Abstract
Production of H2O2 follows a growth phase-dependent pattern that mimics that of many virulence factors of Streptococcus pyogenes. To gain greater insight into mechanisms coupling virulence factor expression to growth phase, we investigated the molecular basis for H2O2 generation and its regulation. Deletion of the gene encoding lactate oxidase (lctO) or culture in the presence of glucose eliminated H2O2 production, implicating carbohydrate regulation of lctO as a key element of growth phase control. In examining known carbohydrate-responsive regulators, deletion of the gene encoding CcpA but not that encoding LacD.1 resulted in both derepression and an uncoupling of lctO transcription from its growth phase pattern. Expanding this analysis to additional virulence factors demonstrated both negative (cfa, encoding CAMP factor) and positive (speB, encoding a cysteine protease) regulation by CcpA and that CcpA mutants were highly cytotoxic for cultured macrophages. This latter property resulted from enhanced transcription of the streptolysin S biogenesis operon. Examination of CcpA-promoter interactions using a DNA pull-down assay mimicking physiological conditions showed direct binding to the promoters of lctO and speB but not those of sagA. CcpA but not LacD.1 mutants were attenuated in a murine model of soft-tissue infection, and analysis of gene expression in infected tissue indicated that CcpA mutants had altered expression of lctO, cfa, and speB but not the indirectly regulated sagA gene. Taken together, these data show that CcpA regulates virulence genes via at least three distinct mechanisms and that disruption of growth phase regulation alters transcriptional patterns in infected tissues.
The restriction of expression of a gene to a specific phase of the bacterial growth cycle is known as growth phase regulation and is a common feature of pathogen gene expression when examined in vitro. This mode of regulation is typically multifactorial, requiring integration of temporal cues linked directly to the growth cycle, with multiple environmental cues, including those that are characteristic of the naïve environment and those that are altered by subsequent bacterial growth (reviewed in reference 67). Of the environmental cues, the former class is composed typically of physical attributes, like oxygen tension, temperature, and specific growth substrates, while the latter class often includes depletion of nutrients and the accumulation of specific quorum-sensing molecules and other metabolites (reviewed in reference 67). It is generally assumed that growth phase-linked patterns of gene expression observed in vitro reflect adaptations that a successful pathogen makes in response to the dynamic host milieu (reviewed in references 55 and 56). However, establishing this link requires identification of specific gene regulatory elements, their hierarchical relationships, and whether the regulatory network responds in a similar pattern in vivo.
Growth phase regulation likely plays a central role in the ability of Streptococcus pyogenes (group A streptococcus) to cause disease. This gram-positive bacterium is the causative agent of numerous diseases of soft tissue ranging from those that are self-limiting (e.g., pharyngitis) to those that are destructive and life-threatening (e.g., necrotizing fasciitis), as well as serious postinfectious sequelae, such as rheumatic fever and acute glomerulonephritis (reviewed in reference 15). Considerable evidence has accumulated to suggest that when examined in vitro, regulation of virtually all of its recognized virulence factors involves a growth phase component. Furthermore, while changes in transcript stability do contribute to growth phase-associated changes (2), most alteration in transcript levels is controlled by regulation of transcription initiation. For example, the CovRS (CsrRS) two-component regulator and the “stand-alone” transcription regulator Mga control expression of 15% and 10%, respectively, of all chromosomal genes, including multiple important virulence-associated surface proteins and toxins (reviewed in references 33 and 43). A prominent characteristic of CovRS and Mga regulation is their growth phase-dependent pattern (33, 43). Both of these regulators also respond to specific environmental signals, including carbon dioxide (Mga [9]), Mg2+, and antimicrobial peptides (CovS [27, 28]). However, as has been noted (2), growth phase control for these regulators is epistatic to the specific signal, such that a temporal pattern of regulation is manifested even when a specific signal is present throughout the growth cycle. Thus, how temporal control is integrated with the processing of specific signals in S. pyogenes is not well understood.
Relatively more progress has been made in understanding the cues that control the timing of expression of growth phase-regulated genes. As a lactic acid bacterium, S. pyogenes has a relatively simple fermentative metabolism, so it is not surprising that nutritional cues have emerged as leading candidates. Examples include both CodY- and RelA-dependent and -independent pathways, which act to couple growth phase regulation to the availability of amino acids as growth substrates (53, 70, 71). Similarly, several mechanisms have been described which could link carbohydrate availability to expression of the Mga regulon. These include the presence of dual phosphotransferase system regulation domains in Mga that function to modulate the activity of regulatory proteins in response to sugar transport and the control of mga transcription itself by the major carbon catabolite repressor protein CcpA (reviewed in reference 33). These observations implicate nutrient availability as an important signal for controlling the timing of growth phase regulation during infection.
The link between the timing of growth phase regulation and carbohydrate availability has been demonstrated more definitively by studies that have directly compared gene expression in infected tissue with that observed during in vitro culture. Comparison of global gene expression between S. pyogenes growth during infection of muscle versus both in vitro biofilm and planktonic growth revealed that the overall pattern of in vivo gene expression most closely resembled that of planktonic culture in medium restricted for carbohydrates (14). Correlations had the highest significance when in vivo expression patterns were compared with in vitro cultures sampled during the early stationary phase of growth (14). Using the gene that encodes the SpeB cysteine protease as a model gene, conditions were identified, including growth phase, that influenced expression of speB transcription in vitro (47). Analyses of global gene expression in response to these conditions identified a set of coregulated genes whose patterns of expression were significantly correlated with that of speB in vivo, suggesting that this cohort of genes was coregulated by the same growth phase-responsive transcriptional program (47).
It was subsequently found that this regulatory program is under the control of LacD.1 (49), an aldolase enzyme that has been adapted to function exclusively as a component of a transcriptional regulatory pathway (48). Mutational analyses suggested that LacD.1 has been adapted to function as a sensor of its substrates, which are the central intermediates of the Embden-Meyerhoff-Parnas glycolytic pathway, implicating LacD.1 as a regulator of global carbon catabolite control (48). This idea was supported by the observation that glucose and certain other carbohydrates are also signals processed by the pathway (48, 49). In most species of firmicutes, the transcriptional regulator CcpA has been recognized as the central regulator of carbon catabolite repression (reviewed in reference 72). However, mutational analyses have shown that the LacD.1 pathway functions independently of the canonical CcpA pathway (48, 49).
Taken together, the data presented above implicate carbohydrate availability as an important signal for the control of growth phase regulation in infected tissue. However, the existence of two pathways for carbon catabolite repression has complicated our understanding of how carbohydrates may regulate the timing of growth phase regulation. At issue is that like LacD.1, the CcpA pathway also monitors the flow of carbon through glycolysis (72). It is not clear why two independent pathways function to process what is essentially the same input signal. Several recent studies have confirmed the role of the S. pyogenes CcpA pathway in global regulation and have suggested that the pathway contributes to virulence (41, 68). However, how this pathway contributes to growth phase regulation, its relationship to the LacD.1 pathway, and how these pathways may interact during infection of tissue is not clear. In the present study, we extended our approach of using model genes for analysis of the CcpA pathway. Our goal was to identify and characterize several suitable model genes for analysis of the contribution of the pathway in growth phase regulation both in vitro and in infected tissue. This analysis revealed at least one model gene for each of the following categories: (i) genes negatively regulated by CcpA, (ii) genes positively regulated by CcpA, (iii) genes regulated indirectly by CcpA, and (iv) genes regulated by both LacD.1 and CcpA. Characterization of regulation of these model genes demonstrated that CcpA plays an important role in linking growth phase regulation and carbohydrate availability during infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The Escherichia coli strain TOP10 (Invitrogen) was used for cloning using standard molecular biology techniques. The S. pyogenes strain HSC5 (29) and various isogenic mutants (see below) were grown in Todd Hewitt broth (THYB) with 0.2% yeast extract (Difco), C medium (51), or RPMI 1640 (Lonza) supplemented with 10% fetal bovine serum (FBS) (Lonza). Routine growth was conducted using sealed culture tubes incubated under static conditions at 37°C. Where indicated, cultures were grown under conditions of enhanced aeration in 125-ml Erlenmeyer flasks containing 10 ml of medium subjected to orbital shaking (225 rpm) at 37°C. Streptococcal strains grown on solid medium containing 1.4% Bacto agar (Difco) were cultured in a sealed jar with a commercial gas generator (GasPak catalogue no. 70304; BBL). Stationary phase was defined as the point when the optical density at 600 nm (OD600) did not change after 20 min. Approximate final OD600 values are noted. When appropriate, antibiotics were added to media at the following concentrations: kanamycin, 500 μg/ml for S. pyogenes and 1 μg/ml for E. coli; and erythromycin, 1 μg/ml for S. pyogenes and 500 μg/ml for E. coli.
DNA and computational techniques.
Plasmid DNA was isolated by standard techniques and used to transform E. coli as described previously (10). Transformation of S. pyogenes was performed by electroporation as previously described (10). Restriction endonucleases, ligases, and polymerases were used according to the manufacturers' recommendations. Chromosomal DNA was purified from S. pyogenes as previously described (10). The fidelity of all plasmid constructs was confirmed by DNA sequencing, which was performed by a commercial service (SeqWright, Houston, TX). All references to genomic loci are based on the genome of strain SF370 (17). Analyses of homology were conducted using BLAST (1, 21). Analysis and identification of CRE sites were carried out using Vector NTI (Invitrogen) and by allowing for one mismatched base pair from the consensus binding site elucidated in Bacillus subtilis.
Measurement of H2O2 and hemolytic titer.
