Abstract
Methylation of tRNA on the four canonical bases adds structural complexity to the molecule, and improves decoding specificity and efficiency. While many tRNA methylases are known, detailed insight into the catalytic mechanism is only available in a few cases. Of interest among all tRNA methylases is the structural basis for nucleotide selection, by which the specificity is limited to a single site, or broadened to multiple sites. General themes in catalysis include the basis for rate acceleration at highly diverse nucleophilic centers for methyl transfer, using S-adenosylmethionine as a cofactor. Studies of tRNA methylases have also yielded insights into molecular evolution, particularly in the case of enzymes that recognize distinct structures to perform identical reactions at the same target nucleotide.
Introduction
RNA modification is a post-transcriptional process by which certain nucleotides are altered after their initial incorporation into an RNA chain. Transfer RNA is the most heavily modified class of RNA molecules. An average tRNA in Escherichia coli, Saccharomyces cerevisiae, and mammalian cytoplasm is modified at approximately 11%, 14%, and 17% of its residues, respectively [1]. These modifications expand the chemical and functional diversity of tRNA, and enhance its structural stability [2]. Their importance is underscored by the large investment of cellular resources required for biosynthesis: in some bacterial genomes, about 1% of the coding genes are devoted to tRNA modification [2]. Additionally, it has become increasingly apparent that incomplete modification can subject tRNA to rapid decay by various quality control surveillance mechanisms [3, 4]. While over 100 different modifications have been so far identified in tRNA, this review will focus on those that arise from methylation to the four canonical nucleotide bases. Methylated tRNA bases are known to help control tRNA folding, to ensure decoding specificity during translation of the genetic code, and to enhance aminoacylation by certain tRNA synthetases. In the great majority of cases, the methyl group is derived from S-adenosyl-methione (SAM), thus linking tRNA modifications to the intermediary metabolism of this cofactor.
A possible rationale that may account for the preferential use of SAM in tRNA methylation reactions is a more favorable thermodynamic driving force for methyl transfer, as compared with other biological methyl donors (such as folate) [5]. The driving force for the SAM-mediated methyl transfer is the electrophilic character of the methyl group attached to the positively charged sulfur atom. As such, SAM is one of the most widely used enzyme cofactors [5], second only to ATP. Methyltransferases (or methylases) that use SAM for methyl transfer reactions have been divided into five structural classes (I–V) based on their active-site folds [6]. Each of these five classes has a distinct α/β topology that forms the SAM binding pocket. Most of the SAM-dependent methylases possess the class I structure, which features the ancient Rossmann fold, consisting largely of a parallel β-sheet with α-helical crossovers. A final β-strand in the class I methylase fold is oriented in an anti-parallel direction relative to the others. Class I methylases dominate among the known enzymes that catalyze tRNA nucleotide base methylation. The remainder of the tRNA methylation reactions is catalyzed by methylases possessing the class IV structure. The key characteristic of this so-called SPOUT family of RNA methyltransferases [7] is a structural core made up of six parallel β-strands, with the final three folded into a rare protein subdomain known as a deep trefoil knot. Another notable feature of the class IV structure is that the active site is assembled at the interface of a homodimer, in which both monomers make substantial and interdigitating contributions to the cofactor binding [8, 9].
Methylation of a specific tRNA nucleotide base is sequence-dependent and varies greatly from species to species for different amino acid acceptors, and even among the three domains of life for the same amino acid acceptor. However, a subset of tRNA nucleotide base methylations is largely conserved in evolution. These include the m5U54 modification (methylation to position C5 in the uridine at position U54) that generates the conserved T54 (also known as ribothymidine) in the T loop of tRNA, and the m1A58 modification (methylation to position N1 in the adenine at position A58) localized in the same loop. These two modifications stabilize a reverse Hoogstein base-pairing interaction between the two bases, and are thus essential for the tertiary folding of the tRNA L-shape. Both modifications are catalyzed by methylases of the class I Rossmann-fold family. Another example of conserved methylation is the m1G37 modification (methylation to position N1 in the guanine at position G37) located at the 3' side of the anticodon. This modification serves an important role to prevent frameshift errors of tRNA on the ribosome [10]. However, while the bacterial enzyme TrmD catalyzing m1G37 formation is a member of the class IV family [8, 9], the eukaryotic and archaeal enzyme Trm5 is a member of the class I family [11–13]. TrmD and Trm5 thus form a pair of analogous enzymes that use distinct and unrelated SAM-binding folds to catalyze the same methylation reaction. The identification of TrmD and Trm5 suggests that the respective SAM-binding folds both evolved early in the process of tRNA maturation. Similarly, another pair of analogous enzymes, consisting of bacterial TrmJ (a member of the class IV family) and eukaryotic and archaeal Trm7 (a member of the class I family), has been identified, which catalyzes formation of 2'-O-methylation to the ribose of C32 and U32 (Cm32 and Um32) [14].
Crystal structures of SAM-dependent methylases that recognize tRNA nucleotide bases are now available for both class I and class IV enzyme families. These structures confirm that enzymes of the same class bind the cofactor in a similar stereochemical environment. In addition, these structures show that the two classes differ consistently in positioning the SAM cofactor: the class I fold binds the cofactor in an extended conformation with the dihedral angle C-4’-C-5’-Sδ-Cγ of 160–180° [12, 13], whereas the class IV-fold binds the cofactor in a bent conformation in which this dihedral angle is ~80° [8, 9]. The difference in the conformation of the cofactor may prove to be important for how each class regulates the rate of methyl transfer in the active site, and the overall steady-state rate of catalytic turnover. This question is largely unexplored at the present. The SAM conformation is also crucial to the mechanism of methyl transfer, which necessarily involves the precise juxtaposition of the electrophilic methyl group with a nucleophilic target atom, resulting in C-methylation, N-methylation, or O-methylation. Of general interest is the question of how a proton associated with the target atom is removed, and whether it is removed before, concurrent with, or after methyl transfer. Crystal structures of substrate-bound complexes have been determined mainly for class I enzymes. This review provides examples of C- and N-methylation by class I enzymes, in which insights into the mechanism of methyl transfer are available from analysis of substrate-bound complexes. These examples will serve as key models for investigation of related methylation reactions, including those that take place on a broad range of ribosomal RNAs and micro and small RNAs.
Methylation at C5 to produce ribothymidine and 5-methylcytosine
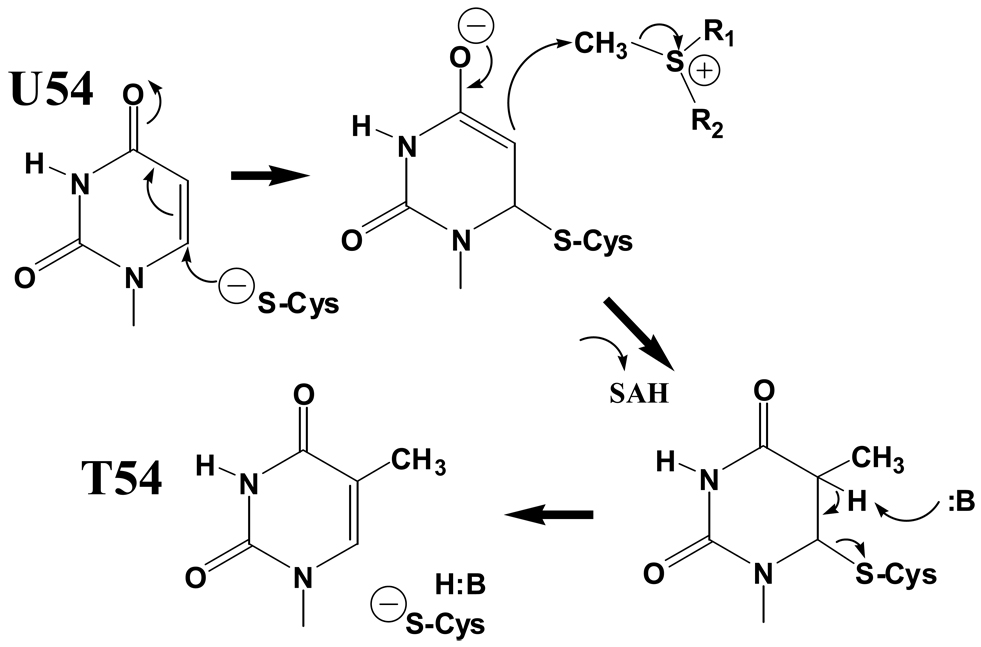
The first tRNA methylase studied in detail was the E. coli tRNA (m5U54)methyltransferase (RUMT; TrmA), which catalyzes SAM-dependent methylation of U54 in nearly all tRNAs, producing ribothymidine at the 5' position of the seven-nucleotide T-loop. A crucial experiment employed to deduce the stereochemical mechanism was use of a substrate analog in which the hydrogen at U54 ring position 5 is substituted with fluorine [15]. The results implicated Michael addition at the C6 ring position by a nucleophilic group on the enzyme to generate a trapped covalent enzyme-tRNA intermediate ([16]; reviewed in [17]; see Figure 1). This trapped complex was used to identify the sulfhydryl group of Cys324 as the catalytic nucleophile [18]. Further experiments involving a chiral methyl SAM cofactor showed that methyl transfer proceeds by direct attack of uracil C5 on the SAM methyl group, leading to the mechanism depicted in Figure 1 [19]. Interestingly, a TrmA-tRNA intermediate complex also accumulates in vivo, perhaps owing to low intracellular levels of SAM [20]. The TrmA mechanism is similar to that proposed for thymidylate synthase and other enzymes catalyzing methyl transfer to C5 of cytosine or uracil, although the identity of the methyl donor may differ (see below) [21]. This body of enzymological work provides an essential framework for predicting the mechanism of any tRNA methylase operating at these positions.
