Abstract
The duplex structure of DNA, with its internal base pairing, protects the nucleobases from chemical damage, but it also poses a barrier to DNA modifying enzymes, including the enzymes that recognize and repair DNA damage. It is known that unpaired, or bulged nucleotides are significantly more accessible, but it is not known whether they might be recognized by nucleotide flipping enzymes. We have investigated this question with human alkyladenine DNA glycosylase (AAG). AAG recognizes a wide variety of structurally disparate lesions, including deoxyinosine (I), which results from the spontaneous oxidative deamination of adenosine, and catalyzes the hydrolysis of the N-glycosidic bond to release the lesion base and initiate the base excision repair pathway. We used single turnover kinetics to characterize the reaction of AAG towards synthetic 25-mer oligonucleotides containing a single I lesion in single-stranded, mismatched, or single nucleotide bulge contexts. We found that AAG has the highest catalytic efficiency towards a lesion that is presented in a single nucleotide bulge. In contrast, AAG has more than 2000-fold reduced catalytic efficiency towards a single stranded I-containing oligonucleotide relative to the duplexes. We observe 20-fold differences in catalytic efficiency for the excision of the presumed biological target (paired with T), relative to alternative pairings such as C that might be formed by the replication of an unrepaired I. Furthermore, a linear free energy relationship shows a strong inverse correlation between duplex stability and catalytic efficiency (slope = −0.6 to −1.0), indicating that gaining access to the base lesion provides a substantial barrier to AAG-catalyzed initiation of DNA repair. The observation that AAG recognizes a single nucleotide bulge as efficiently as a mismatch implies that the recognition of DNA damage is remarkably plastic.
The double-helical structure of DNA protects the nucleobases from oxidative and alkylative chemical damage.1 However, this internal base pairing is also a barrier to the enzymes that recognize and repair DNA damage.2 Enzymes that modify bases in DNA use nucleotide flipping to rotate the target nucleotide out of the duplex into an active site, but the energetic barrier that must be overcome is not fully understood. Unpaired (bulged) nucleotides are more accessible, but it is not known if they are recognized by nucleotide flipping enzymes. We have investigated this question with human alkyladenine DNA glycosylase (AAG). AAG recognizes a wide variety of structurally disparate lesions including deoxyinosine (I), which results from the oxidative deamination of adenosine. AAG catalyzes the hydrolysis of the N-glycosidic bond to release the lesion base and initiate the base excision repair pathway (Figure 1A).3 We used single turnover kinetics to demonstrate that AAG is exquisitely sensitive to the structural context of the I lesion. An inverse correlation is observed between duplex stability and catalytic efficiency, indicating that stable pairing is a barrier to recognition by AAG. Single stranded DNA is a very poor substrate, but a single nucleotide bulge is recognized more efficiently than any other context. These results highlight the intrinsic barrier to nucleotide flipping that DNA repair enzymes face and imply that the recognition of DNA damage by AAG is remarkably plastic.
Figure 1.
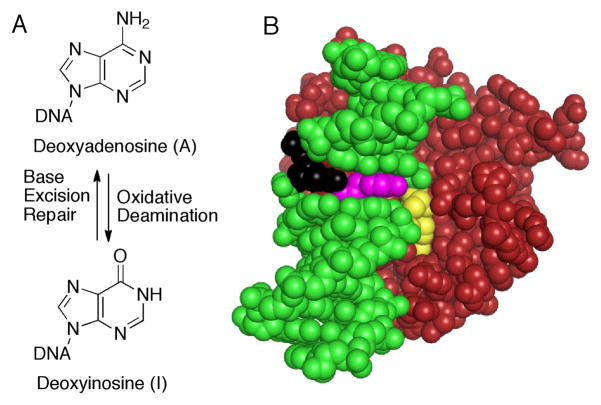
Deamination of A to form I and structure of AAG bound to damaged DNA. (A) Oxidative deamination of A is reversed by AAG and the base excision repair pathway. (B) AAG is red, the intercalating tyrosine (Y162) is magenta, and the DNA is green, except for the lesion (yellow) that is flipped into the active site and the opposing nucleotide (black). This opposing nucleotide is missing in the bulged substrate. Coordinates are from the PDB for AAG bound to 1,N6-ethenoA-DNA; 1EWN.4
Crystal structures of AAG bound to an extrahelical lesion reveal that the DNA is bent, the lesioned base is rotated by ~180° out of the duplex into the active site pocket, and the hole that is left in the DNA is plugged with the phenolic group of Y162 (Figure 1B).4 Intimate protein-DNA contacts in this static structure seem to preclude the possibility that a bulge could be accommodated. However, the barrier to flipping of a bulged nucleotide is much less than for a mismatch. Bulged purines favor an intercalated conformation, but an extrahelical conformation has been observed in a crystal structure.5 If AAG has sufficient flexibility to encompass a bulged nucleotide, then the decreased barrier to flipping would enable more efficient recognition.
