Abstract
OBJECTIVE
The ∼45-cM insulin-dependent diabetes 9 (Idd9) region on mouse chromosome 4 harbors several different type 1 diabetes–associated loci. Nonobese diabetic (NOD) mice congenic for the Idd9 region of C57BL/10 (B10) mice, carrying antidiabetogenic alleles in three different Idd9 subregions (Idd9.1, Idd9.2, and Idd9.3), are strongly resistant to type 1 diabetes. However, the mechanisms remain unclear. This study aimed to define mechanisms underlying the type 1 diabetes resistance afforded by B10 Idd9.1, Idd9.2, and/or Idd9.3.
RESEARCH DESIGN AND METHODS
We used a reductionist approach that involves comparing the fate of a type 1 diabetes–relevant autoreactive CD8+ T-cell population, specific for residues 206–214 of islet-specific glucose 6 phosphatase catalytic subunit–related protein (IGRP206–214), in noncongenic versus B10 Idd9–congenic (Idd9.1 + Idd9.2 + Idd9.3, Idd9.2 + Idd9.3, Idd9.1, Idd9.2, and Idd9.3) T-cell receptor (TCR)–transgenic (8.3) NOD mice.
RESULTS
Most of the protective effect of Idd9 against 8.3-CD8+ T-cell–enhanced type 1 diabetes was mediated by Idd9.1. Although Idd9.2 and Idd9.3 afforded some protection, the effects were small and did not enhance the greater protective effect of Idd9.1. B10 Idd9.1 afforded type 1 diabetes resistance without impairing the developmental biology or intrinsic diabetogenic potential of autoreactive CD8+ T-cells. Studies in T- and B-cell–deficient 8.3-NOD.B10 Idd9.1 mice revealed that this antidiabetogenic effect was mediated by endogenous, nontransgenic T-cells in a B-cell–independent manner. Consistent with this, B10 Idd9.1 increased the suppressive function and antidiabetogenic activity of the FoxP3+CD4+CD25+ T-cell subset in both TCR-transgenic and nontransgenic mice.
CONCLUSIONS
A gene(s) within Idd9.1 regulates the development and function of FoxP3+CD4+CD25+ regulatory T-cells and, in turn, the activation of CD8+ effector T-cells in the pancreatic draining lymph nodes, without affecting their development or intrinsic diabetogenic potential.
Type 1 diabetes in both humans and nonobese diabetic (NOD) mice is the result of a complex T-cell–mediated autoimmune process against the pancreatic β-cells. Putative poorly defined environmental triggers conspire with a constellation of genetic elements scattered throughout the genome to elicit a multifactorial autoimmune response that involves virtually every cell type of the immune system. Genome-wide association studies in humans and mice have identified at least 15 different chromosomal regions harboring candidate type 1 diabetes–associated gene loci, in addition to the HLA and H-2 complexes on human and mouse chromosomes 6 and 17, respectively (1–4).
The ∼45-cM insulin-dependent diabetes 9 (Idd9) region on mouse chromosome 4 has a major effect on type 1 diabetes development (5). NOD mice congenic for the Idd9 region of C57BL/10 (B10) mice are highly resistant to type 1 diabetes (5). However, the underlying mechanisms remain unclear. Initial work correlated protection from type 1 diabetes with recruitment of antidiabetogenic CD30/interleukin (IL)-4–expressing cell types into pancreatic islets (5). Subsequent studies, however, suggested that alternative mechanisms might be at play. For example, using NOD and NOD.B10 Idd9 mice expressing an insulin promoter-driven influenza hemagglutinin transgene, Martinez et al. found that the B10 Idd9 region affords tolerance to hemagglutinin-specific CD8+ T-cells through a mechanism that does not inhibit their activation in the pancreatic lymph nodes (6). Another study suggested that Idd9 controls the recruitment of autoreactive CD4+ T-cells to the pancreas (7). A third study, using NOD mice congenic for the Idd9/11 region, which partially overlaps B10 Idd9, of nonobese resistant mice (8) supported the CD4+ T-cell intrinsic effect(s) of B10 Idd9 reported by Waldner et al. (7). Additional investigations of congenic NOD mice carrying chromosome 4 fragments around the Idd9.1 region of different lengths and donor strains reported effects of Idd9 on invariant natural killer T (iNKT) cell (9), B-cell (10,11), or dendritic cell (DC) biology (12), and even on susceptibility of β-cells to cell death (13). This rather extensive assortment of mechanisms seemingly unrelated to one another clearly indicates that the effects of Idd9 on type 1 diabetes are complex and suggest that the phenotypes are probably determined by more than one gene within the Idd9 region. In fact, it has been established from congenic strain mapping that the B10 Idd9 region contains at least three type 1 diabetes–associated genes: Idd9.1, Idd9.2, and Idd9.3 (5). A congenic strain having the Idd9.2 + Idd9.3 protective subregions (R11) was more susceptible to type 1 diabetes than the strain having all three congenic subregions (R28). However, the R11 strain was more protected from type 1 diabetes than the R35 strain that has a type 1 diabetes–protective B10-derived allele at only Idd9.3.
This study was initiated to dissect mechanisms underlying the type 1 diabetes resistance afforded by B10 Idd9 loci. We used a reductionist experimental approach (14) that involves comparing the fate and diabetogenicity of a type 1 diabetes–relevant autoreactive CD8+ T-cell population in B10 Idd9–congenic versus noncongenic, T-cell receptor (TCR)–transgenic NOD mice. Our data show that the B10-derived Idd9.1 subregion has a powerful suppressive effect on the diabetogenic activity of a prevalent monospecific CD8+ T-cell population without impairing its developmental biology or intrinsic diabetogenic potential. We find that this antidiabetogenic effect is mediated by endogenous, nontransgenic T-cells but not B-cells, and show that it is associated with enhanced regulatory activity of CD4+CD25+ regulatory T-cells not only in TCR-transgenic, but also nontransgenic, NOD.B10 Idd9.1 mice.
RESEARCH DESIGN AND METHODS
Mice.
8.3-NOD and 8.3-NOD.Rag2−/− mice have been described (15). NOD.B10 Idd9R28 (line 1104)–, NOD.B10 Idd9R11 (line 1105)–, and NOD.B10 Idd9.3 (line 1106)–congenic mice were developed as described previously (5). Development of the NOD.B10 Idd9.1 (line 1565) and NOD.B10 Idd9.2 (line 1566) strains and the fine-mapping of Idd9.1 and Idd9.2 will be the subjects of future reports. 8.3-NOD.B10 Idd9R28, 8.3-NOD.B10 Idd9R11, 8.3-NOD.B10 Idd9.1, 8.3-NOD.B10 Idd9.2, and 8.3-NOD.B10 Idd9.3 mice were established by crossing 8.3-NOD mice with NOD.B10 Idd9R28 (line 1104), NOD.B10 Idd9R11 (line 1105), NOD.B10 Idd9.1 (line 1565), NOD.B10 Idd9.2 (line 1566), and NOD.B10 Idd9.3 (line 1106) mice, respectively. The line number associated with each strain refers to the unique line designation for strains bred at Taconic (Germantown, NY). Lines 1565, 1566, and 1106 are available from Taconic through the Emerging Models Program. Lines 1104 and 1105 are no longer extant. Figure 1A shows a map of the congenic strains used in this study in comparison to Idd9-congenic strains described by others (9,16,17). Lines 1565, 1566, and 1106 contain the B10 Idd9.1, Idd9.2, and Idd9.3 regions alone, respectively. The markers that define the boundaries of the lines described here are given in supplementary Table 1 (available in an online appendix at http://diabetes.diabetesjournals.org/content/early/2009/10/14/db09-0648/suppl/DC1), along with the primer sequences of novel markers developed for this study. supplementary Fig. 1 lists the single nucleotide polymorphisms (SNPs) distinguishing B10, B6, and NOD in the Idd9.1 region defined by line 1565. All genes containing polymorphisms are noted. Also noted are the regions of overlap of the B10 Idd9 congenic line with R201 of Ueno et al. (9) and the two Brodnicki et al. (16,17) B6 Idd9/Idd11–congenic lines defining the smallest Idd11 interval published to date. Information on the congenic strains is also available at http://www.t1dbase.org/page/DrawStrains (select the strain of interest). Note that the B10 Idd9 fragment in line 1565 overlaps the distal 30% of the B10 Idd9.1 fragment carried by the R201 strain from Ueno et al., and that Brodnicki et al.'s Idd11-congenic strains were produced using B6 donor mice. The 8.3-NOD.Igμ−/−, 8.3-NOD.B10 Idd9.1/Igμ−/−, and 8.3-NOD.B10 Idd9.1/Rag2−/− mice were established by crossing 8.3-NOD and 8.3-NOD.B10 Idd9.1 with NOD.Igμ−/− and NOD.Rag2−/− mice, respectively, followed by backcrossing 8.3-TCR+, mutant heterozygous F1 mice to 8.3-NOD.B10 Idd9.1 mice and intercrossing 8.3-TCR+, mutant heterozygous B10 Idd9.1 homozygous littermates. All mice were housed in specific pathogen-free conditions.
