Abstract
The NSD (nuclear receptor SET domain-containing) family of histone lysine methyltransferases is a critical participant in chromatin integrity as evidenced by the number of human diseases associated with the aberrant expression of its family members. Yet, the specific targets of these enzymes are not clear, with marked discrepancies being reported in the literature. We demonstrate that NSD2 can exhibit disparate target preferences based on the nature of the substrate provided. The NSD2 complex purified from human cells and recombinant NSD2 both exhibit specific targeting of histone H3 lysine 36 (H3K36) when provided with nucleosome substrates, but histone H4 lysine 44 is the primary target in the case of octamer substrates, irrespective of the histones being native or recombinant. This disparity is negated when NSD2 is presented with octamer targets in conjunction with short single- or double-stranded DNA. Although the octamers cannot form nucleosomes, the target is nonetheless nucleosome-specific as is the product, dimethylated H3K36. This study clarifies in part the previous discrepancies reported with respect to NSD targets. We propose that DNA acts as an allosteric effector of NSD2 such that H3K36 becomes the preferred target.
Introduction
Among the array of posttranslational histone modifications that feature prominently in the regulation of chromatin structure and function is lysine methylation. Histone lysine methyltransferases (HKMTs)3 target specific histone residues, and depending on their substrate specificity and their catalytic properties, the resulting products are mono-, di-, or trimethylated versions of lysine residues. A large body of work has correlated the status of histone lysine methylation as well as the extent of such methylation (mono, di, or tri) with certain cellular processes that include transcriptional regulation. Recent genome-wide studies have mapped the level of transcription as a function of the presence and position of specific states of histone lysine methylation within genes. For example, the levels of trimethylated histone H3 lysine 4 (H3K4me3) were elevated surrounding the transcriptional start sites of genes and positively correlated with transcription, whereas H3K36me3 signals were sharply elevated downstream of the transcriptional start site, peaking at the 3′-end in active genes. On the other hand, the presence of H3K27me3 correlated with transcriptional repression (1).
Lysine 20 of histone H4 is also subject to methylation, but its outcome with respect to transcription is not predictable. Recent studies have demonstrated that monomethylation of this residue (H4K20me1) is important for chromosome organization and compaction (2), yet this mark has also been detected on transcriptionally active genes. A likely explanation for these observations is that a histone modification in isolation might not necessarily be predictive of an outcome. Other modifications, such as methylation itself, acetylation, and/or phosphorylation of residues within the nucleosome(s) (or neighboring nucleosomes constituting a “chromatin domain”) likely contribute to the output of a specific histone modification. Although the extent of methylation serves as recognition sites for binding proteins containing specific domains (chromo, plant homeo domain fingers, malignant brain tumor repeats, Tudor, and others), the amino acids surrounding a specific lysine methylation site also confer specificity. Similarly, other modifications present on other residues in the nucleosome(s) can promote or prevent binding of factors to a specific modified lysine residue. Thus, the substrate specificity and resulting products of a given histone lysine methyltransferase, together with other modifications, are critical to fully understanding its role in chromatin-related functions.
The mammalian NSD family of SET domain-containing methyltransferases includes NSD1, NSD2 (Wolf-Hirschhorn syndrome candidate 1/MMSET (multiple myeloma SET domain)), and NSD3 (Wolf-Hirschhorn syndrome candidate 1 like). The SET domain of NSD proteins shares sequence similarity with SET2, the sole H3K36 methyltransferase in Saccharomyces cerevisiae. SET2 binds to and migrates with elongating RNA polymerase II, catalyzing the formation of H3K36me3 along the coding region of actively transcribed genes (3–7). In yeast, H3K36me3 then recruits complexes containing histone deacetylases that putatively target nucleosomes positioned upstream of the elongating RNA polymerase II and thereby re-establish a chromatin structure that suppresses intragenic transcription initiation (8, 9). Little is known regarding H3K36me1 and H3K36me2. However, a recent study in Drosophila reported that H3K36me2 levels peaked at the 5′-end of the transcribed region and required dMes-4 (the NSD homolog in Drosophila), whereas H3K36me3 accumulated toward the 3′-end of transcribed genes and relied on dHYPB (the SET2 homolog in Drosophila) (10). Studies in Caenorhabditis elegans showed that Mes-4 is responsible for all H3K36me2 in early embryos (11). Given these findings, the NSD methyltransferases would be expected to be H3K36-specific. Yet other reports indicated discrepancies in this regard. For example, NSD1 has been reported to dimethylate H3K36 and H4K20 (12), whereas NSD2 has been reported to methylate H4K20 (13), H3K4 (14), H3K27 (15), and H3K36 (16), and NSD3 has been reported to methylate H3K4 and H3K27 (17).
Of note, disruption of NSD protein integrity or of its proper expression in human cells has been linked to several diseases. Haploinsufficiency of NSD1 leads to Sotos syndrome, a childhood developmental disease characterized by overgrowth and mental retardation (18, 19). Similarly, NSD2 is deleted in Wolf-Hirschhorn syndrome characterized by developmental defects and mental retardation (20, 21). Additionally, chromosome translocation resulting in NUP98 fusion to NSD1 (22, 23) and NSD3 (24) gives rise to acute myeloid leukemia, and chromosome translocation resulting in NSD2 overexpression leads to multiple myeloma (20, 25), whereas reducing its levels suppressed cancer growth (13). Moreover, NSD3 is amplified in breast cancer cell lines and primary breast carcinomas (26). Yet little is currently known regarding the mechanism of action and the functional role(s) of the NSD proteins.
As a first step toward understanding the biological impact of NSD proteins, we characterized their methyltransferase activities. Using recombinant NSD1–3 SET domain-containing proteins, we find that NSD1–3 are highly specific H3K36 dimethylases when nucleosomes are used as substrates. However, in the case of histone octamers, multiple residues are targeted, and this phenomenon is suppressed by the presence of short DNA molecules such that the specificity observed with nucleosomes is rescued. A human complex containing full-length NSD2 and associated polypeptides recapitulates the activity observed with nucleosomes and the recombinant NSD2-SET domain protein.
EXPERIMENTAL PROCEDURES
Antibodies
Antibodies were obtained as follows: anti-H3K36me1 (Upstate; antibody 07-548), anti-H3K36me2 (Cell Signaling; antibody 2901), anti-H3K36me3 (Abcam; ab9050), anti-H3K27me3 (Abcam; antibody ab6002), anti-H4K20me1 (Abcam; antibody ab9051), anti-H4K20me2 (Upstate; antibody 07-367), anti-H4K20me3 (Abcam; antibody ab9053), anti-NSD1 antibody (Bethyl; antibody BL715), and anti-β-tubulin (Abcam; antibody ab6046). Anti-NSD2 is a polyclonal antibody derived from rabbit immunized with recombinant NSD2 (NP_001035889; amino acids 310–540). IgG were purified by protein G-agarose (Invitrogen).
