Abstract
Calcium (Ca2+) is a key second messenger in eukaryotes where it regulates a diverse array of cellular processes in response to external stimuli. An important Ca2+ sensor in both animals and plants is calmodulin (CaM). In addition to evolutionarily conserved CaM, plants possess a unique family of CaM-like (CML) proteins. The majority of these CMLs have not yet been studied, and investigation into their physical properties and cellular functions will provide insight into Ca2+ signal transduction in plants. Here we describe the characterization of CML42, a 191-amino acid Ca2+-binding protein from Arabidopsis. Ca2+ binding to recombinant CML42 was assessed by fluorescence spectroscopy, NMR spectroscopy, microcalorimetry, and CD spectroscopy. CML42 displays significant α-helical secondary structure, binds three molecules of Ca2+ with affinities ranging from 30 to 430 nm, and undergoes a Ca2+-induced conformational change that results in the exposure of one or more hydrophobic regions. Gene expression analysis revealed CML42 transcripts at various stages of development and in many cell types, including the support cells, which surround trichomes (leaf hairs) on the leaf surface. Using yeast two-hybrid screening we identified a putative CML42 interactor; kinesin-interacting Ca2+-binding protein (KIC). Because KIC is a protein known to function in trichome development, we examined transgenic CML42 knockout plants and found that they possess aberrant trichomes with increased branching. Collectively, our data support a role for CML42 as a Ca2+ sensor that functions during cell branching in trichomes.
Introduction
The ability of organisms to sense and respond to environmental perturbation is critical for adaptation and survival. Biochemical pathways involved in response to stimuli often employ second messengers such as Ca2+ ions as key components of the signal transduction machinery. Changes in cytosolic Ca2+ concentrations are evoked by external stimuli and detected by the sensor class of Ca2+-binding proteins (Ca2+ sensors), which directly or indirectly mediate the cellular responses appropriate for the perceived stimulus (1). Ca2+ sensors such as calmodulin (CaM)4 function as relays through the binding and regulation of downstream targets (1–3). In plants, Ca2+ signaling plays a regulatory role in various developmental processes as well as in pathways involved in response to stimuli such as pathogen attack, drought, salinity, and temperature shock (3–5).
Among Ca2+-binding proteins, the EF-hand is the most prevalent structural motif for Ca2+-binding (reviewed in Refs. 6, 7). It is composed of a helix-loop-helix structure where a 12-residue Ca2+-binding loop bridges two α-helices. In the canonical model, Ca2+ is coordinated in the EF-hand Ca2+-binding loop by seven residues arranged in a pentagonal bipyramidal manner, where the carboxylate side chains of aspartate or glutamate residues are particularly important. Typically, EF-hand motifs occur as interacting pairs, which allow for binding of Ca2+ in a positively cooperative manner. In well studied EF-hand proteins, such as CaM and troponin-C (TnC), Ca2+-binding induces significant conformational changes in tertiary structure that facilitates association with downstream target proteins via exposure of a hydrophobic target binding surface. Thus, EF-hand Ca2+ sensors often play key roles in the transduction and amplification of cell signals.
In the model plant Arabidopsis, a survey of the genome predicts ∼250 EF-hand-containing proteins comprising six main phylogenetic groups (I–VI), underscoring the complexity of Ca2+ signaling in plants (8). The majority of these proteins remain unstudied, and many are unique to plants. One group, Group IV, includes several CaMs and some 50 CaM-like proteins (CMLs) whose sequence identity to CaM varies from ∼25% to 75% (9). In plants, CaM has been shown to function in various developmental and stress-response pathways, and a diverse array of CaM targets has been identified that includes transcription factors, ion channels and pumps, metabolic enzymes, signaling proteins such as kinases and phosphatases, and a large number of proteins of unknown function (3, 10, 11). In contrast, the extended family of CMLs, most of which appear to be unique to plants, remain largely uncharacterized at the biochemical and functional levels.
We have been examining the role of several CMLs from Arabidopsis (12, 13). CML42 (At4g20780) and CML43 (At5g44460) are two closely related proteins that display ∼35% identity to CaM. CML43 expression is constitutive in roots but is induced in leaves after pathogen infection and likely plays a role in Ca2+ signaling during pathogen response (12). In the present study, we examined the biochemical properties of CML42 as a Ca2+ sensor and identified a role for CML42 in trichome morphology. Trichomes (leaf hairs) are specialized, epidermal cells that are often branched in many plant species and are thought to play a variety of roles, including herbivore deterrence, UV irradiation protection, and reduction in transpiration (water loss) (14, 15). Due to their uniquely branched structure, trichomes are often studied as a model for cell morphology in plants. Previous work has shown an important role for Ca2+ and CaM in the control of branching in trichomes (16, 17). The present study provides new insight into the biochemical properties of CML42 and demonstrates a novel function for this protein in the Ca2+-mediated regulation of trichome morphology.
EXPERIMENTAL PROCEDURES
Plant Material and Growth Conditions
All experiments with Arabidopsis thaliana were conducted using Columbia ecotype (Col-0, wild type). Seeds were imbibed for 3 days at 4 °C and subsequently transferred to a growth chamber under 16-h light (150 μEinsteins m−2 s−1)/8-h dark photoperiod at 22 °C. A gene knockout T-DNA insertion line (SALK_041400C) for CML42 was provided by the SALK institute (18) and obtained from the Arabidopsis Biological Resource Center, The Ohio State University. Southern blotting confirmed the presence of a single T-DNA insertion in this transgenic line (supplemental Fig. S1).
CML42 Vector Construct Preparation and Subcloning
The initial cloning, subcloning, and expression of recombinant CML42 in Escherichia coli has been previously described (12). The CML42 coding region was further subcloned using standard laboratory protocols to generate the various plasmid constructs used in this work. Where necessary for directional subcloning strategies, PCR primers were designed to introduce appropriate restriction sites. All plasmid constructs were verified by DNA sequencing. To produce recombinant His6-tagged N-terminal region of CML42 (residues 1–106), primers were designed to amplify the N-terminal-encoding cDNA from the ATG start codon to the middle of the interdomain linker region (1–318 bp) using primers NdeI-CML42F (5′-CATATGGAGAGTAACAACAACGAG-3′) and XhoI-NtermCML42R (5′-GCTCGAGTTCTCCTCCTCCACAAGCTC-3′) and subcloned into the pET21a prokaryotic expression vector (Novagen) in-frame with the His6 tag to generate the plasmid construct NtermCML42-pET21a.
Recombinant CML42 Expression and Purification
Recombinant full-length CML42 was expressed in E. coli strain BL21(DE3)pLysS (Stratagene), and recombinant protein was purified using Ca2+-dependent phenyl-Sepharose chromatography as previously described (12), with the exception that bacterial cultures were grown at 30 °C for 4–6 h following the addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 0.5 mm. The bacterial expression of recombinant NtermCML42 was performed as described above for the full-length CML42 except that harvested cells were resuspended in Binding Buffer (25 mm Tris-HCl, pH 7.4, 25 mm NaCl, 40 mm imidazole), and the soluble extract was not supplemented with CaCl2. Recombinant NtermCML42 was purified using Chelating Sepharose Fast Flow resin (Amersham Biosciences) as per the manufacturer's instructions. Protein samples were then applied to a Hi-Load 16/60 Superdex 75 size-exclusion column (Amersham Biosciences) equilibrated with CaM elution buffer (25 mm Tris-HCl, pH 7.5, 2 mm EGTA) containing 1 mm dithiothreitol and eluted in 1-ml fractions using the same buffer. Fractions containing recombinant NtermCML42 were pooled, concentrated using an Amicon Ultra-15 centrifugal filtration (Millipore) device, flash frozen in liquid nitrogen, and stored at −80 °C. Protein samples were exchanged into buffers appropriate for various biophysical analyses by dialysis or via PD-10 Sephadex G-25M desalting columns (Amersham Biosciences). The protein concentrations used for all biophysical analyses were determined using a Bradford reagent (Bio-Rad) assay and known standard concentrations of either CML42 or NtermCML42 as determined by amino acid analysis (Alberta Peptide Institute, Calgary, Alberta, Canada).