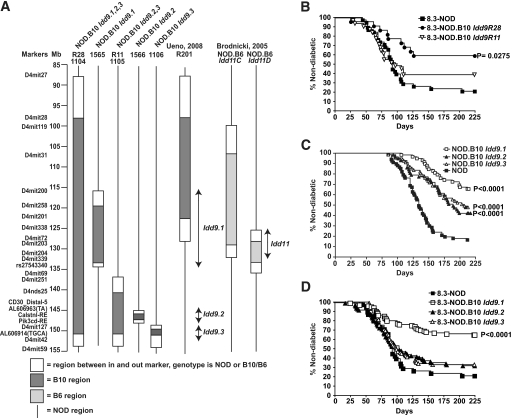
FIG. 1.
Antidiabetogenic effects of different B10 Idd9 intervals on 8.3-CD8+ T-cell–enhanced type 1 diabetes. A: Chromosome 4, B10 Idd9–congenic NOD lines used in this and other studies (9,16). B: Incidence of diabetes in female 8.3-NOD (n = 38), 8.3-NOD.B10 Idd9R28 (n = 22), and 8.3-NOD.B10 Idd9R11 (n = 31) mice. C: Incidence of diabetes in NOD (n = 67) compared with NOD.B10 Idd9.1 (n = 58), NOD.B10 Idd9.2 (n = 57), and NOD.B10 Idd9.3 (n = 67) female mice. D: Incidence of diabetes in female 8.3-NOD (n = 38), 8.3-NOD.B10 Idd9.1 (n = 100), 8.3-NOD.B10 Idd9.2 (n = 94), and 8.3-NOD.B10 Idd9.3 (n = 85) mice. The Kaplan-Meier survival curves were compared with log-rank test.
Peptides and antibodies.
Peptides (NRP-A7 and TUM) were purchased from Mimotopes (Clayton, Victoria, Australia). All monoclonal antibodies (mAbs) were purchased from PharMingen (San Diego, CA), unless indicated otherwise. Anti-mouse glucocorticoid-induced tumor necrosis factor receptor (GITR), anti-mouse/rat FoxP3, and anti–folic receptor 4 (FR4) were from eBiosciences (San Diego, CA). Streptavidin-peridinin-chlorophyll-protein complex was from Becton Dickinson (San Jose, CA).
Preparation of DCs.
Mesenteric lymph node (MLN) and pancreatic lymph node (PLN) DCs were purified from collagenase-digested MLNs and PLNs using anti-CD11c mAb-coated magnetic beads (Miltenyi Biotec). Purified DCs were pulsed with NRP-A7 or TUM peptides (1 μmol/l) for 1 h at 37°C and then used in proliferation and cytokine secretion assays using 8.3-CD8+ T-cells as responders.
Diabetes and insulitis.
Diabetes was monitored by measuring urine glucose with Diastix (Bayer, ON, Canada); animals were considered diabetic after two readings ≥3+. Differences between diabetes survival curves were compared with the Kaplan-Meier log-rank test using Prism software (Graphpad). Insulitis scores were measured on 14 hematoxylin-eosin–stained pancreas sections 150 μm apart, using the following criteria: 0, intact islet; 1, peri-insulitis; 2, up to 25% of the islet infiltrated; 3, 25–50% infiltration; 4, >50% infiltration.
Proliferation and cytokine secretion assays.
Splenic CD8+ T-cells (2 × 104/well) were incubated with NRP-A7/TUM peptide-pulsed (0.0001–1 μmol/l) antigen-presenting cells (APCs; 105 irradiated splenocytes/well) for 2 or 3 days (for cytokine and proliferation assays, respectively) at 37°C in 5% CO2. Cytokines in the supernatants were measured by enzyme-linked immunosorbent assay. The 3-day cultures were pulsed with 1 μCi [3H] thymidine during the last 18 h and harvested. The regulatory activity of CD4+CD25+ T-cells was measured by adding 0.125–1 × 104 CD4+CD25+ T-cells preactivated with anti-CD3 mAb and recombinant (r)IL-2 to CD8+ cell–APC cocultures for 48 h (interferon γ [IFN-γ] content measurements) or 72 h (proliferative activity, assessed by [3H] incorporation during the last 18 h of culture).
Cytotoxic T-lymphocyte differentiation and 51Cr release assays.
Splenic CD8+ T-cells purified from 8.3-NOD and 8.3-NOD.B10 Idd9.1 mice using anti-CD8–coated microbeads (Miltenyi Biotec) (2 × 104 cells/well) were stimulated with NRP-A7–pulsed irradiated NOD splenocytes (105 cells/well) for 3 days and expanded in 0.5 units/ml of rIL-2 (Takeda, Osaka, Japan) for 7–10 days. Cytolytic activity of cytotoxic T-lymphocytes (CTLs) was measured using NRP-A7– or TUM-pulsed (1 μmol/l) RMA-SKd cells.
T-cell transfers.
Splenic CD8+ T-cells were purified using iMAG CD8 beads (BD Bioscience) following the manufacturer's protocols, labeled with carboxyfluorescein succinimidyl ester (CFSE; 2.5 μmol/l), and injected intravenously (107 CD8+ T-cells) into 9- to 11-week-old hosts (NOD or NOD.B10 Idd9.1). Hosts were killed 6 days later and their PLNs and MLNs examined for presence of CD8+ CFSE+ cells. To purify CD4+CD25+ cells, lymph node and/or splenic cells were enriched for CD4+ cells, incubated with anti–CD25-phycoerythrin (PE), and separated using anti-PE mAb-coated beads (Miltenyi Biotec). The purity was >85% for CD4+CD25+ cells. The mice were injected intravenously with 2 × 105 cells and monitored for diabetes for 15 weeks.
Statistical analyses.
Data were compared using log-rank, Mann-Whitney U, or two-way ANOVA tests.
RESULTS
The B10 Idd9 region suppresses CD8+ T-cell–induced diabetes predominantly via Idd9.1.
To determine whether the B10-derived Idd9 region that protects from spontaneous type 1 diabetes can also protect from disease accelerated by the presence of a diabetogenic TCR, we introgressed two copies of the B10 Idd9.1-9.3 and Idd9.2-Idd9.3 intervals (Fig. 1A) into transgenic NOD mice expressing a diabetogenic T-cell receptor, 8.3 (15). This TCR is representative of a large fraction of islet-associated CD8+ T-cells in NOD mice that use highly homologous TCRα chains (15,18,19) and recognize the mimotopes NRP-A7 and NRP-V7 in the context of the major histocompatibility complex (MHC) molecule H-2Kd (20). These T-cells are already a significant component of the earliest NOD islet CD8+ infiltrates (19–21), are pathogenic (15,18), target a peptide from islet-specific glucose 6 phosphatase catalytic subunit–related protein (IGRP206–214, similar to NRP-A7) (22), and are unusually frequent in the periphery (>1/200 CD8+ T-cells) (23).