Accumulation of H2O2 in culture supernatant was measured as previously described (20) and on solid medium modified by the addition of horseradish peroxidase (200 μg/ml; Sigma) and 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS, 3 mg/ml; Sigma) following overnight culture under anaerobic conditions and exposure to ambient air for approximately 1 h at 37°C. Determination of hemolytic titers was conducted following overnight culture in RMPI 1640 with 10% FBS that was subjected to centrifugation, filtered through 0.22-μm filters (Millipore), and then assessed using defibrinated rabbit erythrocytes (Hemostat Labs, CA) as described previously (62). Where indicated, the streptolysin S (SLS)-specific inhibitor trypan blue (Sigma) and streptolysin O (SLO)-specific inhibitor cholesterol (Sigma) were added to undiluted supernatants at concentrations of 50 μg/ml and 25 μg/ml, respectively. Hemolytic titer is represented as the inverse of the greatest dilution in which approximately 50% hemolysis was observed.
Construction of CcpA− and LctO− mutants.
In-frame deletion mutations in the genes encoding CcpA (SPy_0514) and LctO (SPy_0414) were generated using allelic replacement and the PCR primers listed in Table S1 in the supplemental material. The deletion alleles were transferred to the HSC5 chromosome using the allelic replacement vector pJRS233 (19) to create strains CKB207 (CcpA−) and CKB044 (LctO−). The presence of the correct alleles was confirmed by PCR.
Isolation of RNA and real-time RT-PCR.
Total RNA was isolated from strains grown in the media and at the times during the growth cycle indicated in the text, as previously described (6). RNA was subjected to reverse transcription (RT) using Superscript II (Invitrogen) per the manufacturer's instructions. Real-time RT-PCR analysis of cDNA samples was performed using iQ SYBR green Supermix (Bio-Rad) using the primers listed in Table S1 in the supplemental material. Relative transcript levels were determined using the threshold cycle method, with the recA transcript as a standard, as described previously (5). Data presented are the means and standard errors of the mean derived from triplicate determinations of samples prepared from at least two independent experiments. Differences between mean values were tested for significance using the paired Student t test as described previously (5).
Biotinylated DNA pulldowns.
A biotinylated DNA fragment pull-down assay was adapted from the method of Grundling et al. (25) as follows: biotinylated DNA fragments were generated by PCR using 5′ biotinylated primers (Invitrogen) (see Table S1 in the supplemental material) and HSC5 genomic DNA. Unincorporated primers were removed by gel purification of PCR products using a commercial reagent (QIAquick gel extraction kit; Qiagen). A 100-μl aliquot of a solution of streptavidin-coated magnetic beads (Dynabeads M-270 streptavidin; Invitrogen) was then coated with 3 μg of biotinylated DNA per the manufacturer's instructions. A whole-cell soluble protein extract was produced as follows: 40-ml cultures of HSC5 were grown to mid-exponential phase (OD600 of 0.2) in THYB. Cells were harvested by centrifugation (6,000 × g, 5 min, 4°C), and the cell pellet was washed twice with 5 ml of lysis buffer (10 mM Tris-HCl, pH 7.5; 50 mM NaCl) and then resuspended in 1 ml of lysis buffer with EDTA-free complete mini protease inhibitor cocktail (Roche). The resuspended cells were then disrupted using glass beads (G4649; Sigma) in a high-speed reciprocating shaking device (FastPrep 100; MPbiomedicals) at a setting of 6.0 for 40 s. Debris and undisrupted cells were removed by centrifugation (14,000 × g, 5 min, 4°C), and the total protein concentration of the cleared supernatant was determined using a bicinchoninic acid assay (Pierce/Thermo-Fisher). The protein concentration was adjusted to 1.5 mg/ml, glycerol was added to a final concentration of 20%, and the solution was stored at −20°C until use. For the pulldown, an aliquot of DNA-coated beads (200 μl, prepared as described above) was mixed with the protein extract (1 mg) and binding buffer (10 mM Tris-HCl [pH 8], 100 mM KCl, 3 mM MgCl2, 20 mM EDTA, 5% glycerol, 40 μg/ml sheared salmon sperm DNA [Invitrogen], and 10 μg/ml bovine serum albumin [Sigma]) and added to a total volume of 1 ml. Following a 30-min incubation at room temperature with gentle mixing on a rotating mixer, the beads were collected using a magnetic particle separator (catalog number K1585; Invitrogen) for 1 min at room temperature, washed four times with 500 μl binding buffer, resuspended in 30 μl of 1× DNase I buffer (Invitrogen), boiled for 5 min, and then allowed to slowly cool to room temperature. The DNA probes were then removed by incubation with DNase I (2 μl, AMP grade; Invitrogen) for 10 min. Protein sodium dodecyl sulfate (SDS) sample buffer (1×; 50 mM Tris-HCl [pH 6.8], 100 mM dithiothreitol, 2% [wt/vol] SDS, 0.1% [wt/vol] bromophenol blue, 10% glycerol) was then added in a 2× dilution from a 6× stock, and the sample was boiled for an additional 5 min. The magnetic beads were removed with the magnetic particle separator, and the resulting sample was subjected to electrophoresis using a 12% SDS-polyacrylamide gel electrophoresis gel. Protein bands were visualized by staining with Sypro ruby (Molecular Probes/Invitrogen) per the manufacturer's instructions. The band(s) of interest was excised and then identified by matrix-assisted laser desorption ionization-time of flight (mass spectrometry) by the Proteomics Core at Washington University (http://proteomics.wustl.edu/siteman).
Preparation and infection of mouse bone marrow-derived macrophages.
Bone marrow-derived macrophages were prepared as previously described (11). Following 8 days of culture in RPMI 1640 with 20% FBS and 30% L-cell-preconditioned medium, macrophages were harvested by incubation in ice-cold phosphate-buffered saline (PBS) (Cellgro) and then gently scraped. The macrophages were harvested by centrifugation (1,000 × g, 10 min, 4°C), resuspended in RPMI 1640 with 10% FBS, and plated in six-well tissue culture plates (TPP) at 3 × 106 macrophages per well in 4 ml total medium. The macrophages were allowed to settle on the plates for 4 h at 37°C in 5% CO2. The various streptococcal strains were cultured overnight in THYB, diluted to an OD600 of 0.05 in 40 ml RPMI 1640 with 10% FBS, and allowed to grow for an additional 2.5 h. The streptococcal strains were then collected by centrifugation and resuspended in RPMI 1640 with 10% FBS to a final OD600 of 0.87. An aliquot of the bacterial suspension (150 μl) was added to each macrophage-containing well and incubated at 37°C in 5% CO2 for the times indicated in the text. At the conclusion of the infection, the media were removed and each well was washed with 2 ml cold PBS. The PBS was then removed, and cells were stained using a fluorescent reagent to assess viability (Live/Dead for mammalian cells; Invitrogen) per the manufacturer's instructions. Cells were imaged using a Leica DMRE2 fluorescence microscope, and images were analyzed using Openlab software (Improvision). For each tissue culture well, at least 200 macrophages were examined. The data presented represent the means and standard errors of the mean derived from triplicate determinations. Differences between mean values were tested for significance with the paired Student t test.
Subcutaneous infection of mice.
As previously described (6, 8), 5- to 6-week-old female SKH1 hairless mice (Charles River Labs) were injected subcutaneously with 107 CFU of streptococci of the strains indicated in the text, and the areas of the resulting ulcers were quantitated on day 3 postinfection as described previously (6, 8). Briefly, images of resulting lesions were analyzed using MetaMorph (Improvision) image analysis software to determine the area contained by the irregular border of each ulcer. The data presented are representative of at least two independent experiments conducted with 10 mice in each experimental group. Differences in the areas of the resulting ulcers were tested for significance by the Mann-Whitney U test (22). RNA was harvested from selected ulcers and subjected to analysis using real-time RT-PCR to assess expression of various streptococcal genes using the primers listed in Table S1 in the supplemental material and the methods described previously (7).
RESULTS
Lactate oxidase is responsible for the glucose-repressible loss of viability in stationary phase.
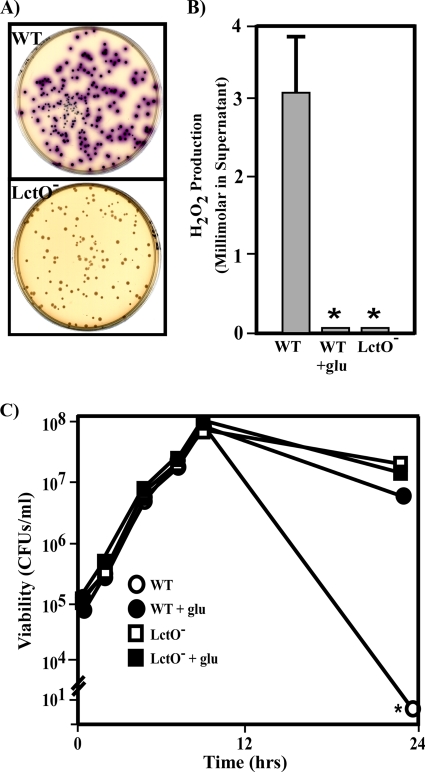
Mutants of S. pyogenes deficient in NADH oxidase (Nox, SPy_1150) produced self-lethal levels of H2O2; however, the addition of glucose both suppressed this lethality and the production of H2O2 (20). It has also been observed that many clinical isolates of S. pyogenes produce auto-intoxicating amounts of H2O2 upon entry into the stationary phase of growth when grown under aerobic conditions (63, 66). These data suggest that peroxide production is both glucose and growth phase regulated. Biochemical analyses of S. pyogenes have suggested that H2O2 is produced by an enzymatic activity consistent with a lactate oxidase (66). Examination of the prototypic S. pyogenes M1 genome revealed a single gene (lctO, SPy_0414) with significant homology to the characterized lactate oxidase gene of Streptococcus iniae (19). This locus is also present in 13/13 sequenced S. pyogenes strains and the wild-type (WT) strains used previously to study Nox (20). An in-frame deletion of lctO was constructed in one of these latter strains (HSC5), and the resulting LctO− mutant (CKB044) was found to produce colonies of normal size compared to HSC5 (WT) when grown anaerobically; however, these colonies failed to produce H2O2 when subsequently exposed to air (Fig. 1A). When grown aerobically in liquid culture, the WT strain rapidly lost viability upon onset of stationary phase, though loss of viability could be suppressed by addition of glucose (Fig. 1C). In contrast, the LctO− strain was protected from a loss of viability in a glucose-independent manner (Fig. 1C) that correlated with failure to produce any detectable amounts of H2O2 in stationary phase, compared to the millimolar amounts produced by the WT strain (Fig. 1B). As previously observed, addition of glucose suppressed the ability of WT bacteria to produce H2O2 (Fig. 1B). The defect in H2O2 production apparent in the LctO− mutant was complemented by an lctO allele expressed constitutively off a plasmid (see Fig. S1 in the supplemental material).