Figure 1.
Mechanism of E. coli tRNA (m5U54) methyltransferase (RUMT, TrmA). Attack of a conserved enzyme cysteine sulfydryl group (Cys324) on C6 leads first to accommodation of the negative charge on O4, allowing for nucleophilic attack by the C5 carbon on the methyl group of SAM, with the sulfonium ion serving as electron sink to produce the SAH product. Base abstraction of a proton from C5 (by Glu358) then leads to elimination of the covalent adduct and regeneration of active enzyme.
The structural basis for the TrmA reaction was recently explained by crystallization and high-resolution structure determination of a trapped covalent complex of E. coli TrmA with the T-arm of tRNAPhe [22]. The complex was trapped by the E358Q TrmA mutant: Glu358 is the catalytic base (:B) that abstracts the proton from C5 in the final catalytic step (Figure 1). Glu358 of TrmA was identified by homology with Glu424 of the homologous E. coli rRNA C5 methylase RumA (the first RNA methyltransferase:RNA cocrystal structure ever determined [23]). The TrmA structure reveals both a trapped covalent adduct of Cys324 with C6 of U54, and a methyl group covalently attached at C5, as predicted by the mechanism. While Glu358 is confirmed as the catalytic base, the S-adenosyl homocysteine (SAH) product is not visualized in the structure, and details of the cofactor binding site in the ternary complex are not available. However, the structures of a binary Pyrococcus abyssi TrmA-SAM complex [24], and of RumA bound in a ternary complex with an [F]5U-substituted rRNA motif and SAH [23], should allow detailed modeling of a fully occupied E. coli TrmA active site.
When bound to TrmA, the T-arm of tRNAPhe adopts a conformation not seen in any NMR structure of the isolated T-arm, suggesting an important induced-fit component to the binding reaction [22]. All the RNA-protein interactions are made with the seven-nucleotide T-loop and flanking two base-pairs, consistent with kinetic measurements indicating that this minimal 11-mer RNA is a competent TrmA substrate [25]. The base of U54 is flipped out of the loop to bind in the TrmA active site, and the T-arm structure is stabilized by entry of the A58 base into the space vacated by the extrusion of U54. These RNA loop stabilization interactions promoted by TrmA in its catalytic complex are remarkably similar to those observed in the local rRNA loop motif that is methylated by RumA [22, 23]. Thus, a consensus RNA-fold for C5 methylation is identified by comparing the cocrystal structures of TrmA and RumA. More generally, comparison of these two complexes provides very detailed insight into how homologous RNA methyltransferases with identical catalytic mechanisms promote highly similar local RNA folding, yet also differentiate to recognize disparate RNA structures farther away from the methylation site.
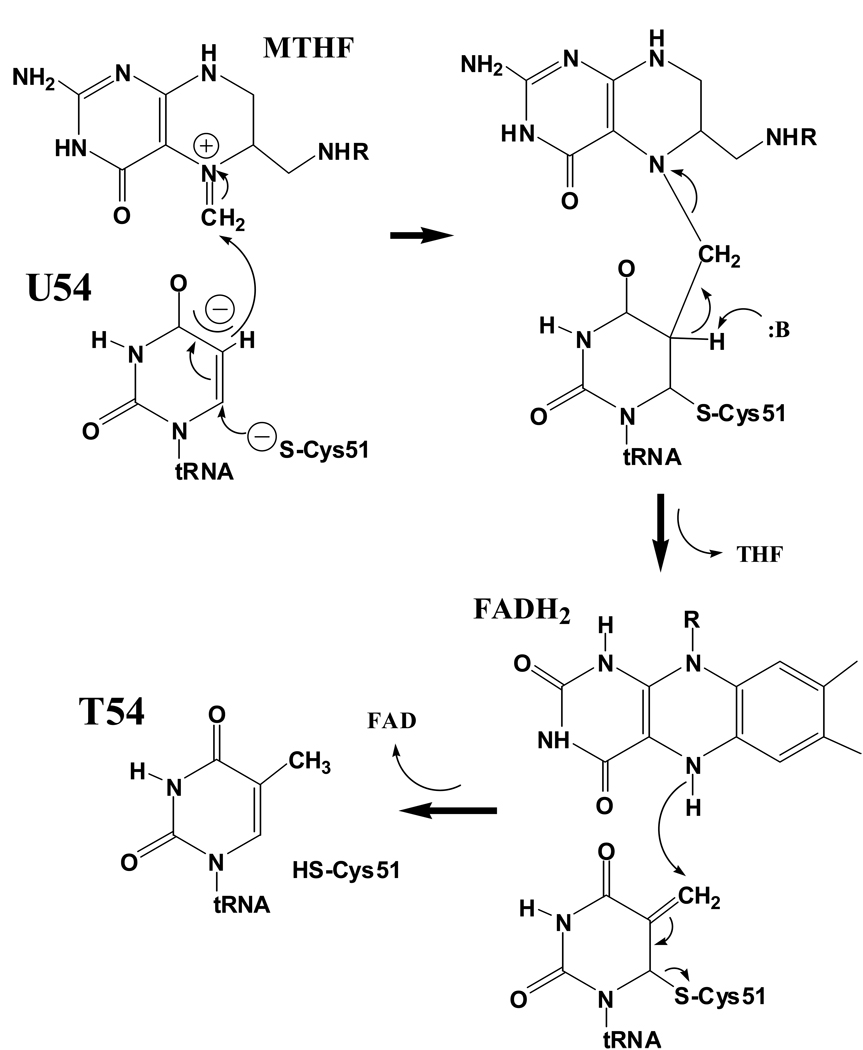
Interestingly, T54 formation in a subset of bacteria is catalyzed not by SAM-dependent TrmA, but by a different enzyme, TrmFO, that instead utilizes folate and FAD as the methyl transfer cofactors [26]. The phylogenetic distributions of TrmA and TrmFO are mutually exclusive. A crystal structure of T. thermophilus TrmFO bound to tetrahydrofolate and FAD showed that the enzyme possesses two domains, both of which share structural similarity with domains of the GidA (MnmG) protein that participates in formation of cmnm5U34 in some tRNAs [27]. However, the relative domain orientations and quaternary organizations of GidA and TrmFO differ significantly, and GidA itself may not possess methylase activity. A stereochemical mechanism for TrmFO has been hypothesized based on the catalytic pathways of TrmA and of the thymidylate synthases ThyX and ThyA - folate-dependent enzymes that methylate C5 of dUMP to form dTMP for DNA synthesis [27]. As in TrmA, the proposed TrmFO pathway initiates with nucleophilic attack on C6 by a conserved enzyme thiolate group. The partially anionic C5 carbon then attacks the methylene group of 5,10-methylenetetrahydrofolate (MTHF), with the iminium ion at N5 of MTHF serving as the electron sink, analogous to the sulfonium of SAM in TrmA (Figure 2). This produces a covalent ternary intermediate that is resolved by base abstraction of the proton from C5 of U54, again analogous to the TrmA mechanism. A final step involves oxidation of FADH2 to FAD, with concomitant reduction of the C5-methylene to a methyl group and regeneration of FADH2 on the enzyme by oxidation of NADPH. This mechanism is consistent with the TrmFO structure, in which the methylene group of MTHF binds adjacent to the redox-active nitrogen of FAD [27]. Further corroboration awaits determination of a tRNA-bound cocrystal structure.
Figure 2.
Proposed mechanism for T. thermophilus TrmFO-catalyzed formation of ribothymidine at position 54 of tRNA. Nucleophilic attack on C6 generates in turn a nucleophilic C5 position that attacks the methylene group of 5,10-methylenetetrahydrofolate (MTHF). The covalent intermediate (upper right) is resolved by base abstraction of a proton at C5, analogous to the TrmA mechanism depicted in Figure 1A, but producing a methylene group at C5. The methylene is reduced to a methyl group by a redox chain involving both FAD and NADP+ cofactors.
