Abstract
The concept of an infectious agent playing a role in cardiovascular disease is slowly gaining attention. Among several pathogens identified, the oral bacterium Streptococcus gordonii has been implicated as a plausible agent. Platelet adhesion and subsequent aggregation are critical events in the pathogenesis and dissemination of the infective process. Here we describe the identification and characterization of a novel cell wall-anchored surface protein, PadA (397 kDa), of S. gordonii DL1 that binds to the platelet fibrinogen receptor GPIIbIIIa. Wild-type S. gordonii cells induced platelet aggregation and supported platelet adhesion in a GPIIbIIIa-dependent manner. Deletion of the padA gene had no effect on platelet aggregation by S. gordonii but significantly reduced (>75%) platelet adhesion to S. gordonii. Purified N-terminal PadA recombinant polypeptide adhered to platelets. The padA mutant was unaffected in production of other platelet-interactive surface proteins (Hsa, SspA, and SspB), and levels of adherence of the mutant to fetuin or platelet receptor GPIb were unaffected. Wild-type S. gordonii, but not the padA mutant, bound to Chinese hamster ovary cells stably transfected with GPIIbIIIa, and this interaction was ablated by addition of GPIIbIIIa inhibitor Abciximab. These results highlight the growing complexity of interactions between S. gordonii and platelets and demonstrate a new mechanism by which the bacterium could contribute to unwanted thrombosis.
Platelets are small anucleate cell fragments of the larger hematopoietic precursor, the megakaryocyte. Being devoid of a nucleus, the platelet has no control of gene expression, but it has limited capabilities in translational protein synthesis. The primary role of platelets in hemostasis is to police the integrity of the endothelium in order to prevent blood loss. Platelets circulate close to the endothelial cell surface as individual entities that ordinarily do not interact with any other cell types (24). In the absence of any stable interactions with endothelial cells, they exist in an antiadhesive state. Upon trauma or injury to vascular endothelium, platelets are rapidly recruited to the site of injury. Recruitment is a highly controlled event that is stimulated by the initial adhesive interaction between the exposed extracellular matrix proteins in damaged endothelium and specific membrane receptors on the platelet (26). Adhesion to the extracellular matrix requires a synergistic function of membrane receptors which ultimately results in platelet activation and aggregation.
Several surface proteins expressed on the surface of the platelet play a role in these adhesive interactions. The initial interaction of platelets with the injured vessel wall occurs between the platelet glycoprotein Ib (GPIb) and immobilized von Willebrand factor (vWf). This interaction initiates the tethering of circulating platelets to the vessel wall (24). Platelets become slowed and roll over vWf in the direction of flow, driven by shear forces experienced by the vasculature (25). A loss of interaction between GPIb and vWf on one side of the platelet leads to the formation of another GPIb-vWf interaction on the other side of the platelet, giving rise to the rolling phenomenon (26). Eventually the platelet will come to a complete stop and end in firm adhesion to the injured part of the vessel. This firm adhesion is mediated by several other membrane receptors, including the platelet integrin GPIIbIIIa, which will have become activated as a result of platelet rolling. Integrin GPIIbIIIa has many ligands, including fibrinogen, vWf, and fibronectin (28).
Recently there have been increased numbers of reports in the literature highlighting the interactions between oral bacteria and platelets (8). Most notably the viridans group streptococci, including Streptococcus sanguinis, Streptococcus oralis, Streptococcus mutans, Streptococcus mitis, Streptococcus parasanguinis, and Streptococcus gordonii (2), are associated with over 30% cases of native valve endocarditis (6). Chronic oral disease such as periodontitis provides a plausible route of entry of bacteria to the circulation, leading to transient bacteremia. If the bacteria evade the immune defenses, then subsequent interaction with platelets is thought to be crucial in pathogenesis of infective endocarditis and disseminated intravascular coagulation (DIC).
Many attempts have been made to identify bacterial proteins recognizing platelets and their reciprocal platelet receptors. A platelet-associated activating peptide present on S. sanguinis that induced platelet aggregation was characterized (9), but the corresponding platelet receptor has not been identified. More recently, it has been shown that a cell wall-linked, serine-rich repeat polypeptide GspB (or Hsa) on S. gordonii (31) is capable of binding to the sialylated N-terminal region of GPIbα on platelets (5, 31). This interaction is critical for supporting S. gordonii adhesion to platelets (15). Inactivation of the hsa gene in S. gordonii reduced platelet adhesion levels by approximately 50% but had no effect on the ability of bacteria to induce platelet aggregation. These results suggested that additional streptococcal factors were involved in supporting platelet adhesion and inducing platelet aggregation.
Progress has also been made in determining the mechanisms through which S. gordonii induces platelet aggregation. The cell surface proteins SspA (172 kDa) and SspB (164 kDa) of S. gordonii belong to the antigen I/II family of adhesins that are expressed by most indigenous strains of oral streptococci (7) These polypeptide adhesins recognize multiple ligands, including salivary agglutinin glycoprotein (gp340), collagen type I, and β1 integrin (23, 12) Deletion of the sspA and sspB genes in S. gordonii does not affect platelet adhesion, but it extends the lag time for streptococci to induce platelet aggregation (15). However, inactivation of the sspA, sspB, and hsa genes together leads to ablation of S. gordonii-induced platelet aggregation, suggesting that the antigen I/II polypeptides and Hsa function synergistically in the aggregation process (10).
There is a growing awareness of the complex mechanisms that bacteria use for survival in the host bloodstream, and until now these three cell surface proteins were thought to be the major determinants by which oral streptococci adhere to and aggregate platelets, leading to thrombosis. This paper describes a new protein with a vWf-A1 like motif (vWA) on the surface of Streptococcus gordonii, designated platelet adherence protein A (PadA). This high-molecular-mass (397-kDa) cell wall-anchored protein may present a novel method by which Streptococcus gordonii can bind to platelets and support platelet adhesion, a step that might contribute to thrombotic vegetations found in blood-borne disease.
MATERIALS AND METHODS
Bacterial strains, plasmids, and cultivation.
The strains and plasmids used in this study are listed in Table S1 in the supplemental material. All Streptococcus strains were stored in 50% glycerol at −80°C and streaked onto Todd-Hewitt (TH) agar plates prior to growth in TH broth. Streptococcus gordonii DL1-Challis and isogenic mutants were cultured at 37°C in an atmosphere of 5% CO2. Escherichia coli was cultured in Luria-Bertani medium aerobically at 37°C for 16 h. For selection and growth of transformed E. coli, ampicillin was added to Luria-Bertani medium at a final concentration of 100 μg/ml, while for S. gordonii, spectinomycin (aad9) or kanamycin (aphA3) was incorporated into Todd Hewitt medium at 250 μg/ml.
Bioinformatic analyses.
