Abstract
MicroRNAs are negative regulators of protein coding genes. The liver-specific microRNA-122 (miR-122) is frequently suppressed in primary hepatocellular carcinomas (HCCs). In situ hybridization demonstrated that miR-122 is abundantly expressed in hepatocytes but barely detectable in primary human HCCs. Ectopic expression of miR-122 in nonexpressing HepG2, Hep3B, and SK-Hep-1 cells reversed their tumorigenic properties such as growth, replication potential, clonogenic survival, anchorage-independent growth, migration, invasion, and tumor formation in nude mice. Further, miR-122-expressing HCC cells retained an epithelial phenotype that correlated with reduced Vimentin expression. ADAM10 (a distintegrin and metalloprotease family 10), serum response factor (SRF), and insulin-like growth factor 1 receptor (Igf1R) that promote tumorigenesis were validated as targets of miR-122 and were repressed by the microRNA. Conversely, depletion of the endogenous miR-122 in Huh-7 cells facilitated their tumorigenic properties with concomitant up-regulation of these targets. Expression of SRF or Igf1R partially reversed tumor suppressor function of miR-122. Further, miR-122 impeded angiogenic properties of endothelial cells in vitro. Notably, ADAM10, SRF, and Igf1R were up-regulated in primary human HCCs compared with the matching liver tissue. Co-labeling studies demonstrated exclusive localization of miR-122 in the benign livers, whereas SRF predominantly expressed in HCC. More importantly, growth and clonogenic survival of miR-122-expressing HCC cells were significantly reduced upon treatment with sorafenib, a multi-kinase inhibitor clinically effective against HCC. Collectively, these results suggest that the loss of multifunctional miR-122 contributes to the malignant phenotype of HCC cells, and miR-122 mimetic alone or in combination with anticancer drugs can be a promising therapeutic regimen against liver cancer.
Introduction
Hepatocellular carcinoma (HCC)4 is the fifth most common cancer, affecting over 500,000 people each year. Although most prevalent in Asia and sub-Saharan Africa, the incidence of HCC in the United States is on the rise, and among men, it is the fastest growing cause of cancer-related death (1). The major risk factors are hepatitis B and C virus infection, alcohol abuse, xenobiotics, primary biliary cirrhosis, diabetes, nonalcoholic fatty liver disease, and genetic disorders like hemochromatosis and α1-antitrypsin deficiency. The high mortality rate is due to its detection at the late stage with limited therapeutic options. The 5-year survival rate for liver cancer is only 5%, and the death rate is predicted to increase significantly in the next decade. Although primary HCCs are the most common form of liver cancer (>90%), liver malignancies can also be caused by metastasis of colon, prostate, or breast carcinomas. The disease is progressive and death usually occurs within 10 months of the initial diagnosis.
Although the molecular mechanism of liver tumorigenesis at the genetic level has been studied well, other potential mechanisms for hepatocarcinogenesis are only beginning to emerge. In recent years, there has been considerable interest in understanding the role of microRNAs in cancer (2, 3). MicroRNAs, which encode small noncoding RNAs of ∼21 nucleotides, are now recognized as a large gene family expressed in plants, animals, and viruses as well as in unicellular algae (4, 5). MicoRNAs act in concert to regulate expression of myriad target proteins either by impeding their translation or destabilizing their mRNAs (6). Most animal microRNAs are evolutionarily conserved and often found in clusters (7). Primary microRNAs with the stem-loop structure are transcribed by RNA polymerase II or RNA polymerase III and are processed both in the nucleus and cytoplasm by RNase III like enzymes such as Drosha and Dicer and associated co-factors to generate mature microRNAs. The mature microRNAs are recognized by specific proteins to form a microRNA-induced silencing complex, which facilitates its interaction with the cognate sequence predominantly located in the 3′-UTR of target mRNAs. Dysregulation of microRNA expression occurs in different diseases including cancer, and certain microRNAs function as oncogenes or tumor suppressors (8).
miR-122 was identified as the most abundant liver-specific microRNA, constituting 70% of total hepatic microRNA while cloning small RNAs from different tissues in mice (9, 10). Expression of this evolutionary conserved microRNA starts during gestation and attains a maximal level in the adult liver. miR-122 facilitates replication (11) and translation (12) of hepatitis C viral RNA and positively regulates cholesterol and triglyceride levels (13, 14). Microarray analysis of hepatic RNA from mice depleted of miR-122 by antagomirs has shown that most of the genes induced upon miR-122 depletion are normally repressed in the liver, implicating dedifferentiation of hepatocytes (15). Its expression has been shown to be regulated by circadian rhythm (16). miR-122 level is very high in mouse and human hepatocytes, but it is either silent or expressed at a very low level in most HCCs and transformed cell lines (9). We are the first to report that miR-122 is down-regulated in the preneoplasic nodules and HCCs developed in rats fed choline deficient diet as well as in primary human HCCs (17). Recently it has been shown that a loss of miR-122 expression correlates with poor prognosis and metastasis of liver cancer (18–20). It is also suppressed in patients with nonalcoholic fatty liver disease, which often leads to liver cancer (21). Thus, it is logical to conceive that the loss of function of miR-122 can predispose hepatocytes to neoplastic transformation. Here, we examined the tumor suppressor properties of miR-122 in vitro in HCC cells in culture and in nude mice and identified its targets that may play a causal role in hepatocarcinogenesis. Furthermore, our data also show that miR-122 potentiates growth inhibitory function of sorafenib in HCC cells.
EXPERIMENTAL PROCEDURES
Cell Culture, Treatment with Sorafenib, and Tissue Procurement
Human HCC cell lines (HepG2, Hep3B, and SK-Hep-1) obtained from ATCC were cultured according to the protocol provided by the supplier. The cells were harvested for RNA isolation, and whole cell extracts were subjected to Western blot analysis. Primary human hepatocellular cancer and adjacent normal tissue samples were obtained from the Cooperative Human Tissue Network at The Ohio State University James Cancer Hospital. Tissue specimens were procured in accordance with The Ohio State University Cancer Internal Review Board guidelines. HepG2-tet-off cells obtained from Clontech were cultured according to the supplier's protocol. HCC cells were treated with different concentrations of sorafenib (purchased from LC Biochemical) dissolved in dimethyl sulfoxide. Primary human dermal microvascular endothelial cells (HDMEC) purchased from Biowhittaker (Walkersville, MD) were maintained in endothelial cell basal medium-2 containing 5% fetal bovine serum and growth supplements.
Generation of stable HCC cell lines expressing miR-122 is described in the supplemental “Materials and Methods.” MicroRNA was isolated from formalin-fixed paraffin-embedded (FFP) blocks using miRVana kit (Ambion) as described (22).
TaqMan RT-PCR for Quantification of miR-122
RNA was isolated from FFP tissue sections as described (23). An aliquot (100 ng) of total RNA was treated with DNase I (2 units) at 37 °C for 15 min followed by heat inactivation of DNase I at 65 °C for 5 min. Ten ng of RNA was assayed using TaqMan MicroRNA kit (Applied Biosystems). miR-122 expression was normalized to 18 S rRNA or RNU6B using the 2−ΔCT method (24). Northern blot analysis was performed as described (25).
Real Time RT-PCR Analysis
ADAM10, SRF, GAPDH mRNA, and 18 S rRNA were measured in cDNA synthesized from DNase I-treated total RNA using the SYBR Green method (26). The primer sequences are provided in the supplemental materials.