FIG. 1.
Endogenous self-lethal H2O2 production is dependent on LctO. (A) WT and LctO− strains were grown on H2O2 indicator solid medium as described in the text. Production of H2O2 is indicated by colorimetric change to purple in the medium. (B) WT and LctO− strains were grown in ThyB or ThyB supplemented with 0.2% glucose (+glu) aerobically. After growth overnight, supernatants were assayed for H2O2 concentration, and data are presented as the means and standard errors of results of two independent experiments carried out in triplicate. Asterisks denote values below the limit of detection (approximately 10 μM). (C) Strains were grown as described in the legend for panel B, samples were taken at indicated times, and appropriate dilutions were plated on solid ThyB medium to count viable CFU. The asterisk indicates a value of less than 100 CFU/ml of culture. Strains: WT, HSC5; LctO−, CKB044.
Glucose and growth phase regulate transcription of lctO.
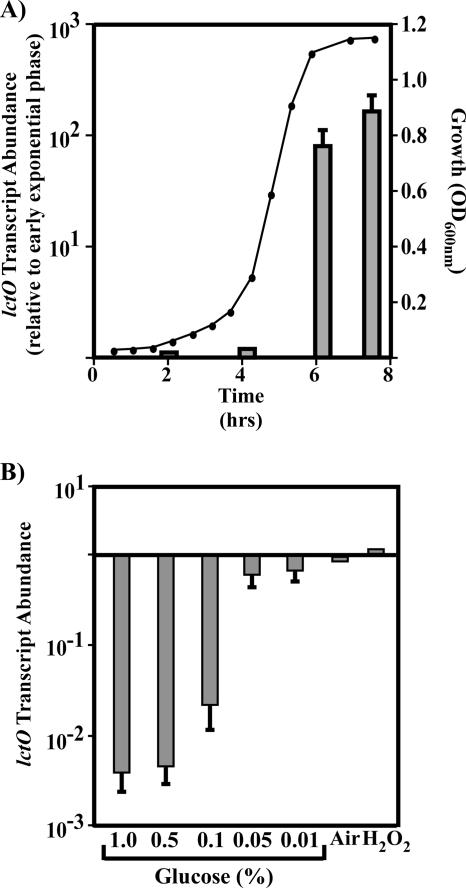
The data presented above suggest that LctO is responsible for H2O2 production and the glucose-inhibited, growth phase-dependent loss of viability in aerobic conditions. To examine the basis of the latter phenomenon, the transcription of lctO in the WT strain was examined using real-time RT-PCR. Relative to early exponential phase (OD600 of 0.05, approximately 2 h postinoculation), the transcript abundance of lctO stays low through mid-exponential-phase growth (OD600 of 0.2, approximately 4 h postinoculation) and then undergoes a large upregulation upon the onset of stationary phase (OD600 of 1.15, approximately 6.5 h postinoculation) (Fig. 2A). In late stationary phase the transcript abundance of lctO was still relatively high (OD600 of 1.15 approximately 7.5 h postinoculation) (Fig. 2A), suggesting that when cultures have reached a maximum OD600, there is a large dynamic shift toward increased transcription of lctO that persists throughout stationary phase. To test if glucose was a signal controlling transcription of lctO, WT streptococci were grown in a carbohydrate-poor medium (C medium). The addition of glucose in exponential phase resulted in a dose-dependent and saturable repressive effect on lctO transcript abundance (Fig. 2B). At low levels of glucose the transcription was relatively high, while the addition of 0.1% or greater amounts of glucose repressed lctO transcript abundance several-hundredfold (Fig. 2B). Further experiments showed that growth while shaking in ambient air (to increase O2 tension) or exposure to oxidative stress in the form of added H2O2 changed lctO transcript abundance less than twofold (Fig. 2B, Air and H2O2).
FIG. 2.
Transcription of lctO is regulated by growth phase and glucose concentration. (A) WT bacteria were grown in C medium plus 0.2% glucose anaerobically to determine transcription of lctO during growth in broth culture. Growth of streptococci was measured by OD600. Total RNA was harvested at the indicated times, and lctO transcript abundance was measured by real-time RT-PCR. Data are presented as the ratios of transcript abundance at the indicated time points compared to transcript levels at the first time point (2 h) and represent the means and standard deviations from a representative experiment carried out with three samples per time point, each analyzed in triplicate. (B) WT bacteria were grown with varied glucose, aeration, and oxidative stress to test the effects on lctO transcription. WT bacteria were grown in C medium supplemented with the indicated amount of glucose, with C medium plus 0.2% glucose shaking in ambient air (Air), or with 100 μM H2O2 (H2O2) added 1 h prior to harvesting. Total RNA was harvested in early exponential phase (OD600 of 0.1) and lctO transcript abundance determined by real-time RT-PCR. Data are presented as the ratios of transcript abundance as noted (compared to C medium plus glucose for glucose analysis and compared to C medium plus 1% glucose for Air and H2O2) and represent the means and standard deviations derived from three samples, each analyzed in triplicate, compared to C medium without glucose (−glu), or for Air and H2O2, compared to C medium plus 0.2% glucose. WT strain, HSC5.
CcpA affects growth phase regulation of lctO in response to carbohydrate signals.
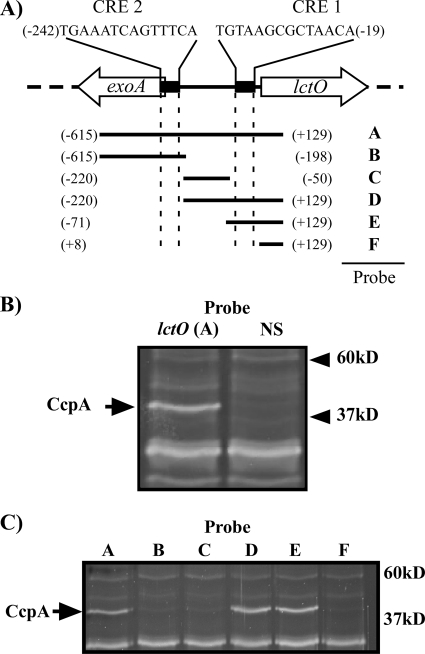
A biochemical approach was used to identify potential growth phase and glucose-responsive regulators of lctO transcription. A biotinylated DNA probe that included the entire intergenic region between lctO and an upstream open reading frame (Fig. 3A, probe A) was incubated with whole-cell lysate prepared from mid-logarithmic cultures; the probe was then recovered by precipitation with streptavidin-coated beads. This analysis consistently revealed a protein band of approximately 37 kDa that was not precipitated with a nonspecific DNA probe, which was identified by matrix-assisted laser desorption ionization-time of flight (mass spectrometry) as the carbon catabolite control protein CcpA (SPy_0514) (Fig. 3B). As a transcriptional repressor, CcpA binds to an operator site termed a catabolite responsive element (CRE), and using the consensus site determined for Bacillus subtilis (73), it was possible to identify two potential CRE sites in the region encompassed by probe A (CRE-1, CRE-2) (Fig. 3A). However, analysis of additional probes that contained various sections of the intergenic region indicated that only those probes containing CRE-1 were capable of coprecipitating CcpA, an observation supported by previous studies of CcpA interaction with CRE elements in S. pyogenes (Fig. 3A and C, probes A, D, and E) (68).
FIG. 3.
CcpA binds to a conserved CRE site in the lctO promoter. (A) Diagram of the lctO locus and DNA probes generated by PCR for biotinylated DNA probe precipitations. Positions of DNA segments are shown as relative to the predicted start of translation of the lctO gene, with the A of the ATG start codon considered +1. Predicted CRE sites are indicated with black rectangles, and sequences of corresponding CRE sites are indicated above each site. Probes were created by PCR using HSC5 genomic DNA as a template and the following primer combinations: A, CK192/CK194; B, CK192/CK303; C, CK304/CK305; D, CK304/CK194; E, CK306/CK194; F, CK307/CK194. Primer sequences are listed in Table S1 in the supplemental material. (B) Biotinylated DNA probes were used to precipitate proteins from a lysate of the WT. The lctO probe A and a control nonspecific fragment (NS) were created by PCR using HSC5 genomic DNA as a template and the primer pair CK195/CK196 (see Table S1 in the supplemental material). As indicated by the arrow, an lctO-specific band was identified as CcpA. The locations of molecular weight standards (in thousands) are indicated at right. (C) Biotinylated DNA probes spanning the lctO locus were used to examine the binding of CcpA to the promoter of lctO. Proteins were precipitated as described in the legend for panel B, with the DNA probes indicated in the legend for panel A. The band identified as CcpA in the legend for panel B is labeled with an arrow. The locations of molecular weight standards (in thousands) are indicated at right. Strain: WT, HSC5.