Open reading frames (ORFs) were identified in the annotated 2,196,662-bp genome of S. gordonii Challis CH1 (GenBank accession number CP000725). Putative signal sequence cleavage sites were determined using SignalP for Gram-positive bacteria (4), and putative functional domains were located via Pfam or BLAST searches (1) for regions of amino acid similarity. Conserved domains (designated by cd or smart numbers) were found in the NCBI Conserved Domain Database and Search Service (21). Putative promoters with −35 and −10 regions were identified with BPROM software available from Softberry (Mount Kisco, NY), and putative transcriptional terminator regions were identified with mfold (37).
DNA manipulations.
Restriction and modifying enzymes (Invitrogen, Roche) were used according to the manufacturer's instructions. Double-stranded PCR products (oligonucleotide primers are listed in Table S2 in the supplemental material) were routinely obtained using AmpliTaq polymerase (GeneAmp, Applied Biosystems). DNA fragments were purified from agarose using the QIAEXII kit (Qiagen). Plasmid DNA was extracted using the Qiagen plasmid mini kit. S. gordonii chromosomal DNA was extracted from TH broth cultures containing 0.5% (wt/vol) glycine by incubating cell pellets with mutanolysin and lysozyme as previously described (30). DNA fragments were cloned by digesting with appropriate restriction enzymes, ligating with T4 DNA ligase into convenient restriction sites in the vector, and then transforming into either CaCl2-competent E. coli DH5α or S. gordonii cells made competent (16) with horse serum.
For Southern hybridization analyses, HindIII-digested S. gordonii chromosomal DNA was electrophoresed on 0.7% (wt/vol) agarose gels and transferred to Hybond-N nylon membranes (Amersham Biosciences) by standard procedures. Double-stranded DNA probes prepared from the aad9 gene or the appropriate upstream amplicon from S. gordonii CH1 chromosomal DNA were labeled with digoxigenin-dUTP (Roche), hybridized to the membranes, washed under stringent conditions, and detected using chemiluminescence.
Construction of S. gordonii mutant strains.
Construction of DL1 strains deficient in expression in each of the genes SGO_2006, SGO_2005, and SGO_2004 was performed by allelic replacement. Internal fragments of the genetic loci at SGO_2005 and SGO_2004 (see Fig. S1 in the supplemental material) were each replaced by a 1,158-bp aad9 spectinomycin resistance gene (accession number M69221) with its own putative promoter and terminator regions. SGO_2006 was disrupted with a 913-bp aad9 gene with the putative promoter but lacking the terminator region to facilitate expression of the downstream ORFs. Plasmids carrying inserts of homologous DNA flanking the aad9 gene were constructed in E. coli. Amplicons homologous to S. gordonii CH1 chromosomal DNA up- and downstream of the region of replacement were amplified by PCR using primers engineered with convenient restriction sites (see Table S2 in the supplemental material).
A 392-bp region upstream of the region of replacement in SGO_2005 was amplified with primers Xho2005usF and Eco2005usR (see Table S2 in the supplemental material) and cloned into the XhoI and EcoRI sites of pGEM-7:spR (17), which carries the aad9 gene with its own putative promoter and terminator. The 752-bp downstream amplicon was generated with primers Hind2005dsF and Sst2005dsR (see Table S2) and cloned into the HindIII and SstI sites of pGEM-7:spR containing the upstream fragment, generating pGEM-7:spR-2005 (see Table S1). The pGEM-7:spRΔ2005 was digested with XhoI and SstI to generate a linear DNA fragment of 2,244 bp, which was purified and transformed into S. gordonii DL1. The DNA fragment was also transformed into S. gordonii UB1545 to generate a double mutant, UB2031, deficient in expression of PadA and Hsa. Transformants were selected on Todd-Hewitt agar medium containing 250 μg/ml spectinomycin. Chromosomal DNA extracts were analyzed by Southern hybridization to confirm the expected allelic replacement (see Fig. S1), which was verified by direct sequencing of PCR amplicons generated by oligonucleotide primers 2005tfJ and Sst2005dsR (see Table S2) that flanked the region of replacement. The premature translational stop codon generated resulted in a putative truncated protein with a molecular mass of approximately 9.6 kDa (see Table S3).
SGO_2004 was disrupted in a similar way, except that the downstream S. gordonii DL1 fragment was first cloned into pGEM-7:spR. A 404-bp downstream amplicon was generated with primers Hind2004dsF and Bam2004dsR (see Table S2 in the supplemental material) and cloned into pGEM-7:spR. The 346-bp upstream fragment was amplified with primers Xho2004usF and Mun2004usR (see Table S2) and was cloned into pGEM-7:spR containing the downstream amplicon to generate pGEM-7:spRΔ2004 (see Table S1). This was linearized by digestion with XhoI and BamHI to a fragment of 1,908 bp. Southern hybridization of Spr transformant DNA confirmed the replacement of 1,454 bp of SGO_2004 with the 1,158-bp aad9 gene, and the replacement was verified by sequencing. For disruption of SGO_2006, primers 5′-(TAGGATC)CATATCATATATAATCTAGAATA-3′ (forward) and 5′-(TACTTAAG)AATATTAAAAAAATTAGACAATAA-3′ (reverse) with engineered sequences (in parentheses) containing restriction sites (underlined) were used to amplify a 913-bp region of the aad9 gene. This was cloned into pBluescript SK II(+) (Invitrogen) to generate pBS:spRt− (see Table S1). A 320-bp fragment, upstream of the region of replacement within SGO_2006, was amplified with primers Sst2006usF and Bam2006usR (see Table S2) and cloned into pBS:spRt−. This amplicon contained a 68-bp intergenic region upstream of the ATG start codon of SGO_2006. A translational stop codon was engineered in the Bam2006usR primer. The 430-bp downstream amplicon (including the 66-bp region between SGO_2006 and SGO_2005, and 321 bp of SGO_2005) was generated with primers Eco1046dsF and Xho2006dsR (see Table S2). The amplicon was cloned into pBS:spRt containing the upstream fragment to generate pBS:spRt−Δ2006 (see Table S1). A linear fragment (1,663 bp) was then transformed into S. gordonii DL1. Southern hybridization of DNA from Spr transformants confirmed the replacement, and the construct was verified by sequencing.
Recombinant protein production.
DNA fragments (Fig. 1) were amplified by PCR using a proofreading enzyme (BioExact; Bioline) with the respective primers (see Table S2 in the supplemental material). The fragments (979 bp, 1,972 bp, and 4,186 bp) were inserted into the ligation-independent cloning vector pET46 (Novagen, Germany,) which incorporates an N-terminal His tag onto the expressed polypeptide. The plasmids were transformed into E. coli JM109, colonies screened by PCR, and purified plasmid constructs then transformed into the E. coli BL21 expression host. Recombinant proteins were expressed and purified as detailed previously (23), except for fragment I (326 amino acid [aa] residues) (Fig. 1) which was insoluble and formed inclusion bodies. To purify this polypeptide, IPTG (isopropyl-β-d-thiogalactopyranoside)-induced cells from 1 liter of culture were collected by centrifugation, incubated with 0. 35 mg lysozyme/ml and 1% Triton X-100, and then ultrasonicated. The extract was treated with 20 μg DNase/ml (37°C, 1 h), and after centrifugation at 15,000 × g for 20 min, the pellet was solubilized in 50 mM HEPES-NaOH (pH 7.5) containing 6 M guanidine-HCl and 25 mM dithiothreitol. The solubilized protein was then diluted into cold folding buffer containing 1 M NDSB201 (34). The remaining guanidine and NDSB were removed by dialysis against 50 mM HEPES-NaOH, pH 7.5. Polyclonal antibodies to fragment I were raised in rabbits (CovalAb UK Ltd., Cambridge).