C5 methylation of cytosine to produce the m5C methylated base occurs in archaea and eukaryotes, but not in eubacteria. The best-characterized tRNA m5C methyltransferase is yeast Trm4, which (in contrast to TrmA and TrmFO) possesses remarkably broad substrate specificity: it is capable of methylating cytosines located at positions 34, 40, 48 and 49 in various yeast tRNAs [28]. Eukaryotic versions of Trm4 possess a C-terminal extension not found in the archaeal enzymes: this extension does not efficiently bind tRNA yet nonetheless significantly enhances the activity of the common core catalytic domain [29]. Sequence analysis indicates that the catalytic domains of m5C RNA methylases are of the class I Rossmann fold type, and possess two conserved cysteine residues residing within “PC” (Pro-Cys) and “TC” (Thr-Cys) motifs, respectively [30]. One conserved cysteine (TC-Cys) is proposed to function as the catalytic nucleophile for addition to C6, as described for TrmA and TrmFO. The other conserved cysteine (PC-Cys) is proposed to function in product release, as suggested by the accumulation of higher molecular weight protein-RNA complexes in cells expressing a Trm4p variant in which the PC motif is replaced by PA (Pro-Ala) [30]. A comprehensive phylogenetic analysis of m5C methyltransferases identified also the highly conserved Lys179 and Asp257 residues, predicted to lie in the active site, as essential to catalysis; this prediction was confirmed by site-directed mutagenesis [31]. The crystal structure of the unliganded homologous rRNA methylase YebU from E. coli confirmed the localization of both cysteines, Lys179, and Asp257 in the active site [32], although a detailed stereochemical mechanism involving all these residues has not yet been formulated. No cocrystal structures of m5C methyltransferases bound to tRNA have yet been reported.
Mechanisms of tRNA methylation at nitrogen
The m6A methylation
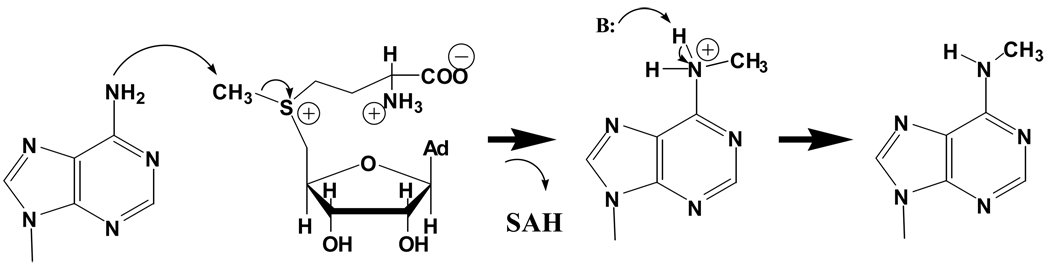
Methylation of the four canonical tRNA bases at nitrogen is known to occur at all nonglycosidic nitrogens except for N3 of adenosine and guanosine and N7 of adenosine. However, despite the large variety of chemical and structural contexts at which these RNA N-methylation events occur, relatively little detailed information is available regarding the stereochemical pathways of the enzyme-catalyzed reactions. Indeed, the best-studied N-methylation reaction in nucleic acids has for some time been that catalyzed by the N6-adenine DNA methyltransferase M.TaqI. The structure of this enzyme bound to DNA and the unreactive cofactor analog 5’-[2-(amino)ethylthio]-5’-deoxyadenosine revealed specific enzyme recognition of an extrahelical target adenine, showing definitively that base-flipping out of duplex nucleic acid occurs for adenine as well as cytosine methylation [33]. In the crystal structure, the N6 of the flipped adenosine points toward the reactive methyl group of the cofactor, and the respective positions of these groups are stabilized by contacts with the conserved active-site motif NPPY. This provides a structural framework for appreciating how the necessary bond-breaking and bond-making steps are facilitated in the active site.
Formulation of a mechanism for M.TaqI was challenging given the very high pKa of 20 for the 6-amino group, the elimination (based on mutational studies) of the NPPY tyrosine as a candidate for general base, and the lack of other obvious candidates for general base in the active site. However, it was noted that the NPPY motif could offer the asparagine amide and a backbone carbonyl oxygen to form hydrogen-bonds with the N6 hydrogens to increase the nucleophilicity of N6 by increasing its electron density. Further, these hydrogen-bond donors are located below the plane of the adenine ring, and might function further to help distort the sp2 character of N6 toward sp3, thus dramatically increasing N6 nucleophilicity by assisting its dissociation from the aromatic system of the adenine ring. Finally, because the pKa of a 6-methyl ammonium ion is sharply decreased as compared with the 6-amino group, even very weak basic groups in the active site (such as the asparagine amide and a backbone oxygen) might function as transient proton acceptors. One or more water molecules in the active site, which are connected to bulk solvent, might also play a role in subsequent proton transfers [34]. Based on these considerations, it appears likely that the lowest-energy transition state for methylation features a substantial sp3 character at N6. This conclusion is supported by a theoretical study based on a model system mimicking the M.TaqI active site [34]. A possible mechanism for methylation to produce m6A is depicted in Figure 3.
Figure 3.
Proposed mechanism for methylation at exocyclic adenine N6 by the DNA methylase M.TaqI – a potential model for tRNA methylases operating at adenine N6, guanine N2, and cytosine N4. Attack on the SAM methyl group is depicted as proceeding via attack by nitrogen in its sp2 hybridization state, because proton removal prior to nucleophilic attack is highly unfavorable. Proton abstraction from the positively charged quaternary nitrogen, by contrast, is relatively facile. See text for details.
The M.TaqI mechanism is important because it provides a model for the function of tRNA methylases that modify exocyclic amines with high pKa values: m6A, m2G, and m4C. The first report of a tRNA methylase that produces the relatively rare m6A modification was only very recently published: the E. coli yfiC gene encodes a methylase that modifies A37 in a tRNAVal isoacceptor [35]. The m4C modification is found in mitochondrial tRNA [36], but the enzyme catalyzing this reaction has not yet been identified. For both m6A and m4C better-studied examples of rRNA methylases are available, but detailed structural and mechanistic work has not yet been reported. Generation of m4C by bacterial DNA modification methylases such as M.BamHI has been the subject of numerous kinetic studies [37], but again a cocrystal structure of an enzyme-DNA complex to provide an interpretive structural framework is lacking.
The m2G and m22G methylation
Although the m2G and m22G modifications have been described at a variety of nucleotide positions, the only examples for which the enzymes have been well-characterized are for methylation at G10, G26, and G27 [38–41]. These three nucleotides are each located near the junction of the D and anticodon arms in tRNA, where the structure deviates from canonical A-form helical geometry owing to a noncontiguous sugar-phosphate backbone and non-Watson-Crick hydrogen-bonding arrangement at the bridging 26–44 pair. The enzyme that catalyzes dimethylation at G26, Trm1, was first characterized from a variety of archaeal and eukaryotic sources, and in several cases has been shown to produce the monomethylated species as a reaction intermediate (reviewed in [38, 39]). In general the enzyme was found to be sensitive to nucleotide alterations in the nearby D-stem as well as in the variable loop, suggesting that these regions of the substrate might be important to site-selectivity. Crystal structures of Pyrococcus horikoshii Trm1 bound to either SAM or SAH were recently reported [39]. The structures revealed an N-terminal class I methylase Rossmann fold catalytic domain that binds the cofactor, as expected, together with a structurally unique 140 amino acid α/β C-terminal domain of unknown function. An additional narrow pocket adjacent to the SAM binding cleft was hypothesized to provide a binding pocket for the G26 base.
Recently, a putative gene encoding a Trm1 homolog was found in the hyperthermophile Aquifex aeolicus, the first example of the enzyme found in the eubacterial domain [38], and its activity confirmed both in vivo and in vitro. Remarkably, it was found that the recombinant enzyme transfers more than two methyl groups per tRNA, and is able to methylate a transcript containing A26. Using LC/MS analysis, it was shown that A. aeolicus Trm1 is able to methylate both G26 and G27 in vitro, and that native tRNACys purified from cells using DNA-affinity chromatography indeed contains both m22G26-m2G27 and m22G26-m22G27. Like yeast Trm4, A. aeolicus Trm1 is a multi-site recognition enzyme [28]. Further, use of variant tRNA transcripts as substrates suggested that recognition by A. aeolicus Trm1 depends on the T-arm structure but not on either the D-arm or variable loop as found for the archaeal and eukayotic enzymes. Thus, Trm1 may have evolved to develop different domain specificity for tRNA recognition.
The enzyme catalyzing m22G10 formation was first identified in the hyperthermophilic archaeon P. abyssi, and is known as Trm11 [40]. Trm11 contains a C-terminal methylase domain of the Rossmann-fold linked to an N-terminal THUMP (thiouridine synthases, RNA methyltransferases, and pseudouridine synthases) domain found in a variety of RNA modifying enzymes. As shown for G26 modification, formation of m22G10 also proceeds through a monomethylated intermediate. The tRNA nucleotide determinants that are required for formation of m22G10 were identified as a G10-U25 base pair in the D-stem, together with a four-nucleotide variable loop. tRNAs containing G10-C25 or a five-nucleotide variable loop were only monomethylated at G10. Interestingly, the tRNA sequence and structural features for monomethylation of G10 are the same as those for dimethylation of G26, suggesting independent evolution of Trm1 and Trm11 toward their distinct tRNA specificities.