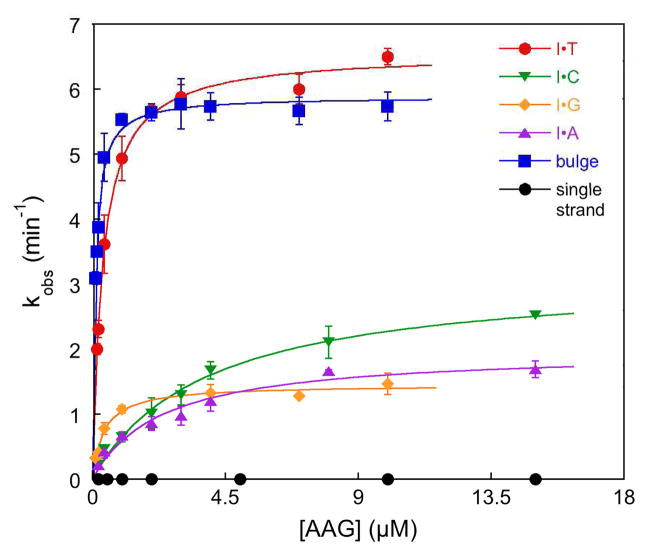
Therefore, we characterized the activity of AAG towards an I lesion in different structural contexts, including a single nucleotide bulge. Product release is rate-limiting for multiple turnover reactions of AAG, requiring the use of single turnover methods.6 The concentration of AAG was varied to determine the maximal single turnover rate constant (kmax) and the half-maximal concentration (K1/2) for each context (Figure 2). Although the individual reaction rates varied by more than 10,000-fold, with half-times that varied from seconds to days, single exponential fits were excellent in all cases (R2>0.97; See Supporting Information for representative data). The rate constant kmax reports on the flipping and N-glycosidic bond hydrolysis steps, and the K1/2 value corresponds to the Kd for substrate dissociation.6 Specificity between competing substrates is given by the ratio kcat/KM, the catalytic efficiency.
Figure 2.
Concentration dependence for single turnover glycosylase activity of AAG. Each data point corresponds to the average and standard deviation from 4–8 individual reactions (See Supporting Information). Lines indicate the best fit to a hyperbolic concentration dependence; kobs = kmax[AAG]/(K1/2 + [AAG]).
The glycosylase activity of AAG is shown in Figure 2 and summarized in Table 1. The bulge is the best context tested, with a catalytic efficiency that is 3-fold better than the biological I·T mismatch. This result indicates that any unfavorable effects of removing the opposing base (Figure 1B) are more than compensated for by an increased ease in flipping the lesion. Further work is needed to evaluate whether the bulge might be more easily bent than the mismatch and whether this could contribute to catalytic recognition. Other mismatches are recognized less efficiently than the I·T mismatch. The single-stranded lesion is even more accessible, and several glycosylases efficiently utilize single-stranded substrates, however we find that AAG has ~2000-fold reduced activity towards this substrate.
Table 1.