Plasmid Construction and Generation of Stable Cell Lines
The miR-122 gene was amplified from mouse genomic DNA using Accuprime Taq polymerase (Invitrogen) and cloned into pBabe (SK-Hep-1), pMSCV-PIG (Hep3B), or p-RevTRE (Clontech) (HepG2) vectors (27). The cells were infected with retroviruses generated in phoenix cells as described (28). After 72 h the cells were selected with 2 μg/ml puromycin. The primers sequences are described in the supplemental data.
The 3′-UTR and the 3′-UTR with the miR-122 site deleted of ADAM10, Igf1R, and SRF were amplified from human lymphocyte DNA and cloned into TA cloning vector (Qiagen). Inserts were retrieved with MluI and NheI and cloned into the same sites of a luciferase reporter vector, pIS0 (29). The correct clones were confirmed by sequencing. The primers sequences are described in the supplemental data.
Transfection of Cells with miR-122 Mimic, Inhibitor (Anti-miR-122), and Respective Negative Control RNAs
HCC cells or HDMEC were transiently transfected with miR-122 mimetic (Dharmacon) or miR-122 inhibitor (Dharmacon) or respective negative control RNAs using Lipofectamine 2000 reagent (Invitrogen) following the manufacturer's protocol. After 24 h the cells were trypsinized, counted, and used for monitoring growth, migration, and invasion as described (30). For miR-122 assay and Western blot analysis cells were harvested for RNA and protein, respectively, after 48 h.
In Situ Hybridization (ISH) and Co-labeling
In situ detection of miR-122 in FFP tissue section with LNA-modified probe (Exicon) was determined as described (23, 24). A scrambled probe was used as the negative control. For co-labeling, the tissue sections were first subjected to ISH followed by immunohistochemical analysis with an anti-SRF antibody following published protocol (31).
Cell Proliferation Assay
Cell proliferation was monitored using cell proliferation reagent kit I (MTT) (Roche Applied Science) as described before (23). All of the experiments were performed in quadruplicate.
Thymidine incorporation assay was measured as described (26, 32). Soft agar assay was performed as described (33). Cell motility assay was performed as described (23). Cell invasion assay described in the supplemental “Materials and Methods.” Matrigel in vitro endothelial tube formation assay was performed as described (30).
Ex Vivo Tumor Growth of SK-Hep-1 Cells
Ex vivo tumor growth of SK-Hep-1 cells was performed as described (34). To image tumor growth in mice first Hep3B cells expressing luciferase were generated by transfecting pCMV-Luciferase and selecting with G418. These cells were next transfected with pMSCV-miR-122 or pMSCV alone and selected with puromycin. Five million of these cells were injected into the flanks of nude mice, and tumor growth was monitored every week using IVIS imaging system (Xenogen Corporation) immediately after injecting 4.2 mg of luciferin (Gold Biotech) in 150 μl of saline.
Western Blot Analysis
Proteins extracted from cells or tumor tissues were immunoblotted with different antibodies following published protocol (24). The catalogue number of the antibodies used is provided in the supplemental text). Protein was estimated using a Bio-Rad protein assay kit with bovine serum albumin as standard. Kodak Imaging software was used to quantify ethidium bromide-stained gels and scanned x-ray films (Western blot data).
Statistical Analysis
Statistical significance of differences between groups was analyzed by unpaired Student's t test, and p ≤ 0.05 was considered to be statistically significant. Paired Student's t test was used to analyze differences in expression of microRNAs and mRNAs levels among tumors and paired nontumor tissues in real time RT-PCR analysis. All real time RT-PCR (assayed in triplicate), Western blotting, and transfection experiments were repeated twice, and reproducible results were obtained. The correlation between miR-122 and Igf1R mRNA levels was analyzed by two-tailed Pearson correlation test. Single and double asterisks denote p ≤ 0.05 and ≤0.01, respectively.
RESULTS
miR-122 Expression Is Significantly Reduced in Primary Human HCCs
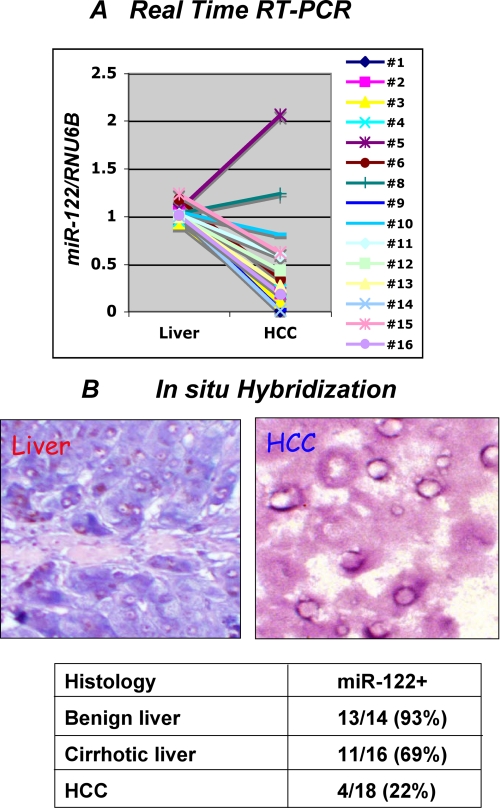
We have shown earlier by Northern blot analysis of RNA isolated from 20 frozen HCC samples that miR-122 is specifically suppressed in tumors (17). Here, we measured miR-122 expression in FFP tissues by TaqMan real time RT-PCR (n = 16) (see “Experimental Procedures” for details). The results showed that miR-122 expression was significantly decreased in 14 HCC samples and increased in two HCC samples compared with the adjacent normal liver (Fig. 1A). The normalized miR-122 level in each sample is presented in supplemental Table S1. We also analyzed miR-122 expression in FFP sections by LNA-ISH that showed its robust expression in benign liver tissues (blue color) but barely detectable in HCCs (Fig. 1B). LNA-ISH with scrambled RNA did not give any signal (data not shown). Analysis of 18 different HCC samples by LNA-ISH showed that miR-122 was detectable only in 22% of HCCs. Notably, miR-122 was also down-regulated (31%) in cirrhotic livers. Thus, the loss of miR-122 expression in the cirrhotic liver might predispose hepatocytes to transformation.
FIGURE 1.
Expression of miR-122 is reduced in primary HCCs. A, expression of miR-122 is significantly down-regulated in primary human HCCs compared with matching liver tissues. TaqMan RT-PCR assay using primer and probe for miR-122 and RNU6B (snRNA U6B) is shown. Each sample was analyzed in triplicate. The results are the means ± S.D. of three independent experiments. B, a representative in situ hybridization data showing miR-122 expression in the liver but not in HCC. Expression of miR-122 was detected by in situ hybridization with LNA-modified antisense miR-122 probe. Tissue sections were hybridized to biotin-labeled oligonucleotide (antisense miR-122 or scrambled), which was captured with alkaline phosphatase conjugated-streptavidin, and the signal (blue) was developed with nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate. The cell nuclei were stained with fast red dye. Quantification of miR-122-positive samples in different benign, cirrhotic, and HCC samples are presented in the lower panel.