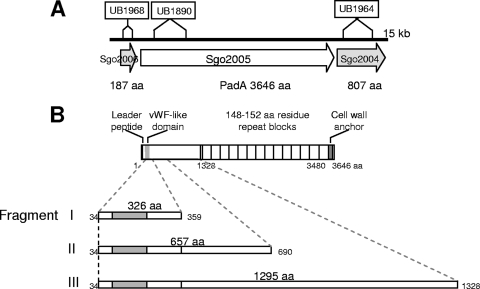
FIG. 1.
Diagrammatic representations of the S. gordonii padA locus and PadA polypeptide. (A) The locus shows sites of sequence replacement with an aad9 cassette (encoding spectinomycin resistance) to generate mutants UB1968 (SGO_2006), UB1980 (padA), and UB1964 (SGO_2004). (B) The primary sequence of PadA comprises a leader peptide (33 aa), a unique N-terminal region (1,295 aa) with a von Willebrand factor-like domain (shaded gray), 14-aa repeat blocks, and a C-terminal cell wall anchor region. For more detailed information about the gene identities at the padA locus and mutant construction, see Fig. S1 in the supplemental material. The complete amino acid sequence of PadA, delineating the features of the N-terminal region (including the vWf-like domain, aa 71 to 229) and the C-terminal amino acid repeat blocks, is provided in Fig. S2 in the supplemental material. Fragments I, II, and III depict N-terminal recombinant polypeptides expressed in E. coli and purified (see Fig. S3 in the supplemental material).
Protein analyses.
Cell wall proteins were extracted from S. gordonii following incubation with mutanolysin (11). These were separated on 8% polyacrylamide gels for 2.5 h at 120 V. Proteins were stained with silver nitrate. For Western immunoblot analysis, proteins were transferred onto Hybond nitrocellulose membranes (Amersham) at 100 V for 1 h. The membranes were probed with rabbit polyclonal antiserum against Streptococcus mutans SpaP polypeptide (cross-reacting with SspA and SspB) at a 1:1,000 dilution, and antibody binding was detected with peroxidase-linked swine anti-rabbit immunoglobulins and ECL (Amersham) (14). For Western immunoblot analysis of PadA, proteins were transferred onto Hybond membranes in cold transfer buffer (0.025 M Tris, 0.192 M glycine) containing10% methanol and 0.1% SDS by electroblotting for 16 h at 50 V. The membranes were then probed with rabbit polyclonal antiserum against S. gordonii PadA N-terminal protein fragment (fragment I) at a 1:250 dilution. Antibody binding was detected as before.
Determination of Hsa expression.
Levels of binding of succinylated wheat germ agglutinin-biotin (sWGA) (EY Laboratories, San Mateo, CA) to immobilized streptococcal cells were used to assess cell surface expression of Hsa as described previously (15).
Bacterial adherence to fetuin.
Fetuin (fetal calf; Sigma) was prepared in 20 mM Na2CO3-20 mM NaHCO3 (pH 9.3) and immobilized (0.4 μg) onto plastic well plates (Immulon) for 16 h at 4°C. Adherence of streptococcal cells (1 × 107 input) was then measured by crystal violet staining as previously described (12).
Platelet preparation.
Whole blood was drawn from the antecubital veins of healthy volunteers who had abstained from taking nonsteroidal anti-inflammatory drugs in the previous 10 days. Informed consent was obtained from all subjects. Nine volumes of blood were added to 1 volume of acid-citrate-dextrose (ACD) or 3.8% sodium citrate. Platelet-rich plasma (PRP) was prepared by centrifugation of anticoagulated whole blood at room temperature at 150 × g for 10 min. Platelets were separated from plasma proteins by gel filtration. PRP was adjusted to pH 6.5 with ACD, and apyrase (1 U/ml) and prostaglandin E1 (1 μM) were added prior to centrifugation at room temperature at 630 × g for 10 min. The resultant supernatant of platelet-poor plasma was removed, and the platelet pellet was suspended in 2 ml modified HEPES-Tyrodes (JNL) buffer (6 mM dextrose, 130 mM NaCl, 9 mM NaCl2, 10 mM Na citrate, 10 mM Tris base, 3 mM KCl, 0.8 mM KH2PO4, and 0.9 mM MgCl2). The platelet suspension was then applied to a chromatograph column containing 5 ml packed Sepharose 2B, which was previously equilibrated with JNL buffer. The resultant platelet fractions were pooled. The platelet concentration was adjusted to 2 × 108 platelets/ml on a Sysmex-100 particle counter (Sysmex, Japan).
Static platelet adhesion assay.
Microtiter plate wells were coated with 100 μl bacteria (optical density at 600 nm [OD600] = 1.0), fibrinogen (20 μg/ml), purified protein fragments (5 μg/ml), or bovine serum albumin (BSA) (1%) and platelet adherence measured by p-nitrophenol phosphate assay as described previously (10). In some experiments, platelets were incubated with inhibitors for 15 min prior to addition to the well plates. In antibody inhibition experiments, S. gordonii cells were incubated for 30 min with antibodies to PadA fragment I diluted at 1:10 or 1:100.
Platelet aggregation.
Platelet aggregation was assessed by monitoring light transmission using a PAP-4 platelet aggregometer (Bio/Data Corp., Horsham, PA). Platelets were tested for normal responses to thrombin receptor-activating peptide (TRAP; 5 μM) or ADP (20 μM). Bacteria were prepared as described previously (10), and 0.05 ml bacterial cells (OD600 = 1.6) were mixed with 0.45 ml platelet rich plasma (PRP). The light transmission of PRP without added bacteria and the light transmission of platelet-poor plasma were defined as 0% and 100% light transmission, respectively, and platelet aggregation was expressed as a final percentage of light transmission after 25 min. Five independent assays were performed for each strain.
Adherence to glycocalicin.
Soluble purified GPIbα (glycocalicin) (a kind gift from Jose Lopez, Puget Sound Blood Center, Seattle, WA) was diluted to 1 μg/ml in phosphate-buffered saline (PBS) and applied to 96-well enzyme-linked immunosorbent assay (ELISA) plates (Costar, Corning, NY) overnight at 4°C. After wells were washed and blocked with 1% BSA, FITC-labeled bacteria (15) were added to coated wells and incubated at 37°C for 45 min. Wells were then washed with PBS twice to remove unbound bacteria, and adherent bacteria were quantified at an excitation wavelength of 490 nm and an emission wavelength of 540 nm using a fluorescence plate reader (FLUOstar Galaxy).