Kinetic Parameters for the AAG-catalyzed hydrolysis of Inosine in Different Structural Contextsa
| Opposing Base | K1/2 (μM) | kmax (min−1) | kmax/K1/2b (M−1s−1) | Relative kmax/K1/2 |
|---|---|---|---|---|
| Bulge (none) | 0.08 | 5.4 | 1.1×106 | 3 |
| T | 0.30 | 6.3 | 3.5×105 | (1) |
| G | 0.41 | 1.5 | 6.2×104 | 0.2 |
| C | 3.8 | 3.3 | 1.4×104 | 0.04 |
| A | 2.3 | 2.0 | 1.4×104 | 0.04 |
| Single strand | 0.34 | 0.0035 | 1.7×102 | 0.0005 |
Data from 23 °C in 50 mM NaMES, pH 6.1, 1 mM EDTA, 1 mM DTT, 0.1 mg/mL BSA. Ionic strength was adjusted to 200 mM with NaCl.
kmax/K1/2 is a measure of the catalytic efficiency, analogous to kcat/KM in steady state kinetics.
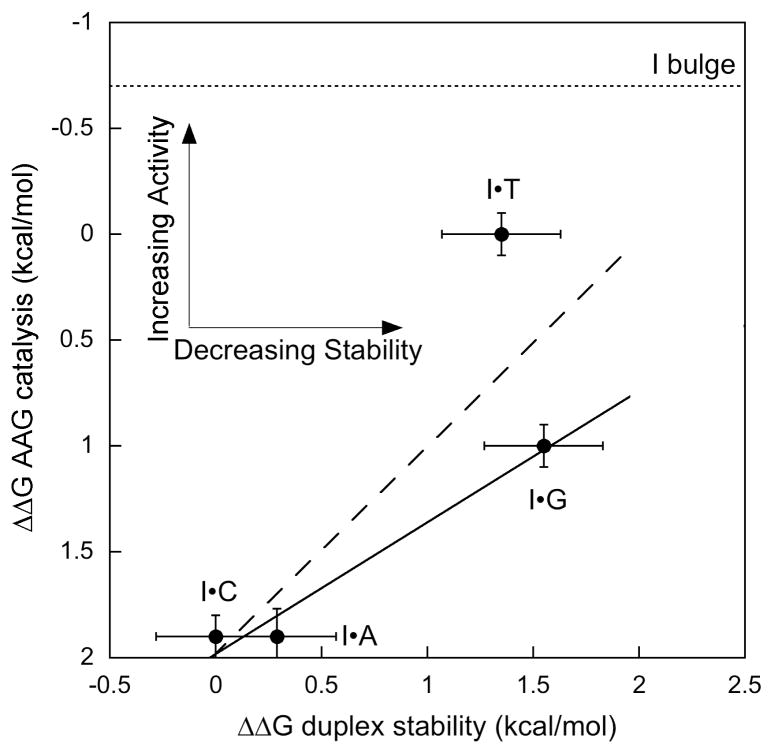
Since AAG does not make specific contacts with the opposing base, the relationship between base pair stability and catalytic efficiency for different mismatches can be assessed. Thermodynamic parameters for duplex stability for inosine paired with each of the normal nucleotides are known7, and serve as a surrogate for base pair stability. The relative free energies (ΔΔG) for catalytic efficiency of the AAG-catalyzed reaction and for duplex stability are presented in Figure 3. A linear fit to all of the mismatches yields a slope of −0.96 (R2 = 0.67), and omission of the I·T substrate gives a slope of −0.59 (R2 = 0.97). This is a limited set of data, but the trend is clear. Previous analysis of the thermodynamics of duplex stability and DNA binding by uracil DNA glycosylase found that disruption of base pairing gives greater accessibility and tighter binding.8 The inverse relationship between duplex stability and efficiency of excision supports the model that AAG and other glycosylases must overcome the barrier provided by base pairing interactions. This behavior maximizes the discrimination between damaged and undamaged nucleotides because undamaged nucleotides have more favorable hydrogen bonding and stacking interactions.
Figure 3.
Linear free energy relationship shows an inverse correlation between duplex stability and glycosylase activity. Differences in free energy (ΔΔG) are from ref 7 for duplex stability and from the equation ΔΔG = −RTln(kcat/KMrel) for AAG activity. Linear fits to all of the mismatches (dashed line; slope = −0.97; R2 = 0.67) or with the exclusion of the T mismatch (solid line; slope = −0.59; R2=0.97) are shown. The relative activity towards a bulge is shown as a dotted line.