8.3-NOD.B10 Idd9R28 mice, carrying the complete B10 Idd9 interval that affords >95% protection from spontaneous type 1 diabetes (5), displayed a significantly reduced frequency of diabetes compared with their noncongenic 8.3-NOD counterparts (∼40 vs. 80%) (Fig. 1B). The shorter B10 Idd9.2-Idd9.3 (Idd9R11) interval, which provides 60% protection from spontaneous type 1 diabetes (5), was also antidiabetogenic, but to a much lesser extent (Fig. 1B). To ascertain whether the higher antidiabetogenic effect of Idd9 was due to additive/synergistic effects of loci contained in different subregions, we developed 8.3-NOD mice congenic for each of the individual Idd9 intervals (Idd9.1, Idd9.2, and Idd9.3); each of the individual Idd9 congenic regions affords non–TCR-transgenic NOD mice protection from spontaneous type 1 diabetes as shown in Fig. 1C. The fine-mapping of the Idd9.1 and Idd9.2 regions is in progress and will be the subject of separate reports (D.B.R. et al. and Hamilton-Williams et al., manuscripts in preparation). The Idd9.3 region is 1.2 Mb with only 15 genes, including the prime candidate Tnfrsf9, which encodes 4–1bb (also known as CD137) (24–26). As shown in Fig. 1D, the incidence of diabetes in 8.3-NOD.B10 Idd9.1 mice was virtually identical to that seen in 8.3-NOD.B10 Idd9R28 mice, suggesting a minor contribution of the B10 Idd9.2 and Idd9.3 alleles on 8.3-CD8+ T-cell–enhanced type 1 diabetes. In agreement with these observations, the diabetes survival curves corresponding to 8.3-NOD.B10 Idd9.2 and 8.3-NOD.B10 Idd9.3 mice were remarkably similar to those seen in 8.3-NOD.B10 Idd9R11 mice. This is in contrast to what occurs in nontransgenic NOD mice, where Idd9.2 and Idd9.3 augment the antidiabetogenic effects of each other and of Idd9.1 (5). Because the Idd9.1 interval is as protective in 8.3-NOD mice as it is in non–TCR-transgenic NOD mice (5), these data suggest that expression of the diabetogenic TCR overwhelms the antidiabetogenic properties of Idd9.2 and Idd9.3, but not the effects of Idd9.1.
The B10 Idd9.1 allele suppresses 8.3-CD8+ T-cell–enhanced diabetes and insulitis without compromising 8.3-CD8+ T-cell development or function.
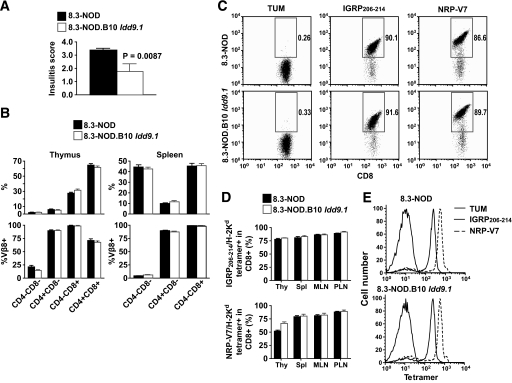
Because the antidiabetogenic effect of the B10 Idd9.1 region in 8.3-NOD.B10 Idd9.1 mice was associated with slower progression of insulitis (Fig. 2A), we asked whether this region's antidiabetogenic activity was mediated by effects on the developmental biology of 8.3-CD8+ T-cells. Cytofluorometric studies indicated that the thymi and spleens of both types of mice contained similar absolute and relative numbers of double-positive CD4+CD8+ thymocytes and/or single-positive CD4+ or CD8+ T-cells, respectively (Fig. 2B and data not shown). In addition, the transgenic CD8+ T-cells of both types of mice expressed comparable levels of the transgenic Vβ8.1+ TCR and CD8 (Fig. 2C), and bound NRP-V7/Kd and IGRP206–214/Kd tetramers (recognized with high and intermediate avidity, respectively) with similar frequency (Fig. 2D) and mean fluorescence intensity (Fig. 2E).
FIG. 2.
Effects of B10 Idd9.1 on insulitis and 8.3-CD8+ T-cell development. A: Insulitis scores of 8.3-NOD (n = 6) and 8.3-NOD.B10 Idd9.1 (n = 5) at 10 weeks of age. B: Flow cytometry profiles of thymocytes (left) and splenocytes (right) in 8.3-NOD (n = 7) and 8.3-NOD.B10 Idd9.1 (n = 6) mice. Values correspond to the means ± standard errors. C: Representative anti-CD8 mAb/IGRP206–214/Kd and anti-CD8 mAb/NRP-V7/Kd tetramer staining profiles of PLN cells. D: Percentages of thymic, splenic MLN, and PLN CD8+ T-cells binding IGRP206–214/Kd and NRP-V7/Kd tetramers. Data (means ± SE) correspond to four mice per strain. E: Representative TUM/Kd (negative control), IGRP206–214/Kd (intermediate avidity), and NRP-V7/Kd (high avidity) tetramer staining profiles of splenic T-cells from 8.3-NOD and 8.3-NOD.B10 Idd9.1 mice.
Functional in vitro assays revealed that the splenic CD8+ T-cells of both types of mice proliferated equally well and secreted similar levels of IFN-γ and IL-2 in response to NRP-A7 peptide stimulation over a range of concentrations (Fig. 3A). Likewise, in vitro–differentiated 8.3-CD8+ T-cells from both strains killed peptide-pulsed target cells with similar efficiency (Fig. 3B).
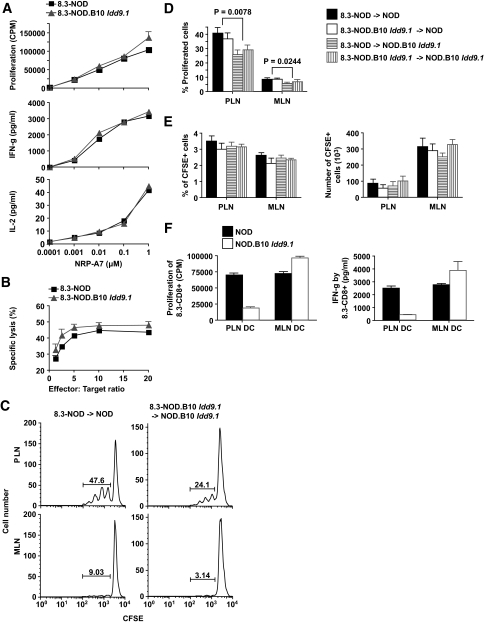
FIG. 3.