Recombinant Protein Expression for NMR Analysis
Recombinant CML42 was uniformly 15N-labeled by growing E. coli BL21(DE3)pLysS harboring the CML42-pET5a construct in M9 minimal media supplemented with 15NH4Cl (1 g/liter) as the sole nitrogen source (Cambridge Isotope Laboratories, Andover, MA). Expression was carried out in a manner similar to that described above for the unlabeled CML42 construct, with the exception that, upon induction with isopropyl-β-d-thiogalactopyranoside, the bacterial cultures were supplemented with 10 ml of 15N-Bio-Express-1000 medium (Cambridge Isotope Laboratories) and allowed to grow for an additional 8–10 h. Purification of the uniformly 15N-labeled recombinant CML42 followed the same protocol as described for the unlabeled protein.
CD Spectroscopy
Far-UV CD spectra of CML42 were acquired from 260 to 179.5 nm on a rapid scanning monochromator fitted with a CD module (RSM 1000, OLIS, Bogart, GA) at room temperature using a 0.1-mm path length cylindrical quartz cuvette. Spectra were collected on samples containing 54–63 μm CML42 in 5 mm Tris-HCl, pH 6.9, 150 mm NaF supplemented with either 5 mm EGTA or 5 mm CaCl2. Samples were manually mixed in situ following every third spectrum collection to counteract apparent photolytic degradation of the proteins. Spectra from twelve scans were averaged and corrected for background by subtracting averaged buffer spectra collected under the same conditions. Molar ellipticity [θ] was calculated according to the formula [θ] = θ × 100/(nlc), where n represents the number of amino acids in the protein, l represents the path length in centimeters, and c represents the concentration in millimolar. Percentage of secondary structure was calculated from the averaged, corrected spectra using CDNN deconvolution software (19).
ITC
Isothermal scanning calorimetry (ITC) experiments were performed on a MicroCal VP-ITC microcalorimeter (MicroCal). In each experiment, 5-μl injections of either 400 μm or 800 μm CaCl2 were made to a 1.5-ml sample cell containing 20 μm CML42 or 20 μm NtermCML42 in 25 mm HEPES, pH 7.5, 100 mm NaCl, with or without 5 mm MgCl2. Experiments were conducted at 30 °C, with 58 injections at 180-s intervals. To ensure that the initial samples remained Ca2+-free, all buffers were prepared in acid-washed plastic. Buffer control runs were conducted to obtain a baseline for each experiment. The heat of dilution/mixing was determined in separate buffer sample and ligand-buffer control experiments; these values were subtracted from the experimental runs. Data were analyzed using Origin 5.1 software (MicroCal) to obtain values for stoichiometry (N) and dissociation constants (Kd). The best fitting model for each experiment was selected based on the experimental N value and minimization of the chi-square values by modifying the binding-type input parameters. All ITC experiments were performed in duplicate.
Fluorescence Spectroscopy
8-Anilino-1-naphthalenesulfonate (ANS) fluorescence spectroscopic studies were performed with 500 μm ANS and 50 μm CML42 in 25 mm HEPES, pH 7.5, 50 mm NaCl. Spectra of the recombinant protein in the presence of 5 mm EGTA or 5 mm CaCl2 were measured at room temperature in a 1.5-ml, 10-mm cuvette on a PerkinElmer Life Sciences LS50B luminescence spectrometer with the following parameters: excitation and emission wavelengths of 380 and 400–600 nm, respectively, and a slit width of 7 nm. A titration experiment was also performed by sequentially adding 2 mm CaCl2, and finally 10 mm EDTA and 10 mm EGTA to an ANS-supplemented apo-CML42 sample. Fluorescence emission readings, in arbitrary intensity units, were taken after vigorous mixing following each addition.
NMR Spectroscopy
Two-dimensional 1H-15N heteronuclear single-quantum coherence (HSQC) NMR spectra of 120 μm 15N-labeled full-length recombinant CML42 were acquired on a Varian INOVA 600-MHz spectrometer equipped with a pulse-field gradient triple-resonance cryoprobe at 25 °C. Samples comprised 10 mm Tris-HCl, pH 6.8, 25 mm KCl, 90% H2O/10% D2O supplemented with either 5 mm EGTA or 5 mm CaCl2. Spectra were also collected for incremental additions of CaCl2 to full-length apo-CML42 to saturating conditions, as determined by the cessation of change in the resonances. The experiments used the enhanced sensitivity pulsed-field gradient approach (20) and comprised a 1024 × 128 real data matrix, which was zero-filled once in each dimension. The proton chemical shifts were referenced to 0.0 ppm by use of the trimethylsilyl resonance of the 2,2-dimethyl-2-silapentane 5-sulfonate signal in the one-dimensional spectrum. Spectra were processed and analyzed by use of NMRPipe (21) and NMRview (22), respectively.
Yeast Two-hybrid Screening
The Matchmaker yeast two-hybrid system (Clontech, Mountain View, CA) was used in this study. The full CML42 coding sequence was excised from the NdeI and BamHI sites of the CML42:pET5a construct (12) and subcloned into the NdeI and BamHI sites of pGBKT7 bait expression vector in-frame with the GAL4 DNA-binding domain (CML42-pGBKT7). The same strategy was used to subclone CML42 into the NdeI and BamHI sites of the “prey” vector (pGADT7) in-frame with the GAL4 activation domain. Competent yeast (strain AH109) cells harboring CML42- pGBKT7 were transformed with a λ-ACT two-hybrid cDNA library (CD4–22, Arabidopsis Biological Resource Center, Columbus OH) according to the Clontech Matchmaker GAL4 Two-Hybrid System 3 User Manual. Transformed cells were screened with high stringency by plating on adenine/tryptophan/leucine/histidine selective dropout minimal medium (SD-LWAH, Clontech) plates. Plates were incubated at 30 °C for a total of 2 weeks to increase the likelihood of detecting weak interactions. Approximately 5 × 105 colonies were screened. Putative positive colonies were replated on SD-LWAH plates four consecutive times to purge plasmids allowing nonspecific reporter activation, and to ensure maintenance of the interaction. Putative interactors were further tested for activation of the MEL1 reporter using an X-galactosidase overlay assay as described in the Clontech Matchmaker manual. Strong interactors exhibited a blue colorimetric reaction within 1–2 h; weaker interactors took up to 10 h to develop. Prey plasmids, representing putative CML42 effectors, were isolated from yeast cultures, and cDNA inserts were sequenced (Genome Quebec Sequencing Platform, Montreal QC) for identification. To further confirm the interactions of the CML42 putative interactor, kinesin-like CaM-binding protein (KCBP)-interacting Ca2+-binding protein (KIC), the KIC cDNA was amplified by PCR using the forward and reverse primers NdeI-KICF (5′-CATATGGAACCAACCGAGAAATCTATG-3′) and SalI-KICR (5′-GTCGACAGGCATAGAAGAGAGATTGTG-3′), respectively, and subcloned into the NdeI and SalI sites of the bait vector (pGBKT7) and tested for the ability to interact with CML42 expressed from the prey vector (pGADT7). A positive control for the yeast two-hybrid assays (FEM-2 and FEM-3) was kindly provided by Dr. Ian Chin-Sang (23).
Expression and Purification of GST-tagged KIC
The full-length KIC coding sequence (556 bp) was amplified by PCR using forward and reverse primers SalI-KICF (5′-GTCGACATGGAACCAACCGAGAAATC-3′) and NotI-KICR (5′-GCGGCCGCTCAAGGCATAGAAGAGAGATTG-3′), respectively, and subcloned into pGEX-4T-3 (Amersham Biosciences) to create an N-terminal GST-tagged KIC fusion protein. This plasmid construct (KIC-pGEX-4T-3) was used for the expression of GST-KIC in E. coli BL21(DE3)pLysS cells as described above for recombinant CML42 and purified by glutathione-affinity chromatography as per manufacturer's instructions (Amersham Biosciences). Purity was assessed by SDS-PAGE following by protein staining.