Construction of Expression Vectors
cDNA of NSD1-SET (NP_071900; amino acids 1849–2094) was cloned into BamHI and XhoI sites of pGEX6P-1 (Amersham Biosciences), and cDNA of NSD2-SET (NP_001035889; amino acids 941–1240) and NSD3-SET (NP_001035889; amino acids 1021–1320) were cloned into the BamHI and SalI sites of pGEX6p-1 (Amersham Biosciences). cDNA for SET2 (Q9BYW2; 1373–2564 amino acids) was cloned into the EcoRI site of pFastBac (Invitrogen). Point mutations of histones H3 and H4 were made based on the Xenopus laevis-like histone H3 expression vector pET3-H3 (AJ556872.1) and the X. laevis-like histone H4 expression vector pET3-H4 (AJ556873.1), respectively, using the QuikChange mutagenesis kit (Stratagene) according to the manufacturer's instructions. All of the constructs were verified by DNA sequencing.
Protein Expression and Purification
GST-SET domain fusion proteins were expressed in Escherichia coli after the addition of 0.5 mm isopropyl β-d-thiogalactopyranoside at 17 °C overnight, and the cells were lysed in lysis buffer (50 mm Tris-HCl, pH 7.9, 500 mm NaCl, 0.2 mm EDTA, 10% glycerol, 1 μg/ml pepstatin A, 1 μg/ml leupeptin, 1 μg/ml aprotinin, and 0.2 mm PMSF). The lysate was cleared by centrifugation, and the supernatant was incubated with glutathione beads (General Electric Healthcare) overnight and washed with 20 column volumes of lysis buffer and eluted with 10 column volumes of lysis buffer plus 20 mm reduced glutathione (Sigma). The eluates were analyzed by CBB staining. The eluates containing the protein of interest were further purified by gel filtration on a Superdex 200 column (General Electric Healthcare) equilibrated with lysis buffer. The fusion proteins were pooled and dialyzed against BC100 (20 mm Tris-HCl, pH 7.9, 100 mm KCl, 0.2 mm EDTA, 10% glycerol, 0.2 mm PMSF, and 1 mm DTT). SET2-C was expressed in baculovirus inset cells and purified by nickel-nitrilotriacetic acid beads (Qiagen) according to the manufacturer's instructions. RAR and RXR were coexpressed using baculovirus and purified by M2 anti-FLAG agarose (Sigma) (27).
Histone Preparation
Recombinant histone octamers were prepared as described previously (28, 29). Briefly, Xenopus histones were expressed in E. coli individually and then pooled at an equal molar ratio in the denatured state and renatured by dialysis. Octamers were then purified from H3-H4 tetramers and H2A-H2B dimers via gel filtration on a Superdex S200 column. HeLa core histones were prepared as described previously (30). Nucleosomes were assembled using pG5E4 plasmid and octamers by step salt dialysis as described (28, 29). Histone H3 premethylated at lysine 36 was prepared by installing methyl-lysine analogs on recombinant histones by a chemical reaction as described previously (31). Briefly, H3K36C-C110A was generated by quick change PCR on a plasmid encoding histone H3 C110A. H3K36C-C110A was expressed in E. coli BL21(DE3) pLysS at 37 °C for 4 h (induction at A600 = 0.6 with 0.2 mm isopropyl β-d-thiogalactopyranoside). Histone-containing inclusion bodies were purified and solubilized in 7 m guanidine hydrochloride, 100 mm NaCl, 10 mm Tris-HCl, pH 8.0, 1 mm DTT, and 1 mm EDTA. After exchanging guanidine hydrochloride for 7 m urea by dialysis, the histones were further purified by sequential anion and cation exchange column chromatography. The histone solutions were loaded onto a 5-ml Q and a SP Sepharose column assembled inline. The Q Sepharose column was then removed, and the protein was eluted from the SP Sepharose column using a linear gradient of NaCl from 100 to 500 mm in the urea buffer (7 m urea, 10 mm Tris-HCl, pH8.0, 0.1 mm EDTA, and 1 mm DTT). Histone-containing fractions were pooled, dialyzed against water supplemented with 5 mm β-mercaptoethanol, and lyophilized. The lyophilized H3K36C-C110A to be modified (5–10 mg) was weighed into 1.5 ml of siliconized microcentrifuge tubes and 950 μl of alkylation buffer (AB) (4 m guanidinium chloride, 1 m Hepes, pH 7.8, and 10 mm d/l-methionine, pH 7.8; the solution was passed through a 0.22-micron filter and purged with argon prior to use) was added. When solubilization of the protein presented a problem, the protein mixture would be sonicated for 10–15 min in a Branson 1510 ultrasonic cleaning bath at ambient temperature. The resultant clear colorless solution was treated with 20 μl of a 1 m DTT solution in AB prepared just prior to use and agitated at 37 °C for 1 h. At the end of this period the reactions were alkylated in the following manner. The histones were treated with the appropriate nitrogen mustard ((ϕK-Me1), 100 μl of a 1 m N-methylaminoethyl chloride hydrochloride solution in AB; (ϕK-Me2), 50 μl of a 1 m 2-(dimethylamine) ethyl chloride hydrochloride solution in AB; and (ϕK-Me3), 100 mg of (2-bromoethyl) trimethylammonium bromide as solid) and agitated in the absence of light at either 25 °C (ϕK-Me2) or 50 °C (ϕK-Me1, ϕK-Me3). After 2 h, the ϕK-Me2 reaction was treated with 10 μl of a 1 m solution of DTT. After 2.5 h, a second portion of alkylating reagent was added to the ϕK-Me2 reaction (50 μl), and a second portion of DTT (10 μl) was added to the ϕK-Me1 and ϕK-Me3 reactions. The mixtures were then agitated in the dark at their respective temperatures for an additional 2.5 h. At the end of this period the reactions were then quenched with 2-mercaptoethanol (50 μl), cooled to room temperature, diluted to 2.5 ml with AB, purified by gel filtration through a PD-10 column (equilibrated with 0.1% BME in 18 Ω water) according to the manufacturer's protocol for centrifugal isolation, and lyophilized. A portion of each (∼0.1 mg) was analyzed by reverse phase high pressure liquid chromatography and MALDI-TOF mass spectrometry to ensure product identity and homogeneity.
Histone Lysine Methyltransferase Assay
A typical methyltransferase assay was carried out in 25-μl reaction mixtures containing 0.35 μm of histone octamers or nucleosomes, 0.15 μm of GST-SET domain fusion proteins, 50 mm Tris-HCl, pH 8.5, 5 mm MgCl2, 0.2 mm DTT, 1 μl of S-adenosyl-[methyl-3H]-l-methionine ([3H[SAM, 50–85 Ci/mmol, 0.55 mCi/ml; PerkinElmer Life Sciences), and 20 μm unlabeled SAM (Sigma), for 1 h at 30 °C. Synthesized short DNA was supplied in some assays at the concentrations indicated in the figure legends and text. The plus strand sequence of the 41-bp DNA was 5′-CTCTCTTTGAGGACACCAACCTGGCGGCCATCCACGCCAAG-3′. The NSD2 complex was used in place of the SET domain in some assays as indicated. The reactions were stopped by the addition of Laemmli sample buffer, and the reaction were products separated by 15% SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The membrane was stained with CBB, sprayed with 3H-ENHANCE (PerkinElmer Life Sciences) and analyzed by fluorography. In some assays, [3H]SAM was replaced with unlabeled SAM (final concentration, 200 μm), and standard Western blot was performed using the antibodies specified.
