a minimum of three nonredundant NF-YC splicing isoforms are present in human cell lines. These variants arise by alternative splicing and are translated into different polypeptides with predicted molecular mass of 37, 48, and 50 kDa. Noteworthy, the region affected by alternative splicing corresponds to the Q-rich transactivation domain of NF-YC, which is reminiscent of the NF-YA situation (31). RT-PCR experiments performed in 13 human cell lines indicate that the three NF-YC mRNAs are simultaneously transcribed in a given cell type, with the 37-kDa mRNA being the most abundant, which explains why this cDNA was originally cloned in human and rat (Fig. 1C). However, the expression of single NF-YC subunits is different at the protein level, with the 37- and 50-kDa isoforms being almost mutually exclusive (Fig. 1D). This indicates strong post-transcriptional mechanisms affecting NF-YC mRNA stability or translation. Furthermore, a strong link between NF-YC and NF-YA subtypes is also evident. Cell lines expressing the 50-kDa NF-YC are specifically enriched for the short NF-YA isoform, whereas the long NF-YA is mostly present in cells containing the 37-kDa NF-YC (Fig. 1D). This is the first demonstration of the presence of a discrete cohort of NF-Y trimer subtypes in vivo, probably reflecting specialized function within the cell line analyzed; all the cell lines of hematopoietic origin, B-Jab, K563, Nalm-6, and U937, express the short NF-YA isoform, as expected (32), and the 50-kDa NF-YC.
Transactivation assays performed on the ΔNp63 promoter confirmed the NF-YC isoform functional specification and the preferential partnership with NF-YA proteins; the 37-kDa NF-YC is more active with the long NF-YA, whereas the short NF-YA isoform is a more potent transactivator with the 50-kDa NF-YC. Interestingly, the polypeptide stretches corresponding to the exon 8bis and exon 9bis are not Q-rich, with exon 9bis coding for a stretch of 104 amino acids particularly rich in Ser (15.1%) and Thr (10.4%) residues. A Ser/Thr-rich domain is present also within NF-YA, just downstream to the Q-rich, and is involved in the modulation of NF-Y transcriptional activity (31). Because 5 of the 16 Ser residues and 2 of the 11 Thr residues are predicted to be bona fide phosphorylation sites, it is reasonable to imagine regulatory functions for the NF-YC Ser/Thr-rich domain, specifically affecting the 48- and 50-kDa isoforms.
NF-YC Alternative Promoters
We identified a region of vertebrate sequence conservation within the first intron of the RefSeq NF-YC. This is clearly a second promoter (P2) because of the following: (i) an mRNA arising from this region is present within the GenBankTM sequence data base; (ii) P2-specific mRNAs are readily observed in RT-PCR (Fig. 4B); (iii) histone marks typical of active chromatin and 5′ promoter regions are present (Fig. 3, A and B); and (iv) P2 possesses promoter activity in transfection experiments (Fig. 2C).
Recent genome-wide analyses indicate that alternative promoter usage is a common event, which occurs at least with the same order of frequency as alternative splicing does, affecting about 50% of human genes. Kimura et al. (25) recently counter-correlated the presence of CpG islands with the occurrence of an alternative promoter in a given gene, concluding that genes that undergo complex transcriptional regulation often contain at least one ubiquitously expressed promoter, along with one or more promoters used for tissue-specific or signal-dependent expression. These general rules seem to be perfectly recapitulated by our results on the NF-YC promoters; P1 is indeed contained within a well conserved CpG island, whereas P2 is not. P1 has a “housekeeping” activity, being used in most cell lines with a similar activity (Fig. 4B, upper panel), whereas P2 is less active in basal conditions and is regulated depending on the cellular context (Fig. 4B, lower panel). RT-PCR experiments also show that P1 transcripts are more abundant.
There are two additional noteworthy features of NF-YC promoters. First, they contain CCAAT boxes, and ChIP experiments confirmed that NF-Y is indeed bound in vivo to both promoters (Fig. 3A), indicating NF-Y self-regulation, mostly on NF-YC P2, as assessed also by transfection experiments (Fig. 5C). Second, because the ATG is present 6 bp after the beginning of exon 2, alternative promoter choice labels nascent transcripts with two different, mutually exclusive, 5′UTR regions. Kimura et al. (25) showed that “mutually exclusive” variations are the most abundant 1st exon pattern resulting from alternative promoter usage. Interestingly, sequence analysis performed with RegRNA (47) identified 23 putative microRNA target sites within exons 1 and 7 within exon 1bis, none of which are in common (supplemental Table S2). Exon 1bis also contains an upstream open reading frame suggesting that alternative transcriptional regulation affects different regulatory levels of the human NF-YC locus, influencing mRNA stability and/or translatability.