miR-122 Inhibits Tumorigenic Properties of HCC Cells
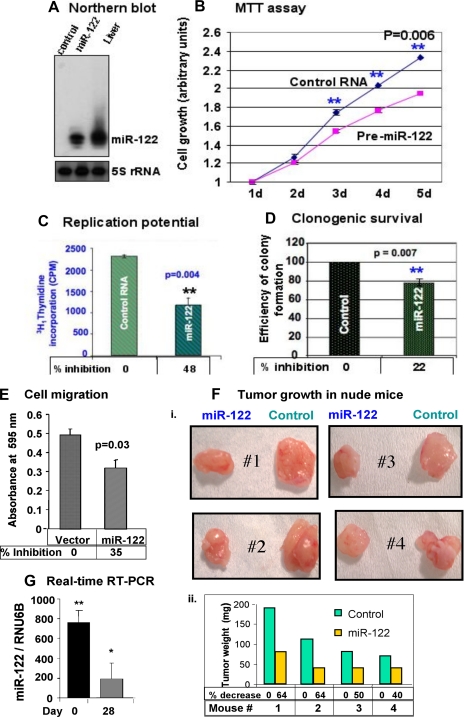
Next, we examined the anti-tumorigenic function of miR-122 in different human HCC cell lines by transfecting miR-122 mimetic or by generating stable cell lines expressing miR-122. miR-122 was detectable only in SK-Hep-1 cells transfected with miR-122 but at a level less than that in the liver (Fig. 2A). Functional studies showed miR-122 expression significantly inhibited growth of these cells, which increased with time (Fig. 2B) and correlated with a 48% reduction in their replication potential (Fig. 2C). Similarly, the ability of single cells to form colonies in soft agar was reduced by 22% upon ectopic expression of miR-122 (Fig. 2D and supplemental Fig. S1). miR-122 expression also impeded migration of SK-Hep-1 cells to serum-containing medium placed at the bottom chamber of a trans-well plate (Fig. 2E). miR-122 also compromised the ability of SK-Hep-1 cells to produce tumors in nude mice (Fig. 2F), another hallmark of cancer cells. Tumor growth decreased by 64, 64, 50, and 40%, respectively, in mice 1–4 compared with the controls. These results demonstrated tumor suppressor properties of miR-122 in SK-Hep-1 cells. A significant drop in miR-122 level in the tumors compared with that in SK-Hep-1 cells before transplantation to the nude mice probably explains the modest inhibition of tumor growth by this microRNA (Fig. 2G).
FIGURE 2.
Expression of ectopic miR-122 inhibited tumorigenic property of SK-Hep-1 cells. A, total RNA (15 μg) from these cells was subjected to Northern blot analysis with 32P-labeled anti-miR-122 or anti-5 S rRNA oligonucleotide as probe. B, cells (1000 cells/well) were seeded in 96-well plate, and cell growth was monitored every 24 h using MTT assay. d denotes days in culture. Each cell type was analyzed in quadruplicate. Absorbance at day 1 was assigned a value of 1. The results are the means ± S.D. of three independent experiments. C, cells (10,000 cells/well of 24-well plate) were serum-starved overnight followed by addition of serum and [3H]thymidine for 2 h, and [3H]thymidine incorporated into DNA was measured in a scintillation counter. Each experiment was performed in triplicate and was repeated twice. D, anchorage-independent growth was inhibited in miR-122-expressing cells. Cells (105 in 60-mm dish) were used for soft agar assay, and colonies formed after 2 weeks were stained with crystal violet and counted. Each sample was analyzed in triplicate. Efficiency of colony formation was determined by arbitrarily assigning the colonies formed in the control as 100. E, miR-122 inhibited cell migration through trans-well inserts (8-μm pore size). Cells (1 × 104) in serum-free medium layered onto the top chamber of a two chamber plate were allowed to migrate to the bottom chamber containing serum-supplemented medium for 48 h at 37 °C. The cells that migrated to the bottom of the insert were suspended in phosphate-buffered saline containing 5% acetic acid and 5% methanol, stained with Hema 3. The color developed was measured at 595 nm. Absorbance of cells that migrated to the bottom chamber containing serum-free medium was used as negative control. The results are the means ± S.D. of three experiments. F, miR-122 inhibited tumor growth in nude mice. Cells (2 × 106) transfected with control RNA or miR-122 mimetic were mixed with 50% Matrigel and injected subcutaneously to the flanks of nude mice. After 4 weeks, the tumors were excised and analyzed. Panel i, photograph of tumors developed in mice. Panel ii, average weight of the tumors developed in each group. G, miR-122 expression in cells before transplantation in nude mice and in tumors developed after 4 weeks by real time RT-PCR. The data were normalized to RNU6B.
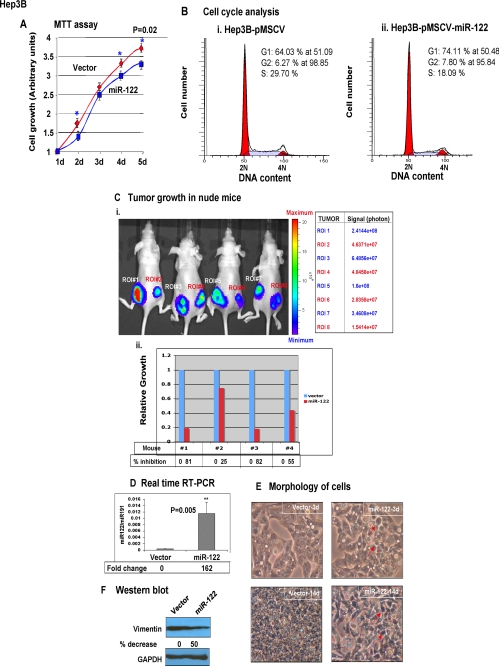
To confirm that the anti-tumorigenic property of miR-122 is not restricted to SK-Hep-1 cells, we tested its functions in two other HCC cell lines. Hep3B cells that do not express detectable miR-122 were transfected with a retroviral vector stably expressing miR-122 (supplemental Fig. S2A). There was a small but significant decrease in the growth of miR-122-expressing cells for 5 days in culture (Fig. 3A) that correlated with an increase in population of cells at the G1 phase (64 and 74% in control and miR-122-expressing cells, respectively) and a decrease in that at the S phase (30 and 18% in control and miR-122-expressing cells, respectively) (Fig. 3B). Similarly, clonogenic survival of Hep3B cells was also significantly compromised (∼50%) in miR-122-expressing cells (supplemental Fig. S2B). Next we tested the ability of miR-122 to impede Hep3B cell growth in nude mice using bioluminescence imaging (Fig. 3C). For this purpose stable cell lines co-expressing firefly luciferase and miR-122 and the empty vector were injected into the right and left flanks, respectively, of nude mice that were imaged every week. A representative photograph of the images of tumors developed in nude mice is shown in Fig. 3C (panel i). Quantification of the luciferase signal showed a reduction in the ability of miR-122-expressing cells to form tumors in all four mice, albeit at different levels (81, 25, 82, and 55% in mice 1–4, respectively) compared with that of controls (Fig. 3C, panel ii).
FIGURE 3.