Glutathione-Sepharose Co-precipitation Assay to Test for in Vitro CML42-KIC Interaction
A glutathione (GSH) Sepharose co-precipitation assay, followed by SDS-PAGE and immunoblotting, was performed to test for an interaction between CML42 and the GST-fused KIC in vitro. Approximately 250 ng of GST-fused KIC was combined with an equimolar quantity of CML42 in a total volume of 200 μl of Interaction Buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1.0 mm CaCl2). Similarly, equimolar amounts of CML42 and purified GST were tested as a negative control. The mixtures were added to 100 μl of GSH-agarose beads (Sigma-Aldrich) pre-equilibrated with Interaction Buffer, and incubated with gentle shaking for 1 h at room temperature. The samples were transferred to empty columns (Bio-Rad) and washed extensively (50 column volumes) with Interaction Buffer. The samples were then returned to microcentrifuge tubes, and the bound proteins were eluted in three 50-μl fractions with GST Elution Buffer (50 mm Tris-HCl, pH 8.0, 10 mm reduced GSH). Aliquots of each assay were subjected to SDS-PAGE followed by Western blot analysis using either anti-GST antiserum or anti-CML42/CML43 antiserum and colorimetric detection with 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium. Assays were also performed in the presence of 1 mm EGTA to test the Ca2+ dependence of the interaction.
CML42 Expression Analysis
CML β-glucuronidase (GUS)-reporter analysis was conducted using transgenic Arabidopsis (Col-0, wild type) plants. The CML42 GUS-reporter construct (CML42::GUS) comprised 0.8 kb (−0.8 kb to 0) of 5′ upstream genomic DNA that was amplified by PCR using the forward and reverse primers HindIII-CML42:pBI121F (5′-TGAAGCTTTCTGGTGTTAGATAGATTG-3′) and XbaI-CML42:pBI121R (5′-CGTCTAGATGTTATCTTGTGTTCTTCTTC-3′), respectively, and subcloned into the HindIII and XbaI sites of the binary plasmid pBI121 (Clontech). This promoter region comprised the full intergenic region between the CML42 start codon and the nearest 5′ upstream gene. Wild-type Arabidopsis plants were transformed via the floral dip method (24). Homozygous T4 generation plants were selected for transgene expression as previously described (13). In situ GUS staining was performed as described (25). Whole plants, seedlings, or specific plant tissue samples were harvested throughout all stages of development. Samples were transferred to 15-ml Falcon tubes or 1.5-ml microcentrifuge tubes containing 90% acetone and incubated at 4 °C for 30 min, followed by extensive washing with water. GUS staining solution (100 mm sodium phosphate buffer, pH 7.0, 10 mm EDTA, 0.1% Triton X-100, 2 mm potassium ferricyanide, 2 mm potassium ferrocyanide, 1 mg/ml 5-bromo-4-chloro-3-indolyl-beta-d-glucuronic acid (X-Gluc, Bioshop Canada Inc., Burlington, Ontario, Canada) was applied, and samples were stained overnight in the dark at 37 °C. Samples were washed with several changes of 70% ethanol, and stored in 70% ethanol to remove remaining plant pigments. Samples were collected and assessed for a minimum of five independent CML::GUS transgenic lines. Sample images were captured with a digital camera connected to a Zeiss DiscoveryV12 (Carl Zeiss Inc.) stereoscope.
Isolation and Analysis of Arabidopsis Trichomes
Arabidopsis plants (wild-type or cml42 knockouts) were grown as described above, and trichomes were isolated from leaves and stained as described (26). For counting trichome branch numbers, samples were mounted on slides and visualized using a Zeiss DiscoveryV12 stereoscope. All cryogenic scanning electron microscopy was performed using a JEOL 6400 scanning electron microscope (JEOL Ltd.) outfitted with an Oxford CT 1500C cryotransfer system and cold stage (Oxford Instruments). Images were captured using a Robinson backscattered electron detector (SPI Inc.) at an accelerating voltage of 6 kV. To examine the CML42 transcript level in cml42 knockout (or wild-type) plants, reverse transcription PCR was performed as described (13) using CML42-specific forward (CML42rtF, 5′-ATGGAGAGTAACAACAACGAG-3′) and reverse (CML42rtR, 5′-GAATCAAGAAGAAGGGATGAC-3′) primers. Actin was used as a control for reverse transcription-PCR analysis and to test for genomic contamination in samples as described (13).
Genetic Rescue of cml42 Mutants with CML42
Total genomic DNA was extracted from wild-type Arabidopsis tissues using the Qiagen Plant DNeasy kit. The full-length CML42 intronless region (576 bp), as well as the genomic (promoter) region upstream of the CML42 ATG translation start site to the predicted nearest-neighbor gene (−894 bp), was amplified by PCR using the pMDCpromoterF primer (5′-TACCTTGTAAAGCTTTATTGGTTTC-3′) and the CML42rtR primer. The 1470-bp PCR product was subsequently cloned into the pMDC99 complementation binary vector via Gateway® LR recombination reaction. The transcription of the transgene CML42 is thus driven by its endogenous promoter in the pMDC99 vector. The pMDC99:CML42 construct was transformed into Agrobacterium tumefaciens (strain LBA4404) and then into Arabidopsis cml42 knockout mutants as described above. Transformants were selected by growth on Murashige and Skoog media (Sigma-Aldrich) plates supplemented with 50 μg/ml hygromycin, and resistant seedlings were transferred to soil for self-pollination and propagation to homozygosity. Total RNA was extracted, and first-strand cDNA was obtained for the CML42-transformed cml42 “rescued” plants as described above. Reverse transcription-PCR, using primers flanking CML42 (CML42rtF and CML42rtR), was performed on cDNA templates to ensure the presence of the CML42 transcript in the CML42-transformed cml42 knockout plants. Genomic DNA was also extracted from these transgenic plants, and PCR was performed using T-DNA-specific forward (SALK Lbc1, 5′-GGACTCTTGTTCCAACATGG-3′) and reverse (CML42rtR) primers to confirm the presence of the T-DNA insert.
RESULTS
CML42 Undergoes Ca2+-induced Changes in Secondary and Tertiary Structure
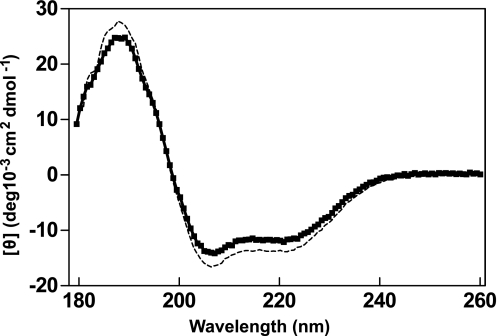
As depicted in Fig. 1, CML42 displays significant sequence identity (∼35%) with CaM, particularly within and adjacent to its three consensus EF-hand motifs (I, III, and IV). However, the sequence of CML42 corresponding to the second EF-hand motif of CaM diverges substantially from canonical architecture, such that an EF-hand-like Ca2+-binding site could not be unambiguously identified. Additionally, CML42 also contains an N-terminal extension of ∼10 residues and longer central linker region when compared with CaM. To assess the impact of Ca2+ binding on the conformation of CML42, we employed far-UV CD and heteronuclear NMR spectroscopy. The far-UV CD spectrum of apo-CML42 reveals a large positive band below 200 nm, with a maximum at 190 nm and local minima at 208 nm and 222 nm indicative of significant α-helical character (Fig. 2). The addition of Ca2+ to CML42 results in a slight decrease in molar ellipticity below 200 nm and a slight increase in the 205–230 nm range suggesting a Ca2+-induced decrease in α-helical content. Deconvolution of the data predicts a 2% decrease in helical content (31% to 29%) for CML42, which is in contrast to the 10% increase observed from CD studies of CaM (27). However, the similar helical content observed in the three-dimensional structures of apo-bound (28, 29) and Ca2+-bound (30) CaM led to the latter observations being attributed to helix reorientation (31). Such a reorientation could account for the modest change in helical content observed for CML42; however, an actual reduction in helical content cannot be ruled out.
FIGURE 1.
Amino acid sequence alignment of Arabidopsis CML42, CML43, and conserved CaM (CaM2). Black shading indicates identical amino acids, gray shading indicates conserved amino acids. Black bars and roman numerals above the residues indicate the CaM2 Ca2+-binding EF-hands. The non-canonical EF hand (II) of CML42 and CML43 is marked with an asterisk. GenBankTM accessions are Q9SVG9, Q9FI19, and NP_850344 for CML42, CML43, and CaM2, respectively.
FIGURE 2.
Ca2+ induces changes in secondary structure in CML42. Secondary structure characteristics of CML42 were monitored by far-UV CD spectroscopy in the presence of saturating (5 mm) CaCl2 (■) or EGTA (dashed line). Each spectrum is representative of at least twelve averaged scans and is reported in molar ellipticity ([θ]).