NF-Y and the DNA-damage Response
NF-Y controls genes involved in cell cycle regulation and response to checkpoint activation (10), and it can directly recruit p53 to specific targets containing multiple CCAAT boxes, usually leading to histone deacetylase complex recruitment and transcriptional repression (41). Here we show that the NF-YC P2 is activated following DNA damage in a p53-dependent way, with kinetics similar to bona fide p53 target genes, CDKN1A and HDM2. P1 activity is almost unaffected. In line with this, the NF-YC 37-kDa isoform is also induced by DNA damage. The 48- and 50-kDa isoforms are unaffected, supporting our previous RT-PCR findings that identified P2 as an isoform-specific promoter. Thus, because P2 transcription seems to become dominant and per se sufficient to affect the level of single NF-YC isoforms (Fig. 5D), it is reasonable to speculate about regulatory mechanisms acting on P2 5′UTR. ChIP experiments demonstrated that p53 is bound to P2 in growing cells, and this binding decreased upon DNA damage, concomitantly to P2 activation. There are no obvious p53 sites in the region, but several well conserved CCAAT boxes are present upstream to the P2 transcription start site, and it is reasonable to imagine NF-Y-p53 complexes bound to P2, mediating regulation (41). Both P2 activity and 37-kDa isoform levels are indeed higher in HCT116/E6 cells, supporting the idea that p53 removal relieves transcriptional repression present in HCT116 cells.
The functional diversification of the isoforms outlined above becomes apparent from the shRNA experiments specifically affecting the 37-kDa isoform, and to a lesser degree the 48-kDa isoform, but not the 50-kDa isoform. The G2/M genes are crucially dependent on NF-Y binding, as exemplified by the fact that GO terms referring to this transition are at the top of the list of genes affected by profiling of genes after NF-YB RNA interference (49); they are normally regulated after knockdown of the 37-kDa isoform, that is, expressed in growing and repressed after DNA damage, indicating that the 50-kDa isoform is specifically targeting them. However, other checkpoints genes are strongly affected by the 37-kDa removal, in two ways. The first is that cell cycle blocking and pro-apoptotic genes are up-regulated under “basal” conditions; this might be either a direct effect of lack of the 37-kDa repression or unbalance with a potentially activating 50-kDa isoform. The second is the lack or decrease in activation of these genes after DNA damage. In this regard, it is important to remember similar experiments of inactivation of NF-YB, in which activation of p53 and a robust proapoptotic response is triggered (49). Therefore, it is possible that an unbalance of a specific NF-YC important for the DNA-damage response is sensed as a potentially threatening condition. Further work along this line of reasoning is required to validate this hypothesis.
In summary, our observations indicate that NF-YC (and NF-YA) splicing isoforms act as tissue/function-specific variants of a widespread and general TF, adding a further layer of complexity in what has been considered a relatively “simple” activator. Further genetic experiments in vivo will be required to fully understand how different NF-Y regulatory networks are established and executed within specialized cellular compartments.
Supplementary Material
Acknowledgments
We thank C. Politi for help with the experiments. We also thank S. Pozzi and M.A. Viganò for helpful discussion and comments on the manuscript.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4 and Tables S1–S2.
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) AK000346.
- ChIP
- chromatin immunoprecipitation
- RT
- reverse transcription
- shRNA
- short hairpin RNA
- UTR
- untranslated region
- TF
- transcription factor
- HFM
- histone-fold motif
- AS
- alternative splicing.