Ectopic expression of miR-122 inhibited tumorigenic properties of Hep3B and HepG2 cells. A, growth of Hep3B cells was measured by MTT assay as described in the legend to Fig. 2. The results are means ± S.D. of three independent experiments. B, analysis of cell cycle profile miR-122-expressing and control Hep3B cells by fluorescence-activated cell sorter analysis. Cells (1 × 106) 72 h post-transfection were fixed overnight at −20 °C in 70% ethanol and washed, and incorporation of propidium iodide in cells treated with RNase A was measured in the FACSCaliber. C, tumor growth in nude mice. Hep3B cells (5 × 106 in 100 μl of phosphate-buffered saline) expressing luciferase alone or along with miR-122 were injected subcutaneously to the left and right flanks, respectively, of nude mice (see “Experimental Procedures” for details). Every week (up to 3 weeks), the mice were injected with luciferin (4.2 mg in 150 μl of saline) to monitor tumor growth by imaging with an IVIS system. The scale denotes the minimum and maximum photon intensity. Panel i, photograph of tumors developed in four mice. Panel ii, luciferase signal, region-of-interest (ROI), captured in each tumor in each mouse is represented. D, real time RT-PCR analysis of miR-122 in stable HepG2-tet-off cells expressing miR-122 or the vector. miR-122 expression was normalized to miR-191. E, morphology of miR-122-expressing HepG2-tet-off cells is distinct from those transfected with the vector. An identical number (1 × 106) of cells seeded in a 100-mm dish was allowed to grow for 14 days and was photographed on days 3 and 14 under a phase contrast microscope. F, Western blot analysis of Vimentin, a marker for mesenchymal cells, and GAPDH in the whole cell extracts. The Vimentin level normalized to that of GAPDH is presented.
Expression of miR-122 in another nonexpressing HCC cell line, HepG2, also resulted in a small but significant reduction in growth in culture and invasion through basement membrane (supplemental Fig. S3). To study the effect of miR-122 on morphology of these cells, we generated stable cell line expressing miR-122 (Fig. 3D). It is notable that the morphology of miR-122-expressing HepG2 cells was distinct from those transfected with the vector. After 3 days in culture both cell types showed epithelial morphology, but cell-cell contact was very prominent in miR-122-expressing cells (Fig. 3E). After 14 days in culture, epithelial morphology was lost in the control cells probably because of overgrowth and transition to mesenchymal phenotype, as evident from the expression of Vimentin, a marker for mesenchymal cells (Fig. 3F). In contrast, miR-122-expressing cells retained epithelial morphology even after 14 days in culture (Fig. 3E) that correlated with significant decrease (50%) in Vimentin expression in these cells (Fig. 3F), implicating the involvement of miR-122 in maintaining epithelial phenotype of HepG2 cells.
Next we investigated whether depletion of miR-122 from Huh-7 cells that express miR-122, albeit at a lower level than the liver (supplemental Fig. S4A), could enhance their tumorigenic properties. Transfection of anti-miR-122 (50 nm) resulted in a ∼50% decrease in its expression (supplemental Fig. S4A) that correlated with facilitated growth (supplemental Fig. S4B), replication potential (supplemental Fig. S4C), migration (supplemental Fig. S4D), and clonogenic survival (supplemental Fig. S4E) of these cells compared with those transfected with control RNA. Taken together, these results demonstrated anti-tumorigenic characteristics of miR-122 in HCC cells.
miR-122 Negatively Regulates Expression of ADAM10 (a Distintegrin and Metalloprotease Family 10), Which Is Up-regulated in Human Primary HCCs
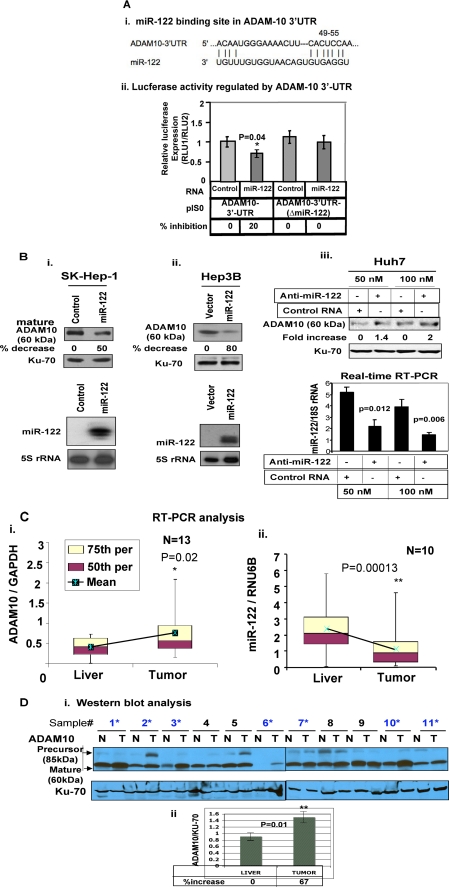
Next, we sought to identify the target mRNAs of miR-122 that might play a role in tumorigenesis. A data base search revealed several growth regulatory mRNAs that harbor conserved miR-122 recognition sites in their 3′-UTR. We focused our attention on ADAM10 (Fig. 4A, panel i) because ADAM family proteins are involved in various biological processes such as cell adhesion, cell fusion, cell migration and invasion, membrane protein shedding, and proteolysis (35, 36).
FIGURE 4.
ADAM10, a target of miR-122, is significantly up-regulated in primary human HCCs. A, panel i, the conserved miR-122 cognate site in 3′-UTR of ADAM10. Panel ii, luciferase activity driven by 3′-UTR of ADAM10 is inhibited by ectopic expression of miR-122. Hep3B cells were co-transfected with firefly luciferase-3′-UTR-(ADAM10) or 3′-UTR of ADAM10 deleted of miR-122 complementary site and miR-122 mimetic or control RNA (50 nm) along with pRL-TK (as an internal control) using Lipofectamine 2000. After 48 h, firefly (RLU-1) and Renilla luciferase (RLU-2) activities were measured using dual luciferase assay kit. The results represented as firefly luciferase normalized to Renilla luciferase are the means ± S.D. of quadruplicate experiments. B, cell extracts were subjected to immunoblot analysis, and the data were normalized to Ku-70. The corresponding expression of miR-122 is presented in the lower panels. Reproducible results were obtained in two independent experiments. C, ADAM10 mRNA and miR-122 were measured in HCCs and matching liver tissues by real time RT-PCR analysis. The data are presented as a box-whisker plot. The horizontal line in each box represents the median value of ADAM10 mRNA or miR-122 normalized to GAPDH or RNU6B, respectively. Boxes represent 50th and 75th percentile range of scores (as indicated), whereas the whiskers represent the highest and lowest values. D, Western blot analysis of ADAM10 in HCC and matching liver extracts. The signal in each lane was quantified using Kodak imaging software. Normalized data are presented in the lower panel.
To demonstrate that miR-122 directly regulates ADAM10 expression by interacting with its 3′-UTR, we co-transfected pIS0 harboring the 3′-UTR of ADAM10 downstream of firefly luciferase reporter along with miR-122 or control RNA. A small but significant decrease (20%, p = 0.04) in luciferase activity was observed specifically in pIS0-ADAM10 following expression of miR-122, which was abrogated when the miR-122 site was deleted from ADAM10–3′-UTR (Fig. 4A, panel ii). These results suggest that miR-122 negatively regulates ADAM10 expression by interacting with its 3′-UTR.
To confirm that miR-122 regulates endogenous ADAM10 protein expression in HCC cells, we measured its level in SK-Hep-1 and Hep3B cells expressing ectopic miR-122 or in Huh-7 cells depleted of endogenous miR-122. The results showed that ADAM10 level was reduced in SK-Hep-1 and Hep3B cells by 50 and 80%, respectively, upon ectopic expression of miR-122 (Fig. 4B). In contrast, its expression increased by 40 and 100%, respectively, in Huh-7 cells transfected with 50 and 100 nm anti-miR-122 (Fig. 4B). The relative miR-122 levels in each cell type are shown in the respective lower panels of Fig. 4B. These data also revealed that the effect of miR-122 on ADAM10 protein was more pronounced than on the heterologous luciferase protein. Notably, the RNA level of ADAM10 and ADAM17, another validated target of miR-122 (20), was also suppressed by miR-122 (supplemental Fig. S5).