The dimethylated m22G10 is not found in eukaryotic tRNAs. In yeast, a heterodimeric enzyme complex catalyzes the monomethylation at m2G10. This complex consists of Trm11 and a second protein designated Trm112, both of which are required for formation of m2G10 in vivo with comparable intracellular abundance. Interestingly, Trm112 also interacts with another tRNA methyltransferase, Trm9, which methylates uridine in the anticodon wobble position of yeast tRNAArg and tRNAGlu [42]. Although a protein complex isolated from yeast extract yields m2G10 synthesis activity, coexpression of Trm11 and Trm112 together in E. coli does not reconstitute the activity. Thus, additional protein factors may be required to form the active complex in yeast cells. Trm11/112 is only one example of a multimeric enzyme that is required for tRNA methylation; other examples include the enzymes responsible for synthesis of m1A58 and m7G46 (see below).
The m1G37 methylation
The formation of endocyclic m1G37 by eukaryal/archaeal Trm5 and bacterial TrmD has been well-characterized. The m1G37 modification is conserved in all three domains of life, and plays an important role in increasing fidelity of ribosomal decoding in both the cytoplasm and mitochondria [43]. Interestingly, the bacterial TrmD and archaeal/eukaryal Trm5 form an example of analogous pairs and also recognize different determinants on tRNA: only Trm5 is sensitive to the integrity of the tertiary tRNA structure [44]. The recently determined ternary complex structure of M. jannaschii Trm5 bound to tRNA and SAM reveals recognition of the tRNA globular hinge domain via a large induced-fit motion of one domain upon binding, explaining the importance of tRNA tertiary conformation to function [13]. Separate crystal structures in complex to tRNACys and to tRNALeu show that the overall orientation of binding is conserved despite the large difference in the tRNA variable arms, but significant differences in the detailed conformations and interactions with the respective anticodon loops are observed.
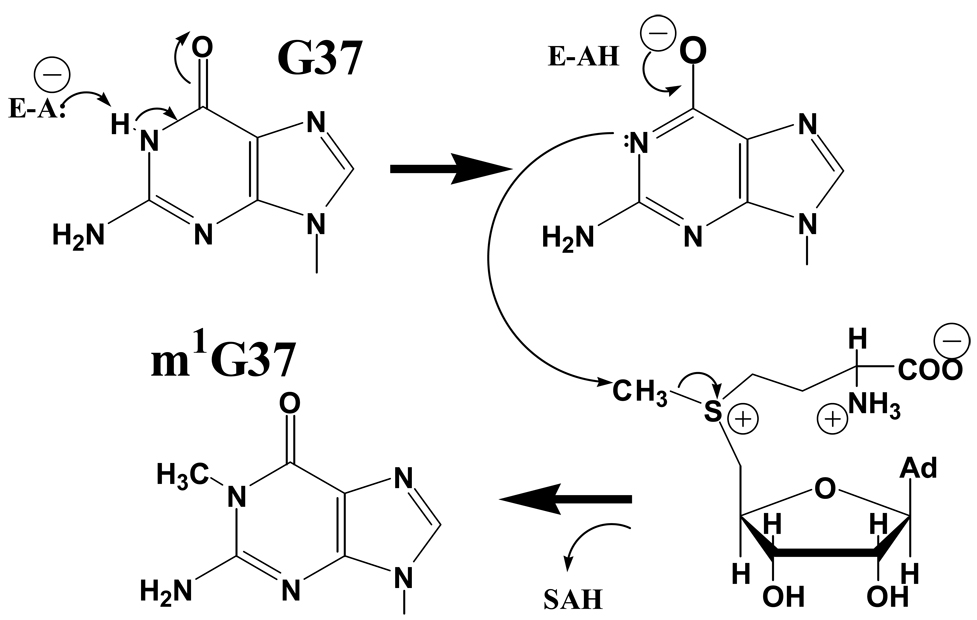
The mechanism of m1G formation has been elucidated for the M. jannaschii Trm5 enzyme. The crystal structure shows a conformation poised for catalysis, in which the N1 of G37 is located 2.8 Å from the methyl group of SAM [13]. However, there is no apparent general base positioned to accept the proton from N1. Further, the positively charged sulfonium also disfavors approach of protonated N1. For these reasons, it appears likely that the crystal lattice has trapped a deprotonated form of G37 in which a lone pair of electrons on nitrogen is poised to attack the SAM methyl group. Measurement of the pH dependence of the single-turnover rate constant for m1G37 formation showed a remarkably steep increase in the rate of methyl transfer as the proton concentration is lowered, up to an asymptote at neutral pH [45]. A plot of log(kobs) versus pH yielded a slope of approximately 2, indicating a mechanism involving two ionizing groups. The dependence of the reaction rate on the most basic group in a doubly ionizing system is consistent with the notion that kobs corresponds to nucleophilic attack of the deprotonated N1 of tRNACys G37 on the methyl group of SAM, with electron transfer to the sulfonium ion (Figure 4). The negative charge developing on O6 is compensated, as shown in the crystal structure, by the conserved guanidinium of Arg145. Because the structure suggests that deprotonation occurs before final docking of G37 in the active site, proton transfer from G37-N1 through two enzyme groups apparently occurs during the induced-fit transition of the tRNA anticodon loop. The identities of the protein groups involved in the double ionization remain to be elucidated.
Figure 4.
Proposed mechanism for M. jannaschii Trm5. Proton abstraction during docking of G37 in the active site (upper left) yields deprotonated G37 as visualized in the crystal structure (upper right). Nucleophilic attack on SAM by deprotonated N1 then leads to product formation.
Mutational analysis of Trm5 shows that replacement of residues playing a role in substrate organization in the active site, such as Asp223, Asn265, or Pro267, have greater effects on the catalytic rate as compared with residues involved more directly in the chemical reaction. For example, while the side chain of Arg145 may serve to stabilize the negative charge on O6 of the G37 base after deprotonation of N1, elimination of the side chain of Arg145 has a lesser effect compared to elimination of the side chains of Asp223, Asn265, or Pro267 [46]. The lesser role of acid-base catalysis for methylation at nitrogen was also suggested previously for catalysis by the DNA N6 adenine methyltransferase M.TaqI, discussed above [33, 34]. A key distinction, however, is that deprotonation of guanine N1 by Trm5 occurs early, and the nucleophilic attack on SAM is by nitrogen in the sp2 hybridization state (Figure 3). In contrast, pKa ~ 20 for adenine N6 suggests that in M.TaqI, deprotonation occurs only after the nitrogen has acquired significant sp3 character [33, 34] (compare Figure 3 and Figure 4]. A pH kinetic analysis of the m6A DNA methylase M.EcoRI showed only a linear four-fold change in kobs over a wide pH range [47]. Enzymatic methylation at exocyclic adenine N6, then, may depend even less on acid-base chemistry to facilitate rate acceleration, than does methylation at endocyclic guanine N1.
The m7G46 methylation
The m7G46 modification is widely present in eukaryotes and bacteria, as well as in some archaea. It occurs in tRNA species that contain a variable loop of the typical size of 4–5 nucleotides. The modified m7G46 base is positively charged when present in the base triple C13-G22-G46 that stabilizes the tRNA tertiary core [48], indicating that the modification can introduce a site-specific electrostatic charge within the tRNA tertiary structure. The m7G46 modification is catalyzed by TrmB in bacteria (the YggH protein) but by the heterodimeric Trm8-Trm82 enzyme in yeast [49]. Interestingly, while the sequences of TrmB can vary significantly among bacterial species, as in the E. coli and Aquifex aeolicus enzymes [50] [51], the mechanism of tRNA recognition appears conserved [51]. The size of the variable loop, conserved sequences in the T stem, and nucleotides flanking the target G46 base each play an important role. In the case of the Trm8-Trm82 heterodimer, however, while Trm8 possesses both the SAM- and tRNA-binding capacity and performs the methyl transfer activity in vivo (albeit reduced) and in vitro, Trm82 does not bind either substrate. Instead, Trm82 functions to stabilize and promote the active conformation of Trm8 [49]. The crystal structure of the apo form of B. subtilis TrmB reveals that, despite the presence of a core catalytic domain consisting of the Rossmann fold, the enzyme exists as a homodimer [52], in contrast to the monomeric form of E. coli TrmB. Gel filtration analysis confirmed the homodimer structure, suggesting that this unusual structure is not a consequence of crystal packing. The dimer interface of B. subtilis TrmB does not involve the Rossmann fold but rather hydrophobic core residues that are unique to the enzyme. The structural diversity among the monomeric E. coli TrmB, the dimeric B. subtilis TrmB, and the heterodimeric yeast Trm8-Trm82 provides an example of how the Rossmann fold can function in different oligomeric contexts.