Effects of B10 Idd9.1 on the function of naive 8.3-CD8+ T-cells. A: Proliferation of, and cytokine (IFN-γ and IL-2) production by, CD8+ splenocytes purified from 8.3-NOD and 8.3-NOD.B10 Idd9.1 mice in response to NRP-A7 peptide–pulsed irradiated NOD splenocytes. Data are representative of two separate experiments. B: CTL differentiation potential and cytolytic activity of in vitro–differentiated 8.3-CTLs from 8.3-NOD and 8.3-NOD.B10 Idd9.1 mice. Naive 8.3-CD8+ T-cells (n = 3 for each strain) were stimulated with NRP-A7 (1 μmol/l)–pulsed APCs for 3 days and cultured in the presence of rhIL-2 for 4 additional days. Cytotoxicity assays were performed using NRP-A7– and TUM-pulsed (1 μmol/l) RMA-SKd cells as targets, at different effector/target ratios. Background responses against TUM-pulsed targets were subtracted. C and D: CFSE-labeled 8.3-CD8+ T-cells were injected into 9- to 11-week-old NOD (n = 9; five 8.3-NOD donors and four 8.3-NOD.B10 Idd9.1 donors) and NOD.B10 Idd9.1 (n = 9; six 8.3-NOD donors and three 8.3-NOD.B10 Idd9.1 donors) hosts and analyzed for dividing (CFSElow) CD8+ T-cells in pancreatic (PLN) and mesenteric (MLN) lymph nodes 6 days later. C: Representative flow cytometry profiles. D: Average quantitative data. E: Percentages and total numbers of 8.3-CD8+ T-cells in the PLNs and MLNs of NOD and NOD.B10 Idd9.1 hosts (the histogram legend is the same as in D). F: Proliferation and secretion of IFN-γ by naive 8.3-CD8+ T-cells in response to NRP-A7–pulsed DCs isolated from the PLNs or MLNs of NOD and NOD.B10 Idd9.1 mice. Bars in A, B, D, E, and F correspond to the standard errors of the means. P values were calculated with Mann-Whitney U test.
Similar results were obtained in vivo. CFSE-labeled 8.3-CD8+ T-cells from 8.3-NOD.B10 Idd9.1 mice proliferated in the pancreatic (but not mesenteric) lymph nodes (PLNs and MLNs, respectively) of wild-type NOD hosts as efficiently as those derived from 8.3-NOD donors (Fig. 3C).
Taken together, these observations indicated that the B10 Idd9.1 allele does not impair the development or function of IGRP206–214-reactive CD8+ T-cells.
CD8+ T-cell–extrinsic inhibition of 8.3-CD8+ T-cell activation by B10 Idd9.1 in vivo.
Whereas CFSE-labeled 8.3-CD8+ T-cells proliferated equally well in vivo regardless of the Idd9.1 genotype of the donor mice, CFSE-labeled 8.3-CD8+ T-cells from both 8.3-NOD and 8.3-NOD.B10 Idd9.1 mice proliferated significantly less in the PLNs of NOD.B10 Idd9.1 hosts than in the PLNs of NOD hosts (Fig. 3C and D), without any obvious effects on the relative or absolute numbers of recruited/retained 8.3-CD8+ T-cells (Fig. 3E). This suggested that B10 Idd9.1 somehow impairs the cross-priming of 8.3-CD8+ T-cells by endogenous autoantigen-loaded dendritic cells. This was not due to interstrain differences in autoantigen loading of DCs in vivo but rather to impaired PLN DC function because whereas ex vivo, NRP-A7–pulsed DCs purified from the MLNs of NOD and NOD.B10 Idd9.1 mice could stimulate naive 8.3-CD8+ T-cells with similar efficiency, NRP-A7–pulsed DCs purified from the PLNs of NOD.B10 Idd9.1 mice could not do so as efficiently as NRP-A7–pulsed DCs purified from the PLNs of NOD mice (Fig. 3F).
Suppression of 8.3-CD8+ T-cell–enhanced diabetes by the B10 Idd9.1 region requires endogenous T- but not B-cells.
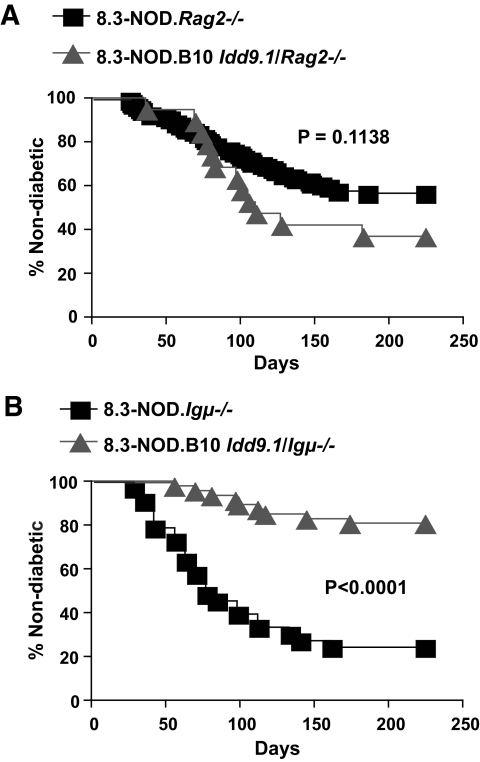
To ascertain whether the CD8+ T-cell–extrinsic effects of B10 Idd9.1 on 8.3-CD8+ T-cell cross-priming in vivo were associated with B10 Idd9.1's antidiabetogenic activity, we introduced a recombination activating gene (Rag2) deficiency into 8.3-NOD and 8.3-NOD.B10 Idd9.1 mice. Although penetrance of type 1 diabetes in 8.3-NOD.Rag2−/− mice is significantly lower than in 8.3-NOD mice (owing to lack of CD4+ T-cell help) (15), the incidence of diabetes in 8.3-NOD.Rag2−/− and 8.3-NOD.B10 Idd9.1/Rag2−/− mice was essentially the same (Fig. 4A), indicating that the RAG-2 deficiency had completely abrogated the antidiabetogenic effects of the B10 Idd9.1 allele. Because these mice export only 8.3-CD8+ T-cells (but not CD4+ T-cells or B-cells) to the peripheral lymphoid organs (15), we reasoned that the protective activity of B10 Idd9.1 was mediated by endogenous T-cells and/or B-cells. To distinguish between these two possibilities, we introduced an immunoglobulin μ heavy chain gene (Igμ) deficiency into 8.3-NOD and 8.3-NOD.B10 Idd9.1 mice and followed both types of B-cell–deficient mice for development of diabetes. B-cell–deficient 8.3-NOD.Igμ−/− mice developed diabetes essentially like B-cell–competent 8.3-NOD mice, indicating that type 1 diabetes development in the 8.3-NOD model, unlike the case in nontransgenic NOD mice, is B-cell independent (Fig. 4B). In contrast, 8.3-NOD.B10 Idd9.1/Igμ−/− mice remained protected from type 1 diabetes despite lacking B-cells, indicating that the protective effects of the B10 Idd9.1 region are B-cell independent and mediated by endogenous T-cells.
FIG. 4.
The antidiabetogenic effects of B10 Idd9.1 are effector T-cell extrinsic and mediated by endogenous T-cells in a B-cell–independent manner. A: Incidence of diabetes in female 8.3-NOD.Rag2−/− (n = 106) and 8.3-NOD.B10 Idd9.1/Rag2−/− mice (n = 19). B: Incidence of diabetes in female 8.3-NOD.Igμ−/− (n = 33) and 8.3-NOD.B10 Idd9.1/Igμ−/− mice (n = 47). The Kaplan-Meier survival curves were compared with log-rank test.
The B10 Idd9.1 region enhances the development and function of FoxP3+CD4+CD25+ regulatory T-cells.