REFERENCES
- 1.Suzuki Y., Tsunoda T., Sese J., Taira H., Mizushima-Sugano J., Hata H., Ota T., Isogai T., Tanaka T., Nakamura Y., Suyama A., Sakaki Y., Morishita S., Okubo K., Sugano S. (2001) Genome Res. 11, 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elkon R., Linhart C., Sharan R., Shamir R., Shiloh Y. (2003) Genome Res. 13, 773–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.FitzGerald P. C., Shlyakhtenko A., Mir A. A., Vinson C. (2004) Genome Res. 14, 1562–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mariño-Ramírez L., Spouge J. L., Kanga G. C., Landsman D. (2004) Nucleic Acids Res. 32, 949–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki Y., Yamashita R., Shirota M., Sakakibara Y., Chiba J., Mizushima-Sugano J., Kel A. E., Arakawa T., Carninci P., Kawai J., Hayashizaki Y., Takagi T., Nakai K., Sugano S. (2004) In Silico Biol. 4, 429–444 [PubMed] [Google Scholar]
- 6.Grskovic M., Chaivorapol C., Gaspar-Maia A., Li H., Ramalho-Santos M. (2007) PLoS Genet. 3, e145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J., Li Z., Brower-Sinning R., John B. (2007) PLoS Comput. Biol. 3, e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ceribelli M., Dolfini D., Merico D., Gatta R., Viganò A. M., Pavesi G., Mantovani R. (2008) Mol. Cell. Biol. 28, 2047–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reed B. D., Charos A. E., Szekely A. M., Weissman S. M., Snyder M. (2008) PLoS Genet. 4, e1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ceribelli M., Alcalay M., Viganò M. A., Mantovani R. (2006) Cell Cycle 5, 1102–1110 [DOI] [PubMed] [Google Scholar]
- 11.Zhu Z., Shendure J., Church G. M. (2005) Genome Res. 15, 848–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tabach Y., Milyavsky M., Shats I., Brosh R., Zuk O., Yitzhaky A., Mantovani R., Domany E., Rotter V., Pilpel Y. (2005) Mol. Syst. Biol. 1, 2005.0022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorn A., Bollekens J., Staub A., Benoist C., Mathis D. (1987) Cell 50, 863–872 [DOI] [PubMed] [Google Scholar]
- 14.Mantovani R. (1998) Nucleic Acids Res. 26, 1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mantovani R. (1999) Gene 239, 15–27 [DOI] [PubMed] [Google Scholar]
- 16.Bellorini M., Lee D. K., Dantonel J. C., Zemzoumi K., Roeder R. G., Tora L., Mantovani R. (1997) Nucleic Acids Res. 25, 2174–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romier C., Cocchiarella F., Mantovani R., Moras D. (2003) J. Biol. Chem. 278, 1336–1345 [DOI] [PubMed] [Google Scholar]
- 18.de Silvio A., Imbriano C., Mantovani R. (1999) Nucleic Acids Res. 27, 2578–2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberati C., di Silvio A., Ottolenghi S., Mantovani R. (1999) J. Mol. Biol. 285, 1441–1455 [DOI] [PubMed] [Google Scholar]
- 20.Coustry F., Maity S. N., Sinha S., de Crombrugghe B. (1996) J. Biol. Chem. 271, 14485–14491 [DOI] [PubMed] [Google Scholar]
- 21.Frontini M., Imbriano C., diSilvio A., Bell B., Bogni A., Romier C., Moras D., Tora L., Davidson I., Mantovani R. (2002) J. Biol. Chem. 277, 5841–5848 [DOI] [PubMed] [Google Scholar]
- 22.Kabe Y., Yamada J., Uga H., Yamaguchi Y., Wada T., Handa H. (2005) Mol. Cell. Biol. 25, 512–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donati G., Imbriano C., Mantovani R. (2006) Nucleic Acids Res. 34, 3116–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Testa A., Donati G., Yan P., Romani F., Huang T. H., Viganò M. A., Mantovani R. (2005) J. Biol. Chem. 280, 13606–13615 [DOI] [PubMed] [Google Scholar]