The contribution of CcpA to the regulation of lctO transcription was determined by analysis of a mutant strain constructed to contain an in-frame deletion allele of ccpA (CKB206). Two major effects were observed with the resulting CcpA− mutant. Whereas the WT strain switched from a state of low lctO transcript abundance in exponential phase to high abundance in stationary phase, transcript levels in the CcpA− strain were similar in both exponential and stationary growth and consistent with the high level seen in stationary phase of the WT strain (Table 1) The increase in lctO transcript abundance upon the onset of stationary phase was no longer apparent in the CcpA− strain; rather, transcript abundance was relatively high and unchanged by growth phase (Table 1). Second, addition of glucose to the culture medium no longer caused a repressive effect on lctO transcription in the CcpA− strain, differing from the WT strain for which transcript abundance was down over 100-fold in the presence of excess glucose (Table 1). Deletion of ccpA led to a loss of repression of lctO transcription and constitutively high transcript levels in the CcpA− mutant. Therefore, we conclude that growth phase and glucose regulation of lctO transcription are due to the direct repressive effects of CcpA.
TABLE 1.
Growth phase and glucose-induced changes in transcript abundance of lctO are CcpA dependent
| Strainc | Relative expressiona |
||
|---|---|---|---|
| Exponentialb |
Stationary | ||
| −Glucose | +Glucose | ||
| WT | 1 | 0.0081 ± 0.002 | 425.8 ± 79.1 |
| CcpA− | 407.5 ± 60.6 | 388.0 ± 53.6 | 448.5 ± 143.3 |
Values are derived from real-time RT-PCR analysis of total cellular RNA and normalized to WT in exponential phase in unsupplemented C medium (−Glucose).
Exponential OD600nm = 0.1; stationary OD600nm = 1.1, in C medium plus glucose. Growth in C medium: unsupplemented (−Glucose) or 0.1% (wt/vol) glucose (+Glucose).
WT, HSC5; CcpA−, CKB206.
CcpA regulates virulence factors CAMP and SpeB in growth phase-dependent and -independent manners, respectively.
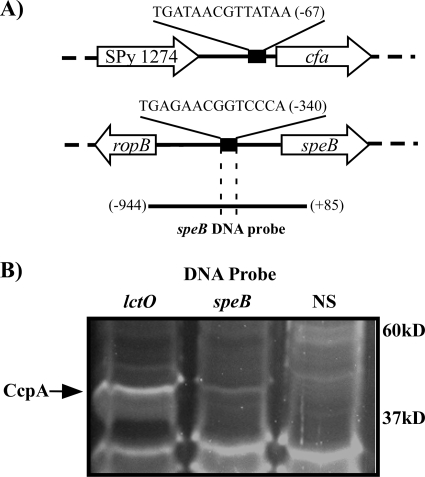
Using the consensus CRE sequence from B. subtilis, we identified two additional genes in group A streptococci likely to be regulated by CcpA. The gene for CAMP factor, cfa (SPy_1273), has a consensus CRE sequence in the promoter region (Fig. 4A) and is a predicted virulence factor found in many pathogenic lactic acid bacteria (46). Similar to those of lctO, cfa transcript levels increase significantly in stationary phase, and the loss of CcpA leads to constitutively high transcript abundance and a loss of growth phase-dependent regulation (Table 2). This suggests that like lctO, cfa is growth phase regulated by CcpA due to repression of cfa transcription in exponential phase and then upregulated due to a release of repression in stationary phase. When CcpA is absent, transcript levels of cfa are constitutively at a level similar to that of stationary-phase transcription in the WT strain (Table 2). The gene for SpeB (SPy_2039), the major secreted cysteine protease of group A streptococci, is preceded by a large intergenic region containing a predicted CRE (Fig. 4A) and is subject to complex transcriptional regulation (47, 57). Unlike that of either lctO or cfa, speB transcript abundance decreased in the absence of CcpA in both phases of growth (Table 2). In the CcpA− strain, transcription of speB was consistently 7- to 10-fold lower than that in WT bacteria in the same stage of growth (Table 2). However, similar to WT bacteria, there was still a large growth phase-dependent upregulation of speB transcription in the CcpA− strain (Table 2). Despite the significant activating effect of CcpA on speB expression, these analyses revealed that CcpA does not function as a growth phase regulator of speB transcription. Thus, CcpA acts as an important regulator of virulence genes but functions with at least two distinct mechanisms: direct repression in response to growth phase and activation of transcription independent of growth phase.
FIG. 4.
CcpA binds predicted CRE sites upstream of CcpA-regulated virulence genes. (A) Diagram of the cfa and speB loci. Predicted CRE sites are indicated with black rectangles; sequences of corresponding CRE sites are indicated above each site. Positions of the DNA features are relative to the start of translation of the cfa and speB genes, with the A of the ATG start codon considered +1. The speB DNA probe is indicated by the line below the speB locus and was generated by PCR using HSC5 genomic DNA as template with the primer pair CK393/CK396 (see Table S1 in the supplemental material). (B) Precipitation of proteins from the WT interacting with the speB promoter region. DNA probes: lctO, probe A (Fig. 3A); speB, as indicated in the legend for panel A; NS, nonspecific (Fig. 3B). The protein band corresponding to CcpA is indicated with an arrow. The positions of molecular weight standards (in thousands) are indicated at right. Strain: WT, HSC5.
TABLE 2.
CcpA affects virulence factor transcript abundance in both growth phase-dependent and -independent manners
| Gene | Relative expressiona |
|||
|---|---|---|---|---|
| Exponentialb |
Stationary |
|||
| WTc | CcpA− | WT | CcpA− | |
| cfa | 1 | 6.76 ± 3.19 | 7.00 ± 3.15 | 7.45 ± 4.35 |
| speB | 1 | 0.14 ± 0.03 | 6975 ± 1294 | 999.7 ± 252.5 |
Values derived from real-time RT-PCR analysis and normalized to these of the WT in exponential phase for each gene.
Strains were grown in C medium plus glucose (0.1% [wt/vol]). Exponential OD600 of 0.1; stationary OD600 of 1.1.
WT, HSC5; CcpA−, CKB206.
CcpA binds the speB promoter.
Since regulation of speB differed from our model gene, lctO, we examined whether CcpA bound directly to the speB promoter. Both in the control lctO fragment used earlier (Fig. 3A, probe A) and in the fragment indicated for the speB promoter (Fig. 4B), CcpA was copreciptated from lysates of the WT strain of streptococci but not by using a nonspecific DNA probe (Fig. 4B). This suggests that direct interaction of CcpA within a promoter region can result in either activation or repression of transcription.
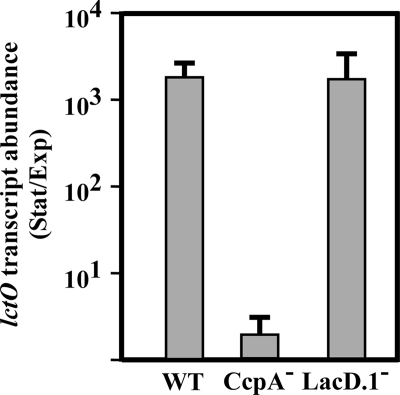
LacD.1 and CcpA do not both regulate lctO.
Complicating the models of speB transcriptional regulation is the recent discovery of a novel carbohydrate sensing regulator, the adapted tagatose aldolase LacD.1 from group A streptococci that also controls transcriptional regulation of speB (49). In contrast to activation by CcpA, the LacD.1 pathway represses speB transcription. Since the mechanism of LacD.1-mediated repression of speB is largely unknown, it was possible that CcpA and LacD.1 acted in a linear pathway (49). To test this hypothesis, we examined the transcription of lctO in the CcpA− strain and a LacD.1− strain in comparison to WT regulation. Deletion of ccpA abolished growth phase-dependent regulation of lctO, whereas inactivation of lacD.1 had no effect on lctO transcription compared to the WT strain (Fig. 5). If CcpA and LacD.1 function in a linear pathway, both deletions should have similar effects on lctO transcription. This suggests that CcpA and LacD.1 are at least partially independent but have opposing transcriptional effects on the speB promoter.
FIG. 5.
Unlike CcpA, LacD.1 does not regulate lctO. WT, CcpA−, and LacD.1− strains were grown in C medium with 0.1% glucose until early exponential phase (OD600 of 0.1) or until stationary phase (maximal OD600 of approximately 1.1 was attained). Total RNA was isolated from these cultures and used for real-time RT-PCR analysis of the lctO transcript. Data are presented as the ratios of transcript abundance in stationary phase to that in exponential phase for each strain and represent the means and standard deviations derived from three samples, each analyzed in triplicate. Strains: WT, HSC5; CcpA−, CKB206; LacD.1−, JL151 (49).
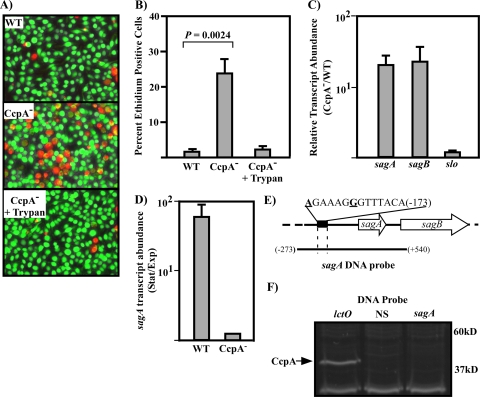
Disruption of the CcpA regulatory pathway renders bacteria more cytotoxic, revealing an indirect mechanism of CcpA regulation of streptolysin S.
