Abstract
In the spring of 2009, a new human influenza A H1N1 virus emerged in Mexico and the United States. The strain was referred to as “swine flu” as it has strong similarities with current circulating swine influenza viruses, although the first outbreak on a swine farm was recorded more than 2 mo following the first human reports. This new strain, designated as pandemic (H1N1) 2009, has shown the ability to spread amongst the human population and can be found on all continents. The way influenza viruses and specifically this influenza A pandemic (H1N1) 2009 virus evolve is described in this manuscript.
Résumé
Le nouveau virus de l’influenza A H1N1 : En équilibre à l’interface des humains et des animaux. Au printemps 2009, un nouveau virus de l’influenza A H1N1 est apparu au Mexique et aux États-Unis. La souche a été appelée «grippe porcine» car elle présente de fortes similarités avec les virus d’influenza porcine en circulation à l’heure actuelle, même si la première éclosion sur une exploitation porcine a été enregistrée plus de 2 mois après les premiers rapports d’infection chez les humains. Cette nouvelle souche, désignée comme virus (H1N1) pandémique 2009, a manifesté la capacité de se propager parmi la population humaine et se trouve sur tous les continents. La façon dont les virus de l’influenza évoluent, particulièrement ce virus (H1N1) de l’influenza A pandémique 2009, est décrite dans ce manuscrit.
(Traduit par Isabelle Vallières)
Introduction
Ever since the H1N1 Spanish flu raged around the world in 1918–1920, people have feared a subsequent outbreak of a similar or greater magnitude. With over 50 million human casualties (1,2), and especially high rates of mortality among young adults aged 18 to 40 y (3,4), the Spanish flu pandemic has set the stage for the preparations underway today. Although it was not the first influenza outbreak (5), it was by far the most devastating and at that time, people were completely unaware of the nature of the disease. The first successful isolation of an influenza virus occurred in 1930 (6,7), and during the last 2 pandemics (the 1957 H2N2 Asian flu and the 1968 H3N2 Hong Kong flu), the scientific world had at least a better understanding of the causative agent (8). This first isolate was a swine influenza virus. The earliest recorded observations of an influenza-like illness in swine coincided with the human influenza pandemic in 1918, and already during that period there were suggestions that human flu and swine flu might be similar diseases (8,9). However, the exact transmission route between species (human-to-pig or pig-to-human) remains unresolved. Until now all pandemics (Spanish, Asian, and Hong Kong flu) have been caused by influenza viruses of avian origin (10), the spread of the pandemic (H1N1) 2009 (pH1N1) virus marks the first known pandemic influenza virus of swine origin. This manuscript describes the characteristics and evolution of influenza viruses and specifically focuses on the pH1N1 virus.
Influenza A viruses are characterized based on the envelope glycoproteins hemagglutinin (H or HA) and neuraminidase (N or NA). So far, the human population has been confronted on an epidemic scale with 3 different HA types: H1, H2, and H3. There is no reason to exclude the possibility that humans can be infected with all other variants, this has already been reported for H5, H7, and H9 (5). Influenza A viruses are members of the Orthomyxoviridae family, which is comprised of enveloped, negative strand RNA viruses. The influenza A genome consists of 8 gene segments [HA, NA, matrix protein (MP), nucleo-protein (NP), Polymerase A (PA), Polymerase B1 and 2 (PB1 and PB2) and the non-structural protein (NS)] coding for 11 different proteins. Sixteen subtypes of HA (H1–H16) and 9 subtypes of NA have been found to date (N1–N9) (11). A new human influenza pandemic will therefore be caused by an influenza virus containing an HA antigenic makeup heretofore unknown to humans. Also, the NA type contributes to antibody generation upon exposure to the immune system. The Hong Kong flu (H3N2) outbreak showed that the neuraminidase (NA) induced a limited protection, as this N2 had some antigenic similarities to the NA type found in the H2N2 Asian flu pandemic (12–17). In light of this, it is not surprising that the emergence of human infections with an H5N1 avian flu (18–20), with a current human mortality rate of 60% (21), led to an international effort to prevent the spread of this virus.