Analysis of ADAM10 RNA level in primary human HCCs showed significant increase (p = 0.02) in tumors compared with matching liver tissues (Fig. 4C). In contrast, miR-122 was down-regulated in the majority of these HCC samples (Fig. 4C). Relative expression of ADAM10 and miR-122 RNA levels are provided in supplemental Tables S2 and S3, respectively. The ADAM10 protein level normalized to Ku-70 was also significantly (p = 0.01) elevated (∼67%) in HCCs compared with matching liver tissues (Fig. 4D). These results suggest that the suppression of miR-122 may be one of the mechanisms involved in the up-regulation of ADAM10 in primary HCCs.
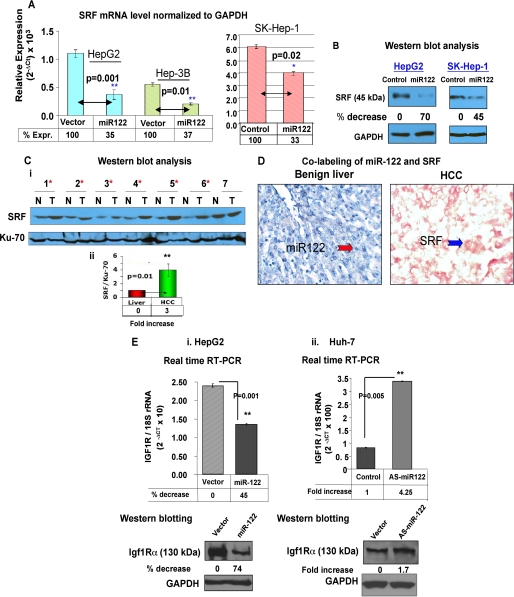
SRF, a Target of miR-122, Is Up-regulated in Primary Human HCCs
SRF is a ubiquitously expressed, pleiotropic transcription factor that activates immediate early genes in response to growth factors and mitogenic stimuli by binding to serum response elements (37). It regulates cell proliferation, differentiation, and cytoskeletal reorganization. We next focused our attention on SRF because it facilitates epithelial mesenchymal transition (38) and angiogenesis of endothelial cells (39). Because 3′-UTR of SRF encompasses a conserved miR-122 cognate site (supplemental Fig. S6A), we attempted to confirm whether SRF was a true target of miR-122. For this purpose, luciferase activity was measured in cells transfected with pIS0-3′-UTR-SRF (wild type) or pIS0-3′-UTR(Δ-miR-122)-SRF along with miR-122 mimic or control RNA. The results showed that miR-122 expression reduced luciferase activity by 35% in cells expressing the wild type but not in cells expressing the mutant reporter (supplemental Fig. S6B). Thus, miR-122 regulates luciferase activity by directly interacting with 3′-UTR of SRF. Real time RT-PCR analysis showed significant reduction in SRF mRNA level in HepG2 (65%), Hep3B (63%), and SK-Hep-1 (33%) cells expressing miR-122 (Fig. 5A). The SRF protein level was also reduced by 70 and 45% in HepG2 and SK-Hep-1 cells, respectively, upon ectopic expression of miR-122 (Fig. 5B).
FIGURE 5.
SRF and Igf1R are targets of miR-122. A, SRF mRNA level in HCC cells transfected with (50 nm) control RNA or mimetic (HepG2, Hep3B, and SK-Hep-1) was measured by real time RT-PCR, and the data were normalized to GAPDH. B, SRF protein level in HCC cells expressing miR-122. Western blot analysis of SRF and GAPDH in extracts of cells transfected with control or miR-122 mimetic (50 nm). C, panel i, Western blot analysis of SRF in HCCs and matching liver tissues. Whole tissue extracts were subjected to immunoblot analysis with SRF and Ku-70 antibodies. The asterisks denote samples showing up-regulation of SRF. Panel ii, SRF level normalized Ku-70 level. D, co-labeling of miR-122 and SRF in FFP sections of primary human HCC. Tissue sections were hybridized to biotinylated and LNA-modified antisense miR-122 probe, which was captured with alkaline phosphatase conjugated-streptavidin, and the signal (blue) was developed with nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate. Next, the section was subjected to immunohistochemistry with anti-SRF antibody using fast red dye as the chromogen. E, expression of Igf1R RNA (upper panels) and protein (lower panels) in cells transfected with miR-122 (in HepG2, panel i) or anti-miR-122 (in Huh-7, panel ii) by real time RT-PCR and Western blot analysis, respectively.
We then assessed the SRF level in primary human HCC samples. Western blot analysis showed an increase in the SRF level in 6 of 7 HCC tissues compared with the matching liver tissues (Fig. 5C). On average, SRF level increased by ∼3-fold in HCCs. SRF and miR-122 levels in tissues were also analyzed by combined LNA-ISH (to detect miR-122) and immunohistochemistry (to detect SRF) (Fig. 5D). As expected, miR-122 was abundantly expressed in the benign liver but not detectable in HCC (Fig. 5D, left panel). In contrast, SRF expression was high in the HCC but negligible in the liver (Fig. 5D, right panel). These results demonstrated reciprocal regulation of miR-122 and its target SRF in primary HCC samples.
miR-122 and Its Target Igf1R Are Reciprocally Regulated in Primary Human HCCs
Igf1R, a receptor tyrosine kinase with high affinity for IGF-1 and IGF-2, stimulates cell growth, survival, differentiation, and proliferation. Signaling through IGF1R regulates initiation, progression, and metastasis of cancer cells as well as resistance to therapy (40, 41). Because Igf1R harbors a conserved miR-122-binding site (supplemental Fig. S7A), we first validated it as target of miR-122. The luciferase activity controlled by Igf1R 3′-UTR was indeed reduced with by ectopic expression of miR-122 (supplemental Fig. S7B). Real time RT-PCR analysis demonstrated that miR-122 expression down-regulated Igf1R expression by 45% in HepG2 cells (Fig. 5E). Conversely, miR-122 depletion in Huh-7 cells resulted in 4.25-fold increase in its level. Western blot analysis confirmed that the Igf1R protein level was negatively regulated by miR-122 in HepG2 and Huh-7 cells (Fig. 5E).
The assessment of Igf1R expression by real time RT-PCR showed a significant increase in HCCs compared with the pair-matched controls (supplemental Fig. S7C). Correlation analysis revealed significant (p = 0.01) inverse correlation of Igf1R and miR-122 in primary tissues (r = −0.584) (supplemental Fig. S7D). These data confirm up-regulation of Igf1R in primary human HCCs and implicate miR-122 as one of the factors involved in its regulation.
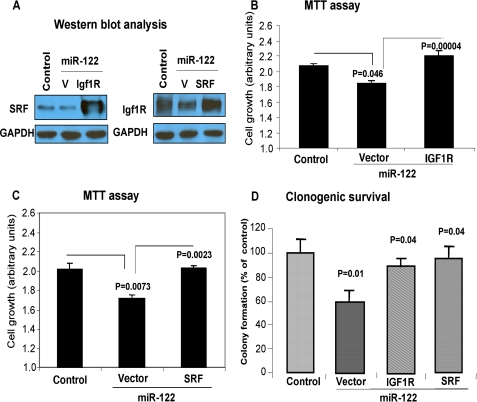
Ectopic Expression of SRF and Igf1R cDNA Can Reverse Growth Inhibitory Property of miR-122
To demonstrate that miR-122 functions, at least in part, are mediated through these targets, we transfected miR-122-expressing HCC cells with expression vectors for SRF or Igf1R lacking respective 3′-UTR. Western blot analysis demonstrated increased expression of these proteins in SK-Hep-1 cells compared with those transfected with the vectors (Fig. 6A). MTT assay showed that overexpression of Igf1R (Fig. 6B) and SRF (Fig. 6C) reversed the inhibitory effect of miR-122 on growth and clonogenic survival (Fig. 6D). Similar results were obtained in HepG2 cells (supplemental Fig. S8).