The m1A58 methylation
The m1A58 modification is found in the highly conserved A58 in the T loop of tRNAs. This modification is rare in bacteria (and is absent from E. coli) but is common in most eukarya and archaea. The bacterial enzymes responsible for this modification are the α4 TrmI tetramers [53, 54], whereas the yeast enzyme consists of the α2β2 Trm6/Trm61 complex [55]. Unlike the functional organization of Trm8-Trm82 for the m7G46 modification, the substrate binding functions of Trm6/Trm61 are segregated: Trm6 binds tRNA, whereas Trm61 binds SAM [56], Thus, both subunits are required to constitute the enzyme activity. In addition, the sequence similarity between Trm6 and Trm61 suggests that the eukaryotic m1A58 methylases evolved by gene duplication, whereas the bacterial enzymes remained as homotetramers. The tetrameric structure of the bacterial TrmI has been confirmed by crystal structural analysis of the Mycobacterium tuberculosis enzyme in complex with SAM [57], and of the T. thermophilus enzyme in complex with SAH [58]. Biophysical analysis with the noncovalent ESI-MS technique demonstrates that the tetramer binds one to two tRNA molecules [58], although the structural origin for this stoichiometry is unclear. Mutational analysis reveals that Asp170 of a DxxxPW motif in T. thermophilus TrmI (analogous to the NPPY motif in m6A methylases) is key to catalysis [58]. Docking of the structure of T. thermophilus TrmI with a substrate analog into the active site suggests a role of Asp170 in stabilizing both the SAM cofactor and the A58 base. In this docking model, Asp170 would be positioned to form a H-bond with the exocyclic N6 of A58, as a possible means for deprotonation of the N6 and activation of the lone electron pair on N1 for attack on the methyl group of SAM. In addition, two catalytically important aromatic residues, Tyr78 and Tyr194, are proposed to stabilize the transition state of methyl transfer [58], analogous to the role of the aromatic Y/F/W residue in the NPPY motif in m6A methylases.
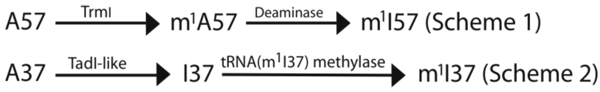
The m1I57 and m1I37 methylations
The m1I-methylation reaction can occur on the modified base inosine I57 in the T loop to synthesize m1I57 as a modification unique to archaeal tRNAs [59], or on the base of inosine I37 in the anticodon loop to synthesize m1I37 unique to eukaryotic tRNAAla (with the IGC anticodon) [60]. Interestingly, the two 1-methylation reactions on inosine proceed by different pathways (schemes 1 and 2 below). The archaea-specific m1I57 methylation is formed in a pathway that first introduces the 1-methylation to the canonical base A57 in tRNA by a specialized SAM-dependent tRNA(m1A57) methylase to synthesize m1A57, which is then converted to m1I57 by a 1-methyladenosine-57 deaminase [59]. In contrast, the eukaryotic tRNAAla-specific m1I37 methylation begins by deamination of A37 by a deaminase to synthesize I37, which is then modified to m1I37 by a methylase [60]. In this pathway, the deaminase would be functionally similar to the deaminase Tad1, which is specific at position 34. Thus, distinct site-specific pathways for synthesis of the m1I-modified base in tRNA appear to exist.
The archaeal enzyme for the synthesis of m1A57 is homologous to the bacterial TrmI. Indeed the enzyme TrmI of the thermophilic archaeon P. abyssi has dual specificity for catalysis of the 1-methylation reaction at both A57 and A58 in tRNA, although the methylation of A57 is more efficient than the methylation of A58 [53]. This suggests the possibility of a sequential methylation mechanism beginning with A57 and continuing to A58. Alternatively, the archaeal TrmI might form a complex with the yet-to-be identified deaminase to proceed with the two-step synthesis of m1I57; upon release from the complex, TrmI might be recruited to catalyze synthesis of m1A58. Because the P. abyssi TrmI can efficiently catalyze m1A58 formation when A57 is replaced by G57 [53], and because inosine is structurally more similar to guanosine than to adenosine, the second possibility may be favored. Possible models for this pathway are the two-step reaction schemes by which certain aminoacyl-tRNAs are synthesized by the action of two consecutive enzymes, which may form a complex to promote the rapid production of the end product [61, 62]. The P. abyssi TrmI exists as an α4 homotetramer, analogous to the oligomeric structure of bacterial TrmI [53]. Interestingly, the dimer-dimer interface of P. abyssi TrmI contains the two conserved cysteine residues C196 and C233 (not present in bacterial TrmI), which appear to enforce the tetrameric structure via inter-subunit disulfide bridges to resist heat denaturation at extreme temperatures. The Rossmann fold is also modified to place the conserved cysteines at subunit interfaces, such that C196 from one monomer and C233 from a neighboring monomer form a disulfide bridge to stabilize a tight-binding dimer. This pair of C196-C233 bridge then interacts with another pair of bridge from the other tight-binding dimer to stabilize the tetramer.
The eukaryotic enzyme responsible for synthesis of m1I37 in the anticodon loop of tRNAAla has not been identified. Limited work has shown that the enzyme can be separated by chromatography from the related eukaryotic Trm5 enzyme that catalyzes synthesis of m1G37 [60]. The two enzymes appear to have overlapping specificity and can both target I37 or G37 in tRNA for 1-methylation, albeit with different efficiencies. It was also shown that the Xenopus laevis oocyte enzyme for synthesis of m1I37 recognizes tRNAAla of different origins [60]. Thus, the sequence determinants in different tRNAAla species that would confer recognition by this enzyme remain elusive. The identification of these determinants may shed light on why only tRNAAla in eukaryotes possesses the m1I37 modification.
Biological significance of tRNA base methylation
Individual methylations to tRNA nucleotide bases generally do not have apparent effects on cell growth. For example, while the m5U54 modification is conserved in evolution, elimination of the responsible TrmA enzyme reveals no functional impact on the normal cell growth rate [63]. This lack of a clear growth effect also holds true for other types of modifications to tRNA nucleotides, such as the thiolation at position 4 to the U8 nucleotide (s4U8) that is commonly present in tRNA species [63]. However, while inactivation of a specific base methylation may have no clear growth effect, inactivation of the methylation in combination with another usually has a strong negative effect on growth. For example, while the m7G46 modification by itself is not critical for cell growth in yeast, elimination of this modification together with another modification, regardless of whether the second modification involves methylation, thiolation, or conversion of uridine to pseudouridine or to dihydrouridine, leads to growth arrest and cell death [4]. This observation suggests that modifications to tRNA nucleotide bases and backbones, including the various methylation reactions, are present to build a network that will improve the overall function of tRNA and in turn will promote cell fitness and viability. When more than one modification is removed, the efficiency of the overall tRNA function is compromised, ultimately leading to a cell growth defect. Recent studies have shown that alteration of tRNA levels within the small margin of 10% can have a large impact on cell growth [64].
The m1G37 modification is a notable exception, however. This modification is essential for growth in bacteria, such as E. coli [65], Salmonella typhimurium [66], and Streptococcus pneumonia [67], and in yeast, such as S. cerevisiae [68]. Earlier genetic studies in bacteria showed that m1G37 is important for promoting aminoacyl-tRNA binding and selectivity to the ribosome A site [69], and for reducing frameshift errors during translation of the genetic code [66, 70]. A structural analysis suggests that the modification imposes constraints on anticodon loop dynamics [71], providing a rationale for the ability of the modification to enforce codon-anticodon pairing on the ribosome. In the yeast S. cerevisiae, the modification prevents mischarging of tRNAAsp by arginyl-tRNA synthetase [72], suggesting that it functions as an anti-determinant against misacylation. Further, the modification is the obligate precursor for subsequent modifications to synthesize the hyper-modified wybutosine base at position 37 in tRNAPhe [73]. The diverse functional roles associated with the m1G37 modification underscore the importance of the modification in bacteria and in yeast. In the archaeal domain, the modification is necessary for efficient tRNA aminoacylation by cysteinyl- and phosphoseryl-tRNA synthetases [62, 74]. Because the efficiency of tRNA aminoacylation is tightly coupled to ribosome function and to cell growth, the m1G37 modification is likely a critical determinant for cellular survival in the archaeal domain as well.