Continuing diversification and host range of influenza viruses
Influenza viruses infecting humans consist of 3 variants: types A, B, and C. The B and C types are almost exclusively found in humans although influenza B can also be found in aquatic mammals (22) and influenza C was shown to be present in some swine herds (23) and caused upper respiratory tract infections (24). The influenza A virus particle is enveloped, and within this envelope 3 membrane proteins are present: HA, NA, and one of the 2 proteins coded by MP the ion channel M2 (24–26). HA and NA appear at the outer surface of the virus and they can induce antibodies with a neutralizing effect (26).
Currently, almost all combinations of HA and NA have been detected in birds, especially gulls and waterfowl (11,26,27). Birds contribute significantly to the spread of influenza viruses through their migration patterns in specific flyways (27). Equines can be only infected with influenza viruses H3N8 and H7N7, although the latter has not been detected in horses in recent years and may have disappeared completely (28). Dogs can also be infected with the H3N8 equine variant (29,30). A variety of influenza viruses have been found in aquatic mammals (H1, H3, H4, H7, and H13 containing variants) (26,31). The avian flu H5N1 outbreak in cats, leopards and tigers demonstrated that it was also possible to infect felids with this strain (32,33). In porcines, the H3N2, H1N1 and reassorted versions of these 2 viruses [H1N2, H3N1, and H1N1 variants containing the swine H3N2 internal genes (rH1N1)] are present (34–37). Recently, a new H1N1 with a human-derived H1 gene (huH1N1) was detected in swine herds and appears to be circulating (36). The classic H1N1 porcine variant is present in both Europe and North America. However, while an avian H1N1 variant introduced in the European swine population has almost completely replaced the classic variant in Europe (38–41), the classic H1N1 still continues to infect swine herds in North America (34,36,37).
As a consequence of the segmentation of the genome, the virus must include all 8 segments during assembly of the viral particle. In the rare event that a second, different, influenza virus enters the same cell, the cell will contain 2 different genes of each segment. The viral particle can include either of the 2 segments leading to a rearrangement (42) of genes. For example: an H1N1 influenza virus infecting a cell already infected with an H3N2 influenza virus might lead to the aforementioned rearrangements resulting in H1N2 or H3N1 viruses. This process is called antigenic shift and represents a dramatic change in antigenic makeup of the viral particle (43). All genes seem to be able to end up in a rearranged influenza virus independently, although whether that is really the case remains to be established (44); it might be that some combinations are less successful or even impossible.
Influenza genes can also change more gradually by means of point mutations which result in alteration of the transcribed proteins (45). These mutations are random errors introduced during the copying of RNA and are part of a regular evolutionary process called antigenic drift (46).
The new pandemic (H1N1) 2009 virus
Recently, a new influenza variant was identified as the source of a cluster of human pneumonia cases in the state of California (USA) and in La Gloria, Mexico (47,48). This variant was soon designated as “swine flu” virus although it was never detected in porcine populations until the occurrence in a swine herd in Alberta, Canada in May 2009 (49), in Argentina in June 2009 (50), and in Australia at the end of July 2009 (51). Later, names like Mexican flu virus, swine-origin H1N1, or pandemic (H1N1) 2009 arose, but the nomenclature remains unclear. Pandemic (H1N1) 2009 abbreviated to pH1N1 will be used herein.
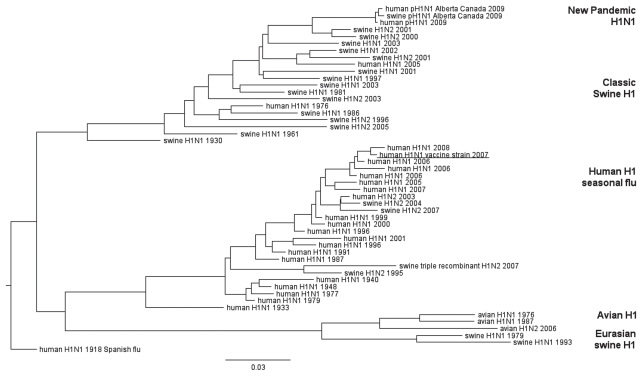
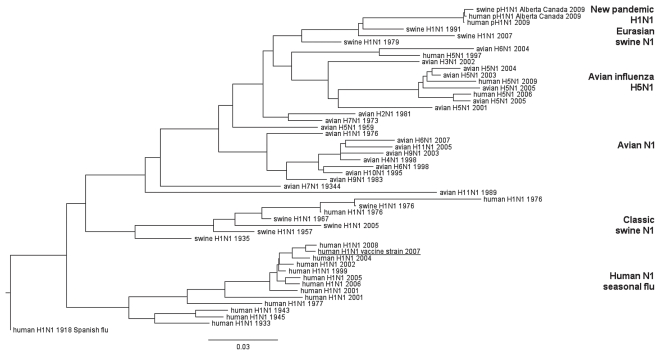
When pH1N1 genes are compared with genes already known in influenza viruses, pH1N1 does not cluster with any of the known and currently circulating human H1N1 seasonal flu viruses. Instead, all gene segments cluster with currently circulating swine viruses. An overview of the phylogenetic trees indicating the relationship of the HA and NA genes is represented in Figures 1 and 2. Phylogenetic trees of other gene segments will be provided upon request.