Expression of GPIIbIIIa by CHO cells.
Glycoprotein Ibiza (GPIIbIIIa)-transfected Chinese hamster ovary (CHO) cells (a kind gift from Niamh Moran and Kelly Aylward, Royal College of Surgeons, Ireland) were maintained in Ham F-12 and Dulbecco's modified Eagle media supplemented with 10% fetal calf serum, 375 μg/ml Geneticin, and 250 μg/ml Zeocin. After 60 h, the cells were harvested with cell dissociation solution and adjusted to 4 × 106 cells. To examine the transfection efficiency, the CHO cells were incubated with 10 μg/ml phycoerythrin (PE)-labeled primary antibody (mouse anti-CD41, clone SZ22) or isotype control antibody for 20 min at room temperature. The samples were then diluted in 1 ml PBS and analyzed on a FACSCalibur flow cytometer (San Jose, CA) using CellQuest Pro software.
To measure adherence of CHO cells to S. gordonii, bacteria (OD600 = 1.0) were immobilized onto a 96-well ELISA plate for 2 h at 37°C. The plate was blocked with 1% BSA for a further 1 h. Mock-transfected (empty vector) or GPIIbIIIa-transfected CHO cells were added to the wells in triplicate and allowed to bind for 45 min at 37°C. Each well was gently washed three times with PBS to remove unbound CHO cells. Bound cells were then lysed with 0.1 ml lysis buffer containing acid phosphatase substrate (0.1 M Na acetate [pH 5.5], 0.1% Triton X-100, 10 mM p-nitrophenol phosphate) and incubated for 2 h at 37°C. The resultant color was read at 405 nm in a microtiter plate reader (Wallac Victor2; Perkin-Elmer).
Fluorescence microscopy.
Platelets were fluoresced with Alexa-Fluor 568 phalloidin for 20 min in the dark at room temperature. Fluorescent platelets were allowed to adhere to immobilized S. gordonii DL1 or UB1890 padA for 45 min in the dark at 37°C. Loose platelets were removed by gentle washing with Tris-buffered saline (TBS), pH 7.2. Slides were mounted for fluorescent visualization using a confocal microscope (LSM 510; Zeiss, United Kingdom). Images of stained cells were acquired using an argon laser at 488 nm.
Statistical analysis.
Statistics were performed using InStat statistical software (GraphPad Software). Data shown are the means ± standard errors of the means (SEM), and comparisons between mean values were performed using the Student paired or unpaired t test.
RESULTS
Structure of the S. gordonii padA locus.
As a result of annotation of the S. gordonii CH1 genome (GenBank accession number CP000725), 20 genes were identified as encoding putative cell wall-anchored polypeptides carrying LPxTG C-terminal motifs (29). One of these genes, originally designated SGO_2005, encoded a polypeptide with a predicted molecular mass of 397 kDa that was particularly interesting in terms of primary sequence structure (see below). It represented the largest open reading frame (ORF) on the S. gordonii chromosome. SGO_2005 formed part of a locus comprising three genes (SGO_2006, SGO_2005, and SGO_2004) (Fig. 1). A more detailed analysis of this locus together with upstream and downstream genes may be found in Fig. S1 in the supplemental material.
SGO_2006 encodes a thioredoxin signature protein (PF00085) with a putative signal sequence cleavage site within the N-terminal region (see Table S3 in the supplemental material). The SGO_2006 polypeptide was determined via BlastP to share 75% identity with S. gordonii CH1 SGO_1267, a thioredoxin family protein. The thioredoxin signature protein contains a conserved domain (cd02966), located at aa 56 to 172 and is a member of the TlpA-like protein family. Since TlpA is essential for biogenesis of cytochrome aa3 (18), SGO_2006 may play a role in cytochrome maturation. A putative promoter was identified 61 bp upstream of the start codon of SGO_2006 (see Fig. S1). No predicted terminator region was found in the 66-bp region between SGO_2006 and SGO_2005. SGO_2005 and SGO_2004 encode cell surface proteins, each with a putative leader peptide (signal sequence) within the N-terminal region (see Table S3) and an LPxTG cell wall anchorage motif followed by a hydrophobic region close to the C terminus.
SGO_2004, located 77 bp downstream of SGO_2005, encodes a protein that, on the basis of Pfam search, contains seven 78-aa G5 domain direct repeats (PF07501) spanning aa 145 to 630, the presence of which is suggestive of binding to N-acetylglucosamine (3). No putative promoters were identified upstream of SGO_2005 or SGO_2004. A putative transcriptional terminator region was identified downstream of SGO_2004 (see Fig. S1 in the supplemental material).
SGO_2005 encodes a protein with 31.5% identity to a putative cell wall-anchored protein in Enterococcus faecalis V583 (AAO80591). SGO_2005 carries a vWf type A (vWA) conserved domain (smart00327) between aa 75 and 229 (Fig. 1; see Fig. S2 in the supplemental material). Since vWf, a glycoprotein found in blood plasma, platelet α-granules, and subendothelial connective tissues, mediates adhesion of platelets to the endothelium (27) (see the introduction), it was hypothesized that SGO_2005 may play a role in binding platelets. Features of SGO_2005 (3,646 aa), later designated PadA (platelet adherence protein A), included a leader peptide (aa 1 to 33), a unique N-terminal region (aa 34 to 1328) containing a vWf-A1 like domain, and a C-terminal region (aa 3486 to 3646) containing a proline-rich sequence (aa 3553 to 3610), the sortase recognition motif LPKTG (aa 3614 to 3618), and charged a C terminus (see Fig. S2). Within the region spanning aa 1329 to 3485 there are 14 repeat blocks of amino acid residues, with the sequences within repeat blocks 1, 13, and 14 more divergent than the rest. Repeat blocks 1 to 12 each contain 148 to 152 amino acid residues, while repeat blocks 13 and 14 contain 176 and 179 amino acid residues, respectively. The C-terminal repeat region appears to have originated through duplication of two blocks each of 755 amino acid residues, aa 1479 to 2230 (RP1) and aa 2231 to 2982 (RP2), each consisting of five 148- to 152-aa repeat blocks (see Fig. S2). There were no significant sequence similarities to these repeat blocks following BlastP search of all the sequenced microbial genomes. The primary sequence of PadA highlighting all the features described above is provided in Fig. S2 in the supplemental material.
Cell wall proteins of S. gordonii isogenic mutants.