Presumably pathogenic bacteria sense and respond to many host-derived signals during infection. We hypothesized that signals encountered during pathogenesis could control either CcpA or LacD.1 activity or both. To test this hypothesis, we infected murine bone marrow-derived macrophages with various streptococcal strains to examine changes in host cell response. Previous studies have shown that interactions with group A streptococci cause macrophages to upregulate many cytokines, such as tumor necrosis factor alpha, interleukin-1, and interleukin-6 (23). However, unlike infection with the WT strain, after infection with the CcpA− strain for 4 h, bone marrow-derived macrophages had rounded and detached from the tissue culture flask surface (not shown). At earlier time points, about 1 hour postinfection, most bone marrow-derived macrophages were viable after interaction with WT streptococci, but significantly more macrophages had lost viability after infection with the CcpA− streptococci (Fig. 6A and B). This suggested that the CcpA− strain was more cytotoxic than the WT strain. In hemolytic assays the supernatant of the CcpA− strain had a hemolytic titer 16-fold higher than that of the WT strain (Table 3).
FIG. 6.
SLS is overproduced in a CcpA− strain due to indirect regulation. (A) Bone marrow-derived macrophages (BMM) were infected with WT and CcpA− for 1 h and then assayed for viability. Viable cells fluoresce green due to calcein acetoxymethyl staining; nonviable cells fluoresce red due to increased permeability of cellular membranes to ethidium homodimer. The SLS-specific inhibitor trypan blue (50 μg/ml) was added as indicated. (B) Quantitation of ethidium-positive (dead) BMMs from infections represented in panel A. At least 200 BMMs were counted in duplicate experiments. Data represents means and standard deviations from two experiments done in triplicate; differences were analyzed for significance using Student's t test. (C) Analysis of cytolysin transcript abundance in WT and CcpA−. Bacteria were grown as indicated for infection of BMMs. Total RNA was harvested and subjected to real-time RT-PCR analysis for the sagA, sagB, and slo transcripts. Data are presented as the ratios of transcript abundance in CcpA− compared to that in WT bacteria for each indicated gene and represent the means and standard deviations derived from two experiments, with three samples in each experimental group, each analyzed in triplicate. (D) Growth phase regulation of sagA in WT and CcpA−. Strains were grown in C medium plus 0.1% glucose until early exponential phase (OD600 of 0.1) or stationary phase (OD600 of approximately 1.1). Total RNA was isolated and used for real-time RT-PCR analysis of sagA transcript abundance as described in the legend for panel C. (E) Diagram of the sagA locus. Positions of DNA features are shown as relative to the predicted start of translation of the sagA gene, with the A of the ATG start codon considered +1. A putative CRE site identified in previous studies (41, 68) is indicated by a black rectangle. The sequence of the putative CRE site is indicated; bold and underlined base pairs differ from the conserved CRE sequence determined from B. subtilis (39). The sagA DNA probe indicated below the diagram was generated by PCR using HSC5 genomic DNA as template and the primer pair CK405/CK371 (for primer sequence, see Table S1 in the supplemental material). (F) Precipitation of proteins interacting with the sagA locus. Biotinylated DNA precipitation of proteins from a WT protein sample (lctO probe A [Fig. 3A]; sagA as indicated in the legend for panel E; NS, nonspecific [Fig. 3B]) was carried out as described previously, and the protein band corresponding to the CcpA band identified previously is indicated with an arrow. The positions of molecular weight standards (in thousands) are indicated at right. Strains: WT, HSC5; CcpA−, CKB206.
TABLE 3.
Inactivation of CcpA leads to increased trypan blue-inhibitable hemolysis
| Strainc | Hemolytic titera |
|||||
|---|---|---|---|---|---|---|
| RPMIb |
ThyB |
|||||
| None | +Chol | +Trypan | None | +Chol | +Trypan | |
| WT | 2 | 2 | NL | 4 | NL | 2 |
| CcpA− | 32 | 16 | NL | 8 | 2 | NL |
Values represent the reciprocal of the dilution of the supernatant in which 50% lysis was observed. NL, no lysis.
Strains were grown in RPMI1640 with 10% FBS or ThyB to an OD600 of 0.1. Supernatant was sterile filtered and used in hemolytic assay. Cholesterol (25 mg/ml) or trypan blue (50 mg/ml) was added as noted.
WT, HSC5; CcpA−, CKB206.
The two best characterized group A streptococcal hemolysins are the cholesterol-dependent cytolysin SLO and the lantibiotic-related peptide hemolysin SLS. Including the SLO-specific inhibitor cholesterol in hemolytic assays did not prevent hemolysis upon incubation with supernatant from the CcpA− strain, suggesting that SLO was not responsible for the increased cytotoxicity observed in macrophage infection (Table 3). Addition of the SLS-specific inhibitor trypan blue to the hemolytic assay prevented hemolysis in the CcpA− supernatant samples (Table 3). This suggested that increased cytotoxicity of the CcpA− strain was due to increased SLS production. When trypan blue was added to bone marrow-derived macrophages during infection with the CcpA− strain, macrophages retained viability comparable to that with infection with WT streptococci (Fig. 6A and B). This confirmed that increased cytotoxicity of the CcpA− mutant was due to overproduction of SLS. To test whether SLS overproduction was due to altered transcriptional regulation, transcript levels of the first two genes in the SLS biogenesis operon (sagA and sagB, SPy_0738 and SPy_0739) were tested as well as the SLO structural gene, slo (SPy_0167). Under the conditions used for infection of macrophages, both the sagA and sagB transcripts were upregulated about 25-fold in the CcpA− strain compared to WT bacteria (Fig. 6C). This result confirms observations from previous studies that inactivation of CcpA leads to an upregulation of sag operon transcription (41, 68) As expected, the slo transcript was not affected by CcpA under these conditions (Fig. 6C). Since both the sagA and sagB transcripts were upregulated to the same degree, it is likely that loss of CcpA regulation results in the entire sag operon being misregulated, not just the pel transcript, which is largely comprised of the intergenic and sagA regions (45).
Examination of the growth phase regulation of transcript abundance in the WT and CcpA− strains revealed that in the WT strain the sagA transcript is upregulated in stationary phase, whereas in the CcpA− strain this upregulation is not apparent (Fig. 6D). Taken with the previous findings, this suggests that the sag operon, like lctO and cfa, is repressed by CcpA in exponential phase and loss of CcpA leads to overproduction of SLS due to a derepression of transcription. Analysis of the sag promoter revealed a poorly conserved CRE sequence located upstream of the start of translation of sagA (Fig. 6E). This CRE site was not found in our earlier searches due to several mutations known to abolish CcpA binding in B. subtilis (38, 39). Using the biotinylated DNA pull-down assay with a DNA fragment from the sag promoter, we were unable to coprecipitate CcpA (Fig. 6E), whereas CcpA was coprecipitated with the lctO DNA fragment using the same protein sample (Fig. 6E). To validate these findings in other serotypes of group A streptococcus, we repeated these DNA pull-down assays using a protein lysate from M1 strain SF370. The results from this pulldown confirm results from our WT strain (see Fig. S2 in the supplemental material). This suggests that regulation of the sag operon by CcpA occurs through an indirect mechanism. Due to the epistatic overproduction of SLS we were unable to determine if host responses by macrophages were altered by CcpA compared to responses to the WT strain.
Regulation by LacD.1 and CcpA in infected tissue is distinct and closely resembles growth-phase regulation of CcpA in vitro.
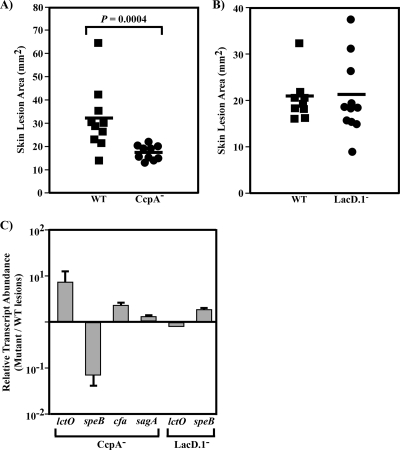
Previous experiments have shown that CcpA functions as an important regulator of virulence genes, acting by at least three different mechanisms: (i) direct repression in response to growth phase, (ii) indirect repression in response to growth phase, and (iii) direct activation independent of growth phase. We analyzed the regulation of lctO, cfa, speB, and sagA in lesions from mice infected with WT, CcpA−, and LacD.1− strains in a subcutaneous infection model to better understand which mechanisms of regulation observed in vitro were similar to patterns of regulation observed during tissue infection. Briefly, immuno competent hairless mice were injected subcutaneously with 107 streptococci. In this model, as with many WT streptococcal strains, a local necrotic lesion forms, enlarging and forming an eschar, but does not progress to invasive disease (8). At 3 days postinjection, when lesions with WT bacteria were maximal, lesions were measured for area, a general indicator of overall strain virulence, and total RNA was extracted from excised lesions and subjected to real-time RT-PCR analysis of these streptococcal genes. Infection by the CcpA− strain leads to significantly smaller lesions than those from the WT strain (Fig. 7A), suggesting that the CcpA− strain is attenuated in this virulence model. Analysis of the transcript levels from these lesions showed that lctO and cfa transcript levels were significantly increased in the CcpA− strain compared to levels from WT lesions and that speB levels are significantly lower than those from WT lesions (Fig. 7B). This closely mirrors the results seen in vitro. Transcript levels of sagA were not significantly different (less than twofold change) between CcpA− and WT lesions (Fig. 7B).