Figure 1.
Genetic relationships of human, swine, and avian influenza viruses for H1.
Names represent viruses all containing an H1 and different neuraminidase types (as indicated). Year of isolation is indicated. The 2007 WHO recommended H1N1 vaccine strain: A/Brisbane/59/2007 (H1N1) is underlined. The 2009 pandemic H1N1 (pH1N1) can be found at the top branch of the tree. Scale bar indicates amino acid substitutions per site.
Figure 2.
Genetic relationships of human, swine, and avian influenza viruses for N1.
Names represent viruses all containing an N1 and different hemagglutinin types (as indicated). Year of isolation is indicated. The 2007 WHO recommended H1N1 vaccine strain: A/Brisbane/59/2007(H1N1) is underlined. The 2009 pandemic H1N1 (pH1N1) can be found at the top branch of the tree. Scale bar indicates amino acid substitutions per site.
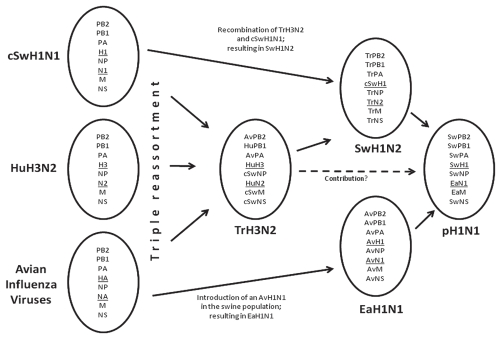
Compared with the genes of other influenza viruses, the pH1N1 seems to be the result of a reassortment of 2, but maybe even 3 different viruses, resulting in a combination of genes not previously reported in humans or porcines (Figure 3). At present, the assumption is that pH1N1 is the product of a reassortment of the genes of 2 viruses, as this is a more likely event than a triple reassortment (3 different viruses). For a reassortment, the viruses have to be in the same cell at the same time. The first contributing virus is the ‘Eurasian’ H1N1 swine influenza virus. This virus donated the NA, and most likely also delivered the MP as it also clusters with the Eurasian swine viruses. The MP gene is a highly conserved gene amongst the influenza A viruses; therefore, it is difficult to type due to its close resemblance with a number of MP genes of other influenza variants. For the same reason the conserved MP gene is used in molecular diagnostics to detect the presence of influenza A viruses in specimens (52,53). Both NA and MP of the Eurasian swine H1N1 virus were originally derived from an avian influenza virus that entered the European swine population at the end of the 1970’s (54). The other genes of pH1N1 were most likely derived from a swine H1N2 virus. The pH1N1 HA gene clusters with swine H1N2 viruses. These swine H1N2 viruses emerged from a rearrangement of classic swine H1N1 and swine triple reassorted H3N2 (trH3N2) genes (55). The H1 gene of swine H1N2 underwent a significant antigenic drift since it was derived from classic swine H1N1, hence the HA of H1N2 can currently be identified as a genetically separated cluster. The swine trH3N2 virus contributed all other gene segments to the swine H1N2. The history of the aforementioned swine trH3N2 virus is interesting and illustrates the ability of influenza viruses to rearrange their genes and cross species barriers. The H3, N2, and PB1 genes of the swine trH3N2 virus were all derived from a human H3N2 virus related to the Hong Kong flu pandemic influenza strain. This contribution could be made by introduction of a human influenza virus into the porcine population. The PB2 and the PA genes of the swine trH3N2 virus show similarities to influenza genes still found in the avian population in North America. The MP, NS, and NP were all derived from a North American classic swine H1N1 virus (56). A triple reassortment of the classic swine H1N1, the human H3N2, and the avian influenza virus resulted in the swine trH3N2 virus. This same swine trH3N2 has also been detected in turkeys (57,58).