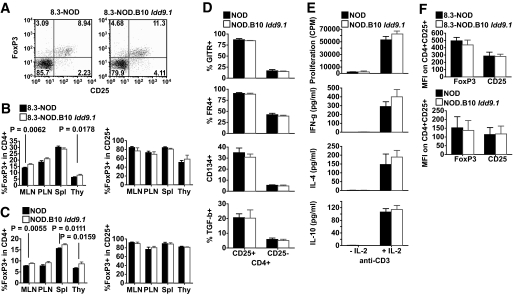
Prompted by these findings and the observation that the protective effect of the B10 Idd9.1 interval of line 1565 was not associated with increased development of thymic iNKT cells (9) (supplementary Fig. 2), we investigated whether B10 Idd9.1 might enhance the development and/or function of FoxP3+CD4+CD25+ regulatory T-cells (Tregs). We compared the relative size of the FoxP3+CD4+ T-cell subset in thymuses, spleens, MLNs, and PLNs of 8.3-NOD and 8.3-NOD.B10 Idd9.1 mice. The lymphoid organs of 8.3-NOD.B10 Idd9.1 mice contained higher percentages (albeit not absolute numbers) of FoxP3+CD4+ T-cells than those of 8.3-NOD mice, although the differences were relatively small and statistically significant only for thymuses and MLNs (Fig. 5A and B, and supplementary Fig. 3A). These differences were not a peculiarity of TCR-transgenic mice, because the thymuses and MLNs of nontransgenic NOD.B10 Idd9.1 mice also contained significantly higher percentages of these T-cells compared with NOD mice (Fig. 5C). As expected, there were no differences in the percentages of FoxP3+ T-cells within the CD4+CD25+ T-cell subsets (Fig. 5A and C). These CD4+CD25+ T-cells had all the hallmarks of CD4+CD25+ Tregs described in other strains: they expressed GITR, FR4, CD134, and transforming growth factor β (TGF-β; Fig. 5D), and produced IFN-γ, IL-4, and IL-10 when stimulated with plate-bound anti-CD3 mAb in the presence, but not absence, of rhIL-2 (Fig. 5E). The CD4+CD25+ T-cells of transgenic and nontransgenic NOD and NOD.B10 Idd9.1 mice expressed similar levels of all these markers, including CD25 and FoxP3 (Fig. 5F and data not shown).
FIG. 5.
Effects of B10 Idd9.1 on the size and phenotype of the regulatory FoxP3+CD4+CD25+ Treg subset. A: Representative FoxP3/CD25 flow cytometry profiles. B: Percentages of FoxP3+ cells in the CD4+ and CD4+CD25+ T-cell subsets in the MLNs, PLNs, spleens (Spl), and thymuses (Thy) of 8- to 11-week-old 8.3-NOD (n = 12) and 8.3-NOD.B10 Idd9.1 (n = 11) mice. Values correspond to the means ± standard errors. P values were calculated with Mann-Whitney U test. C: Percentages of FoxP3+ in the CD4+ and CD4+CD25+ T-cell subsets in the spleens, MLNs, PLNs, and thymuses of 8- to 11-week-old NOD (n = 9 for spleens, MLNs, and PLNs; n = 5 for thymuses) and NOD.B10 Idd9.1 (n = 8 for spleens, MLNs, PLNs; n = 4 for thymuses) mice. D: GITR, FR4, CD134, and surface TGF-β flow cytometry profiles of CD4+CD25+ and CD4+CD25− cells from NOD and NOD.B10 Idd9.1 mice. Values correspond to the means ± standard errors of two to four experiments/strain type. E: Proliferation and cytokine production by CD4+CD25+ cells from NOD and NOD.B10 Idd9.1 mice in response to plate-bound anti-CD3 mAb in the absence/presence of rhIL-2. Values correspond to the means ± standard errors of three independent experiments. F: Comparison of FoxP3 and CD25 mean fluorescence intensities for FoxP3+CD4+CD25+ T-cells from both types of mice.
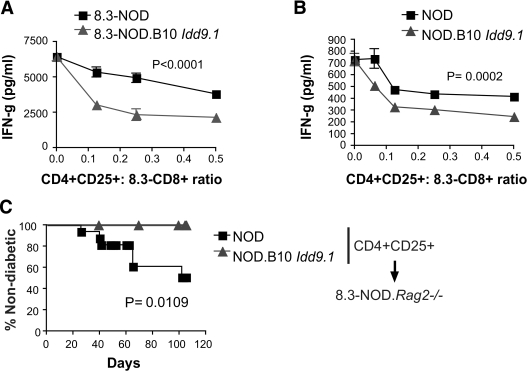
We next asked whether these B10 Idd9.1-associated differences in the relative size of the FoxP3+CD4+CD25+ Treg subset were accompanied by differences in suppressive activity. We first compared the ability of CD4+CD25+ (and CD4+CD25−) T-cells purified from NOD.B10 Idd9.1 and NOD mice to suppress the activation of naive 8.3-CD8+ T-cells from 8.3-NOD mice in response to NRP-A7 peptide–pulsed APCs. The CD4+CD25+ T-cells purified from 8.3-NOD.B10 Idd9.1 or NOD.B10 Idd9.1 mice had significantly higher regulatory activity than the CD4+CD25+ T-cells purified from 8.3-NOD or NOD mice, respectively, at different CD4+CD25+/CD8+ T-cell ratios (Fig. 6A and B, and supplementary Fig. 3B). The CD4+CD25− T-cells of both types of mice were not suppressive in these assays (data not shown), as described previously (27). These strain differences in Treg function were also true in vivo. Whereas 8.3-NOD.Rag2−/− hosts transfused with CD4+CD25+ T-cells from wild-type NOD donors developed spontaneous diabetes essentially like nonmanipulated 8.3-NOD.Rag2−/− mice (15), 8.3-NOD.Rag2−/− hosts transfused with CD4+CD25+ T-cells from NOD.B10 Idd9.1 mice remained diabetes free throughout the follow-up period (Fig. 6C).
FIG. 6.
Effects of B10 Idd9.1 on the in vitro and in vivo suppressive activity of CD4+CD25+ regulatory T-cells. A and B: IFN-γ production by naive 8.3-CD8+ T-cells from 8.3-NOD mice in response to NRP-A7–pulsed APCs in the presence of CD4+CD25+ T-cells from 8.3-NOD and 8.3-NOD.B10 Idd9.1 mice (A) or NOD and NOD.B10 Idd9.1 mice (B), at different Treg/responder ratios. Graphs are representative of five (A) and three (B) independent experiments. C: Incidence of spontaneous diabetes in 8.3-NOD.Rag2−/− hosts (n = 28) transfused with 2 × 105 CD4+CD25+ T-cells from NOD (n = 16) or NOD.B10 Idd9.1 donors (n = 12). P values for A and B were calculated with two-way ANOVA, and with log-rank for C.
Taken together, these data are consistent with the idea that diabetes resistance afforded by the B10 Idd9.1 interval is mediated, at least in part, by enhanced development and function of CD4+CD25+ regulatory T-cells.
DISCUSSION
Analyses of Idd-congenic NOD mice, in which type 1 diabetes–associated chromosomal regions are replaced by the corresponding regions found in nondiabetes-prone strains of mice, such as B6 and B10, have demonstrated the existence of more than 20 non–MHC-linked type 1 diabetes–associated regions/loci (1,28). The nature of the genes that are responsible for these associations has been resolved for only a few Idd regions, including Idd16-β2m (29), Idd3-Il2 (14), Idd5.1-Ctla4 (30,31), and Idd5.2-Nramp1 (30,32,33). Furthermore, with the exception of Idd3 (14), which controls Treg development and function, the mechanisms by which most of these loci contribute to the diabetogenic process remain unclear.