FIG. 7.
Regulation in infected tissue through CcpA, but not LacD.1, resembles regulation in vitro. SKH1 hairless mice were inoculated subcutaneously with the WT and CcpA− (A) or LacD.1− (B), and areas of the resulting lesions were determined at day 3 postinjection. Data shown are numbers of lesion areas from individual mice from a representative experiment. Differences between groups were tested for significance using the Mann-Whitney U test, and P values are reported in the figure. (C) SKH1 hairless mice were infected as described above. On day 3 postinjection, the resulting skin lesions were excised, and total RNA isolated and subjected to real-time RT-PCR analysis of the indicated transcripts. Data presented are ratios of transcript abundance in mutant strains compared to the WT and represent the means and standard deviations derived from two experiments in which three mice for each bacterial strain were analyzed. Strains: WT, HSC5; CcpA−, CKB206; LacD.1−, JL151.
Lesions caused by the LacD.1− strain were not significantly different from those caused by the WT strain (Fig. 7C). In culture conditions, repression through LacD.1 results in a several-hundredfold reduction in speB transcript abundance (49). In samples from mouse lesions, the transcription of the speB gene in LacD.1− samples was only threefold higher than that in WT bacteria from skin lesions (Fig. 7D). This suggests that although certain in vitro conditions lead to LacD.1 activation, these same signals are not active in mouse lesions at this time point.
DISCUSSION
For many pathogens, virulence factor expression is coupled to growth phase by the transduction of extrinsic (environmental) or intrinsic (metabolic signaling molecules) cues to regulators controlling the abundance of virulence factor transcripts. Uncoupling of virulence gene regulation from growth phase through the disruption of signaling pathways or specific regulators can affect pathogen fitness in host tissues (43, 69). The variety of signals and regulators as well as the unique virulence factor repertoires among related pathogens necessitates the identification and characterization of specific regulators and signals to better understand how the pathogenesis of important infectious diseases is controlled by cues linked to growth phase.
In this study we have characterized CcpA as a growth phase-dependent regulator of metabolic and virulence-associated genes and shown that growth phase-dependent regulation through LacD.1 and CcpA are distinguishable in mouse tissue. We have shown that CcpA can regulate transcription with three different mechanisms: direct repression, indirect repression, and direct activation. Our data suggest that the mechanism of CcpA-dependent regulation as observed in vitro has implications for regulation of target genes in infected tissues, thereby validating the study of in vitro signaling for understanding in vivo virulence responses. Furthermore, our results demonstrate a correlation between an altered pattern of virulence gene regulation in tissues and the attenuation of virulence, suggesting that growth phase-associated signals sensed through CcpA are important in pathogenesis.
Growth phase regulation of virulence factor transcription is found among many low-G+C gram-positive pathogens. Conserved regulators such as CodY, quorum-sensing systems, and alternative sigma factors coordinate complex transcriptional programs upon the onset of stationary phase in Staphylococcus aureus, Streptococcus pneumoniae, and Enterococcus faecalis (60, 69). However, while group A streptococci share the CodY response (53), they appear to lack a conserved quorum-sensing system (the signaling peptide SilCR is present in a minority of clinical isolates though enriched in invasive isolates [3, 4, 31]) and possess only one alternate sigma factor, which is not implicated in pathogenesis (58). Through studies of identified virulence factor regulators, it has been shown that CovR/CovS, Mga, RopB, LacD.1, and the vfr locus make significant contributions to the growth phase regulation of the ska, emm, and speB genes (16, 49, 51, 52, 54). However, mutations in CovR/CovS and LacD.1 do not completely uncouple their target genes from growth phase regulation, and the transcriptional activators RopB and Mga and the vfr locus are themselves growth phase regulated, suggesting that the sensing of growth phase signals lies upstream of these regulators. Previous studies of CcpA in growth phase-dependent regulation established that for many largely catabolic genes in B. subtilis and Lactococcus lactis the disruption of CcpA uncoupled the accumulation of transcript from growth phase (50, 75). Since CcpA has been demonstrated to be a catabolite-sensitive regulator in B. subtilis, the signal controlling growth phase is most likely the depletion of glucose sensed through interaction of CcpA with a serine-phosphorylated form of the Hpr protein (34).
Inactivation of ccpA affects virulence and virulence gene expression in several gram-positive pathogens, including S. aureus and many pathogenic streptococci (18, 35, 36, 41, 65, 68). Thus, studies of growth phase regulation through CcpA would likely reveal how regulation observed during infection relates to important signals and pathways elucidated in vitro. Indeed, our findings show that certain aspects of growth phase-dependent regulation observed with culture are similarly manifested in infected tissue. This is the case not only for virulence factor genes, such as cfa and speB, but also for lctO, the gene responsible for H2O2 generation. In S. pneumoniae, the ability to produce H2O2 can serve as a virulence trait or more likely as a colonization factor both by slowing the clearance of bacteria through the inhibition of ciliary beating and by the bacteriostatic and bactericidal activities of H2O2 with other upper respiratory tract bacteria (32, 59, 61). That H2O2 production functions as a colonization factor in a related bacterium may explain why lctO is regulated in S. pyogenes. Like many lactic acid bacteria, group A streptococci are polyauxotrophic, requiring a ready supply of amino acids and cofactors and an easily metabolized organic carbon source for growth. During times of low metabolite availability, the ability to produce H2O2 may inhibit the growth of competing bacteria in the nasopharynx, thereby allowing streptococci to scavenge nutrients, while limiting direct competition. In contrast, in times of high nutrient availability, the repression of lctO production may help to avoid the self-lethal accumulation of H2O2 in the environment.
Coregulation of virulence and metabolic genes in group A streptococci has been previously demonstrated for other regulators. RopB, an rgg family regulator controlling the activation of transcription of the speB gene, has effects on amino acid metabolism (12, 13). The discovery and characterization of LacD.1 as a regulator of speB transcription established a new regulator capable of linking nutritional status to transcription of virulence genes (49). Inactivation of LacD.1 coupled with overexpression of RopB led to the partial uncoupling of speB transcription from growth phase, suggesting a role for LacD.1 as an inhibitor of RopB activity until the onset of stationary phase (49). Our results show that CcpA regulates speB transcription as well, adding a third metabolic sensor. Unlike LacD.1, CcpA appears to activate transcription in a growth phase-independent manner. Activation of transcription is a described function for CcpA in the case of acetate kinase in B. subtilis and many genes in L. lactis as assessed by transcriptome analysis (26, 75). Unique to speB, CcpA-dependent activation is opposed by the inhibitory effects of LacD.1, possibly in response to the same signal. This paradox can be partially explained by measurement of transcript levels in infected tissues. Whereas transcription of speB in lesions by CcpA closely resembles the pattern and magnitude observed in culture, regulation through LacD.1 in lesions follows a similar pattern but does not have a similar magnitude as observed in culture. A possible mechanism for this observation is that both LacD.1 and CcpA are known to respond to multiple signals. LacD.1 was first discovered as a regulator of pH and chloride repressive signals in SpeB production (49), though the mechanism for sensing and regulation in response to these signals is unknown. CcpA has been shown to interact not only with the serine-phosphorylated form of Hpr but also with the metabolite signals glucose-6-phosphate and fructose-1,6-bisphosphate (24) and the cofactor NADP (40). Binding of CcpA to target DNA is not dependent on any of these binding partners; rather, complexing of CcpA with Hpr Ser-P, glucose-6-phosphate, fructose-1,6,-bisphosphate, and NADP affect the affinity of CcpA-CRE interactions through conformational changes in the CcpA dimer (64). Input from any of these alternate signals to either the LacD.1 or CcpA regulatory pathways may lead to a change in regulatory pattern or magnitude of regulation from that observed in vitro. In addition, it is difficult to study the microenvironments or differential expressions among populations of bacteria within an infection. Our data suggest that direct or indirect regulation by CcpA can behave distinctly in infected tissues, perhaps due to the actions of multiple pathways at indirectly regulated promoters. Taken together, these data support a model in which the integration of multiple signals within a regulatory pathway and organization of multiple pathways at specific promoters is important for coordination of a virulence response and pathogen fitness within host tissues.
The mechanism by which CcpA regulates transcription of target genes has best been studied with B. subtilis, in which the CcpA protein has been shown to interact with well-conserved DNA sites in promoter regions, leading to either repression or activation of transcription (34). Studies of CcpA regulation in pathogenesis have generally used genomic-based techniques and focused on the role of CcpA as a transcriptional repressor, as described for regulation of the starch utilization locus of B. subtilis (30). Considerably less is known about the roles and biochemistry of CcpA as an indirect regulator and activator of transcription. What is known about the biochemistry and DNA binding capabilities of CcpA in B. subtilis appears to hold true for group A streptococci and related streptococcal pathogens. In group A streptococcal virulence, two previous studies have suggested that the sag locus is repressed directly by CcpA (41, 68). Our data agree, in that inactivation of CcpA can lead to an overexpression of SLS due to increased transcript abundance of the sag operon. However we have shown here that SLS overexpression in the absence of CcpA does not follow a pattern in tissue similar to that of other directly repressed genes. In contrast to other genes directly repressed by CcpA and within infected tissues, the loss of CcpA has almost no effect on sagA transcription. This may explain why in the murine subcutaneous lesion model, as opposed to infection of macrophages in which we observed hypervirulence, the CcpA mutant shows a significant loss of virulence. Whereas in previous studies many of the results for the link between CcpA and virulence focused on sagA and SLS biogenesis, here our findings suggest that SLS is likely to have only a minor contribution to differences in GAS virulence levels in CcpA− strains in a subcutaneous lesion model of infection.