FIGURE 6.
Growth inhibitory property of miR-122 could be partially reversed by co-expression of SRF or Igf1R lacking 3′-UTR. SK-Hep-1 cells expressing miR-122 or vector were co-transfected with SRF, Igf1R expression vector, or corresponding empty vector. A, 48 h later cell extracts were split for Western blot analysis. B and C, MTT assay (72 h post-transfection). D, clonogenic survival of cells (2 weeks post-transfection).
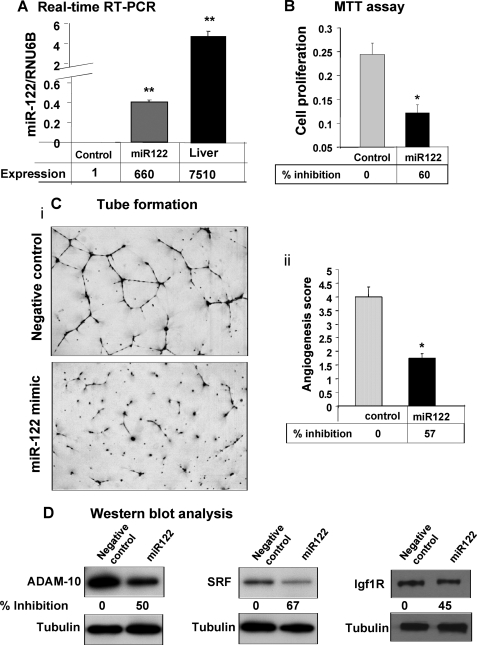
miR-122 Inhibits Angiogenic Potential of Endothelial Cells in Vitro
Formation of new blood vessels or angiogenesis by endothelial cells in the tumor microenvironment is required to sustain tumor growth (42). Angiogenesis is a multi-step process involving endothelial cell proliferation, migration, and capillary tube formation. Because miR-122 inhibited tumor growth in nude mice, we explored whether it directly affects angiogenic function of endothelial cells (HDMEC) in vitro. Transfection of miR-122 or control RNA in HDMEC cells resulted in a significant increase in miR-122 level but at a much lower level than in the liver (Fig. 7A). Growth of these cells was inhibited by 60% because of miR-122 expression (Fig. 7B). The motility of miR-122-expressing cells as measured by lateral migration of cells toward the scratch was significantly impeded (supplemental Fig. S9). Notably, the scratch remained open in cells expressing miR-122 even after 72 h. Similarly, the ability of these cells to form interconnected tubes in Matrigel was compromised (57% decrease) because of expression of miR-122 (Fig. 7C). Decreases in all three targets of miR-122, namely ADAM10, SRF, and Igf1R in endothelial cells (Fig. 7D), upon ectopic miR-122 expression implicate their roles in miR-122-mediated inhibition of angiogenic properties of these cells.
FIGURE 7.
miR-122 inhibits growth and tube formation of endothelial cells in vitro. HDMEC cells transfected with 50 nm of miR-122 or control RNA using Lipofectamine 2000 were trypsinized, counted, and used for different assays after 24 h cells. A, miR-122 level in cells 96 h post-transfection. B, proliferation of HDMEC cells 48 h post-transfection was measured by MTT assay. C, 1.6 × 103 cells in endothelial cell basal medium-2 supplemented with 2% serum were added to a Matrigel-coated well and incubated at 37 °C for 16–18 h. At the end of incubation, the culture medium was aspirated off the Matrigel surface, and the cells were fixed with methanol and stained with Diff-Quick solution II. Each chamber was photographed under microscope, and the total area occupied by endothelial cell derived tubes in each chamber was calculated using NIS-Elements-BS (Nikon) and expressed as an angiogenic score. D, Western blot analysis of cell lysates (72 h post-transfection) with specific antibodies. The levels of ADAM10, SRF, and Igf1R were normalized to tubulin.
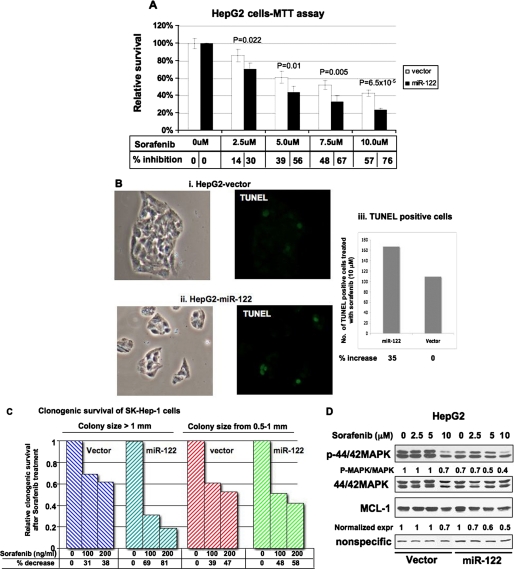
Expression of miR-122 Sensitizes HCC Cells to Sorafenib
Sorafenib is the only oral multi-kinase inhibitor recently approved by the Food and Drug Administration with demonstrated efficacy in enhancing overall survival in patients with advanced HCC (43). Because microRNAs can modulate the sensitivity of cancer cells to chemotherapeutic agents (22, 23), we tested whether miR-122 could sensitize cells to sorafenib. For this purpose we treated growing HCC cell lines expressing miR-122 with the drug and measured cell survival by MTT or colony formation assay. MTT assay showed that the survival of miR-122-expressing HepG2 cells was significantly reduced upon exposure to sorafenib at concentrations ranging from 2.5 to 10 μm compared with control cells (Fig. 8A). Similar results were obtained in Hep3B cells expressing miR-122 (supplemental Fig. S10). Quantification of TUNEL-positive HepG2 cells demonstrated a 35% increase in apoptotic cells expressing miR-122 after treatment with sorafenib (Fig. 8B). Apoptosis was negligible in untreated cells (data not shown). Similarly, clonogenic survival of miR-122-expressing SK-Hep-1 cells was significantly reduced in a dose-dependent manner upon treatment with sorafenib compared with the controls (Fig. 8C).
FIGURE 8.
Survival of miR-122-expressing HCC cells was significantly reduced after sorafenib treatment. A, vector-transfected and miR-122-expressing cells were seeded in a 96-well plate treated with different concentrations of sorafenib for 24 h, and cell survival was measured by MTT assay. Survival of untreated cells was taken as 100%. B, cells treated with 10 μm sorafenib for 48 h were subjected to TUNEL assay using an in situ cell death detection kit (Roche Applied Science), and positive cells in four fields at 10× magnification were counted. Panels i and ii, representative photographs of TUNEL-positive (green) and total number of cells (phase contrast). C, clonogenic survival of miR-122-expressing or vector transfected SK-Hep-1 cells in presence of sorafenib. Cells (500) were plated in a 60-mm dish followed by treatment with the drug 48 h later. Media and drugs were replaced every 72 h. The colonies were stained and counted after 10 days. D, Western blot analysis of cell extracts treated with sorafenib with different antibodies.