The m1A58 modification is another exception. This modification is specifically required to maintain steady-state levels of initiator tRNAMet in S. cerevisiae [55]. Deletion of the Trm6 component of the Trm6-Trm61 heterodimer enzyme responsible for synthesis of m1A58, or mutations of Trm6 that disrupt SAM-binding but do not interfere with formation of the heterodimer, lead to cell death [55]. In addition, all retroviruses utilize m1A58-containing tRNA to prime reverse transcription. In human tRNA3Lys, this modification is required for accurate termination of plus-strand strong-stop DNA synthesis during HIV-1 replication [75], presumably because the N1 methyl group blocks correct base pairing. The A58 to U58 mutation in human tRNA3Lys has been shown to inhibit HIV-1 replication in T cells [76], providing further support for the significance of the m1A58 modification. Interestingly, while m1A58 is essential for yeast viability, reduction in the enzyme activity for the modification is not lethal in a rat mammary adenocarcinoma [77]. This suggests that targeting the m1A58 modification, which is required for HIV replication, may offer new promises as a possible anti-HIV therapy [75, 76].
Also of medical interest is the m1I37 modification unique to eukaryotic tRNAAla. This tRNA harbors the additional base modification of A34 to I34 at the wobble position of the anticodon. The m1I37 and the I34 modifications are both the target of the autoimmune antibodies that are present in the sera of patients afflicted with the inflammatory disease of the PL12 type of polymyositis [78, 79]. Because the unmodified transcript of human tRNAAla does not induce the production of the autoimmune antibodies, it appears that both modifications are the required antigens to specifically stimulate the autoimmune response [60]. Although the native human tRNAAla also contains other modifications in the anticodon loop (such as the 2'-O-methylation of U32 and of G39, and the pseudouridine at position 38), a transcript harboring just the I34 and m1I37 modifications was sufficient to elicit the autoimmune response, suggesting that these two modifications are the dominant factors. The molecular basis for how these two modifications are associated with the autoimmune disease is not known; thus, further studies to elucidate the enzymatic pathways and mechanisms leading to the modifications are warranted. Nonetheless, it is clear that specific methylation to tRNA nucleotide bases can indeed impact human health.
Conclusion and perspective
Table 1 summarizes the diverse methylation reactions that are discussed in this review, together with the well-known enzymes responsible for the reactions. Clearly, while many of these methylation reactions have well-established roles in tRNA structural stability and functional activities, the structural basis for substrate recognition and catalysis remain poorly characterized. Thus, the origins of selectivity for the target base, the juxtaposition and orientation of the target base relative to the methyl group of SAM, the chain(s) of proton transfer that must accompany the process of methyl transfer, and the enzyme residues involved in coordinating proton transfers with electrostatic changes occurring on the target base are not well understood. The importance of these questions is particularly keenly realized in the case of analogous pairs of methylation enzymes that are known to employ different stereochemical pathways to catalyze the same chemical reaction. These questions highlight the need to address how the distinct stereochemistries of two enzymes can be used to overcome the thermodynamic barrier of the same chemical reaction. In addition, the isolation of the pair of the bacterial TrmD and the eukaryotic-archaeal Trm5 for the m1G37 methylation makes a compelling case that a better understanding of the stereochemistry of each enzyme will pave the way to a strong foundation for species-specific targeting of TrmD as a means to develop new anti-bacterial strategies.
Table 1.
Enzyme-catalyzed tRNA base modifications discussed in this review
| Bacterial Enzymes | Eukaryotic Enzymes | Archaeal Enzymes | |
|---|---|---|---|
| m5U54 (riboT54) | E. coli TrmA | P. abyssi TrmA | |
| m5U54 (riboT54) |
T. thermophilus TrmFO |
||
| m5C34, C40, C48, C49 | S. cerevisiae Trm4 | ||
| m6A37 | E. coli yfiC | ||
| m22G26 | A. aeolicus Trm1 | P. horikoshii Trm1 | |
| m22G10 | P. abyssi Trm11 | ||
| m2G10 |
S. cerevisiae Trm11– Trm112 |
||
| m1G37 | E. coli TrmD | S. cerevisiae Trm5 | M. jannaschii Trm5 |
| m7G46 | E. coli TrmB |
S. cerevisiae Trm8– Trm82 |
|
| B. subtilis TrmB | |||
| m1A58 | E. coli TrmI |
S. cerevisiae Trm6– Trm61 |
|
|
M. tuberculosis TrmI |
|||
|
T. thermophilus TrmI |
|||
| m1A57 | P. abyssi TrmI |
This table summarizes enzymes that are discussed in this review. References for each enzyme are cited within the text. Blanks mean that the enzymes are absent or are not well characterized. Abbreviations: E. coli, Escherichia coli; P. abyssi, Pyrococcus abyssi; T. thermophilus. Thermus thermophilus; S. cerevisiae, Saccharomyces cerevisiae; P. horikoshii, Pyrococcus horikoshii; A. aeolicus, Aquifex aeolicus; M. jannaschii, Methanococcus jannaschii; B. subtilis, Bacillus subtilis; M. tuberculocus, Mycobacterium tuberculosis.
To more broadly understand the stereochemistries of methylation enzymes, it is necessary to obtain additional crystal structures of enzymes in complex with tRNA and SAM, and to combine structural analysis with appropriate biochemical tools to address questions regarding reaction mechanisms. The emphasis on the appropriate biochemical tools cannot be overstated. For example, analysis of the Trm5-catalyzed m1G37 methylation reaction has demonstrated that the reaction rate constant measured under steady-state multi-turnover conditions is significantly slower than the rate constant measured under single turnover conditions [46], indicating that in this case the steady-state kinetics does not report on the kinetics of methyl transfer but rather on the events subsequent to methyl transfer. Thus, the development of appropriate kinetic tools for each enzyme and for each specific step of the enzyme-catalyzed reaction must be in place so that the kinetic analysis can be correlated with structural information and interpreted on structural grounds. The development of appropriate kinetic methodologies for each enzyme will remain as one of the major challenges ahead.
It should be noted that while this review focuses only on the methylation reactions that directly target the four canonical bases in tRNA, there are additional methylation reactions that target bases that are already modified. Such methylation reactions on modified bases ultimately generate hyper-modified residues on tRNA. Often these hypermodified bases are localized at positions 34 or 37 in the anticodon loop, where they specifically function to improve codon-anticodon pairing interactions on the ribosome. An example of hyper-modification is found in the biosynthesis pathway of the wybutosine base at position 37 in tRNAPhe. In this pathway, the canonical G37 base is first converted to m1G37, which is then converted to wybutosine in four successive enzymatic reactions, one of which involves methylation to an already highly modified intermediate [73]. Other examples include the methylation to the hyper-modified base N6-isopentenyl-adenosine (i6A) to synthesize ms2i6A, to N6-threonyl-carbamoyl adenosine (t6A) to synthesize ms2t6A, and to 2-thio-uridine (s2U) to synthesize mnm5s2U [80]. All of the methylation reactions that target hypermodified bases involve distinct new SAM-binding enzymes that must have the ability to discriminate against the canonical bases. Obviously, while the elucidation of reaction mechanisms for hypermodifications is necessarily more complex and challenging, it must build upon the mechanistic insights from studies of the primary methylation reactions such as those that are discussed in this review.