Our method of examining interactions with the sagA promoter uses the WT sequence from the sag locus and is nonbiased, in that CcpA is precipitated from a pool of total cellular proteins at protein concentrations less than cellular levels and target DNA concentrations of approximately one to two copies per cell. Using this technique we observed direct CcpA binding to DNA in the lctO and speB promoters, both containing conserved CRE sites, but not to the sagA promoter. In the previous studies of CcpA in streptococcal virulence, direct regulation of the sag operon was observed by use of differing techniques for measurement of binding, which may offer an explanation for these contrasting results. In Shelburne et al. (68), the DNA probe representing the sag operon promoter utilized a CRE site corresponding to the canonical CRE site deduced from studies of B. subtilis (39, 68). The putative CRE site in the sagA promoter differs from the idealized CRE from B. subtilis in several base pairs, including a crucial C-G dyad at the axis of symmetry. It is known from the earlier studies of B. subtilis that this C-G dyad is necessary for CcpA binding; a mutation in either residue leads to a loss of CcpA regulation of the downstream gene (38, 39). It is notable that a poorly conserved putative CRE site in the lctO promoter was also unable to precipitate CcpA. This putative CRE site, termed CRE2, differs at two positions compared to the consensus CRE sequence from B. subtilis and importantly lacks the C-G dyad at the axis of symmetry of the canonical CRE site. Taken together, these findings suggest a specificity of CcpA to DNA containing CRE sites in group A streptococci akin to the specificity described for B. subtilis and may explain why CcpA was unable to be precipitated by the putative CRE site in front of sagA. In Kinkel et al. (41), direct binding of CcpA to the sagA promoter was observed using electrophoretic mobility shift assays to demonstrate protein-DNA complexes (41). Under the conditions used, CcpA was in excess of the DNA probe by 100:1. It has previously been shown that under conditions in which protein is in high excess of target DNA, CcpA can bind to DNA without CRE sites. Furthermore, analysis of CcpA from L. lactis has shown that once CcpA/DNA complexes have been formed, it is difficult to displace CcpA from bound DNA with competing CRE containing unlabeled probe (42). This suggests the importance of the conditions and concentrations under which CcpA is observed to bind DNA since protein to target ratios can affect the specificity of CcpA binding. Our data support indirect regulation of the sag operon; however, it does not rule out the possibility of extremely weak direct interactions. Indirect regulation of the sag operon may suggest a model in which, in infected tissue, another regulatory pathway is epistatic to CcpA, leading to a regulation of the sag operon that differs from patterns observed for genes directly regulated by CcpA.
Interruption of growth phase-dependent regulators has a large impact on virulence and virulence gene regulation in group A streptococci. Inactivation of the CovR (CsrR) gene leads to a marked increase in virulence in mice (44), while inactivation of CcpA leads to attenuation of virulence (68). Due to the importance of these and similar pathways to virulence, defining environmental signals and downstream effects of signaling in vitro is likely to increase our understanding of how pathogens sense signals and adapt while causing an infection. The in vitro signals for CovR/CovS activation include Mg2+ and cationic antimicrobial peptide concentration (27, 28); little is known about the role of either Mg2+ or cathelicidins in growth phase regulation; however, metal homeostasis and adaptation to innate immune responses have been shown to be important in infected tissues (37, 74). CcpA and LacD.1 have been shown to respond to carbohydrate availability (30, 49), an important signal in growth phase regulation. Here we have compared regulation in culture to regulation in infected tissues, showing that the CcpA pathway as defined in vitro is active in virulence; furthermore, our data suggest that integration of multiple signals is important in controlling the activities of both CcpA and LacD.1, possibly preventing the full and antagonistic results of each pathway from affecting speB transcription. Defining the signals and downstream response of the CcpA and LacD.1 pathways in vitro will lead to an understanding of how pathogenic bacteria integrate multiple sensory pathways in virulence.
Supplementary Material
Acknowledgments
This work was supported by Public Health Service Grant AI070759 from the National Institutes of Health.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 19 October 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Barnett, T. C., J. V. Bugrysheva, and J. R. Scott. 2007. Role of mRNA stability in growth phase regulation of gene expression in the group A streptococcus. J. Bacteriol. 189:1866-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bidet, P., C. Courroux, C. Salgueiro, A. Carol, P. Mariani-Kurkdjian, S. Bonacorsi, and E. Bingen. 2007. Molecular epidemiology of the sil streptococcal invasive locus in group A streptococci causing invasive infections in French children. J. Clin. Microbiol. 45:2002-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billal, D. S., M. Hotomi, J. Shimada, K. Fujihara, K. Ubukata, R. Sugita, and N. Yamanaka. 2008. Prevalence of streptococcus invasive locus (sil) and its relationship with macrolide resistance among group A Streptococcus strains. J. Clin. Microbiol. 46:1563-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenot, A., K. Y. King, and M. G. Caparon. 2005. The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes. Mol. Microbiol. 55:221-234. [DOI] [PubMed] [Google Scholar]
- 6.Brenot, A., K. Y. King, B. Janowiak, O. Griffith, and M. G. Caparon. 2004. Contribution of glutathione peroxidase to the virulence of Streptococcus pyogenes. Infect. Immun. 72:408-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenot, A., B. F. Weston, and M. G. Caparon. 2007. A PerR-regulated metal transporter (PmtA) is an interface between oxidative stress and metal homeostasis in Streptococcus pyogenes. Mol. Microbiol. 63:1185-1196. [DOI] [PubMed] [Google Scholar]
- 8.Bunce, C., L. Wheeler, G. Reed, J. Musser, and N. Barg. 1992. Murine model of cutaneous infection with gram-positive cocci. Infect. Immun. 60:2636-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caparon, M. G., R. T. Geist, J. Perez-Casal, and J. R. Scott. 1992. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J. Bacteriol. 174:5693-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caparon, M. G., and J. R. Scott. 1991. Genetic manipulation of pathogenic streptococci. Methods Enzymol. 204:556-586. [DOI] [PubMed] [Google Scholar]
- 11.Celada, A., P. W. Gray, E. Rinderknecht, and R. D. Schreiber. 1984. Evidence for a gamma-interferon receptor that regulates macrophage tumoricidal activity. J. Exp. Med. 160:55-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaussee, M. A., E. A. Callegari, and M. S. Chaussee. 2004. Rgg regulates growth phase-dependent expression of proteins associated with secondary metabolism and stress in Streptococcus pyogenes. J. Bacteriol. 186:7091-7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaussee, M. S., G. A. Somerville, L. Reitzer, and J. M. Musser. 2003. Rgg coordinates virulence factor synthesis and metabolism in Streptococcus pyogenes. J. Bacteriol. 185:6016-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho, K. H., and M. G. Caparon. 2005. Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes. Mol. Microbiol. 57:1545-1556. [DOI] [PubMed] [Google Scholar]
- 15.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Federle, M. J., K. S. McIver, and J. R. Scott. 1999. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J. Bacteriol. 181:3649-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giammarinaro, P., and J. C. Paton. 2002. Role of RegM, a homologue of the catabolite repressor protein CcpA, in the virulence of Streptococcus pneumoniae. Infect. Immun. 70:5454-5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibello, A., M. D. Collins, L. Dominguez, J. F. Fernandez-Garayzabal, and P. T. Richardson. 1999. Cloning and analysis of the l-lactate utilization genes from Streptococcus iniae. Appl. Environ. Microbiol. 65:4346-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson, C. M., T. C. Mallett, A. Claiborne, and M. G. Caparon. 2000. Contribution of NADH oxidase to aerobic metabolism of Streptococcus pyogenes. J. Bacteriol. 182:448-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gish, W., and D. J. States. 1993. Identification of protein coding regions by database similarity search. Nat. Genet. 3:266-272. [DOI] [PubMed] [Google Scholar]
- 22.Glantz, S. A. 2002. Primer of biostatistics, 5th ed. McGraw-Hill, New York, NY.