Mass Spectrometric Analysis
The methyltransferase assay was carried out as described above using unlabeled SAM, and the reaction mixtures were separated by SDS-PAGE and stained with CBB. The histones were excised from SDS-PAGE gels, destained, and then digested in-gel at 37 °C for 5 h with chymotrypsin (for H3; Roche Applied Science), Arg-C (for H4; Roche Applied Science), or Asp-N (for H4; Roche Applied Science) at 20 ng/μl. The digestion buffer for chymotrysin and Arg-C was 100 mm Tris-HCl, pH 8.0, and 10 mm CaCl2. The digestion buffer for Asp-N was 50 mm sodium phosphate, pH 8.0. The resulting peptides were extracted and dried under vacuum. Reverse phase C18 ZipTip microcolumns (Millipore) were used to desalt the samples. 10–25% of the desalted digests were submitted for the MALDI-TOF MS or tandem MS analyses using either a Waters (Milford, MA) MALDI Q-TOF Ultima or a Bruker (Billerica, MA) Autoflex MALDI-TOF mass spectrometer using standard operating parameters. The signals from 500 to 1000 laser shots were summed into each mass spectrum.
Stable Cell Line and FLAG Affinity Purification
HT1080 cells stably expressing FLAG- and Myc-tagged NSD2 (NP_001035889) were established with the murine stem cell virus retroviral system using puromycin selection. Nuclear extract from 1 × 109 cells was incubated overnight at 4 °C with M2 anti-FLAG-agarose (Sigma) equilibrated in the same buffer as nuclear extract (20 mm Tris-HCl, pH 7.9, 1.5 mm MgCl2, 0.42 m NaCl, 25% glycerol, 0.2 mm PMSF, and Roche Applied Science protease inhibitor mixture). The resin was washed with excess amounts of wash buffer (20 mm Tris-HCl, pH 7.9, 350 mm KCl, 0.2 mm EDTA, 10% glycerol, 0.01% Nonidet P-40, and 0.2 mm PMSF), and the bound proteins were eluted with wash buffer plus 0.2 mg/ml 3×FLAG peptide (Sigma). The eluates were analyzed by Western blot with anti-FLAG or anti-NSD2 antibodies followed by silver staining.
Gel Mobility Shift Assay
DNA oligos and proteins were incubated in 10 mm Tris-HCl, pH 7.5, 0.1 mm EDTA, 5 mm DTT, 5% glycerol, and 100 mm NaCl in a total volume of 20 μl at 4 °C for 30 min. The products of the reaction were analyzed by 5% native polyacrylamide gel electrophoresis (37.5:1) using 5 mm Tris-HCl, pH 7.5, and 50 mm glycine. The gel was stained with SYBR Gold (Invitrogen) for DNA detection and then with CBB for protein detection. The 41-bp dsDNA and the NSD2-SET domain used in the HKMT assay were used for the gel shift assay. RAR/RXR and oligo containing the RAR-RXR-specific DNA-binding sequence were used as positive controls (27). The plus strand sequence for the RAR/RXR oligo is 5′-GCAATTAAAGATGAACTTTGGGTGAACTAATTTGTCTG-3′.
siRNA Knockdown
HeLa cells were transfected with siRNA oligos at a 10 nm final concentration by Lipofectamine RNAiMAX (Invitrogen) and analyzed after 72 h. The upper strand sequences of the siRNA oligos against NSD2 (Qiagen) were as follows: 5′-AACGGCCAGAACAAGCUCUUA-3′ and 5′-AGGGATCGGAAGAGTCTTCAA-3′. The upper strand sequence of the control siRNA oligo was 5′-UAACGACGCGACGACGUAATT-3′. Nuclear extracts of the treated cells were analyzed by Western blot using anti-NSD2 antibody. Histones were acid-extracted following the protocol from Abcam. The levels of histone modifications were analyzed by Western blot using the antibodies specified.
RESULTS
The NSD Family of Proteins Targets H3K36 for Methylation
Three highly related NSD proteins exist in human cells, and each contains a SET domain at the C terminus that is homologous to that of SET2 (Fig. 1A, left panel). Toward analyzing NSD substrate specificity, each SET domain was independently fused to GST, expressed in bacteria, purified (Fig. 1A, right panel, and see “Experimental Procedures”), and analyzed for HKMT activity. The well characterized HKMTs, SET2 and PR-Set7 that target H3K36me3 and H4K20me1, respectively, served as controls. With recombinant nucleosomes as substrate, the SET domains of NSD1–3 and SET2 targeted H3 (Fig. 1B). Of note, the lower signal detected in the NSD2 and SET2 HKMT assay performed with recombinant nucleosomes was not directed toward H4 but toward a slower migrating species that we have identified to be a proteolyzed form of H3 by Western blot (see below). In contrast to recombinant nucleosomes in which only H3 was targeted, when recombinant octamers were used as substrate the NSD1-SET domain targeted H3, H2A/H2B, and H4, whereas the NSD2-SET domain mainly targeted H4 with very weak activity on H3 (Fig. 1B). NSD3 and SET2 activity remained specific for H3.
FIGURE 1.
The NSD SET domain methylates H3K36 on nucleosomes. A, the left panel shows the schematic of human NSD1–3 domain organization. NID denotes the nuclear receptor interaction domain (12). PWWP denotes the PWWP domain which contains the proline-tryptophan-tryptophan-proline motif in the consensus amino acid sequence. The region of the SET domain protein expressed and used in the following enzymatic assays is indicated above the sequences. All of the proteins include the pre-SET, SET, and post-SET domain and flanking regions. The right panel shows CBB staining of the purified GST-NSD1–3 SET domain fusion proteins and the SET2 C-terminal fragment separated on SDS-PAGE. B, HKMT assay on nucleosomes (Nuc) and octamers (Oct) with enzymes analyzed in A using [3H]SAM as the methyl donor. The protein concentrations used for NSD1–3 SET were 0.05, 0.1, 0.2, and 0.4 μm; those for SET2-C were 0.01, 0.02, 0.04, and 0.08 μm. Nucleosomes and octamers contained recombinant, unmodified histones, and their final concentrations in the reactions were 0.35 μm unless otherwise noted. The assay mixtures were separated by SDS-PAGE. The lower panel shows CBB staining of the histones, and the upper panel shows results with fluorography. The asterisk denotes a proteolyzed form of H3 (see text). C, HKMT assays were performed on wild type (WT) or H3K36A nucleosomes using enzymes described in A; bovine serum albumin (BSA) and full-length recombinant PR-SET7 served as controls. The final concentrations for NSD1–3 SET in these and subsequent reactions were 0.15 μm unless otherwise noted.
We then determined the exact lysine targeted by the different NSD-SET domain-containing proteins using recombinant nucleosomes reconstituted with H3 polypeptides carrying a single substitution at particular methylation sites. As controls, we included PR-Set7 and SET2. NSD-SET domain-containing proteins displayed activity toward nucleosomal H3 in the wild type case as did SET2, as expected (Fig. 1C, left panel). Although the specific activity varied among the different NSD polypeptides, none of the NSD-SET domain-containing proteins nor SET2 displayed activity in the case of nucleosomes carrying an H3K36 to A mutation (Fig. 1C). This substrate was successfully targeted for methylation by PR-SET7, however. Other lysine to alanine substitutions such as H3K4A, H3K9A, or H3K27A did not affect the activity of the NSD proteins (data not shown; see below). These results collectively suggest that the NSD-SET domain-containing proteins specifically methylate nucleosomal H3K36. However, with recombinant octamers as substrate, these SET domain-containing proteins exhibited disparate specificities.