Figure 3.
The origin of pandemic (H1N1) 2009
The triple reassortment event between an avian influenza virus (Av), a human H3N2 (HuH3N2), and a classic swine H1N1 (cSwH1N1) that lead to the swine triple reassorted H3N2 (TrH3N2) virus is indicated on the left. The introduction and spread of an AvH1N1 into the swine herd resulted in the Eurasian H1N1 (EaH1N1). TrH3N2 reassorted with cSwH1N1 which resulted in SwH1N2. Eventually a reassortment event of EaH1N1 and SwH1N2 lead to the new pandemic (H1N1) 2009 (pH1N1). The origin of the respective genes is indicated in the oval shapes, hemagglutinin and neuraminidase genes are underlined.
Eventually, the swine H1N2 contributed together with the H1N1 Eurasian viruses to the pH1N1 (59–63). Although this seems a likely sequence of events, it should still be regarded as a theoretical genesis of pH1N1 deducted from publicly known sequences. It cannot be excluded, however, that the actual reassortment events were different. This reassortment event is not unique, and indeed swine H1-containing influenza viruses are regularly infecting humans (64), but this extensive spread within the human population is extremely rare. The results of evaluations of available swine and human sequences show that all genes of the pH1N1 virus were derived from swine influenza viruses, but they were derived from geographically widely distributed ancestors (62).
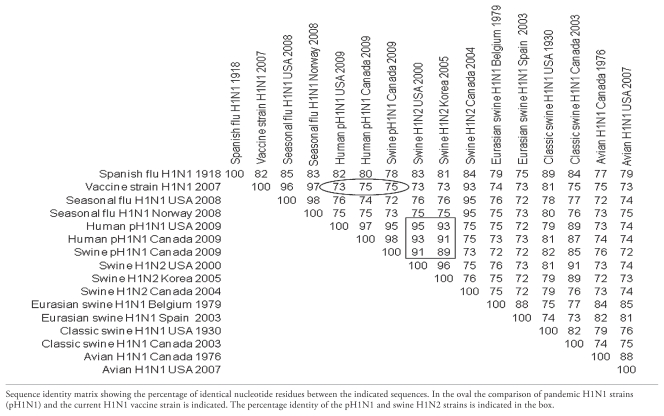
The divergence between the pH1N1 virus and the circulating human seasonal flu H1N1 variants is significant. Because only 73% of the pH1N1 HA gene is genetically similar to the H1N1 vaccine strain A/Brisbane/59/2007(H1N1) (65) the introduction of pH1N1 in the human population may even be described as a “pseudo-antigenic shift” (5) (Table 1). The currently applied seasonal H1N1 influenza vaccine will therefore likely provide only limited protection to humans. As such, given that pH1N1 has appeared in the fall of 2009 an updated vaccine that protects the human population is currently applied (66).
Table 1.
Hemagglutinin gene sequence comparison of pandemic (H1N1) 2009 and avian, human, and swine H1 genes
The situation is slightly different concerning protection of porcine herds. As the HA of the pH1N1 closely resembles the HA of an H1N2 swine virus, application of a vaccine containing a recent strain of a swine H1N2 may provide some protection against introduction and/or influenza virus-related pathology in a swine herd.
It is unclear why there are no reports of an earlier infection of a swine herd than the outbreak recorded in Alberta. Serological evaluations of human and porcine populations and detailed back-tracing of the outbreak may lead to an answer. But these studies can be time consuming, costly, and ultimately unsuccessful. The only way to avoid unexpected confrontations with new viruses is an intensive surveillance of influenza viruses around the world and in a variety of animal species. The establishment of open access databases such as The Global Initiative on Sharing Avian Influenza Data (GISAID) and the Influenza Virus Resource (NCBI IVR) (67,68) are recent initiatives contributing to a better understanding of influenza virus evolution. Not only should the HA and NA be characterized, but the other 6 gene segments that can prove valuable for backtracing and evolutionary studies, as this pH1N1 example illustrates, should also be routinely sequenced.