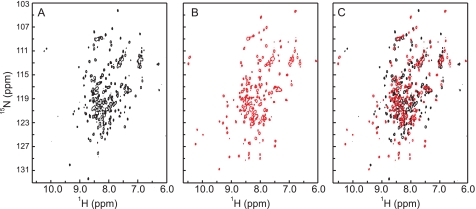
To obtain greater insight into the degree of Ca2+-induced structural changes in CML42, two-dimensional 1H-15N HSQC NMR spectra of uniformly 15N-labeled recombinant CML42 were collected in the absence or presence of Ca2+ (Fig. 3, A–C). In the absence of Ca2+, the 1H-15N HSQC spectrum of CML42 displays good chemical shift dispersion of 1H-15N correlation resonances, with approximately the number of resonances expected for the 186 non-proline amino acid residues comprising CML42 (Fig. 3A). The large number of resonances located in the central region of the spectrum (7.5–8.5 ppm) suggests a protein fold comprising substantial random coil and α-helical structure; an observation commensurate with the CD data described above. A subset of well resolved resonances with backbone HN chemical shifts of >8.5 ppm is indicative of extended or β-structure within CML42 and is consistent with the short strands that would be expected to immediately follow the Ca2+-binding loops within the EF-hand motifs. Addition of Ca2+ to saturating levels results in an increase in dispersion of backbone HN resonances, including the appearance of a larger number of HN resonances of >8.5 ppm (Fig. 3B). Overlays of the spectra of apo- and Ca2+-bound CML42 (Fig. 3C) illustrate that a substantial number of resonances change position upon addition of Ca2+ and suggest that CML42 undergoes a Ca2+-induced conformational change.
FIGURE 3.
Two-dimensional 1H-15N HSQC NMR spectra of uniformly 15N-labeled CML42. Spectra were collected in the absence (A) or presence (B) of Ca2+. The overlay of the two spectra (C) indicates the extent of the Ca2+-induced conformational changes. Spectra were recorded on 0.12 mm 15N-CML42 at 600-MHz frequency in the presence of 5 mm EGTA or 5 mm CaCl2.
One particularly notable feature of the apo- and Ca2+-bound spectra of CML42 that relates to Ca2+ binding involves the resonances located at 10–10.5 ppm in the 1H dimension and 111–113 ppm in the 15N dimension. Resonances found in this region are typical of the backbone HN resonances of the hinge position 6 of the EF-hand Ca2+-binding loop (typically a Gly residue) in the presence of Ca2+ (32–35). Although the identity of these resonances has yet to be confirmed, the presence of two backbone HN resonances in this region of the Ca2+-free spectrum (Fig. 3A) suggests that the regions around the hinge position 6 of two of the EF-hand motifs are in a preformed conformation similar to that of an EF-hand in the Ca2+-bound form. These resonances undergo a Ca2+-induced change in chemical shift (Fig. 3, B and C) and a third resonance appears in the same region under saturating Ca2+ concentrations, which suggests that at least three functional EF-hands exist within CML42.
CML42 Displays Ca2+ Sensor Properties
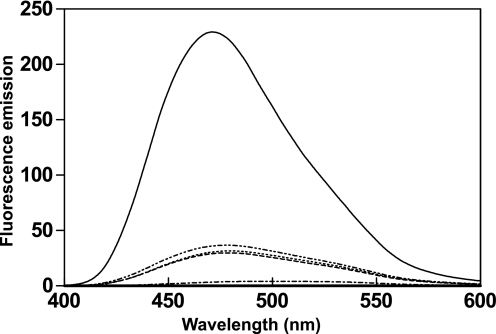
We previously demonstrated that CML42 displays an electrophoretic mobility shift in the presence of Ca2+ (12), typical of a Ca2+ sensor. Additional features of EF-hand Ca2+ sensors include the Ca2+-induced exposure of a hydrophobic surface responsible for target recognition and Ca2+ affinities tuned to detect Ca2+ fluxes within a cellular environment. To probe the effects of Ca2+ binding on surface hydrophobicity of CML42, ANS-based fluorescence was employed (Fig. 4). ANS is a hydrophobic compound that displays marked changes in its fluorescence emission profile, including a blue shift and an increase in intensity, when in contact with hydrophobic regions of proteins. A blue shift in ANS fluorescence, from 504 to 478.5 nm, occurs in the presence apo-CML42 with a relatively modest 7.2-fold increase in intensity, whereas Ca2+-CML42 caused a blue shift from 504 to 470 nm and a 55.4-fold increase in fluorescence, when compared with the emission spectrum of ANS alone. Addition of EDTA and EGTA returned fluorescence emissions to near-apo levels, illustrating the reversibility of Ca2+ binding and of the exposure of hydrophobic surface(s) in CML42 (Fig. 4).
FIGURE 4.
Changes in exposed hydrophobicity of CML42 in the presence of Ca2+. ANS fluorescence emission spectra (400–600 nm) were used to monitor the exposed hydrophobic surfaces of CML42. Scans were recorded for the unbound state (ANS and CML42 alone) (dashed line), and then sequentially following the addition of 5 mm MgCl2 (dotted line), 2 mm CaCl2 (solid line), and 5 mm each EGTA and EDTA (dash-dot-dot-dash line). ANS fluorescence in the absence of CML42 is also shown (dash-dot-dash line). The y axis represents fluorescence emission in arbitrary units.
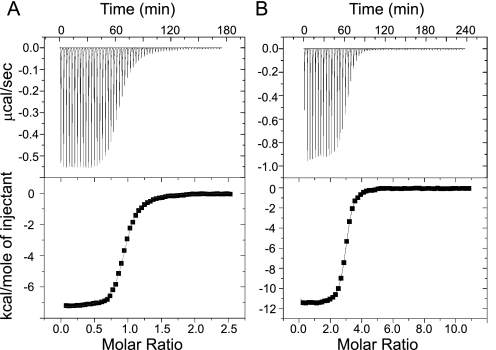
ITC studies were conducted to investigate the thermodynamic parameters of the CML42-Ca2+ interaction. Representative experimental ITC curves and the derived binding isotherms for Ca2+ binding to the isolated N-terminal domain of CML42 (NtermCML42) comprising residues 1–106 and full-length CML42 are presented in Fig. 5. Data for the NtermCML42-Ca2+ interaction was modeled to a single binding site based on the derived stoichiometry (N) of 1 with a dissociation constant (Kd) of 340 nm, although data for full-length CML42 best fit to a sequential 3-site binding model with Kd values of 300 nm, 7 nm, and 250 nm (Table 1). The latter model is also consistent with the experimental stoichiometry (N) of 3, and overall these observations are consistent with the sequence- based prediction that NtermCML42 comprises only one Ca2+-binding site (Fig. 1) (9). The CML42-Ca2+ interaction displays a negative enthalpy change (ΔH), indicative of an exothermic binding event (Fig. 5 and Table 1). This ΔH can be attributed almost entirely to Ca2+-induced conformational changes in the N- and C-terminal domains of CML42, because the ΔH associated with actual Ca2+ binding/desolvation can be approximated to be zero (36). Ca2+ affinities decreased by 20-fold for NtermCML42 and up to 4-fold for full-length when assessed in the presence of saturating amounts of Mg2+ (Table 1). These latter changes in Ca2+ affinity in this study are similar to the Mg2+-based changes in Ca2+ affinity seen for CaM (36). Despite the observed changes in Ca2+ affinity by CML42 associated with the presence of Mg2+, the values for two of the EF-hand Ca2+-binding sites in CML42 remain within the range expected for Ca2+ sensors (∼10−6-10−7 m) (6).
FIGURE 5.
ITC analysis of Ca2+ binding to NtermCML42 (A) and full-length CML42 (B). Representative data curves of NtermCML42-Ca2+ (residues 1–106) and full-length CML42-Ca2+ are presented. The top panels show the calorimetric titration. The lower panels show the corresponding integrated binding isotherm modeled to a single binding site for NtermCML42 and three sites for full-length CML42.
TABLE 1.