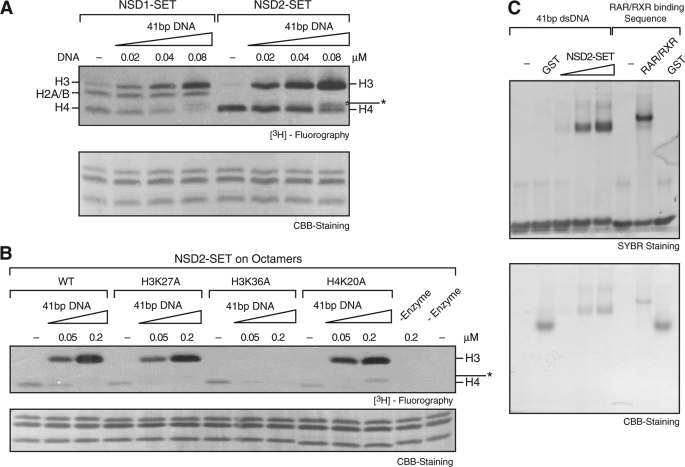
DNA Stimulates Activity and Alters NSD1 and NSD2 Specificity
We next investigated whether the presence of DNA, a discriminating feature of nucleosomes versus octamers, influences the disparate catalysis/specificity exhibited by NSD1 and NSD2. We observed that different single- or double-stranded polynucleotides stimulated NSD1 and NSD2 activity when octamers were used as substrate (data not shown; see below). The observed stimulation appeared to be DNA sequence independent and was not due to the formation of nucleosomes, because the size of the DNA fragments utilized precluded this. We selected a double-stranded 41-bp DNA fragment for further studies. As shown above (Fig. 1B), using octamers in the absence of nucleic acid, NSD1 or NSD2 displayed activity toward H3, H2A/H2B, and H4 or primarily H4, respectively (Fig. 2A). Upon addition of increasing amounts of short DNA fragment, NSD1 and NSD2 activity toward H4 was eliminated, and that toward H3 was increased in a DNA-dose dependent manner (Fig. 2A). NSD1 activity toward H2A/H2B required higher DNA concentrations to be inhibited. We next analyzed the residue targeted by NSD2 as a function of the presence of DNA using octamers carrying independently an alanine substitution of H3K27, H3K36, or H4K20 as substrate (Fig. 2B). In agreement with the findings obtained above in the case of nucleosomes, NSD2 displayed activity toward octamers containing H3K36 in the presence of DNA (Fig. 2B). Interestingly, in the absence of DNA, NSD2 activity directed toward H4 in octamers was not affected by mutant H4K20A, demonstrating that NSD2 targeted a different H4 residue, in contrast with previous studies (Ref. 13 and see below). We concluded that NSD2 targets primarily nucleosomal H3K36 and that it also targets H3K36 contained within octamers in the presence of short DNA. In the absence of DNA, NSD2 activity is mainly directed toward octamer-containing H4 independent of its Lys20 residue.
FIGURE 2.
Short dsDNA stimulates NSD2-SET HKMT activity on H3K36 and inhibits activity on H4. A and B, increasing amounts of 41 bp dsDNA were added to HKMT assays containing octamers and NSD1-SET or NSD2-SET (A) or NSD2-SET with WT octamers or octamers bearing various lysine to alanine point mutations (B). Wild type (WT) octamers were used in the minus (−) enzyme controls. The bottom panels show CBB staining of histones. C, 0.4 μm 41 bp dsDNA was incubated alone or with 6 μm GST or with 0.3, 1.0, or 2.0 μm NSD2-SET domain. As positive control, RAR-RXR dimers were incubated with 0.4 μm oligo containing the RAR-RXR-specific DNA binding sequence. The products of the reaction were resolved by 5% native polyacrylamide gel electrophoresis. The upper panel shows the SYBR Gold stained DNA, and the lower panel shows CBB-stained proteins.
We next analyzed whether the 41-bp DNA fragment binds directly to the enzyme using a gel mobility shift assay. Indeed the NSD2-SET domain shifted the 41-bp dsDNA (Fig. 2C), whereas GST protein alone did not. As a positive control, RAR and RXR dimers shifted oligonucleotide containing their cognate DNA-binding site. We also observed NSD2-SET binding to single-stranded DNA (data not shown). Our data demonstrate that DNA affected NSD2-SET activity by binding to the enzyme.
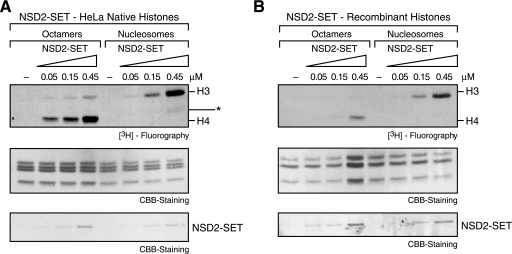
A possible caveat to the experiments described above (Fig. 2B) is that recombinant histones lack posttranslational modification(s) that might affect the activity of NSD proteins. To address this issue, histones were isolated from HeLa cells, used to reconstitute octamers and nucleosomes, and then compared with recombinant species with respect to NSD2 activity. Interestingly, the apparent specific activity of NSD2 was stimulated with HeLa-derived histones relative to recombinant ones, in the context of octamers (Fig. 3, compare A and B). A similar trend was observed with nucleosomes reconstituted with HeLa histones versus recombinant ones, yet the stimulation was not as pronounced (Fig. 3). Of note, histone specificity was not affected by the use of recombinant versus HeLa purified histones. NSD2 preferred H4 in the case of octamers and H3 in the case of nucleosomes (Fig. 3). We concluded that native histones (isolated from HeLa cells) might contain one or more modifications that stimulate NSD2 activity, yet these putative modifications did not affect its specificity for the histone polypeptides.
FIGURE 3.
HKMT assay comparing octamers and nucleosomes assembled from HeLa native histones, bearing natural histone modifications (A) or recombinant histones without modifications (B). The amount of enzyme was titrated as indicated. The middle and bottom panels show CBB staining of histones and enzyme, respectively. The asterisk denotes a proteolyzed form of H3.
NSD2 Is Specific for H3K36me2 Catalysis
The SET domains of the NSD family of proteins are highly similar and highly related to the SET domain of SET2 (Fig. 4A) that trimethylates H3K36. This similarity led us to analyze the extent (mono, di, or tri) of H3K36 methylation performed by the NSD proteins. We and others have established that SET2 could carry out all levels of methylation at H3K36 in vitro (4) yet functions as a trimethylase in vivo (4, 10, 33). Using highly specific antibodies directed toward di- and trimethylated versions of H3K36 (see “Experimental Procedures”) in Western blot analyses, we confirmed these previous studies and observed that SET2 carried out primarily trimethylation of nucleosomal H3K36, with negligible H3K36me2 in vitro (Fig. 4B). On the other hand, the product of the reaction only reached H3K36me2 in the case of NSD2 (Fig. 4B).
FIGURE 4.