To understand the underlying mechanism, we measured the level of phospho-MAPK, a target of sorafenib, in HepG2 cells treated with different concentrations of sorafenib. The results showed that the p-MAPK level was high in control (vector transfected) cells and was marginally reduced only after treatment with 10 μm sorafenib (Fig. 8D). In contrast, the basal level of p-MAPK was at least 30% less in miR-122-expressing cells, which decreased further with increasing concentrations of sorafenib. Similarly, expression of anti-apoptotic Mcl-1 gradually decreased with increasing concentrations of sorafenib in miR-122-expressing cells. In contrast, it was reduced by 30% in control cells only after treatment with 10 μm sorafenib (Fig. 8D). Taken together, these results reveal that miR-122 expression promotes growth inhibitory property of sorafenib toward HCC cells.
DISCUSSION
Ever since Lagos-Quintana et al. (10) cloned miR-122 from the liver, lots of attention has been focused on trying to understand the functions of this developmentally regulated liver-specific microRNA (for review, see Ref. 44). It was one of the microRNAs that could be effectively depleted in the liver by systemic administration of modified anti-microRNA. Its depletion resulted in decrease in serum cholesterol and triglyceride, implicating its positive regulation of lipid metabolism (13, 14). This observation generated lots of excitement because of its potential therapeutic efficacy against hypercholesterolemia. We serendipitously identified miR-122 as a potential tumor suppressor while profiling microRNAs in a rodent model of nonalcoholic steatohepatitis-induced hepatocarcinogenesis (17). Extending this study to humans showed that the loss of miR-122 correlates with a poor prognosis and metastasis in primary human HCC patients (18–20). Furthermore, its decrease also in human nonalcoholic steatohepatitis patients (21) suggests that its loss of expression may be a prognostic marker for different liver disease including HCCs. Interestingly miR-122 can be a potential biomarker of liver damage because it is elevated in the serum of mice treated with acetaminophen (45).
A remarkable decrease in this liver-specific microRNA in HCCs suggested to us that this most abundant microRNA might play a role in maintaining hepatic functions, aberrations in which may lead to dedifferentiation of hepatocytes leading to hepatocellular carcinoma. In the present study we sought to examine anti-tumorigenic properties in vitro and ex vivo of miR-122 in HCC cells. Our results showed that miR-122 inhibits all characteristic properties of cancer cells such as clonogenic survival, anchorage-independent growth, migration, invasion, epithelial-mesenchymal transition, and the ability to form tumors in nude mice. These results support the notion that miR-122 functions as a tumor suppressor in the liver. In fact, activation of genes that are normally suppressed in the liver in mice depleted of endogenous miR-122 implicated its role in maintaining differentiation state of the liver. Furthermore, our results also demonstrate that miR-122 directly inhibits angiogenic properties of endothelial cells in vitro, implicating its potential application in anticancer therapy.
Among the predicted targets of miR-122 are factors involved in differentiation, cell cycle progression, inflammation, transcription, protein biosynthesis, cholesterol, and carbohydrate metabolism (reviewed in Ref. 21). Our study identified three key targets of miR-122 namely, ADAM10, Igf1R, and SRF, which play key roles in tumorigenesis in various cancers. In fact, antibodies and small molecule inhibitors against Igf1R are currently being tested as therapeutic regimen against different malignancies in preclinical and clinical trial.
It is known that certain microRNAs impart drug resistance (22) or sensitivity (23) to cancer cells. We were curious to examine whether miR-122 potentiates sensitivity of HCC cells to any chemotherapeutic agent. Until recently the overall survival rate of HCC patients was very poor because of late stage of detection and unavailability of effective drugs. Sorafenib is the only drug that improved overall survival of HCC patients (43). Because it is a broad spectrum inhibitor of different Ser/Thr and Tyr kinases including Igf1R, a target of miR-122, we tested whether miR-122 can synergize its inhibitory function. Promising results with HCC cells in culture provide the rationale for testing a combination therapy of miR-122 mimetic and sorafenib in animal models.
During our preparation of this manuscript, another group published a paper demonstrating tumor suppressor function of miR-122 in an orthotopic mouse model of HCC by targeting ADAM17 (20). ADAM family proteins ADAM10 and ADAM17 are involved growth factor, cytokine, and Notch signaling as well as cell-cell adhesion by virtue of their ectodomain sheddase activities (35, 36). Down-regulation of ADAMs can explain tumorigenic properties of HCCs because they are key regulators of epidermal growth factor receptor- and G protein-coupled receptor-induced migration and invasion in different tumors. Intense search is being pursued to develop effective inhibitors of ADAM10 and ADAM17 protease activities. Targeting of two ADAM family members by miR-122 suggests its potential therapeutic application in regulating these proteases.
SRF is a MADS (Mcm1, Ag, Defa, and Srf) box transcription factor that regulates multiple genes involved in cell growth, differentiation, migration, cytoskeletal organization, energy metabolism, and myogenesis (38). It has recently been reported that SRF is up-regulated specifically in high grade HCCs, and its expression facilitates migration, invasion, and mesenchymal transition of HCC cells (46). SRF is a downstream target of vascular endothelial growth factor signaling and is essential for VEGF-induced angiogenesis of endothelial cells. Our study has shown that miR-122 can directly inhibit angiogenesis in vitro with concomitant suppression of all three targets, namely ADAM10, SRF, and Igf1R.
All of the functional studies elucidating tumor suppressor properties of miR-122 are based on HCC cells in culture or ex vivo in mice. The true biological functions of miR-122 and its role in hepatocarcinogenesis can be addressed using knock-out mice. Studies along these lines are in progress.
Supplementary Material
Acknowledgments
We thank Drs. John Taylor and David Bartel for providing Huh-7 cells and pIS0, respectively. We thank Drs. Samson T. Jacob, Sarmila Majumdar, and Tasneem Motiwala for critically reading the manuscript. We also thank Dr. Sarmila Majumder for assistance with the TUNEL assay.
This work was supported, in whole or in part, by National Institutes of Health Grants CA122694 and CA086978.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Figs. S1–S10, and Tables S1–S3.
- HCC
- hepatocellular carcinoma
- miR-122
- microRNA-122
- SRF
- serum response factor
- LNA-ISH
- in situ hybridization with locked nucleic acid modified antisense probe
- TUNEL
- deoxynucleotidyltransferase-mediated dUTP nick end labeling
- Igf1R
- insulin-like growth factor 1 receptor
- UTR
- untranslated region
- HDMEC
- human dermal microvascular endothelial cell(s)
- FFP
- formalin-fixed paraffin-embedded
- RT
- reverse transcription
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- MAPK
- mitogen-activated protein kinase.