Thermodynamic parameters for calcium binding to CML42
| Construct | Binding sites used for modeling | Kda | ΔHb |
|---|---|---|---|
| μm | kcal/mol | ||
| NtermCML42 | 1 | 0.34 ± 0.02 | −7.31 ± 0.04 |
| NtermCML42 + Mg2+c | 1 | 7.14 ± 0.66 | −6.49 ± 0.15 |
| CML42 | 3 | 0.30 ± 0.09 | −11.87 ± 1.12 |
| 0.007 ± 0.002 | −7.78 ± 1.17 | ||
| 0.25 ± 0.02 | −9.90 ± 0.14 | ||
| CML42 + Mg2+ | 3 | 0.35 ± 0.02 | −10.79 ± 0.21 |
| 0.03 ± 0.01 | −12.65 ± 0.24 | ||
| 0.43 ± 0.01 | −10.44 ± 0.08 |
a The errors indicate the standard deviation from duplicate measurements.
b The reproducibility of ΔH for duplicate measurements was 0.3 kcal/mol or better.
c 5 mm MgCl2 was present in the samples.
CML42 Interacts with KIC, a Ca2+-binding Protein Associated with Trichome Branching
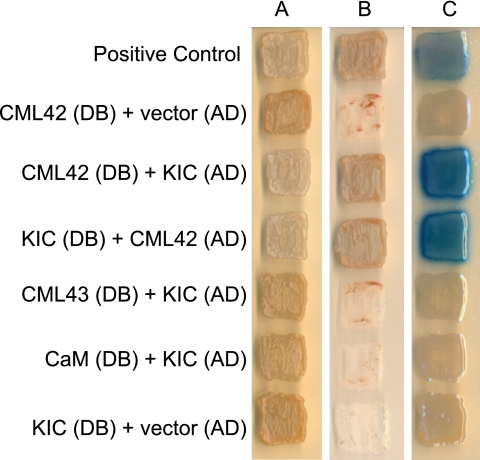
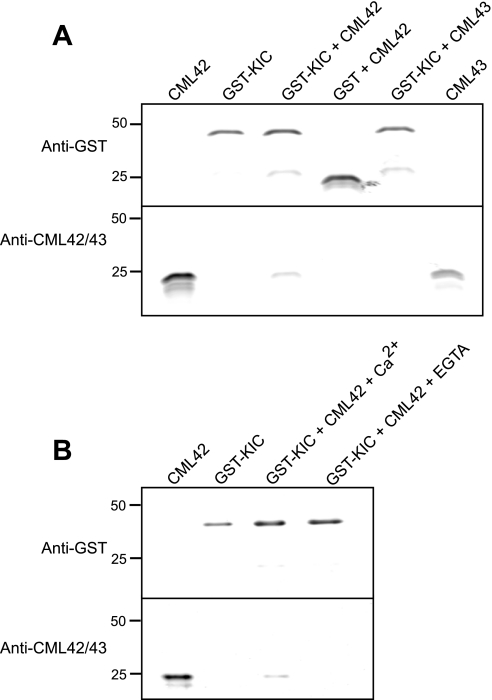
CaM exerts its effect as a Ca2+ sensor by interacting with a variety of downstream targets. To identify possible effectors of CML42, we conducted a yeast two-hybrid screen using CML42 as bait (Fig. 6). Three positive interactors were identified, and all represented a Ca2+-binding, caltractin/centrin-like protein (At2g46600) termed KIC (kinesin-interacting Ca2+-binding protein), which has been shown to play a role in regulating trichome branching through interaction with a specific kinesin, KCBP (kinesin calmodulin-binding protein), which is itself a CaM target (17). Reciprocal testing of CML42 and KIC as respective activation-domain (prey) and DNA-binding (bait) fusion proteins confirmed their interaction in the yeast two-hybrid system. Notably, neither CaM nor the CML42 homologue CML43 were able to associate with KIC in our yeast two-hybrid analyses indicating specificity of the CML42-KIC interaction (Fig. 6). To corroborate the yeast two-hybrid result and examine the effect of Ca2+ on CML42-KIC interaction, a co-precipitation assay with bacterially expressed recombinant GST-KIC and CML42 was performed (Fig. 7). CML42 co-precipitated with GST-KIC in a Ca2+-dependent manner but did not interact with GST alone. This interaction was specific to CML42, because CML43 did not co-precipitate with GST-KIC. These in vitro results support the yeast two-hybrid data and suggest that CML42-KIC interaction is both specific and Ca2+-dependent.
FIGURE 6.
Interaction between CML42 and KIC in the yeast two-hybrid system. Yeast cells containing the indicated combinations of CML42, KIC, CaM, and CML43 in the “bait” DNA-binding domain (DB) vector or “prey” activation-domain (AD) vector, as indicated, were grown on selective media and tested for interaction. Panels A, B, and C, respectively, show growth on media selective for the presence of both vectors (controls), dropout media selective for activation of the reporter genes indicating interaction of “bait” and “prey” fusion proteins, and reporter β-galactosidase activity (blue coloration) on media selective for bait and prey vectors. As a positive control for the two-hybrid analysis, two known interacting proteins (FEM2 and FEM-3 (39)) were also tested.
FIGURE 7.
Pulldown assays show CML42 and KIC interact in vitro in a Ca2+-dependent manner. A, in the presence of 1 mm Ca2+, recombinant GST-KIC or GST (negative control) were incubated with recombinant CML42 or CML43 as indicated above each lane. Glutathione-agarose beads were used to pull down GST or GST-KIC interacting proteins. Following washing, the proteins were eluted, electrophoresed in replicate gels, and blotted to nitrocellulose membranes, and immunoblotting was performed using either anti-GST (upper panels) or anti-CML42/CML43 antiserum (lower panels). Pure recombinant CML42 or CML43 were run in outer lanes as positional markers for immunodetection. B, similar pulldown assays and immunodetection as described in A were repeated in the presence of 1 mm CaCl2 or 1 mm EGTA as indicated. The electrophoretic mobility of molecular weight markers (kilodaltons) are shown on the left for both A and B.
CML42 Is Widely Expressed among Tissues and Is Found in Trichome Support Cells
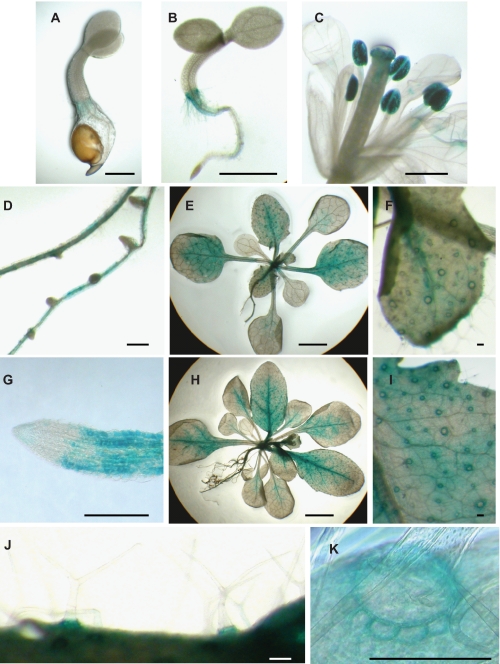
To investigate the transcript expression patterns of CML42 in Arabidopsis, transgenic plants containing fusion constructs comprising the CML42 promoter region and the GUS reporter enzyme (constructs CML42:: GUS) were tested for GUS activity throughout development (Fig. 8). The results shown represent GUS-staining patterns in at least five independent transgenic Arabidopsis lines. CML42::GUS was expressed at the hypocotyl-root interface beginning shortly after germination (Fig. 8, A and B). In floral tissue, GUS activity was detected in pollen grains, and diffuse expression appeared in the anther and filament of the stamens (Fig. 8C), whereas in roots expression was restricted to mature tissue and was absent from root meristems and the root cap (Fig. 8, D and G). In leaves, expression was present throughout young developing rosette (Fig. 8E) and cauline leaves (not shown) and receded to veins and the petiole as the leaves matured (Fig. 8H). Expression in leaves was localized primarily to the ring of cells surrounding the developing trichomes (Fig. 8, E, F, and H–K). Expression in these cells, termed trichome support (or socket) cells, began in the third rosette leaf and was present in all subsequent rosette and cauline leaves (Fig. 8, E and H). Notably, weak expression was detectable in the trichomes themselves (Fig. 8J). Expression ceased in trichome support cells at the distal edge of the expanding leaf blade, but continued in the younger leaf tissue (Fig. 8H). Control plants transformed with the empty pBI121 plasmid showed no detectable GUS activity (data not shown).
FIGURE 8.