NSD2-SET domain dimethylates H3K36 on nucleosomes. A, a schematic representation of NSD2 and SET2 domain organization is shown on top, and the sequence alignment of the SET domains of NSD1–3 and SET2 is shown on the bottom. Identical residues are replaced by dots, similar residues are shown in black, and others are in red. The active site residues described in previous SET domain structures are highlighted in yellow. PWWP, PWWP domain containing proline-tryptophan-tryptophan-proline motif in consensus amino acid sequence. B, HKMT assays were performed with NSD2-SET and SET2-C on wild type (WT) and H3K36A nucleosomes using unlabeled SAM, and the product of the reaction was analyzed by Western blot using anti-H3K36me3 (upper panel) and anti-H3K36me2 (lower panel) antibodies. C, recombinant histones bearing methyl analogs mimicking different degrees of methylation on H3K36 were generated by chemical reaction as described under “Experimental Procedures” and analyzed by Western blot using anti-H3K36me2 and anti-H3K36me3 antibodies. D, premethylated histones were assembled into nucleosomes and used as substrates in HKMT assays with NSD2-SET (left panel) and SET2-C (right panel). Me1, monomethyl-H3ϕK36; Me2, dimethyl-H3ϕK36; Me3, trimethyl-H3ϕK36. E, MALDI-TOF mass spectra for chymotyptic digests of H3 (top panel), H3 methylated by NSD2 (middle panel), and H3 methylated by SET2-C (bottom panel). HKMT assays were performed on nucleosomes, and the products of the reaction were separated by SDS-PAGE. The H3 bands were excised and digested in-gel using chymotrypsin, and the resulting peptides were analyzed using MALDI-TOF MS. The calculated protonated monoisotopic masses of peptide 21–41 carrying zero to three methylations are 2195.25, 2209.27, 2223.28, and 2237.30, respectively. The asterisk denotes a proteolyzed form of H3.
We next tested the capacity of NSD2 to methylate a chemically modified H3 already carrying a mono-, di-, or trimethyl group at Lys36, in the context of recombinant nucleosomes. These species were generated by cysteine specific alkylation of H3K36C-C110A with mono-, di-, and trimethylated nitrogen mustards as described (Ref. 29 and “Experimental Procedures”). This reaction generated specifically methylated pseudo-lysines that have been demonstrated to be a close analog of methylated lysines (29). The antibody specificity was analyzed anew using nucleosomes reconstituted with chemically synthesized di- and trimethylated H3K36 (Fig. 4C). Indeed the antibodies directed toward H3K36me2 preferentially recognized H3 on nucleosomes reconstituted with chemically synthesized H3K36me2, in Western blot analysis. Also, when chemically synthesized H3K36me3 was used to reconstitute nucleosomes, the antibodies directed toward this mark specifically detected H3K36me3, whereas H3K36me2-specific antibodies were ineffectual. Next, we performed HKMT assays using nucleosomes carrying either wild type H3 or the chemically modified H3 species and found that the NSD2-SET domain was capable of methylating the unmodified and monomethylated H3 but was inactive on nucleosomes carrying either a di- or trimethylated H3K36 (Fig. 4D, left panels). On the other hand, SET2 methylated either unmodified H3 or H3K36me2, but not nucleosomes reconstituted with H3K36me3 (Fig. 4D, right panels). To further evaluate the extent of NSD2-mediated methylation of nucleosomes, we analyzed the products by mass spectrometry when the reaction was performed with recombinant unmodified nucleosomes, SAM, and NSD2 (Fig. 4E). In agreement with the results presented above, NSD2 carried out mono- and dimethylation of nucleosomal H3, whereas SET2 carried out all levels of methylation. Taken together with the results presented above, we concluded that NSD2 is a dimethylase, whereas SET2 can carry out trimethylation.
To verify our in vitro observation that NSD2 dimethylated nucleosomal H3K36, we knocked down NSD2 using RNA interference and analyzed the resulting level of H3K36 methylation. Two different siRNA oligos against NSD2 were used, and both led to an efficient knockdown of NSD2 but had no effect on NSD1 protein accumulation (Fig. 5A). In agreement with the results obtained in vitro, the total amount of H3K36me2 was reduced in these two cell lines relative to cells treated with control siRNA (Fig. 5). This was specific for H3K36me2 because siRNA against NSD2 was ineffectual with respect to the other histone modifications analyzed (Fig. 5B). Thus, NSD2 carried out H3K36 dimethylation in vitro and in vivo.
FIGURE 5.
NSD2 carried out dimethylation of H3K36 in vivo. siRNA silencing experiment was carried out in HeLa cells using two different siRNA oligos against NSD2 and a control (Ctr) siRNA oligo. A, nuclear extracts of the siRNA-treated cells were prepared and analyzed by Western blot using antibodies against NSD1, NSD2, and β-tubulin. Histones were acid-extracted from the treated cells and analyzed by Western blot using the antibodies specified. B, histones isolated from cells treated with NSD2- or control (Ctr)-siRNA were loaded in two amounts, and the methylation levels on H3K36 and H4K20 were analyzed by Western blot using specific antibodies.
Full-length NSD2 Targets H3K36 for Dimethylation
To extend our studies in vivo, a cDNA encoding full-length NSD2 with a FLAG and Myc TAG at its N terminus was transfected into HT1080 cells. Stably transfected cells were then harvested, and the full-length NSD2 was isolated (Fig. 6A) and assayed for activity (Fig. 6B). Consistent with our findings above, the HKMT activity of full-length NSD2 was directed toward unmodified and monomethylated H3K36 contained in nucleosomes (Fig. 6B). Moreover, when the full-length NSD2 was analyzed for its activity on octamers, low levels of activity toward H3 and H4 were detectable, but consistent with our results using the NSD2-SET domain, the addition of DNA suppressed the activity toward H4 and stimulated the activity toward H3 (Fig. 6C). We concluded that full-length NSD2 isolated from transfected cells and the NSD2-SET domain display the same histone specificity.
FIGURE 6.
NSD2 complex dimethylates H3K36 on nucleosomes or octamers plus DNA. A, NSD2 complex was purified from HT1080 cells stably expressing FLAG- and Myc-tagged NSD2 by anti-FLAG resin. As a control, a similar purification from a stable cell line with empty vector was carried out, side by side (Mock). The input (IN), flow though (FL), and eluate (E) from the FLAG purification were separated by SDS-PAGE and analyzed by Western blots (left panel) using anti-FLAG M2 or anti-NSD2 antibodies as indicated at the bottom and by silver staining (right panel). B and C, HKMT assay performed with the isolated NSD2 complex and nucleosomes composed of wild type (WT), H3K36A, or premethylated H3K36 as indicated (B, top panel) or with octamers as a function of increasing amounts of 41-bp dsDNA (C, top panel). The asterisk denotes a proteolyzed form of H3. CBB staining of histones is shown (bottom panels).
NSD2 Mediates Methylation of Histone H4 Lysine 44
An unresolved issue with respect to NSD2 specificity involves its methylation of H4. As shown above, this activity was observed on octamers but eliminated in the presence of short DNA and was undetectable with nucleosomes. Moreover, our results did not support previous findings that H4K20 is the target site (13). Instead, we found that an alanine substitution of Lys20 (H4K20A) did not affect NSD2-mediated H4 methylation. To determine the H4 residue(s) targeted, we reconstituted octamers with tail-less core histones (schematic shown in Fig. 7A, left panels) and used these as substrate for NSD2-SET in HKMT assays. NSD2-SET was still able to methylate H4 devoid of residues 1–19, and this activity was drastically reduced by the addition of short DNA, as previously observed with full-length octamers (Fig. 7A, right panels). To map the residue target by NSD2-SET, HKMT assays were performed as above, but the reaction products were separated by SDS-PAGE, and the H4 polypeptide was excised, digested with Arg-C or Asp-N, and subjected to mass spectrometric analysis. The site of methylation mapped to lysine 44. Both mono- and dimethylation were observed (Fig. 7B, top panel). Additionally, an H4 peptide containing lysine 20 was not methylated (Fig. 7B, bottom panel).