- 23.Goldmann, O., M. von Kockritz-Blickwede, C. Holtje, G. S. Chhatwal, R. Geffers, and E. Medina. 2007. Transcriptome analysis of murine macrophages in response to infection with Streptococcus pyogenes reveals an unusual activation program. Infect. Immun. 75:4148-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gosseringer, R., E. Kuster, A. Galinier, J. Deutscher, and W. Hillen. 1997. Cooperative and non-cooperative DNA binding modes of catabolite control protein CcpA from Bacillus megaterium result from sensing two different signals. J. Mol. Biol. 266:665-676. [DOI] [PubMed] [Google Scholar]
- 25.Grundling, A., L. S. Burrack, H. G. Bouwer, and D. E. Higgins. 2004. Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proc. Natl. Acad. Sci. USA 101:12318-12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grundy, F. J., D. A. Waters, S. H. Allen, and T. M. Henkin. 1993. Regulation of the Bacillus subtilis acetate kinase gene by CcpA. J. Bacteriol. 175:7348-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gryllos, I., J. C. Levin, and M. R. Wessels. 2003. The CsrR/CsrS two-component system of group A Streptococcus responds to environmental Mg2+. Proc. Natl. Acad. Sci. USA 100:4227-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gryllos, I., H. J. Tran-Winkler, M. F. Cheng, H. Chung, R. Bolcome III, W. Lu, R. I. Lehrer, and M. R. Wessels. 2008. Induction of group A Streptococcus virulence by a human antimicrobial peptide. Proc. Natl. Acad. Sci. USA 105:16755-16760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanski, E., P. A. Horwitz, and M. G. Caparon. 1992. Expression of protein F, the fibronectin-binding protein of Streptococcus pyogenes JRS4, in heterologous streptococcal and enterococcal strains promotes their adherence to respiratory epithelial cells. Infect. Immun. 60:5119-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henkin, T. M., F. J. Grundy, W. L. Nicholson, and G. H. Chambliss. 1991. Catabolite repression of alpha-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacl and galR repressors. Mol. Microbiol. 5:575-584. [DOI] [PubMed] [Google Scholar]
- 31.Hidalgo-Grass, C., M. Ravins, M. Dan-Goor, J. Jaffe, A. E. Moses, and E. Hanski. 2002. A locus of group A Streptococcus involved in invasive disease and DNA transfer. Mol. Microbiol. 46:87-99. [DOI] [PubMed] [Google Scholar]
- 32.Hirst, R. A., K. S. Sikand, A. Rutman, T. J. Mitchell, P. W. Andrew, and C. O'Callaghan. 2000. Relative roles of pneumolysin and hydrogen peroxide from Streptococcus pneumoniae in inhibition of ependymal ciliary beat frequency. Infect. Immun. 68:1557-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hondorp, E. R., and K. S. McIver. 2007. The Mga virulence regulon: infection where the grass is greener. Mol. Microbiol. 66:1056-1065. [DOI] [PubMed] [Google Scholar]
- 34.Hueck, C. J., and W. Hillen. 1995. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the gram-positive bacteria? Mol. Microbiol. 15:395-401. [DOI] [PubMed] [Google Scholar]
- 35.Iyer, R., N. S. Baliga, and A. Camilli. 2005. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 187(24):8340-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jankovic, I., R. Brèuckner, and U. T. G. Mikrobielle Genetik. 2002. Carbon catabolite repression by the catabolite control protein CcpA in Staphylococcus xylosus. J. Mol. Microbiol. Biotechnol. 4(3):309-314. [PubMed] [Google Scholar]
- 37.Janulczyk, R., S. Ricci, and L. Bjorck. 2003. MtsABC is important for manganese and iron transport, oxidative stress resistance, and virulence of Streptococcus pyogenes. Infect. Immun. 71:2656-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim, J. H., and G. H. Chambliss. 1997. Contacts between Bacillus subtilis catabolite regulatory protein CcpA and amyO target site. Nucleic Acids Res. 25:3490-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim, J. H., Z. T. Guvener, J. Y. Cho, K. C. Chung, and G. H. Chambliss. 1995. Specificity of DNA binding activity of the Bacillus subtilis catabolite control protein CcpA. J. Bacteriol. 177:5129-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim, J. H., M. I. Voskuil, and G. H. Chambliss. 1998. NADP, corepressor for the Bacillus catabolite control protein CcpA. Proc. Natl. Acad. Sci. USA 95(16):9590-9595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinkel, T. L., and K. S. McIver. 2008. CcpA-mediated repression of streptolysin S expression and virulence in the group A streptococcus. Infect. Immun. 76:3451-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kowalczyk, M., B. Borcz, D. Plochocka, and J. Bardowski. 2007. In vitro DNA binding of purified CcpA protein from Lactococcus lactis IL1403. Acta Biochim. Pol. 54:71-78. [PubMed] [Google Scholar]
- 43.Kreikemeyer, B., K. S. McIver, and A. Podbielski. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11:224-232. [DOI] [PubMed] [Google Scholar]
- 44.Levin, J. C., and M. R. Wessels. 1998. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol. Microbiol. 30:209-219. [DOI] [PubMed] [Google Scholar]
- 45.Li, Z., D. D. Sledjeski, B. Kreikemeyer, A. Podbielski, and M. D. Boyle. 1999. Identification of pel, a Streptococcus pyogenes locus that affects both surface and secreted proteins. J. Bacteriol. 181:6019-6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu, G. Y., and V. Nizet. 2004. Extracellular virulence factors of group B Streptococci. Front Biosci. 9:1794-1802. [DOI] [PubMed] [Google Scholar]
- 47.Loughman, J. A., and M. Caparon. 2006. Regulation of SpeB in Streptococcus pyogenes by pH and NaCl: a model for in vivo gene expression. J. Bacteriol. 188:399-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loughman, J. A., and M. G. Caparon. 2007. Comparative functional analysis of the lac operons in Streptococcus pyogenes. Mol. Microbiol. 64:269-280. [DOI] [PubMed] [Google Scholar]
- 49.Loughman, J. A., and M. G. Caparon. 2006. A novel adaptation of aldolase regulates virulence in Streptococcus pyogenes. EMBO J. 25:5414-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lulko, A. T., G. Buist, J. Kok, and O. P. Kuipers. 2007. Transcriptome analysis of temporal regulation of carbon metabolism by CcpA in Bacillus subtilis reveals additional target genes. J. Mol. Microbiol. Biotechnol. 12:82-95. [DOI] [PubMed] [Google Scholar]
- 51.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 17:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma, Y., A. E. Bryant, D. B. Salmi, E. McIndoo, and D. L. Stevens. 2009. vfr, a novel locus affecting cysteine protease production in Streptococcus pyogenes. J. Bacteriol. 191:3189-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malke, H., K. Steiner, W. M. McShan, and J. J. Ferretti. 2006. Linking the nutritional status of Streptococcus pyogenes to alteration of transcriptional gene expression: the action of CodY and RelA. Int. J. Med. Microbiol. 296:259-275. [DOI] [PubMed] [Google Scholar]
- 54.McIver, K. S., and J. R. Scott. 1997. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J. Bacteriol. 179:5178-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mekalanos, J. J. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 174:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller, J. F., J. J. Mekalanos, and S. Falkow. 1989. Coordinate regulation and sensory transduction in the control of bacterial virulence. Science 243:916-922. [DOI] [PubMed] [Google Scholar]
- 57.Neely, M. N., W. R. Lyon, D. L. Runft, and M. Caparon. 2003. Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. J. Bacteriol. 185:5166-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Opdyke, J. A., J. R. Scott, and C. P. Moran, Jr. 2001. A secondary RNA polymerase sigma factor from Streptococcus pyogenes. Mol. Microbiol. 42:495-502. [DOI] [PubMed] [Google Scholar]
- 59.Pericone, C. D., K. Overweg, P. W. Hermans, and J. N. Weiser. 2000. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect. Immun. 68:3990-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Podbielski, A., and B. Kreikemeyer. 2004. Cell density-dependent regulation: basic principles and effects on the virulence of Gram-positive cocci. Int. J. Infect. Dis. 8:81-95. [DOI] [PubMed] [Google Scholar]
- 61.Regev-Yochay, G., K. Trzcinski, C. M. Thompson, M. Lipsitch, and R. Malley. 2007. SpxB is a suicide gene of Streptococcus pneumoniae and confers a selective advantage in an in vivo competitive colonization model. J. Bacteriol. 189:6532-6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ruiz, N., B. Wang, A. Pentland, and M. Caparon. 1998. Streptolysin O and adherence synergistically modulate proinflammatory responses of keratinocytes to group A streptococci. Mol. Microbiol. 27:337-346. [DOI] [PubMed] [Google Scholar]
- 63.Saito, M., S. Ohga, M. Endoh, H. Nakayama, Y. Mizunoe, T. Hara, and S. Yoshida. 2001. H(2)O(2)-nonproducing Streptococcus pyogenes strains: survival in stationary phase and virulence in chronic granulomatous disease. Microbiology 147:2469-2477. [DOI] [PubMed] [Google Scholar]
- 64.Schumacher, M. A., G. Seidel, W. Hillen, and R. G. Brennan. 2007. Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors glucose 6-phosphate and fructose 1,6-bisphosphate. J. Mol. Biol. 368:1042-1050. [DOI] [PubMed] [Google Scholar]
- 65.Seidl, K., M. Stucki, M. Ruegg, C. Goerke, C. Wolz, L. Harris, B. Berger-Bächi, and M. Bischoff. 2006. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 50:1183-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seki, M., K. Iida, M. Saito, H. Nakayama, and S. Yoshida. 2004. Hydrogen peroxide production in Streptococcus pyogenes: involvement of lactate oxidase and coupling with aerobic utilization of lactate. J. Bacteriol. 186:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seshasayee, A. S., P. Bertone, G. M. Fraser, and N. M. Luscombe. 2006. Transcriptional regulatory networks in bacteria: from input signals to output responses. Curr. Opin. Microbiol. 9:511-519. [DOI] [PubMed] [Google Scholar]
- 68.Shelburne, S. A., III, D. Keith, N. Horstmann, P. Sumby, M. T. Davenport, E. A. Graviss, R. G. Brennan, and J. M. Musser. 2008. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc. Natl. Acad. Sci. USA 105:1698-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sonenshein, A. L. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 8:203-207. [DOI] [PubMed] [Google Scholar]
- 70.Steiner, K., and H. Malke. 2000. Life in protein-rich environments: the relA-independent response of Streptococcus pyogenes to amino acid starvation. Mol. Microbiol. 38:1004-1016. [DOI] [PubMed] [Google Scholar]
- 71.Steiner, K., and H. Malke. 2001. relA-independent amino acid starvation response network of Streptococcus pyogenes. J. Bacteriol. 183:7354-7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Warner, J. B., and J. S. Lolkema. 2003. CcpA-dependent carbon catabolite repression in bacteria. Microbiol. Mol. Biol. Rev. 67:475-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weickert, M. J., and G. H. Chambliss. 1990. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 87:6238-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weston, B. F., A. Brenot, and M. G. Caparon. 2009. The metal homeostasis protein Lsp of Streptococcus pyogenes is necessary for acquisition of zinc and virulence. Infect. Immun. 77:2840-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zomer, A. L., G. Buist, R. Larsen, J. Kok, and O. P. Kuipers. 2007. Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:1366-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.