CML42 is expressed broadly in Arabidopsis and is found in trichome support cells. Images showing GUS activity in CML42::GUS transgenic plants are presented of: A, 2-day-old seedling; B, 5-day-old seedling; C, fully developed flower; D, mature (35-day-old) roots; E, 3-week-old plant; F, developing rosette leaf of 3-week-old plant; G, root tip of mature (35-day-old) root; H, 4-week-old plant; I, base of expanded rosette leaf of 4-week-old plant; J, trichomes on a rosette leaf; K, support cells surrounding a trichome on a rosette leaf. Images are representative of staining in six independent transgenic lines. Scale bars: 0.5 mm (A–C); 2 mm (E and H); 250 μm (F and I); and 50 μm (D, G, J, and K).
Transgenic Plants Lacking CML42 Show Altered Trichome Branching
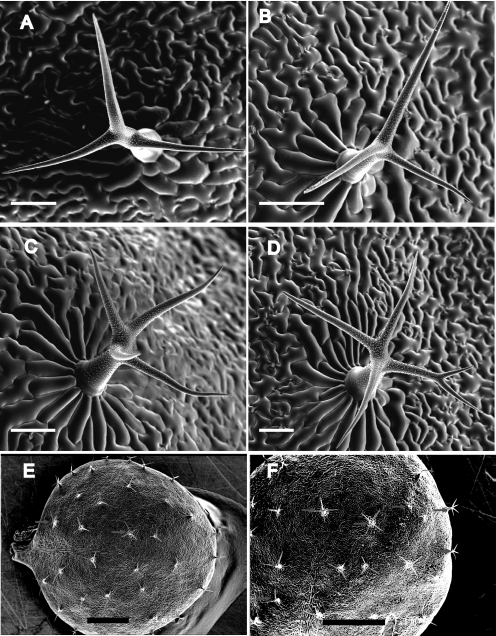
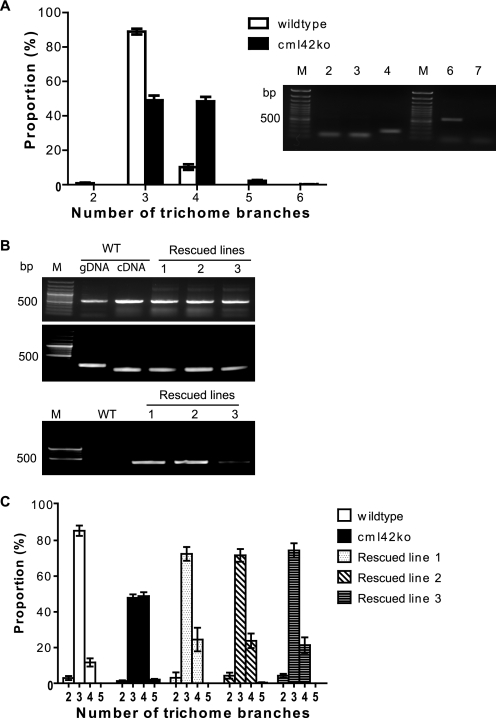
Given the interaction of CML42 with KIC (Figs. 6 and 7), a protein involved in the Ca2+-signaling pathways regulating trichome branching (17), and the strong expression of CML42 in trichome support cells (Fig. 8), we investigated the phenotype of transgenic knockout plants lacking CML42. These plants (cml42) were indistinguishable from wild-type plants with respect to all aspects of growth, development, and phenotype examined (data not shown), with the notable exception that cml42 knockout plants displayed abnormal trichome morphology with increased branches (Fig. 9). Whereas wild-type plants predominantly possessed three-branched trichomes (Fig. 10A), cml42 plants had a statistically significant (t test, p < 0.005) increase in trichomes with four or more branches. This phenotype co-segregated with the T-DNA insertion in the CML42 gene, and, using DNA hybridization blots, we confirmed that these knockout plants possess only a single T-DNA insertion (supplemental Fig. S1). Moreover, we genetically “rescued” the wild-type phenotype by expressing wild-type CML42 under its endogenous promoter in cml42 knockout plants (Fig. 10C). These data unequivocally implicate CML42 in the regulation of trichome branching. We also obtained transgenic knockout plants with a T-DNA insertion in KIC from ABRC, confirmed the absence of KIC transcripts in homozygous lines, but did not observe any alteration in their trichome morphology (data not shown).
FIGURE 9.
Transgenic knockout plants, cml42, show abnormal trichome branching. Representative cryo scanning electron micrograph images are presented of: A, a three-branched trichome from a wild-type plant; B–D, three-, four-, and five-branched trichomes, respectively, from a cml42 knockout plant. E and F, aerial views of a typical rosette leaf from wild-type and cml42 knockout plants, respectively, where arrows indicate trichomes with more than three branches. Scale bars: A–D = 100 μm; E and F = 1 mm.
FIGURE 10.
cml42 knockout plants have increased trichome branching and can be rescued by the wild-type CML42 gene. A, number of trichome branches (mean ± S.E.) of wild-type (open bars) compared with cml42 knockout (black bars) plants. Asterisks indicate statistically significant differences (t test, p < 0.005) between wild-type and cml42 knockout plants. Inset shows RT-PCR detection of CML42 transcript in leaf tissue. Actin controls are shown in lanes 2 and 3 for wild-type and cml42 knockout plants, respectively; lane 4 shows actin amplified from genomic DNA (intron present) derived from a cml42 knockout plant; lanes 6 and 7 show CML42 transcript amplification from wild-type and cml42 knockout plants, respectively; lanes M contain DNA ladders. B, RT-PCR (top panel) was conducted to examine the presence of CML42 transcript in wild-type (WT) and three independent transgenic lines of CML42-transformed cml42 knockout plants (rescued lines 1–3). Actin was used as a control (middle panel). C, PCR was also performed (lower panel) on genomic DNA to confirm that the CML42 rescued lines were from the cml42 knockout genetic background. The presence of a T-DNA insert (primers SALK LBc1 and CML42rtR) in the endogenous CML42 gene was detected only in the rescued lines. DNA marker (M) lanes are shown on the left of each panel. C, number of trichome branches (mean ± S.E.) in wild-type plants (open bars) compared with cml42 knockout plants (solid bars) and the three independent rescued lines described in B.
DISCUSSION
CML42 Is a Putative Ca2+ Sensor
During signal transduction, Ca2+ sensors undergo significant conformational changes induced by Ca2+-binding to at least one of their EF-hand motifs (6, 37). This structural rearrangement allows these proteins to elicit a cellular response, typically via hydrophobic interaction with target proteins. The broad range of Ca2+ affinities of Ca2+ sensors (typically from 10−4 to 10−7 m) reflects their diversity of function in responding to transient increases in [Ca2+]cyt (6, 38). CML42 displays characteristics reminiscent of a relay Ca2+ sensor, including the physical properties associated with Ca2+-induced conformational changes and Ca2+ affinity. Substantial changes in 1H-15N HSQC NMR spectra, upon the addition of Ca2+ (Fig. 5), reversible, Ca2+-dependent exposure of one or more hydrophobic regions as measured by ANS fluorescence (Fig. 2) and interaction with hydrophobic phenyl-Sepharose resin (“Experimental Procedures”), collectively illustrate the significant Ca2+-induced conformational changes in CML42. These properties are indicative of Ca2+ sensors rather than Ca2+-buffering proteins, the latter of which undergo only very minor structural changes and typically have a very high affinity for Ca2+ (6, 37, 38). The regulatory domains of Ca2+ sensors usually comprise two canonical EF-hand Ca2+-binding motifs, as seen for CaM and TnC (30, 37, 39). However, there are cases where only one canonical EF-hand is present, as seen for the S100 family (40). Three functional EF-hand motifs were confirmed by ITC (Table 1), and the NMR data (Fig. 5) suggest that two of the EF-hand Ca2+-binding sites adopt a Ca2+-ready conformation in apo-CML42, as illustrated by the presence of two HN resonances with HN and 15N chemical shifts typical for backbone amide groups of the invariant Gly at position 6 of the EF-hand loop in the Ca2+-bound state (33–35). Addition of Ca2+ resulted in the appearance of a third resonance, which is consistent with binding of Ca2+ to a third EF-hand motif. The fact that addition of excess chelator returned the spectrum to its apo-state (data not shown) suggests that the abovementioned resonances in the apo-state are not due to exogenous Ca2+ in the initial NMR sample.