The current study aimed to elucidate mechanisms underlying the resistance to type 1 diabetes afforded by B10 Idd9 (5), to help pave the way toward eventual identification of the responsible gene. We compared the fate and diabetogenicity of a type 1 diabetes–relevant autoreactive CD8+ T-cell population in B10 Idd9–congenic versus noncongenic TCR-transgenic NOD mice. We found that this reductionist system, which has enabled us to solve Idd3 (14), is not informative to dissect mechanisms underlying Idd9.2- and Idd9.3-linked suppression, presumably because the large size of the transgenic CD8+ T-cell population born by these mice largely overwhelms the corresponding antidiabetogenic mechanisms. This, however, was not the case for Idd9.1. Our data show that Idd9.1 regulates the suppressive activity and antidiabetogenic potential of FoxP3+CD4+CD25+ Tregs. Tregs arising in B10 Idd9.1–congenic NOD mice were significantly more suppressive and antidiabetogenic than those arising in NOD mice. Because FoxP3+CD4+ T-cells suppress diabetogenic T-cell responses, at least in part, by inhibiting DCs in the pancreatic draining lymph nodes (27), these observations are consistent with the suboptimal ability of autoantigen-loaded (in vivo) and peptide-pulsed (ex vivo) PLN DCs from NOD.B10 Idd9.1 mice to support 8.3-CD8+ T-cell activation, both in vitro and in vivo; selective suppression of PLN DCs in these mice may result from cognate suppression of autoantigen-loaded DCs, which predominantly (if not exclusively) reside in the PLNs, by local autoregulatory CD4+CD25+ Tregs. Accordingly, we propose that B10 Idd9.1–“enhanced” Tregs contribute to B10 Idd9.1–linked type 1 diabetes inhibition by suppressing the activation of CD8+ (and possibly other) effector T-cells in the pancreatic draining lymph nodes, without affecting their developmental biology or intrinsic diabetogenic potential. These data might explain the observations of Chamberlain et al. in mice expressing a rat insulin promoter-driven tumor necrosis factor α transgene, in which B10 Idd9.1 somehow suppressed the ability of CD8+ T-cells to respond to islet-infiltrating APCs (34). The results are also compatible with the DC phenotype described by O'Keeffe et al. (12), with the caveat that this congenic strain, unlike the one described here, was produced using B6, as opposed to B10, donor mice and that B10 and B6 DNAs differ substantially in the distal region of chromosome 4 (supplementary Fig. 1). These observations, however, do not exclude, and are compatible with, possible effects of Idd9.1 polymorphisms on other lymphocyte populations. For example, it has been reported that a B10 chromosome 4 region partially overlapping the B10 Idd9.1 fragment studied here restores impaired iNKT cell development and an iNKT cell–related DC phenotype described in NOD mice (9). Our congenic line 1565, however, bears a B10 chromosome 4 fragment that overlaps only the distal 30% of the B10 chromosome 4 fragment present in Ueno et al.'s R201 line. Furthermore, line 1565 does not display Ueno et al.'s iNKT cell phenotype, suggesting that this phenotype is encoded in a gene that is not in the overlapping region. Also, our work does not exclude possible effects of genes within Idd9.1, Idd9.2, and/or Idd9.3 on peripheral tolerance of autoreactive T-cell clones other than those expressing the 8.3-TCR (35), such as for example CD8+ T-cells recognizing IGRP206–214 with higher avidity (36).
It is intriguing that the effects of the B10 Idd9.1 allele on Treg function and type 1 diabetes susceptibility observed in this study are remarkably similar to those we have described recently for B6 Idd3, where Treg development and function are enhanced by increased transcriptional activity of Il2 (14). Accordingly, it is tempting to suspect that regulatory lymphocytes, including the FoxP3+CD4+CD25+ subset, may be common targets of type 1 diabetes–associated chromosomal regions. This hypothesis is compatible with what we know about the type 1 diabetes gene associations in humans. In addition to the IL2-IL21 region on human chromosome 4q27 (2,4,37), and IL2RA on chromosome 10p15.1, which encodes CD25, the high-affinity receptor for IL-2 expressed by activated T-cells, and FoxP3+CD4+CD25+ Tregs (37–40), several other candidate human type 1 diabetes–associated loci play a role in Treg development and function. For example, the type 1 diabetes susceptibility gene CTLA4 on chromosome 2q33.2 encodes a negative regulator of T-cell–mediated immunity (31,41,42). The susceptibility allele generates reduced levels of an alternatively spliced transcript that encodes the soluble form of CTLA-4, which has been correlated with variations in the peripheral frequency of CD4+CD25+ Tregs, which constitutively express CTLA-4 (43). PTPN22 on chromosome 1p13.2, associated with susceptibility to a number of autoimmune diseases, including type 1 diabetes (40, 44–46), encodes lymphoid tyrosine-phosphatase (LYP), another negative regulator of T-cell activation. A nonsynonymous polymorphism (R620W) abolishes the binding of Csk, a negative regulatory kinase, to LYP. Consequently, the R620W LYP variant results in a gain of function that presumably enables LYP to more effectively suppress T-cell signaling compared with the non–type 1 diabetes–associated variants. It has been suggested that PTPN22 polymorphisms contribute to autoimmune disease susceptibility by impairing negative selection of autoreactive thymocytes and by decreasing the number and function of CD4+CD25+ Tregs (47,48). Another locus possibly implicated in autoimmune disease susceptibility by modulating T-cell signaling is PTPN2 on chromosome 18p11.21, a nonreceptor type 2 tyrosine phosphatase that is a negative regulator of T-cell signaling (2).
The fine-mapping of Idd9.1 has not progressed sufficiently to productively comment on candidate genes present in the region. The B10-derived congenic interval present in the Idd9.1 strain used in this study (line 1565) has 329 annotated genes in this 18.5-Mb segment. The Idd11 region as defined by B6-derived congenic strains (16) is an ∼8-Mb region that is wholly contained within the Idd9.1 interval defined by the congenic strain used in this study. However, B6 and B10 are not identical by descent throughout the overlapping region (http://phenome.jax.org); from SNP analyses, B6 and B10 differ at 32.3% of the SNPs within the 8-Mb Idd9.1/Idd11 interval (supplementary Fig. 1). Notably, within this 8 Mb, there are small regions that appear to be identical-by-descent for B6 and B10, including ∼200 kb surrounding Lck, a previously highlighted candidate gene in the Idd9.1 region (16), where the ancestral haplotype shared by the B6 and B10 strains differs from that present in NOD mice (supplementary Fig. 1). Further fine-mapping and gene expression studies are required to define the genes that are responsible for Idd9.1- and Idd11-mediated type 1 diabetes protection and to determine whether the protective alleles are indeed shared by the B6 and B10 strains in this region.
In conclusion, we have shown that the B10 Idd9.1 locus affords diabetes resistance, at least in part, by promoting the function of CD4+CD25+ regulatory T-cells without altering the developmental biology of effector CD8+ T-cells.
Supplementary Material
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (to P.S.). J.Y. was supported by fellowships from the Canadian Diabetes Association and the Juvenile Diabetes Research Foundation (JDRF). P.S. is a Scientist of the Alberta Heritage Foundation for Medical Research (AHFMR). The JMDRC is supported by the Diabetes Association (Foothills). L.S.W. is supported by grants from the JDRF and the Wellcome Trust (WT) and is a JDRF/WT Principal Research Fellow. Support for this work was also provided by National Institute of Allergy and Infectious Disease (NIAID) Grant AI 070351 from the National Institutes of Health. The Cambridge Institute for Medical Research (CIMR) is in receipt of a Wellcome Trust Strategic Award (079895). The availability of NOD congenic mice through the Taconic Farms Emerging Models Program has been supported by grants from the Merck Genome Research Institute, NIAID, and the JDRF.
No potential conflicts of interest relevant to this article were reported.