Methods used for figures 1, 2 and table 1
Sequences of H1N1 2009 influenza viruses and all other viruses described in this paper and in Figures 1 and 2 and Table 1 were downloaded from the databases GISAID and NCBI IVR (67,68). Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 4 (69) and Bioedit 7.0.9.0 (70). The phylogenetic trees were modified using FigTree v1.2.2 (71).
Strains used for Table 1: Spanish flu H1N1 1918 = A/South Carolina/1/1918 (H1N1); Vaccine strain H1N1 2007 = A/Brisbane/59/2007 (H1N1); Seasonal flu H1N1 USA 2008 = A/New York/05/2008 (H1N1); Seasonal flu H1N1 Norway 2008 = A/Norway/76/2008 (H1N1); Human pH1N1 USA 2009 = A/California/04/2009 (H1N1); Human pH1N1 Canada 2009 = A/Canada-AB/RV1532/2009 (H1N1); Swine pH1N1 Canada 2009 = A/swine/Alberta/OTH-33-1/2009 (H1N1); Swine H1N2 USA 2000 = A/Swine Indiana/P12439/00 (H1N2); Swine H1N2 Korea 2005 = A/swine/Korea/JL04 2005 (H1N2); Swine H1N2 Canada 2004 = A/swine/Ontario/48235/04 (H1N2); Eurasian swine H1N1 Belgium 1979 = A/swine/Belgium/WVL1/1979 (H1N1); Eurasian swine H1N1 Spain 2003 = A/swine/Spain/51915/2003 (H1N1); Classic swine H1N1 USA 1930 = A/swine/Iowa/15/1930 (H1N1); Classic swine H1N1 Canada 2003 = A/swine/Alberta/56626/03 (H1N1); Avian H1N1 Canada 1976 = A/duck/Alberta/35/76 (H1N1); Avian H1N1 USA 2007 = A/mallard/Minnesota Sg-00121/2007 (H1N1)
For detailed versions of phylogenies of genes of pH1N1 please contact the corresponding author. CVJ
Footnotes
Use of this article is limited to a single copy for personal study. Anyone interested in obtaining reprints should contact the CVMA office ( hbroughton@cvma-acmv.org) for additional copies or permission to use this material elsewhere.
References
- 1.Palese P. Influenza: Old and new threats. Nat Med. 2004;10 (12 Suppl):S82–87. doi: 10.1038/nm1141. [DOI] [PubMed] [Google Scholar]
- 2.Johnson NP, Mueller J. Updating the accounts: Global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull Hist Med. 2002;76:105–115. doi: 10.1353/bhm.2002.0022. [DOI] [PubMed] [Google Scholar]
- 3.Taubenberger JK, Morens DM. 1918 Influenza: The mother of all pandemics. Emerg Infect Dis. 2006;12:15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray CJL, Lopez AD, Chin B, Feehan D, Hill KH. Estimation of potential global pandemic influenza mortality on the basis of vital registry data from the 1918–20 pandemic: A quantitative analysis. The Lancet. 2006;368:2211. doi: 10.1016/S0140-6736(06)69895-4. [DOI] [PubMed] [Google Scholar]
- 5.Gatherer D. The 2009 H1N1 influenza outbreak in its historical context. J Clin Virol. 2009;45:174–178. doi: 10.1016/j.jcv.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Shope RE. The etiology of swine influenza. Science. 1931;73:214–215. doi: 10.1126/science.73.1886.214. [DOI] [PubMed] [Google Scholar]
- 7.Shope RE. Swine influenza: I. Experimental transmission and pathology. J Exp Med. 1931;54:349–359. doi: 10.1084/jem.54.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taubenberger JK, Reid AH, Janczewski TA, Fanning TG. Integrating historical, clinical and molecular genetic data in order to explain the origin and virulence of the 1918 Spanish influenza virus. Philos Trans R Soc Lond B Biol Sci. 2001;356:1829–1839. doi: 10.1098/rstb.2001.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shope RE. The incidence of neutralizing antibodies for swine influenza virus in the sera of human beings of different ages. J Exp Med. 1936;63:669–684. doi: 10.1084/jem.63.5.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459:931. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fouchier RA, Munster V, Wallensten A, et al. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol. 2005;79:2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox NJ, Subbarao K. Global epidemiology of influenza: Past and present. Annu Rev Med. 2000;51:407–421. doi: 10.1146/annurev.med.51.1.407. [DOI] [PubMed] [Google Scholar]