Acknowledgements
The authors acknowledge support by NIH grants GM081601 to YMH and GM053763 to JJP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bjork GR, Ericson JU, Gustafsson CE, Hagervall TG, Jonsson YH, Wikstrom PM. Transfer RNA modification. Annu Rev Biochem. 1987;56:263–287. doi: 10.1146/annurev.bi.56.070187.001403. [DOI] [PubMed] [Google Scholar]
- 3.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 5.Cantoni GL. Biological methylation: selected aspects. Annu Rev Biochem. 1975;44:435–451. doi: 10.1146/annurev.bi.44.070175.002251. [DOI] [PubMed] [Google Scholar]
- 6.Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anantharaman V, Koonin EV, Aravind L. SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J Mol Microbiol Biotechnol. 2002;4:71–75. [PubMed] [Google Scholar]
- 8.Ahn HJ, Kim HW, Yoon HJ, Lee BI, Suh SW, Yang JK. Crystal structure of tRNA(m1G37)methyltransferase: insights into tRNA recognition. Embo J. 2003;22:2593–2603. doi: 10.1093/emboj/cdg269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elkins PA, Watts JM, Zalacain M, van Thiel A, Vitazka PR, Redlak M, Andraos-Selim C, Rastinejad F, Holmes WM. Insights into catalysis by a knotted TrmD tRNA methyltransferase. J Mol Biol. 2003;333:931–949. doi: 10.1016/j.jmb.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 10.Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Bjork GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. Embo J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christian T, Evilia C, Williams S, Hou YM. Distinct origins of tRNA(m1G37) methyltransferase. J Mol Biol. 2004;339:707–719. doi: 10.1016/j.jmb.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 12.Goto-Ito S, Ito T, Ishii R, Muto Y, Bessho Y, Yokoyama S. Crystal structure of archaeal tRNA(m(1)G37)methyltransferase aTrm5. Proteins. 2008;72:1274–1289. doi: 10.1002/prot.22019. [DOI] [PubMed] [Google Scholar]
- 13.Goto-Ito S, Ito T, Kuratani M, Bessho Y, Yokoyama S. Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation. Nat Struct Mol Biol. 2009;16:1109–1115. doi: 10.1038/nsmb.1653. [DOI] [PubMed] [Google Scholar]
- 14.Purta E, van Vliet F, Tkaczuk KL, Dunin-Horkawicz S, Mori H, Droogmans L, Bujnicki JM. The yfhQ gene of Escherichia coli encodes a tRNA:Cm32/Um32 methyltransferase. BMC molecular biology. 2006;7:23. doi: 10.1186/1471-2199-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci U S A. 1984;81:6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santi DV, Hardy LW. Catalytic mechanism and inhibition of tRNA (uracil-5-)methyltransferase: evidence for covalent catalysis. Biochemistry. 1987;26:8599–8606. doi: 10.1021/bi00400a016. [DOI] [PubMed] [Google Scholar]
- 17.Kealey JT, Gu X, Santi DV. Enzymatic mechanism of tRNA (m5U54)methyltransferase. Biochimie. 1994;76:1133–1142. doi: 10.1016/0300-9084(94)90042-6. [DOI] [PubMed] [Google Scholar]
- 18.Kealey JT, Santi DV. Identification of the catalytic nucleophile of tRNA (m5U54)methyltransferase. Biochemistry. 1991;30:9724–9728. doi: 10.1021/bi00104a022. [DOI] [PubMed] [Google Scholar]
- 19.Kealey JT, Lee S, Floss HG, Santi DV. Stereochemistry of methyl transfer catalyzed by tRNA (m5U54)-methyltransferase--evidence for a single displacement mechanism. Nucleic Acids Res. 1991;19:6465–6468. doi: 10.1093/nar/19.23.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urbonavicius J, Jager G, Bjork GR. Amino acid residues of the Escherichia coli tRNA(m5U54)methyltransferase (TrmA) critical for stability, covalent binding of tRNA and enzymatic activity. Nucleic Acids Res. 2007;35:3297–3305. doi: 10.1093/nar/gkm205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivanetich KM, Santi DV. 5,6-dihydropyrimidine adducts in the reactions and interactions of pyrimidines with proteins. Prog Nucleic Acid Res Mol Biol. 1992;42:127–156. doi: 10.1016/s0079-6603(08)60575-9. [DOI] [PubMed] [Google Scholar]
- 22.Alian A, Lee TT, Griner SL, Stroud RM, Finer-Moore J. Structure of a TrmA-RNA complex: A consensus RNA fold contributes to substrate selectivity and catalysis in m5U methyltransferases. Proc Natl Acad Sci U S A. 2008;105:6876–6881. doi: 10.1073/pnas.0802247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee TT, Agarwalla S, Stroud RM. A unique RNA Fold in the RumA-RNA-cofactor ternary complex contributes to substrate selectivity and enzymatic function. Cell. 2005;120:599–611. doi: 10.1016/j.cell.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 24.Walbott H, Leulliot N, Grosjean H, Golinelli-Pimpaneau B. The crystal structure of Pyrococcus abyssi tRNA (uracil-54, C5)-methyltransferase provides insights into its tRNA specificity. Nucleic Acids Res. 2008;36:4929–4940. doi: 10.1093/nar/gkn437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu XR, Santi DV. The T-arm of tRNA is a substrate for tRNA (m5U54)-methyltransferase. Biochemistry. 1991;30:2999–3002. doi: 10.1021/bi00226a003. [DOI] [PubMed] [Google Scholar]
- 26.Urbonavicius J, Skouloubris S, Myllykallio H, Grosjean H. Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria--evolutionary implications. Nucleic Acids Res. 2005;33:3955–3964. doi: 10.1093/nar/gki703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishimasu H, Ishitani R, Yamashita K, Iwashita C, Hirata A, Hori H, Nureki O. Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase. Proc Natl Acad Sci U S A. 2009;106:8180–8185. doi: 10.1073/pnas.0901330106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motorin Y, Grosjean H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. Rna. 1999;5:1105–1118. doi: 10.1017/s1355838299982201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walbott H, Auxilien S, Grosjean H, Golinelli-Pimpaneau B. The carboxyl-terminal extension of yeast tRNA m5C methyltransferase enhances the catalytic efficiency of the amino-terminal domain. J Biol Chem. 2007;282:23663–23671. doi: 10.1074/jbc.M703818200. [DOI] [PubMed] [Google Scholar]
- 30.King MY, Redman KL. RNA methyltransferases utilize two cysteine residues in the formation of 5-methylcytosine. Biochemistry. 2002;41:11218–11225. doi: 10.1021/bi026055q. [DOI] [PubMed] [Google Scholar]
- 31.Bujnicki JM, Feder M, Ayres CL, Redman KL. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. 2004;32:2453–2463. doi: 10.1093/nar/gkh564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hallberg BM, Ericsson UB, Johnson KA, Andersen NM, Douthwaite S, Nordlund P, Beuscher AEt, Erlandsen H. The structure of the RNA m5C methyltransferase YebU from Escherichia coli reveals a C-terminal RNA-recruiting PUA domain. J Mol Biol. 2006;360:774–787. doi: 10.1016/j.jmb.2006.05.047. [DOI] [PubMed] [Google Scholar]
- 33.Goedecke K, Pignot M, Goody RS, Scheidig AJ, Weinhold E. Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nat Struct Biol. 2001;8:121–125. doi: 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- 34.Newby ZE, Lau EY, Bruice TC. A theoretical examination of the factors controlling the catalytic efficiency of the DNA-(adenine-N6)-methyltransferase from Thermus aquaticus. Proc Natl Acad Sci U S A. 2002;99:7922–7927. doi: 10.1073/pnas.122231499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golovina AY, Sergiev PV, Golovin AV, Serebryakova MV, Demina I, Govorun VM, Dontsova OA. The yfiC gene of E. coli encodes an adenine-N6 methyltransferase that specifically modifies A37 of tRNA1Val(cmo5UAC) Rna. 2009;15:1134–1141. doi: 10.1261/rna.1494409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H, Rother K. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 2009;37:D118–D121. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bheemanaik S, Reddy YV, Rao DN. Structure, function and mechanism of exocyclic DNA methyltransferases. The Biochemical journal. 2006;399:177–190. doi: 10.1042/BJ20060854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Awai T, Kimura S, Tomikawa C, Ochi A, Ihsanawati, Bessho Y, Yokoyama S, Ohno S, Nishikawa K, Yokogawa T, Suzuki T, Hori H. Aquifex aeolicus tRNA (N2,N2-guanine)-dimethyltransferase (Trm1) catalyzes transfer of methyl groups not only to guanine 26 but also to guanine 27 in tRNA. J Biol Chem. 2009;284:20467–20478. doi: 10.1074/jbc.M109.020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ihsanawati, Nishimoto M, Higashijima K, Shirouzu M, Grosjean H, Bessho Y, Yokoyama S. Crystal structure of tRNA N2,N2-guanosine dimethyltransferase Trm1 from Pyrococcus horikoshii. J Mol Biol. 2008;383:871–884. doi: 10.1016/j.jmb.2008.08.068. [DOI] [PubMed] [Google Scholar]
- 40.Armengaud J, Urbonavicius J, Fernandez B, Chaussinand G, Bujnicki JM, Grosjean H. N2-methylation of guanosine at position 10 in tRNA is catalyzed by a THUMP domain-containing, S-adenosylmethionine-dependent methyltransferase, conserved in Archaea and Eukaryota. J Biol Chem. 2004;279:37142–37152. doi: 10.1074/jbc.M403845200. [DOI] [PubMed] [Google Scholar]