To determine the function of PadA in adherence properties of S. gordonii, isogenic mutants were generated in strain DL1 by allelic replacement of 1,960 bp within the N-terminal coding region of padA (nucleotides 479 to 2438) with the 1158-bp aad9 gene encoding spectinomycin resistance (Spr). A number of SGO_2005 mutants were generated from three independent transformation experiments. We included three of these independent mutants in various experiments, and they all were shown to have identical phenotypes. One that was representative of these, designated UB1890, was used throughout (Fig. 1; see Table S1 in the supplemental material). S. gordonii surface protein Hsa has been previously shown to play a critical role in inducing platelet aggregation (15); therefore, a double mutant (UB2031) lacking expression of both Hsa and PadA was generated by allelic exchange with the same DNA construct fragment as used to generate the padA mutant UB1890 (see Table S1). Mutants were also generated by allelic replacements in downstream gene SGO_2004 and in upstream gene SGO_2006, the latter replacement being with aad9 without a transcriptional terminator so as to avoid possible polar effects on downstream padA gene expression (Fig. 1).
The levels of expression of mRNAs from SGO_2005 (padA) and SGO_2004 (downstream of padA) were measured, by real-time quantitative PCR, for strains DL1, UB1968 Δ(SGO_2006), UB1890 Δ(SGO_2005), and UB1964 Δ(SGO_2004) in the mid-exponential phase of growth. There were no significant differences in SGO_2005 expression between DL1, UB1968, and UB1964, confirming that the aad9 (terminator-less) replacement into SGO_2006 (UB1968) did not affect expression levels of padA (see Table S4 in the supplemental material). As expected, there was reduced expression of SGO_2005 (padA) in UB1890 (see Table S4). Residual mRNA expression levels in UB1890 may be as a result of aad9 read-through. Expression levels of SGO_2004 were reduced in UB1890, showing that the allelic replacement within SGO_2005 had a downstream effect (see Table S4), while in UB1968 expression of SGO_2004 was unaffected.
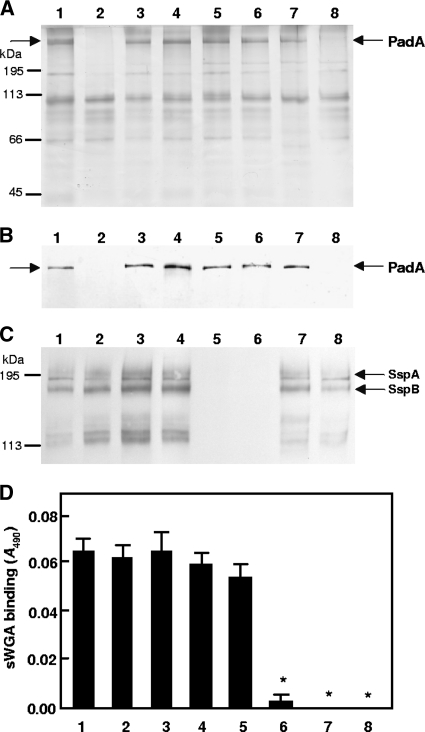
Cell wall protein profiles of wild-type and mutant strains were compared by SDS-PAGE and staining with silver nitrate. S. gordonii DL1 extracts contained a range of polypeptides, as described previously (13), and within the high-molecular-mass protein region of the gel a prominent band at ∼390 kDa, corresponding to PadA, was evident (Fig. 2A). This band was absent in extracts of strain UB1890 ΔpadA (Fig. 2A, lane 2) and UB2031 ΔpadA hsa (Fig. 2A, lane 8), but it was present in extracts from isogenic mutants UB1964 (SGO_2004::aad9) and UB1968 (SGO_2006::aad9) (Fig. 2A, lanes 3 and 4). Polyclonal antibodies generated to the N-terminal region fragment I of PadA (Fig. 1) reacted with the 390-kDa band (Fig. 2B) except in those strains carrying the padA mutation (Fig. 2B, lanes 2 and 8), confirming the identity of PadA.
FIG. 2.
Phenotypic properties of S. gordonii mutants deficient in production of proteins encoded by padA, sspAB, or hsa. (A) SDS-PAGE of proteins extracted from the cell wall of S. gordonii and stained with silver nitrate; (B) corresponding Western blot reacted with polyclonal antibody to PadA fragment I (Fig. 1), which was specific for PadA; (C) corresponding Western blot reacted with polyclonal antibody to S. mutans antigen I/II (cross-reactive with SspA and SspB); (D) cell surface Hsa present on immobilized S. gordonii cells as detected by sWGA-biotin and horseradish peroxidase (HRP)-streptavidin. *, P < 0.0001. Lanes and bars: 1, wild-type DL1; 2, UB1890 padA::aad9; 3, UB1964 SGO_2004::aad9; 4, UB1968 SGO_2006::aad9; 5, UB1360 Δ(sspA sspB)::aad9; 6, UB1552 hsa::aphA3 Δ(sspAsspB)::aad9; 7, UB1545 hsa::aphA3; 8, UB2031 hsa::aphA3 padA::aad9. Positions of molecular mass protein markers are indicated in panels A and C. Error bars indicate SEM.
The antigen I/II family genes sspA and sspB have been shown to play a role in inducing platelet aggregation (15). To determine if mutations in the sspA and sspB genes or in the hsa gene, singly or in combination, affected PadA production, cell wall extracts of strains UB1360 Δ(sspA sspB), UB1545 hsa, and UB1552 Δ(sspA sspB) hsa were compared for presence of PadA by SDS-PAGE and staining of proteins with silver nitrate. None of these mutants were shown to be affected in PadA production (Fig. 2A, lanes 5 to 7).
Mutants UB1890 and UB2031, in which the padA gene was deleted, were unaffected in expression of SspA and SspB as determined by Western immunoblot analysis (Fig. 2C, lanes 2 and 8). They were also unaffected in surface expression of Hsa as determined by biotinylated succinylated wheat germ agglutinin assay (Fig. 2D, lanes 2 and 8).
PadA does not mediate adherence to fetuin.
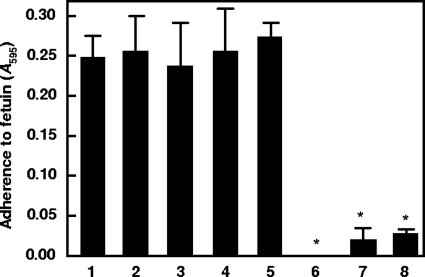
To investigate the ability of PadA to bind to sialylated residues on host receptors, we immobilized fetuin, a highly sialylated human plasma protein, onto a 96-well ELISA plate and measured adhesion of the wild-type strain DL1 and the various mutants constructed. Wild-type S. gordonii DL1 cells adhered to fetuin (Fig. 3), and isogenic mutant strains UB1890 padA, UB1964 (SGO_2004::aad9), UB1968 (SGO_2006::aad9), and UB1360 ΔsspA sspB were all similar to strain DL1 in their levels of adherence to fetuin (Fig. 3). However, deletion of the hsa gene resulted in a significant reduction in adhesion, as seen in the UB1545 hsa mutant (Fig. 3, bar 7) (P < 0.0001), the UB1552 hsa Δ(sspA sspB) mutant, and the UB2031 hsa padA mutant (Fig. 3, bar 8) (P < 0.0001). These results confirm that Hsa is a major mediator of sialic acid recognition in S. gordonii and show that the padA mutation has no effect on recognition of sialic acid by streptococci.