- 13.Kilbourne ED. Perspectives on pandemics: A research agenda. J Infect Dis. 1997;176 (Suppl 1):S29–31. doi: 10.1086/514171. [DOI] [PubMed] [Google Scholar]
- 14.Schulman JL, Kilbourne ED. Independent variation in nature of hemagglutinin and neuraminidase antigens of influenza virus: Distinctiveness of hemagglutinin antigen of Hong Kong-68 virus. Proc Natl Acad Sci U S A. 1969;63:326–333. doi: 10.1073/pnas.63.2.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stuart-Harris C. Epidemiology of influenza in man. Br Med Bull. 1979;35 (1):3–8. doi: 10.1093/oxfordjournals.bmb.a071538. [DOI] [PubMed] [Google Scholar]
- 16.Monto AS, Kendal AP. Effect of neuraminidase antibody on Hong Kong influenza. Lancet. 1973;1:623–625. doi: 10.1016/s0140-6736(73)92196-x. [DOI] [PubMed] [Google Scholar]
- 17.Viboud C, Grais RF, Lafont BA, Miller MA, Simonsen L. Multinational impact of the 1968 Hong Kong influenza pandemic: Evidence for a smoldering pandemic. J Infect Dis. 2005;192:233–248. doi: 10.1086/431150. [DOI] [PubMed] [Google Scholar]
- 18.Wuethrich B. An avian flu jumps to people. Science. 2003;299:1504. doi: 10.1126/science.299.5612.1504. [DOI] [PubMed] [Google Scholar]
- 19.Cauthen AN, Swayne DE, Schultz-Cherry S, Perdue ML, Suarez DL. Continued circulation in China of highly pathogenic avian influenza viruses encoding the hemagglutinin gene associated with the 1997 H5N1 outbreak in poultry and humans. J Virol. 2000;74:6592–6599. doi: 10.1128/jvi.74.14.6592-6599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Jong JC, Claas ECJ, Osterhaus ADME, Webster RG, Lim WL. A pandemic warning? Nature. 1997;389:554. doi: 10.1038/39218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.World Health Organisation (WHO) Cumulative number of confirmed human cases of avian influenza A/(H5N1) Reported to WHO. [Last accessed November 17, 2009]. Available from: http://www.who.int/csr/disease/avian_influenza/country/cases_table_2009_07_01/en/index.html.
- 22.Osterhaus AD, Rimmelzwaan GF, Martina BE, Bestebroer TM, Fouchier RA. Influenza B virus in seals. Science. 2000;288:1051–1053. doi: 10.1126/science.288.5468.1051. [DOI] [PubMed] [Google Scholar]
- 23.Yuanji G, Fengen J, Ping W, Min W, Jiming Z. Isolation of influenza C virus from pigs and experimental infection of pigs with influenza C virus. J Gen Virol. 1983;64:177–182. doi: 10.1099/0022-1317-64-1-177. [DOI] [PubMed] [Google Scholar]
- 24.Lamb RA, Krug RM. Orthomyxoviridae: The viruses and their replication. 4 ed. Philadelphia: Lippincott Williams and Wilkins; 2001. pp. 1487–1531. [Google Scholar]
- 25.Cheung TK, Poon LL. Biology of influenza a virus. Ann N Y Acad Sci. 2007;1102:1–25. doi: 10.1196/annals.1408.001. [DOI] [PubMed] [Google Scholar]
- 26.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Mol Biol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus AD, Fouchier RA. Global patterns of influenza a virus in wild birds. Science. 2006;312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 28.Paillot R, Hannant D, Kydd JH, Daly JM. Vaccination against equine influenza: Quid novi? Vaccine. 2006;24:4047–4061. doi: 10.1016/j.vaccine.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 29.Dubovi EJ, Njaa BL. Canine influenza. Vet Clin North Am Small Anim Pract. 2008;38:827–835. viii. doi: 10.1016/j.cvsm.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Crawford PC, Dubovi EJ, Castleman WL, et al. Transmission of equine influenza virus to dogs. Science. 2005;310:482–485. doi: 10.1126/science.1117950. [DOI] [PubMed] [Google Scholar]
- 31.Webby RJ, Webster RG, Richt JA. Influenza viruses in animal wildlife populations. Curr Top Microbiol Immunol. 2007;315:67–83. doi: 10.1007/978-3-540-70962-6_4. [DOI] [PubMed] [Google Scholar]