- 41.Purushothaman SK, Bujnicki JM, Grosjean H, Lapeyre B. Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA. Mol Cell Biol. 2005;25:4359–4370. doi: 10.1128/MCB.25.11.4359-4370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol Cell. 2007;28:860–870. doi: 10.1016/j.molcel.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee C, Kramer G, Graham DE, Appling DR. Yeast mitochondrial initiator tRNA is methylated at guanosine 37 by the Trm5-encoded tRNA (guanine-N1-)-methyltransferase. J Biol Chem. 2007;282:27744–27753. doi: 10.1074/jbc.M704572200. [DOI] [PubMed] [Google Scholar]
- 44.Christian T, Hou YM. Distinct determinants of tRNA recognition by the TrmD and Trm5 methyl transferases. J Mol Biol. 2007;373:623–632. doi: 10.1016/j.jmb.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoffmann K, Christian T, Hou YM, Perona JJ. Mechanism of N-methylation by the archaeal/eukaryal tRNA m1G37 methylase. 2009 Submitted. [Google Scholar]
- 46.Christian T, Evilia C, Hou YM. Catalysis by the second class of tRNA(m1G37) methyl transferase requires a conserved proline. Biochemistry. 2006;45:7463–7473. doi: 10.1021/bi0602314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mashhoon N, Reich NO. Investigation of ionizable residues critical for sequence-specific enzymatic DNA modification: protein modification and steady-state and pre-steady-state kinetic pH analyses of EcoRI DNA methyltransferase. Biochemistry. 1994;33:7113–7119. doi: 10.1021/bi00189a014. [DOI] [PubMed] [Google Scholar]
- 48.Agris PF, Sierzputowska-Gracz H, Smith C. Transfer RNA contains sites of localized positive charge: carbon NMR studies of [13C]methyl-enriched Escherichia coli and yeast tRNAPhe. Biochemistry. 1986;25:5126–5131. doi: 10.1021/bi00366a022. [DOI] [PubMed] [Google Scholar]
- 49.Alexandrov A, Martzen MR, Phizicky EM. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. Rna. 2002;8:1253–1266. doi: 10.1017/s1355838202024019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Bie LG, Roovers M, Oudjama Y, Wattiez R, Tricot C, Stalon V, Droogmans L, Bujnicki JM. The yggH gene of Escherichia coli encodes a tRNA (m7G46) methyltransferase. J Bacteriol. 2003;185:3238–3243. doi: 10.1128/JB.185.10.3238-3243.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okamoto H, Watanabe K, Ikeuchi Y, Suzuki T, Endo Y, Hori H. Substrate tRNA recognition mechanism of tRNA (m7G46) methyltransferase from Aquifex aeolicus. J Biol Chem. 2004;279:49151–49159. doi: 10.1074/jbc.M408209200. [DOI] [PubMed] [Google Scholar]
- 52.Zegers I, Gigot D, van Vliet F, Tricot C, Aymerich S, Bujnicki JM, Kosinski J, Droogmans L. Crystal structure of Bacillus subtilis TrmB, the tRNA (m7G46) methyltransferase. Nucleic Acids Res. 2006;34:1925–1934. doi: 10.1093/nar/gkl116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roovers M, Wouters J, Bujnicki JM, Tricot C, Stalon V, Grosjean H, Droogmans L. A primordial RNA modification enzyme: the case of tRNA (m1A) methyltransferase. Nucleic Acids Res. 2004;32:465–476. doi: 10.1093/nar/gkh191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varshney U, Ramesh V, Madabushi A, Gaur R, Subramanya HS, RajBhandary UL. Mycobacterium tuberculosis Rv2118c codes for a single-component homotetrameric m1A58 tRNA methyltransferase. Nucleic Acids Res. 2004;32:1018–1027. doi: 10.1093/nar/gkh207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Bjork GR, Tamame M, Hinnebusch AG. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes & development. 1998;12:3650–3662. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ozanick SG, Bujnicki JM, Sem DS, Anderson JT. Conserved amino acids in each subunit of the heteroligomeric tRNA m1A58 Mtase from Saccharomyces cerevisiae contribute to tRNA binding. Nucleic Acids Res. 2007;35:6808–6819. doi: 10.1093/nar/gkm574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gupta A, Kumar PH, Dineshkumar TK, Varshney U, Subramanya HS. Crystal structure of Rv2118c: an AdoMet-dependent methyltransferase from Mycobacterium tuberculosis H37Rv. J Mol Biol. 2001;312:381–391. doi: 10.1006/jmbi.2001.4935. [DOI] [PubMed] [Google Scholar]
- 58.Barraud P, Golinelli-Pimpaneau B, Atmanene C, Sanglier S, Van Dorsselaer A, Droogmans L, Dardel F, Tisne C. Crystal structure of Thermus thermophilus tRNA m1A58 methyltransferase and biophysical characterization of its interaction with tRNA. J Mol Biol. 2008;377:535–550. doi: 10.1016/j.jmb.2008.01.041. [DOI] [PubMed] [Google Scholar]
- 59.Grosjean H, Constantinesco F, Foiret D, Benachenhou N. A novel enzymatic pathway leading to 1-methylinosine modification in Haloferax volcanii tRNA. Nucleic Acids Res. 1995;23:4312–4319. doi: 10.1093/nar/23.21.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grosjean H, Auxilien S, Constantinesco F, Simon C, Corda Y, Becker HF, Foiret D, Morin A, Jin YX, Fournier M, Fourrey JL. Enzymatic conversion of adenosine to inosine and to N1-methylinosine in transfer RNAs: a review. Biochimie. 1996;78:488–501. doi: 10.1016/0300-9084(96)84755-9. [DOI] [PubMed] [Google Scholar]
- 61.Bailly M, Blaise M, Lorber B, Becker HD, Kern D. The transamidosome: A dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagines biosynthesis. Mol Cell. 2007;26:228–239. doi: 10.1016/j.molcel.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 62.Zhang CM, Liu C, Slater S, Hou YM. Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys) Nat Struct Mol Biol. 2008;15:507–514. doi: 10.1038/nsmb.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bjork GR. Genetic dissection of synthesis and function of modified nucleosides in bacterial transfer RNA. Prog Nucleic Acid Res Mol Biol. 1995;50:263–338. doi: 10.1016/s0079-6603(08)60817-x. [DOI] [PubMed] [Google Scholar]
- 64.Marshall L, Kenneth NS, White RJ. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- 65.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bjork GR, Wikstrom PM, Bystrom AS. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science. 1989;244:986–989. doi: 10.1126/science.2471265. [DOI] [PubMed] [Google Scholar]
- 67.O'Dwyer K, Watts JM, Biswas S, Ambrad J, Barber M, Brule H, Petit C, Holmes DJ, Zalacain M, Holmes WM. Characterization of Streptococcus pneumoniae TrmD, a tRNA methyltransferase essential for growth. J Bacteriol. 2004;186:2346–2354. doi: 10.1128/JB.186.8.2346-2354.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bjork GR, Jacobsson K, Nilsson K, Johansson MJ, Bystrom AS, Persson OP. A primordial tRNA modification required for the evolution of life? Embo J. 2001;20:231–239. doi: 10.1093/emboj/20.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li J, Esberg B, Curran JF, Bjork GR. Three modified nucleosides present in the anticodon stem and loop influence the in vivo aa-tRNA selection in a tRNA-dependent manner. J Mol Biol. 1997;271:209–221. doi: 10.1006/jmbi.1997.1176. [DOI] [PubMed] [Google Scholar]
- 70.Hagervall TG, Tuohy TM, Atkins JF, Bjork GR. Deficiency of 1-methylguanosine in tRNA from Salmonella typhimurium induces frameshifting by quadruplet translocation. J Mol Biol. 1993;232:756–765. doi: 10.1006/jmbi.1993.1429. [DOI] [PubMed] [Google Scholar]
- 71.Stuart JW, Koshlap KM, Guenther R, Agris PF. Naturally-occurring modification restricts the anticodon domain conformational space of tRNA(Phe) J Mol Biol. 2003;334:901–918. doi: 10.1016/j.jmb.2003.09.058. [DOI] [PubMed] [Google Scholar]
- 72.Putz J, Florentz C, Benseler F, Giege R. A single methyl group prevents the mischarging of a tRNA. Nat Struct Biol. 1994;1:580–582. doi: 10.1038/nsb0994-580. [DOI] [PubMed] [Google Scholar]
- 73.Noma A, Kirino Y, Ikeuchi Y, Suzuki T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. Embo J. 2006;25:2142–2154. doi: 10.1038/sj.emboj.7601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hauenstein SI, Hou YM, Perona JJ. The homotetrameric phosphoseryl-tRNA synthetase from Methanosarcina mazei exhibits half-of-the-sites activity. J Biol Chem. 2008;283:21997–22006. doi: 10.1074/jbc.M801838200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Renda MJ, Rosenblatt JD, Klimatcheva E, Demeter LM, Bambara RA, Planelles V. Mutation of the methylated tRNA(Lys)(3) residue A58 disrupts reverse transcription and inhibits replication of human immunodeficiency virus type 1. Journal of virology. 2001;75:9671–9678. doi: 10.1128/JVI.75.20.9671-9678.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Renda MJ, Bradel-Tretheway B, Planelles V, Bambara RA, Dewhurst S. Inhibition of HIV type 1 replication using lentiviral-mediated delivery of mutant tRNA(Lys3)A58U. AIDS research and human retroviruses. 2004;20:1324–1334. doi: 10.1089/aid.2004.20.1324. [DOI] [PubMed] [Google Scholar]
- 77.Salas CE, Uschmann BD, Leboy PS. Methyl-accepting RNA in 13762 mammary adenocarcinoma correlated with low adenine methyltransferase levels. Cancer research. 1982;42:5004–5009. [PubMed] [Google Scholar]
- 78.Bunn CC, Mathews MB. Autoreactive epitope defined as the anticodon region of alanine transfer RNA. Science. 1987;238:1116–1119. doi: 10.1126/science.2446387. [DOI] [PubMed] [Google Scholar]
- 79.Bunn CC, Mathews MB. Two human tRNA(Ala) families are recognized by autoantibodies in polymyositis sera. Molecular biology & medicine. 1987;4:21–36. [PubMed] [Google Scholar]
- 80.Limbach PA, Crain PF, McCloskey JA. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]