FIG. 3.
Adherence of S. gordonii DL1 and isogenic mutants to fetuin. Bacterial cells (2 × 107) were incubated with immobilized fetuin (0.5 μg), and adherence was measured by crystal violet staining assay (A595). Bars: 1, wild-type DL1; 2, UB1890 padA::aad9; 3, UB1964 SGO_2004::aad9; 4, UB1968 SGO_2006::aad9; 5, UB1360 Δ(sspAsspB)::aad9; 6, UB1552 hsa::aphA3 Δ(sspAsspB)::aad9; 7, UB1545 hsa::aphA3; 8, UB2031 hsa::aphA3 padA::aad9. *, P < 0.0001. Error bars indicate SEM.
PadA mediates interactions of S. gordonii with human platelets.
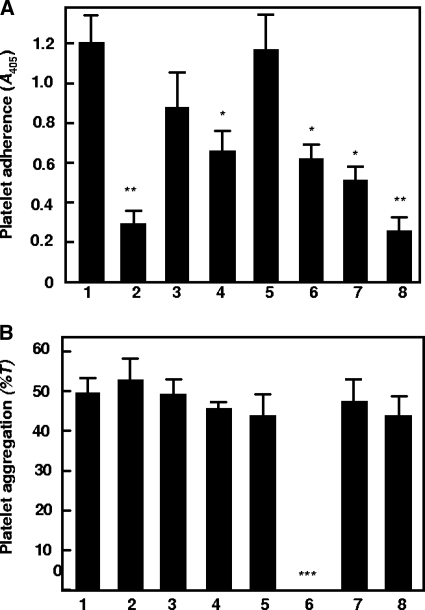
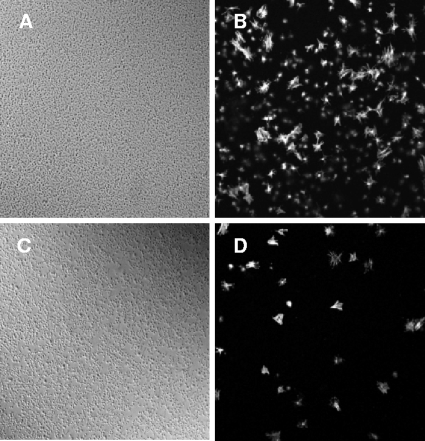
Platelet adhesion to immobilized bacteria is thought to be an initial step leading to thrombus formation. Hsa protein plays an essential role in supporting platelet adhesion (12). Disruption of the hsa gene resulted in approximately 50% reduced levels of platelet adhesion, whereas deletion of the sspA and sspB genes had no effect. Platelet adhesion to immobilized S. gordonii is calcium independent and occurs in the absence of plasma proteins (15), suggesting that there is a direct interaction between S. gordonii and a specific platelet receptor. Deletion of the padA gene resulted in significantly reduced (P < 0.001) platelet adhesion (Fig. 4A, bar 2). Although levels of platelet adhesion to the isogenic mutants UB1964 (SGO_2004::aad9) and UB1968 (SGO_2006::aad9) were also reduced, only the latter reduction was significant (P < 0.05) (Fig. 4A). Consistent with previous findings, platelet adhesion to UB1360 Δ(sspA sspB) remained unaffected, while platelet adhesion to UB1545 Δhsa and to the double mutant UB1552 Δ(sspA sspB hsa) was reduced (Fig. 4A, bars 6 and 7) (P < 0.05). The double mutant UB2031 Δhsa ΔpadA was reduced in platelet adhesion to a similar level as UB1890 (Fig. 4A, bar 8). Thus, both Hsa and PadA are involved in mediating platelet adhesion. However, since platelet adhesion to UB2031 was not completely abrogated, it is suggested that there may be factors in addition to Hsa and PadA that play a role in supporting adhesion, thus highlighting the complexity of the adhesion interaction. The requirement for PadA in platelet adherence was also confirmed microscopically. Fluorescently labeled platelets adhered in high numbers to immobilized cells of S. gordonii DL1 (Fig. 5A and B). However, levels of adherence of platelets to S. gordonii UB1890 padA were at least 10-fold less than those to S. gordonii DL1 (Fig. 5C to D).
FIG. 4.
Interactions of S. gordonii DL1 and isogenic mutants with human platelets. (A) Platelets were incubated with immobilized S. gordonii cells and platelet adherence measured by acid phosphatase activity (A405) as described in Materials and Methods. (B) Platelet-rich plasma (PRP) was incubated with bacterial cells in an aggregometer and aggregation expressed as a final percentage of light transmission (T). Bars: 1, wild-type DL1; 2, UB1890 padA::aad9; 3, UB1964 SGO_2004::aad9; 4, UB1968 SGO_2006::aad9; 5, UB1360 Δ(sspAsspB)::aad9; 6, UB1552 hsa::aphA3 Δ(sspAsspB)::aad9; 7, UB1545 hsa::aphA3; 8, UB2031 hsa::aphA3 padA::aad9. ***, P < 0.0001; **, P < 0.001; *, P < 0.05. Error bars indicate SEM.
FIG. 5.
Interaction of S. gordonii and recombinant PadA polypeptides with platelets. (B and D) Fluorescently labeled platelets are shown attached to immobilized S. gordonii DL1 wild-type bacteria (B) or to S. gordonii UB1890 padA mutant bacteria (D). (A and C) Phase-contrast microscopic images of slides coated with strain DL1 (A) or UB1890 (C).
To further characterize the adhesive interactions between S. gordonii and platelets, the effects of some platelet receptor inhibitors were investigated. Platelet adhesion to immobilized S. gordonii DL1 cells was reduced by 49% ± 1% (n = 3) by preincubation of platelets with monoclonal antibody MB45 against the α chain of GPIb. Furthermore, platelet adhesion to immobilized strain DL1 cells was also significantly reduced (54% ± 1%; n = 3) by addition of Abciximab (ReoPro), a potent and specific inhibitor of GPIIbIIIa (data not shown). Preincubation of platelets with AN51 antibody against GPIb receptor, together with Abciximab, reduced levels of platelet adhesion by 45% ± 3% (n = 3) (data not shown). These observations support the evidence that S. gordonii interacts directly with platelets through recognition of receptors GPIb and GPIIbIIIa.
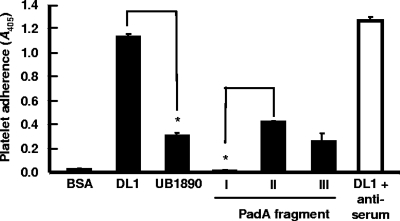
Platelets adhered to immobilized fragments II and III of PadA containing the complete N terminus and vWA domain; however, they failed to adhere to PadA fragment I containing the N-terminal 326 amino acid residues (Fig. 6). Antibodies to fragment I did not block interactions of S. gordonii DL1 with platelets (Fig. 6). It is possible that platelets do not bind fragment I because of conformational differences between the native protein and the purified protein fragment.