FIGURE 7.
NSD2-SET domain can methylate H4K44 on octamers. A, schematic of tail-less histones (left panel). ΔH3 lacks amino acid 1–26, ΔH4 lacks amino acid 1–19, ΔH2A lacks amino acid 1–12, and ΔH2B lacks amino acid 1–23. The tail-less histones were assembled into octamers, and the HKMT assay was carried out on wild type (WT) or tail-less octamers (right panel). 41-bp dsDNA was added in some reactions as indicated. The reactions without enzyme were included as negative controls. B, mapping of the H4 methylation site by mass spectrometry. MALDI-TOF mass spectra for Arg-C digests (upper panel) and Asp-N digests (lower panel) of H4 methylated by NSD2-SET. The peptide identified as H4 (amino acids 42–54) exhibited mono- and dimethylation (upper panel), whereas the peptide identified as H4 (amino acids 1–23) was unmethylated (lower panel). The calculated protonated monoisotopic masses of peptide 41–55 carrying zero to two methylations are 1677.92, 1691.94, and 1705.95, respectively. Tandem mass spectra of precursor ions with m/z of 1691.94 and 1705.95 confirmed the mono- and dimethylation on Lys44 (data not shown). Unmodified peptide 1–23 has a calculated protonated monoisotopic mass of 2360.43. C, HKMT assay with octamers composed of WT H4 or H4K44A (left panel) or H4K44Q (middle panel). The enzyme was titrated at two different amounts, and 41-bp dsDNA was added to the assays as indicated. HKMT assays performed with WT H4, H4K44A, H4K44Q, or H4K44E octamers assembled onto nucleosomes (right panel).
That lysine 44 was the H4 substrate targeted by NSD2 was confirmed using octamers reconstituted with H4 containing a substitution of Lys44 with alanine (H4K44A) (Fig. 7C, left panel). The activity directed toward H4 was eliminated, but importantly, this did not affect the activity directed toward H3 in the presence of DNA (Fig. 7C, left panel). Similar results were observed when nucleosomes reconstituted with H4K44A were used as substrate (Fig. 7C, right panel), demonstrating that NSD2-mediated methylation of H4K44 is not required for its methylation of H3K36. In agreement with studies obtained with yeast SET2 that carries out H3K36me (34), the substitution of H4K44 with glutamine (H4K44Q) led to a severe reduction in NSD2-mediated methylation of H3K36 using either octamers in the presence of DNA (Fig. 7C, middle panel) or nucleosomes (Fig. 7C, right panel). Similarly a substitution of H4K44E also decreased NSD2 activity (Fig. 7C, right panel). We concluded that NSD2 methylates H4K44 present in octamers and that Lys44 can affect methylation of nucleosomal H3K36, but this occurs in a manner independent of Lys44 itself being methylated.
DISCUSSION
We showed here that NSD1, -2, and -3 that comprise the mammalian NSD family of proteins are histone methyltransferases that target H3K36 on nucleosomes. NSD2 mono- and dimethylates H3K36 on nucleosomes but mono- and dimethylates H4K44 on octamers. In the case of octamers, the addition of short double-stranded or single-stranded DNA stimulates the activity on H3 and inhibits the activity on H4.
Previous studies indicated that members of the NSD family of proteins methylate H3K4 (14, 17), H3K36 (12, 16), H3K27 (15, 17), and H4K20 (12, 13). These discrepancies in the reported NSD target sites might be attributable to the differential activity exhibited by NSD proteins on octamers versus nucleosomes, as shown here. The activity we scored on nucleosomes is strong and specific as evidenced by the lack of detectable activity when the nucleosomal substrates contained a substitution of H3K36 to alanine (K36A). However, the activity on octamers is less specific. Activity is detectable on octamers containing H3K36A, suggesting that other residues, such as H3K4 and/or H3K27 may be viable target sites under this condition. Consistent with this possibility, the activity observed on H3K4 and/or H3K27 by previous groups was directed at peptides or individual histones (13, 15, 17). The relevance of such activity is unclear at this point given that we find it to be weak and nonspecific on octamers, relative to nucleosomes. In the case of the discrepancy involving the H4 target site, the previous studies reported H4K20 methylation using antibodies, but our findings point only to H4K44 based on mass spectrometric analyses and comparison of mutant histone substrates. The likely explanation for the disparate results regarding NSD2 specificity (and other HKMTs) is the use of suboptimal substrates and/or the use of mischaracterized antibodies. Given the technology now available with which specific residues on histones can be chemically methylated to different extents (31), these modified proteins can now be used to characterize the specificity of the antibodies in future studies.
Does H3K36me2 Have a Function Distinct from That of H3K36me3?
Our results showed that the human NSD2 SET domain and the NSD2 complex purified from an NSD2 stable cell line were solely capable of mono- and dimethylation of H3K36 on nucleosomes. Knockdown of NSD2 in HeLa cells led to reduced levels of H3K36me2, but not of H3K36me1 or H3K36me3. On the other hand, human Set2 carried out mono-, di-, and trimethylation in vitro (4). Its knockdown abolished H3K36me3 in mouse fibroblasts but did not affect H3K36me1 or H3K36me2 levels (4, 33). These findings are in agreement with those reported in Drosophila. Down-regulation of the NSD homolog, dMES-4, by RNA interference led to a significant reduction in both H3K36me2 and H3K36me3 levels, whereas down-regulation of the Set2 homolog, dHYPB, led to a complete loss of H3K36me3 (10). It is unclear why dMES-4 knockdown affected not only the levels of H3K36me2 as was the case with NSD2, but also those of H3K36me3. It is possible that dMES-4 deficiency leads to a global reduction of transcription and hence indirectly a global down-regulation of H3K36me3 (10).
Although SET2 is responsible for all H3K36 methylation in yeast, yeast strains harboring mutations in genes affecting transcription elongation such as Δspt6, Δctk1, and Δpaf1 specifically abolished trimethylation while preserving dimethylation, suggesting that a level of regulation between di- and trimethylation states exists and is coupled to the efficiency of elongation by RNA polymerase II (35, 36). For comparison, H3K4me3 localizes at the 5′-end of genes, whereas H3K4me1-2 is enriched within the coding region toward the 3′-end. Only H3K4me3 showed a strong correlation with active transcription (37). As for the activities that recognize and bind histone marks, both H3K36me2 and H3K36me3 appear sufficient to recruit the Rpd3s histone deacetylase complex and inhibit internal initiation by RNA polymerase II (35). However, whereas H3K4me3 recruits the HAT complex NuA3 to the promoter, H3K4me2 recruits the Set3 histone deacetylase complex to the 5′-transcribed region. The PHD finger of Set3 exhibits preferential binding to H3K4me2, relative to H3K4me1/3 (38).