REFERENCES
- 1.El-Serag H. B., Rudolph K. L. (2007) Gastroenterology 132, 2557–2576 [DOI] [PubMed] [Google Scholar]
- 2.Visone R., Petrocca F., Croce C. M. (2008) Gastroenterology 135, 1866–1869 [DOI] [PubMed] [Google Scholar]
- 3.Braconi C., Patel T. (2008) Hepatology 47, 1807–1809 [DOI] [PubMed] [Google Scholar]
- 4.Bartel D. P. (2009) Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim V. N., Han J., Siomi M. C. (2009) Nat. Rev. Mol. Cell Biol. 10, 126–139 [DOI] [PubMed] [Google Scholar]
- 6.Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008) Nat. Rev. Genet. 9, 102–114 [DOI] [PubMed] [Google Scholar]
- 7.Kim V. N. (2005) Nat. Rev. Mol. Cell Biol. 6, 376–385 [DOI] [PubMed] [Google Scholar]
- 8.Calin G. A., Croce C. M. (2006) Nat. Rev. Cancer 6, 857–866 [DOI] [PubMed] [Google Scholar]
- 9.Chang J., Guo J. T., Jiang D., Guo H., Taylor J. M., Block T. M. (2008) J. Virol. 82, 8215–8223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T. (2002) Curr. Biol. 12, 735–739 [DOI] [PubMed] [Google Scholar]
- 11.Jopling C. L. (2008) Biochem. Soc. Trans. 36, 1220–1223 [DOI] [PubMed] [Google Scholar]
- 12.Henke J. I., Goergen D., Zheng J., Song Y., Schüttler C. G., Fehr C., Jünemann C., Niepmann M. (2008) EMBO J. 27, 3300–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esau C., Davis S., Murray S. F., Yu X. X., Pandey S. K., Pear M., Watts L., Booten S. L., Graham M., McKay R., Subramaniam A., Propp S., Lollo B. A., Freier S., Bennett C. F., Bhanot S., Monia B. P. (2006) Cell Metab. 3, 87–98 [DOI] [PubMed] [Google Scholar]
- 14.Krützfeldt J., Rajewsky N., Braich R., Rajeev K. G., Tuschl T., Manoharan M., Stoffel M. (2005) Nature 438, 685–689 [DOI] [PubMed] [Google Scholar]
- 15.Krützfeldt J., Kuwajima S., Braich R., Rajeev K. G., Pena J., Tuschl T., Manoharan M., Stoffel M. (2007) Nucleic Acids Res. 35, 2885–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gatfield D., Le Martelot G., Vejnar C. E., Gerlach D., Schaad O., Fleury-Olela F., Ruskeepää A. L., Oresic M., Esau C. C., Zdobnov E. M., Schibler U. (2009) Genes Dev. 23, 1313–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kutay H., Bai S., Datta J., Motiwala T., Pogribny I., Frankel W., Jacob S. T., Ghoshal K. (2006) J. Cell. Biochem. 99, 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coulouarn C., Factor V. M., Andersen J. B., Durkin M. E., Thorgeirsson S. S. (2009) Oncogene, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fornari F., Gramantieri L., Giovannini C., Veronese A., Ferracin M., Sabbioni S., Calin G. A., Grazi G. L., Croce C. M., Tavolari S., Chieco P., Negrini M., Bolondi L. (2009) Cancer Res. 69, 5761–5767 [DOI] [PubMed] [Google Scholar]
- 20.Tsai W. C., Hsu P. W., Lai T. C., Chau G. Y., Lin C. W., Chen C. M., Lin C. D., Liao Y. L., Wang J. L., Chau Y. P., Hsu M. T., Hsiao M., Huang H. D., Tsou A. P. (2009) Hepatology 49, 1571–1582 [DOI] [PubMed] [Google Scholar]
- 21.Cheung O., Puri P., Eicken C., Contos M. J., Mirshahi F., Maher J. W., Kellum J. M., Min H., Luketic V. A., Sanyal A. J. (2008) Hepatology 48, 1810–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller T. E., Ghoshal K., Ramaswamy B., Roy S., Datta J., Shapiro C. L., Jacob S., Majumder S. (2008) J. Biol. Chem. 283, 29897–29903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nasser M. W., Datta J., Nuovo G., Kutay H., Motiwala T., Majumder S., Wang B., Suster S., Jacob S. T., Ghoshal K. (2008) J. Biol. Chem. 283, 33394–33405 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Datta J., Kutay H., Nasser M. W., Nuovo G. J., Wang B., Majumder S., Liu C. G., Volinia S., Croce C. M., Schmittgen T. D., Ghoshal K., Jacob S. T. (2008) Cancer Res. 68, 5049–5058 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S. T., Patel T. (2007) Gastroenterology 133, 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghoshal K., Datta J., Majumder S., Bai S., Kutay H., Motiwala T., Jacob S. T. (2005) Mol. Cell. Biol. 25, 4727–4741 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. (2003) Cell 115, 787–798 [DOI] [PubMed] [Google Scholar]
- 28.Datta J., Majumder S., Kutay H., Motiwala T., Frankel W., Costa R., Cha H. C., MacDougald O. A., Jacob S. T., Ghoshal K. (2007) Cancer Res. 67, 2736–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis B. P., Burge C. B., Bartel D. P. (2005) Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 30.Kumar P., Gao Q., Ning Y., Wang Z., Krebsbach P. H., Polverini P. J. (2008) Mol. Cancer Ther. 7, 2060–2069 [DOI] [PubMed] [Google Scholar]
- 31.Nuovo G. J., Elton T. S., Nana-Sinkam P., Volinia S., Croce C. M., Schmittgen T. D. (2009) Nat. Protoc. 4, 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai S., Datta J., Jacob S. T., Ghoshal K. (2007) J. Biol. Chem. 282, 27171–27180 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Motiwala T., Kutay H., Ghoshal K., Bai S., Seimiya H., Tsuruo T., Suster S., Morrison C., Jacob S. T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13844–13849 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Motiwala T., Majumder S., Ghoshal K., Kutay H., Datta J., Roy S., Lucas D. M., Jacob S. T. (2009) J. Biol. Chem. 284, 455–464 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Duffy M. J., McKiernan E., O'Donovan N., McGowan P. M. (2009) Clin. Cancer Res. 15, 1140–1144 [DOI] [PubMed] [Google Scholar]
- 36.Maretzky T., Reiss K., Ludwig A., Buchholz J., Scholz F., Proksch E., de Strooper B., Hartmann D., Saftig P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9182–9187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarnawski A., Pai R., Chiou S. K., Chai J., Chu E. C. (2005) Biochem. Biophys. Res. Commun. 334, 207–212 [DOI] [PubMed] [Google Scholar]
- 38.Franco C. A., Li Z. (2009) Cell Adh. Migr. 3, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chai J., Jones M. K., Tarnawski A. S. (2004) FASEB J. 18, 1264–1266 [DOI] [PubMed] [Google Scholar]
- 40.Tanaka S., Arii S. (2009) Cancer Sci. 100, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Höpfner M., Huether A., Sutter A. P., Baradari V., Schuppan D., Scherübl H. (2006) Biochem. Pharmacol. 71, 1435–1448 [DOI] [PubMed] [Google Scholar]
- 42.Finn R. S., Zhu A. X. (2009) Exp. Rev. Anticancer Ther. 9, 503–509 [DOI] [PubMed] [Google Scholar]
- 43.Llovet J. M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J. F., de Oliveira A. C., Santoro A., Raoul J. L., Forner A., Schwartz M., Porta C., Zeuzem S., Bolondi L., Greten T. F., Galle P. R., Seitz J. F., Borbath I., Häussinger D., Giannaris T., Shan M., Moscovici M., Voliotis D., Bruix J. (2008) N. Engl. J. Med. 359, 378–390 [DOI] [PubMed] [Google Scholar]
- 44.Girard M., Jacquemin E., Munnich A., Lyonnet S., Henrion-Caude A. (2008) J. Hepatol. 48, 648–656 [DOI] [PubMed] [Google Scholar]
- 45.Wang K., Zhang S., Marzolf B., Troisch P., Brightman A., Hu Z., Hood L. E., Galas D. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4402–4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park M. Y., Kim K. R., Park H. S., Park B. H., Choi H. N., Jang K. Y., Chung M. J., Kang M. J., Lee D. G., Moon W. S. (2007) Int. J. Oncol. 31, 1309–1315 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.