FIG. 6.
Adherence of platelets to immobilized recombinant PadA fragments I, II, and III (Fig. 1) and effect of preincubation of S. gordonii DL1 cells with antiserum to the N-terminal fragment I of PadA on subsequent adherence to platelets (see Materials and Methods). *, P < 0.001. Error bars indicate SEM.
Platelet aggregation by S. gordonii cells involves the concerted activities of Hsa, SspA, and SspB proteins (15). All S. gordonii strains, including UB1890 padA, were found to aggregate platelets, except for strain UB1552 hsa Δ(sspA sspB), as expected (Fig. 4B, bar 6). Aggregation occurred in an all-or-nothing manner. Increasing the concentration of bacteria reduced the lag time for first commencement of aggregation but not the maximum level of aggregation. Overall, the data suggest that PadA is important for platelet adhesion and that it does not play a role in inducing platelet aggregation.
S. gordonii PadA does not recognize purified GPIbα.
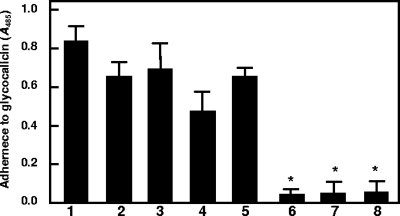
PadA contains a short amino acid residue stretch similar in conformation to the A1 domain of vWf. The A1 domain of vWf contains the recognition site for platelet GPIbα. Therefore, we tested the ability of S. gordonii strains to adhere to glycocalicin (soluble GPIbα). S. gordonii wild-type DL1, UB1890 padA, UB1964 (SGO_2004::aad9), and UB1360 Δ(sspA sspB) all bound glycocalicin to similar degrees (Fig. 7). Strain UB1968 (SGO_2006::aad9) showed partly reduced adherence of glycocalicin (Fig. 7, bar 4). However, all strains that were disrupted in hsa, viz., UB1545, UB1552, and UB2031, showed abrogated binding of glycocalicin (Fig. 7). The latter results are consistent with previous reports in the literature demonstrating that Hsa binds to GPIbα (5, 15). These observations suggest that although PadA contains a region similar to the vWf A1 domain, the PadA protein does not recognize GPIbα.
FIG. 7.
Adherence of S. gordonii DL1 and isogenic mutants to purified glycocalicin. Human platelet glycocalicin (the external portion of platelet receptor GPIb) was immobilized onto plastic wells and attachment of FITC-labeled bacteria quantified at excitation and emission wavelengths of 490 nm and 540 nm, respectively, using a fluorescence plate reader. Bars: 1, wild-type DL1; 2, UB1890 padA::aad9; 3, UB1964 SGO_2004::aad9; 4, UB1968 SGO_2006::aad9; 5, UB1360 Δ(sspAsspB)::aad9; 6, UB1552 hsa::aphA3 Δ(sspAsspB)::aad9; 7, UB1545 hsa::aphA3; 8, UB2031 hsa::aphA3 padA::aad9. *, P < 0.001. Error bars indicate SEM.
S. gordonii PadA binds to platelet receptor GPIIbIIIa.
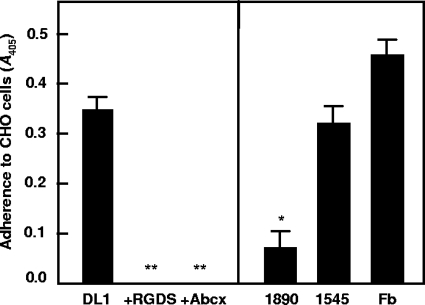
Several domains on vWf, including the C1 domain (19) and the A1 domain (22), have been shown to bind to GPIIbIIIa. Chinese hamster ovary (CHO) cells naturally express the beta 3 subunit (GPIIIa); however, they lack the alpha IIb subunit (GPIIb) to make up the GPIIbIIIa complex. Stable transfection of CHO cells with the gene encoding the GPIIb subunit resulted in a functional GPIIbIIIa receptor being expressed on the cell surface, as determined by fluorescence-activated cell sorter (FACS) analysis and Western immunoblot analysis (see Fig. S4 in the supplemental material). Binding of mock-transfected (empty vector) CHO cells to S. gordonii wild-type or mutant cells was at basal levels, whereas GPIIb-transfected cells adhered to strain DL1 cells at similar levels to those for fibrinogen, the natural receptor for GPIIbIIIa (Fig. 8). Addition of the integrin-blocking peptide RGDS or of Abciximab (ReoPro) completely abrogated binding of CHO cells expressing GPIIbIIIa to S. gordonii DL1 (Fig. 8). Strain UB1890 padA cells were >80% reduced in binding GPIIbIIIa (P < 0.01), while UB1545 hsa cells were unaffected in binding (Fig. 8). Taken collectively, these data suggest that PadA represents a novel bacterial polypeptide that interacts specifically with platelet integrin receptor GPIIbIIIa.
FIG. 8.
Adherence of CHO cells expressing GPIIbIIIa to S. gordonii wild-type and relevant isogenic mutant strains. (A) Adherence to strain DL1 cells and effects of integrin inhibitor RGDS and GPIIbIIIa inhibitor Abciximab (Abcx) on adherence; (B) Adherence to strains UB1890 padA and UB1545 hsa and to fibrinogen (Fb). **, P < 0.001; *, P < 0.01. Error bars indicate SEM.
DISCUSSION
It is now well established that a range of viridians group oral streptococci, including S. gordonii, have the ability to adhere to and induce platelet aggregation. Under normal hemostatic conditions, platelet adhesion is a very separate event from platelet aggregation, because the protein-protein interactions involved in mediating platelet adhesion are distinct from the interactions that mediate platelet aggregation. Many attempts to decipher the mechanisms through which oral streptococci support platelet adhesion or induce platelet aggregation have been made, and now a picture similar to that of normal hemostasis is emerging.
In the present study we have identified a novel protein on the surface of S. gordonii which we have designated PadA. Disruption of the padA gene had no effect on Streptococcus-induced platelet aggregation but significantly reduced platelet adhesion. This reduction in platelet adhesion was not related to any secondary effect on Hsa production, since Hsa expression levels in cell wall protein profiles, and levels of adherence to sialic acid substrates, were unaffected in the padA mutant strain. We do not describe complementation of the padA mutant strain, because it has not yet been possible to clone the entire 11-kb coding region into a plasmid. However, a number of padA mutants obtained from three independent transformation experiments showed similar phenotypes. All the evidence, including direct binding of the PadA N-terminal region to platelets, indicates that the mutant phenotype is a direct result of padA disruption.