Of the three identified functional EF-hand motifs, one appears to play a structural role based on its high affinity for Ca2+ (3 nm), whereas the other two fall into the sensory category as they display affinities relevant in magnitude to [Ca2+]cyt transients that occur in the cell during signaling events (3, 6, 8) (Table 1). The Ca2+ affinities of these latter two sites are comparable to those for other Ca2+ sensors (e.g. CaM) (36), and the Ca2+-binding properties of CML42 are also reminiscent of TnC, a mammalian EF-hand Ca2+-sensing protein involved in muscle contraction. TnC contains a C-terminal structural domain with two high affinity Ca2+-binding sites (Kd ≈ 10−7 m) that are constitutively Ca2+-bound, and an N-terminal regulatory domain containing two lower affinity sites (Kd ≈ 10−5 m) that specifically bind two Ca2+ ions upon increases in [Ca2+]cyt (32, 41). The Mg2+-induced change in Ca2+ affinity observed for full-length CML42 is similar to the effects observed in CaM (36) and TnC (41); this modest effect allows the EF-hands of CML42 to be classified as Ca2+-specific sites and not Ca2+/Mg2+ sites (6). Interestingly, the Ca2+ affinity of isolated N-terminal CML construct (NtermCML42) was substantially more affected by the presence of Mg2+, where a 20-fold change was observed (340 nm to 7 μm). This deviation may arise from a difference in the inherent stability of the isolated NtermCML42 construct relative to the full-length protein; an observation consistent with differential scanning calorimetric results (data not shown). In line with such a rationale, is the possibility that, unlike CaM and TnC, whose two domains are independent of one another and separated by a flexible linker, the two domains of CML42 may interact more extensively in the absence of a target protein, such as in the case for recoverin (42, 43) and sarcoplasmic Ca2+-binding proteins (44). While detailed structural studies are ongoing, the data presented in this study indicate that CML42 is a bona fide Ca2+ sensor.
CML42 Is Involved in Trichome Branching via Interaction with KIC
Genetic analysis of transgenic (knockout) plants lacking CML42 clearly implicate CML42 in the regulation of trichome branching (Figs. 9 and 10), and these data are supported by our demonstration that CML42 interacts in vitro with KIC, a known negative regulator of trichome branching (17). Trichomes are specialized epidermal structures that develop on the surfaces of leaves and other shoot-derived tissues. The physiological function of trichomes has not been clearly elucidated, however, they likely serve to increase the boundary layer between the epidermal tissue and the environment to reduce water loss and afford protection against insect herbivory or pathogen attack (14, 15). Arabidopsis trichomes are large, predominantly three-branched, unicellular structures that provide an ideal model for studies of polarized cell growth, proliferation, differentiation, intercellular communication, and morphogenesis. To date, about 30 genes are known to regulate aspects of trichome initiation and morphogenesis (15). In our studies, transgenic knockout plants lacking CML42 expression show a statistically significant increase in trichome branch number, where trichomes display a predominance of four branches (versus three in wild type) and in many cases five or six branches (Fig. 9). At least six genes have been shown to regulate trichome branching, and many of these, such as KIC, are involved in microtubule function (14, 15). KIC is a 15-kDa, acidic Ca2+-binding protein possessing one EF-hand but lacking any other identifiable sequence motifs (17). A recent study implicated KIC in trichome morphogenesis through its interaction with KCBP, a kinesin-like CaM interactor (17). Importantly, we have demonstrated the binding specificity of KIC for CML42, as both CaM and CML43 (which shows ∼75% sequence identity to CML42) failed to associate in vitro with KIC. The identification of this target protein suggests a role for CML42 in processes important to trichome development. KIC interaction with KCBP is Ca2+-dependent and results in the inhibition of KCBP binding to microtubules (15, 17). It is noteworthy that KIC interacts with the CaM-binding domain of KCBP and inhibits the microtubule-stimulated ATPase activity of KCBP at a lower [Ca2+]cyt than CaM (17). Although the cellular role of KIC-KCBP interaction remains unclear, KCBP is recognized as a modulator of trichome growth and development (14–16). Although initially identified as a CaM-binding protein (45), genetic screening for trichome mutants identified KCBP as the product of the Arabidopsis ZWICHEL (ZWI) gene (16). Mutations in ZWI result in trichomes with a short stalk and a reduced number of branches, thereby establishing KCBP as a positive regulator of trichome branching (16, 46). Previous studies have demonstrated genetic interactions between KCBP and positive (ANGUSTIFOLIA or FURCA1) and negative (suppressor of zwi3 (SUZ)) trichome developmental regulators (47, 48). Notably, overexpression of KIC results in a mild zwi-like phenotype (reduced trichome branching), indicating disruption of KCBP interaction with microtubules and further illustrating the role of KIC as a negative regulator of KCBP, and thus trichome morphogenesis (17). The abnormal trichome-branching phenotype of cml42 is reminiscent of the Arabidopsis mutant suz2, which has been identified as a potential bypass suppressor that negates the requirement for KCBP and FURCA1 in trichome branching (46). The SUZ2 gene has not yet been identified, however, preliminary mapping studies have indicated that it is located on chromosome IV ∼9 centimorgans from the AG1 CAPS marker (46), which is consistent with the location of the CML42 gene (data not shown). Based on genetic analysis, it has been hypothesized that the protein products of SUZ2 and KCBP physically interact (46). Given the mapping location of SUZ2, the similar phenotype of suz2 and cml42 mutants, and the possibility that CML42 could interact with KCBP through a complex with KIC, it is possible that the SUZ2 gene is indeed CML42, and future studies should address this issue.
Ultimately, what emerges is a complex picture of the Ca2+-mediated regulation of trichome branching given that three EF-hand proteins, CaM, KIC, and CML42, all potentially impact KCBP function. Moreover, the Ca2+ concentration range at which KIC and CaM modulate KCBP activity in vitro (14) is comparable to the affinity of CML42 for Ca2+. Not surprisingly, there likely exists considerable functional overlap in the pathways controlling trichome branching, and this is reflected in the phenotype of various mutants (including cml42) where only a fraction of trichomes show aberrant morphology (14, 15). In contrast to cml42 mutants, KIC knockouts (kic transgenics lacking KIC) did not show altered trichome morphology (data not shown) suggesting that KIC is not essential for normal trichome development. It should be noted that, although we clearly detected CML42::GUS expression in support cells, we observed only weak expression in trichomes themselves (Fig. 8). Although it is intriguing to consider that CML42 in support cells may be directly influencing the branching pattern of the adjacent trichome, we speculate that the level of CML42 expression in trichomes was near the limits of detection in our GUS assay given that a recent transcriptome analysis of trichomes observed transcripts for CML42 as well as for KIC and KCBP (49). Interestingly, in addition to their clear involvement in trichome morphogenesis KCBP (ZWI) (45), KIC (17), and CML42 (Fig. 8) are broadly expressed in plant tissues, particularly in flowers, leaves, and roots. Furthermore, S-tagged KIC co-precipitated native KCBP from extracts of Arabidopsis flowers, pollen, and young seedlings (17). Taken together, these data raise the possibility that the CML42-KIC-KCBP relationship may not be exclusive to trichomes and their support cells and that perhaps these proteins play a role in Ca2+-mediated regulation of cell division, cytoskeletal rearrangement, or other “housekeeping” functions. It is worth noting that, to the best of our knowledge, CML42 is the most evolutionarily divergent member (35% identity to CaM) of the CML family in Arabidopsis (∼50 members) to have been biochemically and functionally examined and for which a downstream target has been identified. The paucity of data on CML function and target interaction has been a major hindrance to advancing our understanding of this gene family. Given the importance of Ca2+ signaling in plants, as additional studies on CMLs emerge, it will be interesting to see what other physiological roles these unique Ca2+-binding proteins are involved in.
Supplementary Material
Acknowledgments
We thank Kim Munro (Protein Function Discovery Facility, Queen's University) for technical assistance in the biophysical studies, Jonathan Plett (Biology, Queen's University) for assistance with the transmission electron microscopy imaging, and Dr. Ian Chin-Sang (Biology, Queen's University) for the gift of the yeast two-hybrid feminization-1/2 constructs.
This work was supported in part by National Science and Engineering Research Council (NSERC) of Canada Grants RGPIN-23887-05 (to W. S.) and RGPGP-312316 (to S. P. S.). Operation and maintenance of equipment within the Protein Function Discovery Facility was supported by Canadian Institutes of Health Research (CIHR) Resource Grant PRG-80162.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- CaM
- calmodulin
- CML
- calmodulin-like
- KIC
- kinesin-interacting calcium-binding protein
- ITC
- isothermal titration calorimetry
- ANS
- 8-anilino-1-naphthalenesulfonate
- HSQC
- heteronuclear single-quantum coherence
- KCBP
- kinesin-like calmodulin-binding protein
- GUS
- β-glucuronidase
- TnC
- troponin C
- GST
- glutathione S-transferase.