- 25.Kimura K., Wakamatsu A., Suzuki Y., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Ishii S., Sugiyama T., Saito K., Isono Y., Irie R., Kushida N., Yoneyama T., Otsuka R., Kanda K., Yokoi T., Kondo H., Wagatsuma M., Murakawa K., Ishida S., Ishibashi T., Takahashi-Fujii A., Tanase T., Nagai K., Kikuchi H., Nakai K., Isogai T., Sugano S. (2006) Genome Res. 16, 55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singer G. A., Wu J., Yan P., Plass C., Huang T. H., Davuluri R. V. (2008) BMC Genomics 9, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwan T., Benovoy D., Dias C., Gurd S., Provencher C., Beaulieu P., Hudson T. J., Sladek R., Majewski J. (2008) Nat. Genet. 40, 225–231 [DOI] [PubMed] [Google Scholar]
- 28.Pan Q., Shai O., Lee L. J., Frey B. J., Blencowe B. J. (2008) Nat. Genet. 40, 1413–1415 [DOI] [PubMed] [Google Scholar]
- 29.Sultan M., Schulz M. H., Richard H., Magen A., Klingenhoff A., Scherf M., Seifert M., Borodina T., Soldatov A., Parkhomchuk D., Schmidt D., O'Keeffe S., Haas S., Vingron M., Lehrach H., Yaspo M. L. (2008) Science 321, 956–960 [DOI] [PubMed] [Google Scholar]
- 30.Xin D., Hu L., Kong X. (2008) PLoS ONE 3, e2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X. Y., Hooft van Huijsduijnen R., Mantovani R., Benoist C., Mathis D. (1992) J. Biol. Chem. 267, 8984–8990 [PubMed] [Google Scholar]
- 32.Zhu J., Zhang Y., Joe G. J., Pompetti R., Emerson S. G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11728–11733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bellorini M., Zemzoumi K., Farina A., Berthelsen J., Piaggio G., Mantovani R. (1997) Gene 193, 119–125 [DOI] [PubMed] [Google Scholar]
- 34.Sinha S., Maity S. N., Lu J., de Crombrugghe B. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 1624–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinha S., Maity S. N., Seldin M. F., de Crombrugghe B. (1996) Genomics 37, 260–263 [DOI] [PubMed] [Google Scholar]
- 36.Chen F., Ogawa K., Liu X., Stringfield T. M., Chen Y. (2002) Biochem. J. 364, 571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castrignanò T., D'Antonio M., Anselmo A., Carrabino D., D'Onorio, De Meo A., D'Erchia A. M., Licciulli F., Mangiulli M., Mignone F., Pavesi G., Picardi E., Riva A., Rizzi R., Bonizzoni P., Pesole G. (2008) Bioinformatics 24, 1300–1304 [DOI] [PubMed] [Google Scholar]
- 38.Santos-Rosa H., Schneider R., Bannister A. J., Sherriff J., Bernstein B. E., Emre N. C., Schreiber S. L., Mellor J., Kouzarides T. (2002) Nature 419, 407–411 [DOI] [PubMed] [Google Scholar]
- 39.Roh T. Y., Cuddapah S., Zhao K. (2005) Genes Dev. 19, 542–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miao F., Natarajan R. (2005) Mol. Cell. Biol. 25, 4650–4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imbriano C., Gurtner A., Cocchiarella F., Di Agostino S., Basile V., Gostissa M., Dobbelstein M., Del Sal G., Piaggio G., Mantovani R. (2005) Mol. Cell. Biol. 25, 3737–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang J., Xie Z., Glover B. J. (2005) New Phytol. 165, 623–631 [DOI] [PubMed] [Google Scholar]
- 43.Siefers N., Dang K. K., Kumimoto R. W., Bynum W. E., 4th, Tayrose G., Holt B. F., 3rd (2009) Plant Physiol. 149, 625–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhattacharya A., Deng J. M., Zhang Z., Behringer R., de Crombrugghe B., Maity S. N. (2003) Cancer Res. 63, 8167–8172 [PubMed] [Google Scholar]
- 45.Lipscombe D. (2005) Curr. Opin. Neurobiol. 15, 358–363 [DOI] [PubMed] [Google Scholar]
- 46.Ge Y., Jensen T. L., Matherly L. H., Taub J. W. (2002) Biochim. Biophys. Acta 1579, 73–80 [DOI] [PubMed] [Google Scholar]
- 47.Huang H. Y., Chien C. H., Jen K. H., Huang H. D. (2006) Nucleic Acids Res. 34, W429–W434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borrelli S., Testoni B., Callari M., Alotto D., Castagnoli C., Romano R. A., Sinha S., Vigano A. M., Mantovani R. (2007) BMC Mol. Biol. 8, e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benatti P., Basile V., Merico D., Fantoni L. I., Tagliafico E., Imbriano C. (2008) Nucleic Acids Res. 36, 1415–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.