In higher eukaryotes, few studies have gauged the distribution of H3K36me2 on chromatin in vivo. Chromatin immunoprecipitation combined with microarray studies in Drosophila showed that H3K36me2 levels peak around the 5′-end of active genes, downstream of transcription start sites, whereas H3K36me3 levels peak around the 3′-end of active genes (10). In HeLa cells, H3K36me2 peaks immediately downstream of the transcriptional start site of the PABPC1 gene and then drops 0.5 kb further downstream, whereas H3K36me3 peaks at ∼20 kb downstream of this transcriptional start site (39). NSD1 and NSD2 have been shown to interact with nuclear receptors such as the androgen receptor (14, 40) and/or the retinoid acid receptor (12) and have been implicated in transcriptional activation (14, 40) and repression (12, 13, 41). This suggests that in contrast to SET2 that interacts with RNA polymerase II and carries out H3K36me3 during elongation, the NSD family of proteins might function during the early events of transcription such as initiation, promoter escape, and/or polymerase pausing. Although the available data is still limited regarding the distribution of H3K36me2 versus H3K36me3 on genes, it is tempting to postulate that H3K36me2 is likely to aid in the recruitment of factors other than the Rpd3s complex that regulate earlier events in the transcription process.
DNA Triggers a Switch from H4K44 to H3K36 Methylation
Here we showed that NSD2 methylates H4K44 in vitro and that short single-stranded or double-stranded DNA (41 bp) that are unable to form nucleosomes inhibited NSD2 activity on H4K44 and stimulated its activity on H3K36. Interestingly, an interaction between SET2 and histone H4 is required for H3K36 methylation in yeast (34), and H4K44 is a critical residue mediating this interaction. H4K44Q and H4K44E mutants eliminated H3K36 methylation by SET2 in vivo and in vitro (34). We observed similar effects of H4K44 with respect to NSD2 activity toward methylation of H3K36, yet we also found that NSD2 methylates H4K44 on octamers in vitro and that this activity is inhibited by the presence of DNA. The sequences surrounding H3K36 (GGVKK) and H4K44 (GGVKR) are similar, and this might pertain to how NSD2 targets these two residues in vitro. H4K44 is located within the L1 loop of histone H4 and contacts the DNA within the nucleosome, suggesting that the presence of DNA could potentially inhibit H4K44 methylation through its binding to and masking of the residue (29, 42). Yet we found that the SET domain of NSD2 binds directly to short dsDNA, and this is consistent with DNA having an allosteric affect on the enzyme. Whether or not H4K44 is methylated in vivo is presently not clear. H4K44 monomethylation has been detected using liquid chromatography-MS in the case of histones isolated from bovine calf thymus, although the peptide sequence was not confirmed by tandem mass spectrometry (43). Because DNA is a strong inhibitor of NSD2-mediated H4K44 methylation and H4K44me has not been unequivocally demonstrated in vivo, we suggest that methylation at this site is likely a biochemical artifact caused by the similarity of the amino acids surrounding H3K36 and H4K44. Yet, as with SET2 in yeast, H4K44 is important for targeting NSD2-mediated H3K36 recognition. However, this study indicates that in addition to the importance of H4K44, the DNA itself modulates NSD2 catalysis and its specificity for H3K36me2. If NSD2 methylates H4K44 in vivo, this function may be related to histone deposition and/or to the response to DNA damage, which remains to be studied.
That nucleic acid (RNA or DNA) regulates the activity of HKMTs is not unique to NSD2. Indeed studies have demonstrated that the motif flanking the SET domain (pre- or post-SET) can bind to DNA (44). Additionally, whereas PR-Set7 is a nucleosomally specific HKMT that carries out H4K20 monomethylation, studies have indicated that PR-Set7 can exhibit activity toward octamers in the presence of plasmid DNA (32), although the possibility exists that nucleosomes might have formed given the size of the DNA used (26, 27). We have recapitulated this effect with short DNA fragments.4 It is possible that DNA binding to the SET domain of NSD2 (and other HKMTs) might trigger a conformational change within the SET domain that regulates specificity and enhances catalysis.
Acknowledgments
We thank Dr. Lynne Vales for critical reading of the manuscript. We also thank Drs. Eric Campos, Guohong Li, Gang Li, Raphael Margueron, Hisanobu Oda, and Shengjiang Tu for comments and discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants 4R37GM037120–24 (to D. R.), S10 RR017990, P30 NS050276, and P30CA016087 (to T. A. N.) and GM063716 (to R. M. X.). This work was also supported by a grant from the Howard Hughes Medical Institute (to D. R.) and Basic Research Program of China Grant 2009CB825501 (to R. M. X.).
P. Trojer and D. Reinberg, unpublished results.
- HKMT
- histone lysine methyltransferase
- H3K36
- histone H3 lysine 36
- H4K44
- histone H4 lysine 44
- H3K4Me3
- trimethylated histone H3 lysine 4
- CBB
- Coomassie Brilliant Blue
- ds
- double-stranded
- oligo
- oligonucleotide
- GST
- glutathione S-transferase
- PMSF
- phenylmethylsulfonyl fluoride
- DTT
- dithiothreitol
- AB
- alkylation buffer
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- SAM
- S-adenosyl-[methyl-3H]-l-methionine
- MS
- mass spectrometry
- siRNA
- small interfering RNA.