The I·T mismatch deviates by ~1 kcal/mol relative to the other mismatches, raising the possibility that AAG recognizes the I·T wobble pair geometry independent of its effects on duplex stability. Indeed, I·G forms a less stable duplex than I·T and yet it is removed with 5-fold lower catalytic efficiency. Deamination of A in DNA generates an I·T pair, and AAG-initiated repair will restore the correct sequence. Incorporation of dIMP or replication of an I lesion predominantly forms an I·C pair.9 Under these scenarios the activity of AAG would make a permanent mutation. The much lower efficiency of AAG towards I·C leaves open the possibility that another DNA repair pathway corrects replicative events.
By quantifying the energetic differences in the catalytic efficiency of AAG with different mismatches, we obtained strong evidence that base pairing provides a barrier to base excision. The fact that a bulge is recognized with the same efficiency as a mismatch indicates considerable flexibility in DNA recognition. This also has biological ramifications, because DNA polymerases can slip on repetitive or damaged templates to generate bulged structures. Initiation of base excision repair could correct nascent +1 frameshifts, but would make −1 frameshifts permanent. This ability of AAG to act on bulged nucleotides may explain the observation that an increased level of AAG expression is correlated with increased frequency of frameshift mutations.10
Supplementary Material
Acknowledgments
This work was supported by a grant from the NIH (CA122254). D.L. was supported in part by an NIH training grant at the Chemistry/Biology Interface.
Footnotes
Supporting Information Available: Materials and methods and supporting results and discussion. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lindahl T. Nature. 1993;362:709–15. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Roberts RJ, Cheng X. Annu Rev Biochem. 1998;67:181–98. doi: 10.1146/annurev.biochem.67.1.181. [DOI] [PubMed] [Google Scholar]
- 3.Stivers JT, Jiang YL. Chem Rev. 2003;103:2729–59. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 4.Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T. Proc Natl Acad Sci USA. 2000;97:13573–8. doi: 10.1073/pnas.97.25.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Patel DJ, Kozlowski SA, Marky LA, Rice JA, Broka C, Itakura K, Breslauer KJ. Biochemistry. 1982;21:445–51. doi: 10.1021/bi00532a004. [DOI] [PubMed] [Google Scholar]; (b) Joshua-Tor L, Rabinovich D, Hope H, Frolow F, Appella E, Sussman JL. Nature. 1988;334:82–4. doi: 10.1038/334082a0. [DOI] [PubMed] [Google Scholar]
- 6.(a) Abner CW, Lau AY, Ellenberger T, Bloom LB. J Biol Chem. 2001;276:13379–87. doi: 10.1074/jbc.M010641200. [DOI] [PubMed] [Google Scholar]; (b) Baldwin MR, O’Brien PJ. Biochemistry. 2009;48:6022–33. doi: 10.1021/bi900517y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watkins NE, Jr, SantaLucia J., Jr Nucleic Acids Res. 2005;33:6258–67. doi: 10.1093/nar/gki918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krosky DJ, Schwarz FP, Stivers JT. Biochemistry. 2004;43:4188–95. doi: 10.1021/bi036303y. [DOI] [PubMed] [Google Scholar]
- 9.(a) Yasui M, Suenaga E, Koyama N, Masutani C, Hanaoka F, Gruz P, Shibutani S, Nohmi T, Hayashi M, Honma M. J Mol Biol. 2008;377:1015–23. doi: 10.1016/j.jmb.2008.01.033. [DOI] [PubMed] [Google Scholar]; (b) Zhang H, Bren U, Kozekov ID, Rizzo CJ, Stec DF, Guengerich FP. J Mol Biol. 2009;392:251–69. doi: 10.1016/j.jmb.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofseth LJ, Khan MA, Ambrose M, Nikolayeva O, Xu-Welliver M, Kartalou M, Hussain SP, Roth RB, Zhou X, Mechanic LE, Zurer I, Rotter V, Samson LD, Harris CC. J Clin Invest. 2003;112:1887–94. doi: 10.1172/JCI19757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.