We thank S. Bou, S. Thiessen, M. Deuma-Myers, H. Metselaar, M. DeCrom, and T. Irvine for technical assistance with mouse breeding, genotyping, and animal care; L. Kennedy and L. Robertson for flow cytometry; C. Gwozd for histology; Y. Yang for aGalCer/Cd1 tetramers; and S. Tsai, A. Shameli, and Y. Yang for helpful discussions.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Maier LM, Wicker LS: Genetic susceptibility to type 1 diabetes. Curr Opin Immunol 2005;17:601–608 [DOI] [PubMed] [Google Scholar]
- 2.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, Lowe CE, Szeszko JS, Hafler JP, Zeitels L, Yang JH, Vella A, Nutland S, Stevens HE, Schuilenburg H, Coleman G, Maisuria M, Meadows W, Smink LJ, Healy B, Burren OS, Lam AA, Ovington NR, Allen J, Adlem E, Leung HT, Wallace C, Howson JM, Guja C, Ionescu-Tirgoviste C, Simmonds MJ, Heward JM, Gough SC, Dunger DB, Wicker LS, Clayton DG: Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet 2007;39:857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper JD, Smyth DJ, Smiles AM, Plagnol V, Walker NM, Allen JE, Downes K, Barrett JC, Healy BC, Mychaleckyj JC, Warram JH, Todd JA: Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet 2008;40:1399–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyons PA, Hancock WW, Denny P, Lord CJ, Hill NJ, Armitage N, Siegmund T, Todd JA, Phillips MS, Hess JF, Chen SL, Fischer PA, Peterson LB, Wicker LS: The NOD Idd9 genetic interval influences the pathogenicity of insulitis and contains molecular variants of Cd30, Tnfr2, and Cd137 Immunity 2000;13:107–115 [DOI] [PubMed] [Google Scholar]
- 6.Martinez X, Kreuwel HT, Redmond WL, Trenney R, Hunter K, Rosen H, Sarvetnick N, Wicker LS, Sherman LA: CD8+ T cell tolerance in nonobese diabetic mice is restored by insulin-dependent diabetes resistance alleles. J Immunol 2005;175:1677–1685 [DOI] [PubMed] [Google Scholar]
- 7.Waldner H, Sobel RA, Price N, Kuchroo VK: The autoimmune diabetes locus Idd9 regulates development of type 1 diabetes by affecting the homing of islet-specific T cells. J Immunol 2006;176:5455–5462 [DOI] [PubMed] [Google Scholar]
- 8.Chen YG, Scheuplein F, Osborne MA, Tsaih SW, Chapman HD, Serreze DV: Idd9/11 genetic locus regulates diabetogenic activity of CD4 T-cells in nonobese diabetic (NOD) mice. Diabetes 2008;57:3273–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueno A, Wang J, Cheng L, Im JS, Shi Y, Porcelli SA, Yang Y: Enhanced early expansion and maturation of semi-invariant NK T cells inhibited autoimmune pathogenesis in congenic nonobese diabetic mice. J Immunol 2008;181:6789–6796 [DOI] [PubMed] [Google Scholar]
- 10.Silveira PA, Chapman HD, Stolp J, Johnson E, Cox SL, Hunter K, Wicker LS, Serreze DV: Genes within the Idd5 and Idd9/11 diabetes susceptibility loci affect the pathogenic activity of B cells in nonobese diabetic mice. J Immunol 2006;177:7033–7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rolf J, Motta V, Duarte N, Lundholm M, Berntman E, Bergman ML, Sorokin L, Cardell SL, Holmberg D: The enlarged population of marginal zone/CD1d(high) B lymphocytes in nonobese diabetic mice maps to diabetes susceptibility region Idd11. J Immunol 2005;174:4821–4827 [DOI] [PubMed] [Google Scholar]
- 12.O'Keeffe M, Brodnicki TC, Fancke B, Vremec D, Morahan G, Maraskovsky E, Steptoe R, Harrison LC, Shortman K: Fms-like tyrosine kinase 3 ligand administration overcomes a genetically determined dendritic cell deficiency in NOD mice and protects against diabetes development. Int Immunol 2005;17:307–314 [DOI] [PubMed] [Google Scholar]
- 13.Hill NJ, Stotland A, Solomon M, Secrest P, Getzoff E, Sarvetnick N: Resistance of the target islet tissue to autoimmune destruction contributes to genetic susceptibility in Type 1 diabetes. Biol Direct 2007;2:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamanouchi J, Rainbow D, Serra P, Howlett S, Hunter K, Garner VE, Gonzalez-Munoz A, Clark J, Veijola R, Cubbon R, Chen SL, Rosa R, Cumiskey AM, Serreze DV, Gregory S, Rogers J, Lyons PA, Healy B, Smink LJ, Todd JA, Peterson LB, Wicker LS, Santamaria P: Interleukin-2 gene variation impairs regulatory T cell function and causes autoimmunity. Nat Genet 2007;39:329–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verdaguer J, Schmidt D, Amrani A, Anderson B, Averill N, Santamaria P: Spontaneous autoimmune diabetes in monoclonal T cell nonobese diabetic mice. J Exp Med 1997;186:1663–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brodnicki TC, Fletcher AL, Pellicci DG, Berzins SP, McClive P, Quirk F, Webster KE, Scott HS, Boyd RL, Godfrey DI, Morahan G: Localization of Idd11 is not associated with thymus and NKT cell abnormalities in NOD mice. Diabetes 2005;54:3453–3457 [DOI] [PubMed] [Google Scholar]
- 17.Brodnicki TC, McClive P, Couper S, Morahan G: Localization of Idd11 using NOD congenic mouse strains: elimination of Slc9a1 as a candidate gene. Immunogenetics 2000;51:37–41 [DOI] [PubMed] [Google Scholar]
- 18.Verdaguer J, Yoon JW, Anderson B, Averill N, Utsugi T, Park BJ, Santamaria P: Acceleration of spontaneous diabetes in TCR-beta-transgenic nonobese diabetic mice by beta-cell cytotoxic CD8+ T cells expressing identical endogenous TCR-alpha chains. J Immunol 1996;157:4726–4735 [PubMed] [Google Scholar]
- 19.DiLorenzo TP, Graser RT, Ono T, Christianson GJ, Chapman HD, Roopenian DC, Nathenson SG, Serreze DV: Major histocompatibility complex class I-restricted T cells are required for all but the end stages of diabetes development in nonobese diabetic mice and use a prevalent T cell receptor alpha chain gene rearrangement. Proc Natl Acad Sci U S A 1998;95:12538–12543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson B, Park BJ, Verdaguer J, Amrani A, Santamaria P: Prevalent CD8(+) T cell response against one peptide/MHC complex in autoimmune diabetes. Proc Natl Acad Sci U S A 1999;96:9311–9316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amrani A, Verdaguer J, Serra P, Tafuro S, Tan R, Santamaria P: Progression of autoimmune diabetes driven by avidity maturation of a T-cell population. Nature 2000;406:739–742 [DOI] [PubMed] [Google Scholar]
- 22.Lieberman SM, Evans AM, Han B, Takaki T, Vinnitskaya Y, Caldwell JA, Serreze DV, Shabanowitz J, Hunt DF, Nathenson SG, Santamaria P, DiLorenzo TP: Identification of the beta cell antigen targeted by a prevalent population of pathogenic CD8+ T cells in autoimmune diabetes. Proc Natl Acad Sci U S A 2003;100:8384–8388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trudeau JD, Kelly-Smith C, Verchere CB, Elliott JF, Dutz JP, Finegood DT, Santamaria P, Tan R: Prediction of spontaneous autoimmune diabetes in NOD mice by quantification of autoreactive T cells in peripheral blood. J Clin Invest 2003;111:217–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cannons JL, Chamberlain G, Howson J, Smink LJ, Todd JA, Peterson LB, Wicker LS, Watts TH: Genetic and functional association of the immune signaling molecule 4–1BB (CD137/TNFRSF9) with type 1 diabetes. J Autoimmun 2005;25:13–20 [DOI] [PubMed] [Google Scholar]