- 32.Keawcharoen J, Oraveerakul K, Kuiken T, et al. Avian influenza H5N1 in tigers and leopards. Emerg Infect Dis. 2004;10:2189–2191. doi: 10.3201/eid1012.040759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leschnik M, Weikel J, Mostl K, et al. Subclinical infection with avian influenza A (H5N1) virus in cats. Emerg Infect Dis. 2007;13:243–247. doi: 10.3201/eid1302.060608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olsen CW, Brown IH, Easterday BC, Van Reeth K. Swine influenza. In: Straw BE, Zimmerman JJ, d’Allaire S, Taylor DJ, editors. Diseases of Swine. 9 ed. Oxford: Blackwell Publ; 2006. pp. 469–482. [Google Scholar]
- 35.Thacker E, Janke B. Swine influenza virus: Zoonotic potential and vaccination strategies for the control of avian and swine influenzas. J Infect Dis. 2008;197 (Suppl 1):S19–24. doi: 10.1086/524988. [DOI] [PubMed] [Google Scholar]
- 36.Vincent AL, Ma W, Lager KM, Janke BH, Richt JA. Swine influenza viruses a North American perspective. Adv Virus Res. 2008;72:127–154. doi: 10.1016/S0065-3527(08)00403-X. [DOI] [PubMed] [Google Scholar]
- 37.Webby RJ, Rossow K, Erickson G, Sims Y, Webster R. Multiple lineages of antigenically and genetically diverse influenza A virus co-circulate in the United States swine population. Virus Research. 2004;103:67. doi: 10.1016/j.virusres.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Hinshaw VS, Alexander DJ, Aymard M, et al. Antigenic comparisons of swine-influenza-like H1N1 isolates from pigs, birds and humans: An international collaborative study. Bull World Health Organ. 1984;62:871–878. [PMC free article] [PubMed] [Google Scholar]
- 39.Scholtissek C, Burger H, Bachmann PA, Hannoun C. Genetic relatedness of hemagglutinins of the H1 subtype of influenza A viruses isolated from swine and birds. Virology. 1983;129:521–523. doi: 10.1016/0042-6822(83)90194-0. [DOI] [PubMed] [Google Scholar]
- 40.Campitelli L, Donatelli I, Foni E, et al. Continued evolution of H1N1 and H3N2 influenza viruses in pigs in Italy. Virology. 1997;232:310–318. doi: 10.1006/viro.1997.8514. [DOI] [PubMed] [Google Scholar]
- 41.Dunham EJ, Dugan VG, Kaser EK, et al. Different evolutionary trajectories of European avian-like and classical swine H1N1 influenza A viruses. J Virol. 2009;83:5485–5494. doi: 10.1128/JVI.02565-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cox NJ, Bender CA. The molecular epidemiology of influenza viruses. Seminars in Virology. 1995;6:359. [Google Scholar]
- 43.Young JF, Palese P. Evolution of human influenza A viruses in nature: Recombination contributes to genetic variation of H1N1 strains. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:6547–6551. doi: 10.1073/pnas.76.12.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bean WJ, Cox NJ, Kendal AP. Recombination of human influenza A viruses in nature. Nature. 1980;284:638. doi: 10.1038/284638a0. [DOI] [PubMed] [Google Scholar]
- 45.Laver WG, Gerhard W, Webster RG, Frankel ME, Air GM. Antigenic drift in type A influenza virus: Peptide mapping and antigenic analysis of A/PR/8/34 (HON1) variants selected with monoclonal antibodies. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:1425–1429. doi: 10.1073/pnas.76.3.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webster RG, Kendal AP, Gerhard W. Analysis of antigenic drift in recently isolated influenza A (H1N1) viruses using monoclonal antibody preparations. Virology. 1979;96:258. doi: 10.1016/0042-6822(79)90189-2. [DOI] [PubMed] [Google Scholar]
- 47.Swine influenza A (H1N1) infection in two children — Southern California, March–April 2009. MMWR Morb Mortal Wkly Rep. 2009;58:400–402. Various authors. [PubMed] [Google Scholar]
- 48.Fraser C, Donnelly CA, Cauchemez S, et al. Pandemic potential of a strain of influenza A (H1N1): Early findings. Science. 2009;324:1557–1561. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.World Animal Health Information Database (WAHID) [Last accessed November 17, 2009]. Available from: http://www.oie.int/wahis/public.php?page=single_report&pop=1&reportid=8065.