REFERENCES
- 1.Clapham D. E. (2007) Cell 131, 1047–1058 [DOI] [PubMed] [Google Scholar]
- 2.Berridge M. J., Bootman M. D., Roderick H. L. (2003) Nat. Rev. Mol. Cell Biol. 4, 517–529 [DOI] [PubMed] [Google Scholar]
- 3.Bouché N., Yellin A., Snedden W. A., Fromm H. (2005) Annu. Rev. Plant Biol. 56, 435–466 [DOI] [PubMed] [Google Scholar]
- 4.Lecourieux D., Ranjeva R., Pugin A. (2006) New Phytol. 171, 249–269 [DOI] [PubMed] [Google Scholar]
- 5.Gong D., Guo Y., Schumaker K. S., Zhu J. K. (2004) Plant Physiol. 134, 919–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gifford J. L., Walsh M. P., Vogel H. J. (2007) Biochem. J. 405, 199–221 [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharya S., Bunick C. G., Chazin W. J. (2004) Biochim. Biophys. Acta 1742, 69–79 [DOI] [PubMed] [Google Scholar]
- 8.Day I. S., Reddy V. S., Ali G. S., Reddy A. S. (2002) Genome Biol. 3, 56.1–56.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCormack E., Braam J. (2003) New Phytol. 159, 585–598 [DOI] [PubMed] [Google Scholar]
- 10.Yang T., Poovaiah B. W. (2003) Trends Plant Sci. 8, 505–512 [DOI] [PubMed] [Google Scholar]
- 11.Reddy V. S., Ali G. S., Reddy A. S. (2002) J. Biol. Chem. 277, 9840–9852 [DOI] [PubMed] [Google Scholar]
- 12.Chiasson D., Ekengren S. K., Martin G. B., Dobney S. L., Snedden W. A. (2005) Plant Mol. Biol. 58, 887–897 [DOI] [PubMed] [Google Scholar]
- 13.Vanderbeld B., Snedden W. A. (2007) Plant Mol. Biol. 64, 683–697 [DOI] [PubMed] [Google Scholar]
- 14.Ishida T., Kurata T., Okada K., Wada T. (2008) Annu. Rev. Plant Biol. 59, 365–386 [DOI] [PubMed] [Google Scholar]
- 15.Schellmann S., Hülskamp M. (2005) Int. J. Dev. Biol. 49, 579–584 [DOI] [PubMed] [Google Scholar]
- 16.Oppenheimer D. G., Pollock M. A., Vacik J., Szymanski D. B., Ericson B., Feldmann K., Marks M. D. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6261–6266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy V. S., Day I. S., Thomas T., Reddy A. S. (2004) Plant Cell 16, 185–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., Stevenson D. K., Zimmerman J., Barajas P., Cheuk R., Gadrinab C., Heller C., Jeske A., Koesema E., Meyers C. C., Parker H., Prednis L., Ansari Y., Choy N., Deen H., Geralt M., Hazari N., Hom E., Karnes M., Mulholland C., Ndubaku R., Schmidt I., Guzman P., Aguilar-Henonin L., Schmid M., Weigel D., Carter D. E., Marchand T., Risseeuw E., Brogden D., Zeko A., Crosby W. L., Berry C. C., Ecker J. R. (2003) Science 301, 653–657 [DOI] [PubMed] [Google Scholar]
- 19.Böhm G., Muhr R., Jaenicke R. (1992) Protein Eng. 5, 191–195 [DOI] [PubMed] [Google Scholar]
- 20.Kay L. E., Keiffer P., Saarinen T. (1992) J. Am. Chem. Soc. 114, 10663–10665 [Google Scholar]
- 21.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 22.Johnson B. A. (2004) Methods Mol. Biol. 278, 313–352 [DOI] [PubMed] [Google Scholar]
- 23.Chin-Sang I. D., Spence A. M. (1996) Genes Dev. 10, 2314–2325 [DOI] [PubMed] [Google Scholar]
- 24.Clough S. J., Bent A. F. (1998) Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 25.Jefferson R. A., Kavanagh T. A., Bevan M. W. (1987) EMBO J. 6, 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X., Oppenheimer D. G. (2004) Plant Cell Physiol 45, 221–224 [DOI] [PubMed] [Google Scholar]
- 27.Martin S. R., Bayley P. M. (1986) Biochem. J. 238, 485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang M., Tanaka T., Ikura M. (1995) Nat. Struct. Biol. 2, 758–767 [DOI] [PubMed] [Google Scholar]
- 29.Finn B. E., Evenäs J., Drakenberg T., Waltho J. P., Thulin E., Forsén S. (1995) Nat. Struct. Biol. 2, 777–783 [DOI] [PubMed] [Google Scholar]
- 30.Babu Y. S., Bugg C. E., Cook W. J. (1988) J. Mol. Biol. 204, 191–204 [DOI] [PubMed] [Google Scholar]
- 31.Manning M. C. (1989) J. Pharm. Biomed. Anal. 7, 1103–1119 [DOI] [PubMed] [Google Scholar]
- 32.Krudy G. A., Brito R. M., Putkey J. A., Rosevear P. R. (1992) Biochemistry 31, 1595–1602 [DOI] [PubMed] [Google Scholar]
- 33.Slupsky C. M., Reinach F. C., Smillie L. B., Sykes B. D. (1995) Protein Sci. 4, 1279–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith S. P., Shaw G. S. (1997) J. Biomol. NMR 10, 77–88 [DOI] [PubMed] [Google Scholar]
- 35.Ikura M., Kay L. E., Bax A. (1990) Biochemistry 29, 4659–4667 [DOI] [PubMed] [Google Scholar]
- 36.Gilli R., Lafitte D., Lopez C., Kilhoffer M., Makarov A., Briand C., Haiech J. (1998) Biochemistry 37, 5450–5456 [DOI] [PubMed] [Google Scholar]
- 37.Ikura M. (1996) Trends Biochem. Sci. 21, 14–17 [PubMed] [Google Scholar]
- 38.Reddy V. S., Reddy A. S. (2004) Phytochemistry 65, 1745–1776 [DOI] [PubMed] [Google Scholar]
- 39.Herzberg O., James M. N. (1988) J. Mol. Biol. 203, 761–779 [DOI] [PubMed] [Google Scholar]
- 40.Kligman D., Hilt D. C. (1988) Trends Biochem. Sci. 13, 437–443 [DOI] [PubMed] [Google Scholar]
- 41.Potter J. D., Gergely J. (1975) J. Biol. Chem. 250, 4628–4633 [PubMed] [Google Scholar]
- 42.Tanaka T., Ames J. B., Harvey T. S., Stryer L., Ikura M. (1995) Nature 376, 444–447 [DOI] [PubMed] [Google Scholar]
- 43.Ames J. B., Ishima R., Tanaka T., Gordon J. I., Stryer L., Ikura M. (1997) Nature 389, 198–202 [DOI] [PubMed] [Google Scholar]
- 44.Vijay-Kumar S., Cook W. J. (1992) J. Mol. Biol. 224, 413–426 [DOI] [PubMed] [Google Scholar]
- 45.Reddy A. S., Safadi F., Narasimhulu S. B., Golovkin M., Hu X. (1996) J. Biol. Chem. 271, 7052–7060 [DOI] [PubMed] [Google Scholar]
- 46.Krishnakumar S., Oppenheimer D. G. (1999) Development 126, 3079–3088 [DOI] [PubMed] [Google Scholar]
- 47.Folkers U., Kirik V., Schöbinger U., Falk S., Krishnakumar S., Pollock M. A., Oppenheimer D. G., Day I., Reddy A. S., Jürgens G., Hülskamp M., Reddy A. R. (2002) EMBO J. 21, 1280–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo D., Oppenheimer D. G. (1999) Development 126, 5547–5557 [DOI] [PubMed] [Google Scholar]
- 49.Marks M. D., Betancur L., Gilding E., Chen F., Bauer S., Wenger J. P., Dixon R. A., Haigler C. H. (2008) Plant J. 56, 483–492 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.