- 25.Irie J, Wu Y, Kachapati K, Mittler RS, Ridgway WM: Modulating protective and pathogenic CD4+ subsets via CD137 in type 1 diabetes. Diabetes 2007;56:186–196 [DOI] [PubMed] [Google Scholar]
- 26.Irie J, Wu Y, Sass DA, Ridgway WM: Genetic control of anti-Sm autoantibody production in NOD congenic mice narrowed to the Idd9.3 region. Immunogenetics 2006;58:9–14 [DOI] [PubMed] [Google Scholar]
- 27.Serra P, Amrani A, Yamanouchi J, Han B, Thiessen S, Utsugi T, Verdaguer J, Santamaria P: CD40 ligation releases immature dendritic cells from the control of regulatory CD4+CD25+ T cells. Immunity 2003;19:877–889 [DOI] [PubMed] [Google Scholar]
- 28.Serreze DV, Leiter EH: Genes and cellular requirements for autoimmune diabetes susceptibility in nonobese diabetic mice. Curr Dir Autoimmun 2001;4:31–67 [DOI] [PubMed] [Google Scholar]
- 29.Hamilton-Williams EE, Serreze DV, Charlton B, Johnson EA, Marron MP, Mullbacher A, Slattery RM: Transgenic rescue implicates beta2-microglobulin as a diabetes susceptibility gene in nonobese diabetic (NOD) mice. Proc Natl Acad Sci U S A 2001;98:11533–11538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wicker LS, Chamberlain G, Hunter K, Rainbow D, Howlett S, Tiffen P, Clark J, Gonzalez-Munoz A, Cumiskey AM, Rosa RL, Howson JM, Smink LJ, Kingsnorth A, Lyons PA, Gregory S, Rogers J, Todd JA, Peterson LB: Fine mapping, gene content, comparative sequencing, and expression analyses support Ctla4 and Nramp1 as candidates for Idd5.1 and Idd5.2 in the nonobese diabetic mouse. J Immunol 2004;173:164–173 [DOI] [PubMed] [Google Scholar]
- 31.Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KM, Smith AN, Di Genova G, Herr MH, Dahlman I, Payne F, Smyth D, Lowe C, Twells RC, Howlett S, Healy B, Nutland S, Rance HE, Everett V, Smink LJ, Lam AC, Cordell HJ, Walker NM, Bordin C, Hulme J, Motzo C, Cucca F, Hess JF, Metzker ML, Rogers J, Gregory S, Allahabadia A, Nithiyananthan R, Tuomilehto-Wolf E, Tuomilehto J, Bingley P, Gillespie KM, Undlien DE, Ronningen KS, Guja C, Ionescu-Tirgoviste C, Savage DA, Maxwell AP, Carson DJ, Patterson CC, Franklyn JA, Clayton DG, Peterson LB, Wicker LS, Todd JA, Gough SC: Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 2003;423:506–511 [DOI] [PubMed] [Google Scholar]
- 32.Dai YD, Marrero IG, Gros P, Zaghouani H, Wicker LS, Sercarz EE: Slc11a1 enhances the autoimmune diabetogenic T-cell response by altering processing and presentation of pancreatic islet antigens. Diabetes 2009;58:156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kissler S, Stern P, Takahashi K, Hunter K, Peterson LB, Wicker LS: In vivo RNA interference demonstrates a role for Nramp1 in modifying susceptibility to type 1 diabetes. Nat Genet 2006;38:479–483 [DOI] [PubMed] [Google Scholar]
- 34.Chamberlain G, Wallberg M, Rainbow D, Hunter K, Wicker LS, Green EA: A 20-Mb region of chromosome 4 controls TNF-alpha-mediated CD8+ T cell aggression toward beta cells in type 1 diabetes. J Immunol 2006;177:5105–5114 [DOI] [PubMed] [Google Scholar]
- 35.Hamilton-Williams EE, Martinez X, Lyman M, Hunter K, Wicker LS, Sherman LA: The use of Idd congenic mice to identify checkpoints of peripheral tolerance to islet antigen. Ann N Y Acad Sci 2007;1103:118–127 [DOI] [PubMed] [Google Scholar]
- 36.Han B, Serra P, Yamanouchi J, Amrani A, Elliott JF, Dickie P, Dilorenzo TP, Santamaria P: Developmental control of CD8 T cell-avidity maturation in autoimmune diabetes. J Clin Invest 2005;115:1879–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lowe CE, Cooper JD, Brusko T, Walker NM, Smyth DJ, Bailey R, Bourget K, Plagnol V, Field S, Atkinson M, Clayton DG, Wicker LS, Todd JA: Large-scale genetic fine mapping and genotype-phenotype associations implicate polymorphism in the IL2RA region in type 1 diabetes. Nat Genet 2007;39:1074–1082 [DOI] [PubMed] [Google Scholar]
- 38.Vella A, Cooper JD, Lowe CE, Walker N, Nutland S, Widmer B, Jones R, Ring SM, McArdle W, Pembrey ME, Strachan DP, Dunger DB, Twells RC, Clayton DG, Todd JA: Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. Am J Hum Genet 2005;76:773–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qu HQ, Montpetit A, Ge B, Hudson TJ, Polychronakos C: Toward further mapping of the association between the IL2RA locus and type 1 diabetes. Diabetes 2007;56:1174–1176 [DOI] [PubMed] [Google Scholar]
- 40.Smyth DJ, Plagnol V, Walker NM, Downes K, Yang JHM, Cooper JD, Howson JMM, Stevens H, McManus R, Wijmenga C, Heap G, Dubois P, Clayton DG, Hunt KA, van Heel DA, Todd JA: Shared, distinct and opposing genetic factors in type 1 diabetes and celiac disease. N Engl J Med 2008;359:2767–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nistico L, Buzzetti R, Pritchard LE, Van der Auwera B, Giovannini C, Bosi E, Larrad MT, Rios MS, Chow CC, Cockram CS, Jacobs K, Mijovic C, Bain SC, Barnett AH, Vandewalle CL, Schuit F, Gorus FK, Tosi R, Pozzilli P, Todd JA: The CTLA-4 gene region of chromosome 2q33 is linked to, and associated with, type 1 diabetes: Belgian Diabetes Registry. Hum Mol Genet 1996;5:1075–1080 [DOI] [PubMed] [Google Scholar]
- 42.Marron MP, Raffel LJ, Garchon HJ, Jacob CO, Serrano-Rios M, Martinez Larrad MT, Teng WP, Park Y, Zhang ZX, Goldstein DR, Tao YW, Beaurain G, Bach JF, Huang HS, Luo DF, Zeidler A, Rotter JI, Yang MC, Modilevsky T, Maclaren NK, She JX: Insulin-dependent diabetes mellitus (IDDM) is associated with CTLA4 polymorphisms in multiple ethnic groups. Hum Mol Genet 1997;6:1275–1282 [DOI] [PubMed] [Google Scholar]
- 43.Atabani SF, Thio CL, Divanovic S, Trompette A, Belkaid Y, Thomas DL, Karp CL: Association of CTLA4 polymorphism with regulatory T cell frequency. Eur J Immunol 2005;35:2157–2162 [DOI] [PubMed] [Google Scholar]
- 44.Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M, Eisenbarth GS, Comings D, Mustelin T: A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet 2004;36:337–338 [DOI] [PubMed] [Google Scholar]
- 45.Vang T, Miletic AV, Bottini N, Mustelin T: Protein tyrosine phosphatase PTPN22 in human autoimmunity. Autoimmunity 2007;40:453–461 [DOI] [PubMed] [Google Scholar]
- 46.Smyth D, Cooper JD, Collins JE, Heward JM, Franklyn JA, Howson JM, Vella A, Nutland S, Rance HE, Maier L, Barratt BJ, Guja C, Ionescu-Tirgoviste C, Savage DA, Dunger DB, Widmer B, Strachan DP, Ring SM, Walker N, Clayton DG, Twells RC, Gough SC, Todd JA: Replication of an association between the lymphoid tyrosine phosphatase locus (LYP/PTPN22) with type 1 diabetes, and evidence for its role as a general autoimmunity locus. Diabetes 2004;53:3020–3023 [DOI] [PubMed] [Google Scholar]
- 47.Vang T, Congia M, Macis MD, Musumeci L, Orru V, Zavattari P, Nika K, Tautz L, Tasken K, Cucca F, Mustelin T, Bottini N: Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet 2005;37:1317–1319 [DOI] [PubMed] [Google Scholar]
- 48.Siggs OM, Miosge LA, Yates AL, Kucharska EM, Sheahan D, Brdicka T, Weiss A, Liston A, Goodnow CC: Opposing functions of the T cell receptor kinase ZAP-70 in immunity and tolerance differentially titrate in response to nucleotide substitutions. Immunity 2007;27:912–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.