- 50.World Animal Health Information Database (WAHID) [Last accessed November 17, 2009]. Available from: http://www.oie.int/wahis/public.php?page=single_report&pop=1&reportid=8227.
- 51.World Animal Health Information Database (WAHID) [Last accessed 8/1/2009]. Available from: http://www.oie.int/wahis/public.php?page=single_report&pop=1&reportid=8332.
- 52.Fouchier RAM, Bestebroer TM, Herfst S, Van Der Kemp L, Rimmelzwaan GF, Osterhaus ADME. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J Clin Microbiol. 2000;38:4096–4101. doi: 10.1128/jcm.38.11.4096-4101.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cherian T, Bobo L, Steinhoff MC, Karron RA, Yolken RH. Use of PCR-enzyme immunoassay for identification of influenza A virus matrix RNA in clinical samples negative for cultivable virus. J Clin Microbiol. 1994;32:623–628. doi: 10.1128/jcm.32.3.623-628.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pensaert M, Ottis K, Vandeputte J, Kaplan MM, Bachmann PA. Evidence for the natural transmission of influenza A virus from wild ducts to swine and its potential importance for man. Bull World Health Organ. 1981;59:75–78. [PMC free article] [PubMed] [Google Scholar]
- 55.Karasin AI, Olsen CW, Anderson GA. Genetic characterization of an H1N2 influenza virus isolated from a pig in Indiana. J Clin Microbiol. 2000;38:2453–2456. doi: 10.1128/jcm.38.6.2453-2456.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou NN, Senne DA, Landgraf JS, et al. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol. 1999;73:8851–8856. doi: 10.1128/jvi.73.10.8851-8856.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi YK, Lee JH, Erickson G, et al. H3N2 influenza virus transmission from swine to turkeys, United States. Emerg Infect Dis. 2004;10:2156–2160. doi: 10.3201/eid1012.040581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang Y, Lee CW, Zhang Y, et al. Isolation and characterization of H3N2 influenza A virus from turkeys. Avian Dis. 2005;49:207–213. doi: 10.1637/7288-101304R. [DOI] [PubMed] [Google Scholar]
- 59.Garten RJ, Davis CT, Russell CA, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Investigative team. Novel Swine-Origin Influenza AVIT. Emergence of a Novel Swine-Origin Influenza A (H1N1) Virus in Humans. N Engl J Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 61.Trifonov V, Khiabanian H, Greenbaum B, Rabadan R. The origin of the recent swine influenza A(H1N1) virus infecting humans. Euro Surveill. 2009;14 [PubMed] [Google Scholar]
- 62.Smith GJ, Vijaykrishna D, Bahl J, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 63.Human/Swine A/H1N1 Influenza Origins and Evolution. [Last accessed November 17, 2009]. Available from: http://tree.bio.ed.ac.uk/groups/influenza/
- 64.Shinde V, Bridges CB, Uyeki TM, et al. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005–2009. N Engl J Med. 2009;360:2616–2625. doi: 10.1056/NEJMoa0903812. [DOI] [PubMed] [Google Scholar]
- 65.World Health Organization (WHO) Recommendations for influenza vaccines. [Last accessed November 17, 2009]. Available from: http://www.who.int/csr/disease/influenza/vaccinerecommendations/en/
- 66.Collin N, de Radigues X, Kieny MP. New influenza A(H1N1) vaccine: How ready are we for large-scale production? Vaccine. 2009;27:5184–5186. doi: 10.1016/j.vaccine.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 67.National Center for Biothechnology Information (NCBI) Influenza Virus Resource. [Last accessed November 17, 2009]. Available from: http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html.
- 68.The Global Initiative on Sharing Avian Influenza Data (GISAID) [Last accessed November 17, 2009]. Available from: http://platform.gisaid.org/
- 69.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 70.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 71.Rambaut A. Molecular evolution, phylogenetics and epidemiology Web site: Figtree v1.2.2. [Last accessed November 17, 2009]. Available from: http://tree.bio.ed.ac.uk/software/figtree/