REFERENCES
- 1.Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 2.Oda H., Okamoto I., Murphy N., Chu J., Price S. M., Shen M. M., Torres-Padilla M. E., Heard E., Reinberg D. (2009) Mol. Cell. Biol. 29, 2278–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krogan N. J., Kim M., Tong A., Golshani A., Cagney G., Canadien V., Richards D. P., Beattie B. K., Emili A., Boone C., Shilatifard A., Buratowski S., Greenblatt J. (2003) Mol. Cell. Biol. 23, 4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan W., Xie J., Long C., Erdjument-Bromage H., Ding X., Zheng Y., Tempst P., Chen S., Zhu B., Reinberg D. (2009) J. Biol. Chem. 284, 15701–15707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J., Moazed D., Gygi S. P. (2002) J. Biol. Chem. 277, 49383–49388 [DOI] [PubMed] [Google Scholar]
- 6.Xiao T., Hall H., Kizer K. O., Shibata Y., Hall M. C., Borchers C. H., Strahl B. D. (2003) Genes Dev. 17, 654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kizer K. O., Phatnani H. P., Shibata Y., Hall H., Greenleaf A. L., Strahl B. D. (2005) Mol. Cell. Biol. 25, 3305–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrozza M. J., Li B., Florens L., Suganuma T., Swanson S. K., Lee K. K., Shia W. J., Anderson S., Yates J., Washburn M. P., Workman J. L. (2005) Cell 123, 581–592 [DOI] [PubMed] [Google Scholar]
- 9.Keogh M. C., Kurdistani S. K., Morris S. A., Ahn S. H., Podolny V., Collins S. R., Schuldiner M., Chin K., Punna T., Thompson N. J., Boone C., Emili A., Weissman J. S., Hughes T. R., Strahl B. D., Grunstein M., Greenblatt J. F., Buratowski S., Krogan N. J. (2005) Cell 123, 593–605 [DOI] [PubMed] [Google Scholar]
- 10.Bell O., Wirbelauer C., Hild M., Scharf A. N., Schwaiger M., MacAlpine D. M., Zilbermann F., van Leeuwen F., Bell S. P., Imhof A., Garza D., Peters A. H., Schübeler D. (2007) EMBO J. 26, 4974–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bender L. B., Suh J., Carroll C. R., Fong Y., Fingerman I. M., Briggs S. D., Cao R., Zhang Y., Reinke V., Strome S. (2006) Development 133, 3907–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang N., vom Baur E., Garnier J. M., Lerouge T., Vonesch J. L., Lutz Y., Chambon P., Losson R. (1998) EMBO J. 17, 3398–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marango J., Shimoyama M., Nishio H., Meyer J. A., Min D. J., Sirulnik A., Martinez-Martinez Y., Chesi M., Bergsagel P. L., Zhou M. M., Waxman S., Leibovitch B. A., Walsh M. J., Licht J. D. (2008) Blood 111, 3145–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang H. B., Choi Y., Lee J. M., Choi K. C., Kim H. C., Yoo J. Y., Lee Y. H., Yoon H. G. (2009) FEBS Lett. 583, 1880–1886 [DOI] [PubMed] [Google Scholar]
- 15.Kim J. Y., Kee H. J., Choe N. W., Kim S. M., Eom G. H., Baek H. J., Kook H., Kook H., Seo S. B. (2008) Mol. Cell. Biol. 28, 2023–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nimura K., Ura K., Shiratori H., Ikawa M., Okabe M., Schwartz R. J., Kaneda Y. (2009) Nature 460, 287–291 [DOI] [PubMed] [Google Scholar]
- 17.Kim S. M., Kee H. J., Eom G. H., Choe N. W., Kim J. Y., Kim Y. S., Kim S. K., Kook H., Kook H., Seo S. B. (2006) Biochem. Biophys. Res. Commun. 345, 318–323 [DOI] [PubMed] [Google Scholar]
- 18.Douglas J., Hanks S., Temple I. K., Davies S., Murray A., Upadhyaya M., Tomkins S., Hughes H. E., Cole T. R., Rahman N. (2003) Am. J. Hum. Genet. 72, 132–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurotaki N., Imaizumi K., Harada N., Masuno M., Kondoh T., Nagai T., Ohashi H., Naritomi K., Tsukahara M., Makita Y., Sugimoto T., Sonoda T., Hasegawa T., Chinen Y., Tomita Ha H. A., Kinoshita A., Mizuguchi T., Yoshiura Ki K., Ohta T., Kishino T., Fukushima Y., Niikawa N., Matsumoto N. (2002) Nat. Genet. 30, 365–366 [DOI] [PubMed] [Google Scholar]
- 20.Stec I., Wright T. J., van Ommen G. J., de Boer P. A., van Haeringen A., Moorman A. F., Altherr M. R., den Dunnen J. T. (1998) Hum. Mol. Genet. 7, 1071–1082 [DOI] [PubMed] [Google Scholar]
- 21.Wright T. J., Ricke D. O., Denison K., Abmayr S., Cotter P. D., Hirschhorn K., Keinänen M., McDonald-McGinn D., Somer M., Spinner N., Yang-Feng T., Zackai E., Altherr M. R. (1997) Hum. Mol. Genet. 6, 317–324 [DOI] [PubMed] [Google Scholar]
- 22.Wang G. G., Cai L., Pasillas M. P., Kamps M. P. (2007) Nat. Cell Biol. 9, 804–812 [DOI] [PubMed] [Google Scholar]
- 23.Jaju R. J., Fidler C., Haas O. A., Strickson A. J., Watkins F., Clark K., Cross N. C., Cheng J. F., Aplan P. D., Kearney L., Boultwood J., Wainscoat J. S. (2001) Blood 98, 1264–1267 [DOI] [PubMed] [Google Scholar]
- 24.Rosati R., La Starza R., Veronese A., Aventin A., Schwienbacher C., Vallespi T., Negrini M., Martelli M. F., Mecucci C. (2002) Blood 99, 3857–3860 [DOI] [PubMed] [Google Scholar]
- 25.Chesi M., Nardini E., Lim R. S., Smith K. D., Kuehl W. M., Bergsagel P. L. (1998) Blood 92, 3025–3034 [PubMed] [Google Scholar]
- 26.Angrand P. O., Apiou F., Stewart A. F., Dutrillaux B., Losson R., Chambon P. (2001) Genomics 74, 79–88 [DOI] [PubMed] [Google Scholar]
- 27.Dilworth F. J., Fromental-Ramain C., Remboutsika E., Benecke A., Chambon P. (1999) Proc. Natl. Acad. Sci. U. S. A. 96, 1995–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luger K., Rechsteiner T. J., Richmond T. J. (1999) Methods Enzymol. 304, 3–19 [DOI] [PubMed] [Google Scholar]
- 29.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 30.Orphanides G., LeRoy G., Chang C. H., Luse D. S., Reinberg D. (1998) Cell 92, 105–116 [DOI] [PubMed] [Google Scholar]
- 31.Simon M. D., Chu F., Racki L. R., de la Cruz C. C., Burlingame A. L., Panning B., Narlikar G. J., Shokat K. M. (2007) Cell 128, 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang J., Feng Q., Ketel C. S., Wang H., Cao R., Xia L., Erdjument-Bromage H., Tempst P., Simon J. A., Zhang Y. (2002) Curr. Biol. 12, 1086–1099 [DOI] [PubMed] [Google Scholar]
- 33.Edmunds J. W., Mahadevan L. C., Clayton A. L. (2008) EMBO J. 27, 406–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du H. N., Fingerman I. M., Briggs S. D. (2008) Genes Dev. 22, 2786–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li B., Jackson J., Simon M. D., Fleharty B., Gogol M., Seidel C., Workman J. L., Shilatifard A. (2009) J. Biol. Chem. 284, 7970–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Youdell M. L., Kizer K. O., Kisseleva-Romanova E., Fuchs S. M., Duro E., Strahl B. D., Mellor J. (2008) Mol. Cell. Biol. 28, 4915–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shilatifard A. (2008) Curr. Opin. Cell Biol. 20, 341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim T., Buratowski S. (2009) Cell 137, 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gregory G. D., Vakoc C. R., Rozovskaia T., Zheng X., Patel S., Nakamura T., Canaani E., Blobel G. A. (2007) Mol. Cell. Biol. 27, 8466–8479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X., Yeh S., Wu G., Hsu C. L., Wang L., Chiang T., Yang Y., Guo Y., Chang C. (2001) J. Biol. Chem. 276, 40417–40423 [DOI] [PubMed] [Google Scholar]
- 41.Todoerti K., Ronchetti D., Agnelli L., Castellani S., Marelli S., Deliliers G. L., Zanella A., Lombardi L., Neri A. (2005) Br. J. Haematol. 131, 214–218 [DOI] [PubMed] [Google Scholar]
- 42.Luger K., Richmond T. J. (1998) Curr. Opin. Struct. Biol. 8, 33–40 [DOI] [PubMed] [Google Scholar]
- 43.Zhang L., Freitas M. A. (2004) Int. J. Mass Spectrom. 234, 213–225 [Google Scholar]
- 44.Krajewski W. A., Nakamura T., Mazo A., Canaani E. (2005) Mol. Cell. Biol. 25, 1891–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]