PadA contains a short stretch of amino acid residues which displays only weak homology to either the A1 or C1 domain of vWf (see Fig. S5 in the supplemental material). These domains in vWf are essential for the interaction with platelet GPIbα and GPIIbIIIa, respectively. GPIb recognizes the A1 domain of vWf at Arg1334 (under shear conditions). GPIIbIIIa recognizes the C1 domain of vWf at Arg/Gly/Asp 2507/2508/2509 (RGD sequence).
We initially investigated whether PadA was binding to the vWf receptor, GPIbα. This is the second most abundant platelet receptor, with roughly 25,000 copies per platelet. It is part of the leucine-rich repeat family of proteins and contains binding sites for the vWf A1 domain, thrombin, P-selectin, and many other proteins. The crucial importance of GPIb and vWf in hemostasis is due to the fact that at higher shear rates (arterial shear) (25), GPIb interacting with the vWf A1 domain is able to efficiently capture platelets from flowing blood at sites of vessel wall injury, firmly adhere, and initiate thrombus formation. Firm platelet adhesion, which occurs only when the platelets are slowed down, is an integrin-mediated process. Previously we have shown that S. gordonii DL1 binds to purified GPIb (glycocalicin), an interaction that is dependent on Hsa. Recent work has characterized this interaction and demonstrated that Hsa binds specifically to the N-linked sialic acid residues on GPIbα. This interaction was further characterized (15), and it was found that under conditions of shear, platelets rolled along immobilized bacteria in an interaction that was characteristic of platelets rolling on immobilized vWf at sites of vascular damage. Disruption of the hsa gene in S. gordonii DL1 led to abrogation of platelet rolling, thereby preventing firm adhesion (15).
Disruption of the padA gene did not affect binding levels of glycocalicin (Fig. 7) suggesting that the weak homology seen between the N-terminal region of PadA and the vWF A1 domain is not significant in a GPIb interaction. However, platelets adhered to purified protein fragments II and III of PadA containing the vWf A1-like domain, although this does not appear to be mediated through an interaction with GPIb. The possibility that C-terminal repeats of PadA also interact with platelets is currently under study. Inactivation of the gene SGO_2006 upstream of padA (Fig. 1) led to reduced binding of glycocalicin and of platelets by strain UB1968. The reason for this is not clear, because expression of padA (see Table S4 in the supplemental material) and levels of PadA protein (Fig. 2) produced by UB1968 were similar to those for the wild-type strain DL1. However, it might be that the gene product of SGO_2006 modulates activity of Hsa, and this possibility is also under investigation.
PadA, unlike Hsa, does not appear to recognize sialic acid residues on the platelet glycoprotein GPIb. Fetuin is a highly sialyated human glycoprotein, with well-characterized carbohydrate chains that include sialic acid linked in both α(2,3) and α(2,6). This protein has been extensively used in previous work to simulate binding of Hsa and GspB to platelets (5, 33) Only mutants deficient in Hsa production showed reductions in binding to fetuin and glycocalicin.
GPIIbIIIa is the most abundant receptor on the platelet surface, with over 80,000 copies per platelet (35). It represents 3% of total platelet protein and 20% of the platelet membrane protein mass. GPIIbIIIa can bind multiple ligands, including extracellular matrix proteins fibrinogen, fibronectin, vitronectin, thrombospondin, and vWf. Binding to these ligands results in firm platelet adhesion at sites of thrombus formation. GPIIbIIIa recognizes the short peptide sequence RGD, which is present in many GPIIbIIIa ligands. Both fibrinogen and vWf, ligands that support platelet adhesion under physiological shear, contain RGD sequences (present in the α-chain and C1 domain, respectively). Platelet adhesion to immobilized strain DL1 cells was significantly reduced by addition of an RGD-containing peptide (100%, n = 3) or Abciximab (54% ± 1%, n = 3), a potent and specific inhibitor of GPIIbIIIa (data not shown), further suggesting a role for integrin and more specifically GPIIbIIIa in interaction with S. gordonii.
CHO cells naturally express the integrin beta 3 subunit (GPIIIa); however, they lack the alpha IIb (GPIIb) subunit to make up the GPIIbIIIa complex. CHO cells stably transfected with the gene encoding the GPIIb subunit resulted in a functional GPIIbIIIa receptor being expressed on the surface. These cells, stably transfected with GPIIbIIIa, were utilized to further investigate the interactions with S. gordonii. The mock-transfected cells (empty vector) were not bound by S. gordonii, but the CHO cells expressing GPIIbIIIa were (Fig. 8). Deletion of padA, but not disruption of hsa, reduced binding of GPIIbIIIa-expressing CHO cells by >80%. Preincubation of CHO GPIIbIIIa cells with a monoclonal antibody against GPIIbIIIa (Abciximab) or the RGD mimetic inhibitor eptifibatide (integrelin) led to complete abrogation of GPIIbIIIa binding by streptococci, emphasizing the specific nature of the interaction between PadA and GPIIbIIIa. This clearly demonstrates new evidence for a direct interaction between a bacterial protein and GPIIbIIIa. Furthermore it also suggests that another, as-yet-unidentified binding site, other than the RGD binding site, exists on GPIIbIIIa.
Although it has been shown that Staphylococcus aureus clumping factor A and fibronectin binding protein A bound to GPIIbIIIa, this occurred via a fibrinogen and specific antibody bridge (20). In addition, it was demonstrated that S. gordonii DL1 bound specifically to GPIbα and to GPIIb, but not to GPIIIa, on affinity blots. However, the bacterial protein responsible for binding to GPIIb was not identified (32). The results presented in this paper strongly suggest that this protein is PadA.
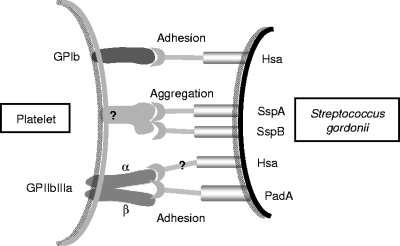
Our results indicate that there are multiple interactions involved in supporting platelet adhesion (Fig. 9). We suggest that platelets roll along immobilized S. gordonii cells, an interaction that primarily involves platelet GPIb interacting with S. gordonii Hsa. This interaction requires a high on-off rate through which the loss of GPIb receptor interactions at one side of the platelet and the generation of GPIb receptor interactions at the other side of the platelet lead to a typical rolling phenomenon. This slows the platelet down long enough to trigger a second interaction that leads to firm adhesion. Firm adhesion is completed when the platelet integrin GPIIbIIIa interacts with S. gordonii PadA through a much slower interaction. Our previous results demonstrated that deletion of Hsa from the surface of S. gordonii prevents any interaction with the platelet (15). This may be explained because in the absence of the platelet rolling phenomenon, the platelet cannot slow down long enough for firm attachment to be mediated by S. gordonii PadA. Thus, S. gordonii beguiles platelets to engage in normal hemostasis. The discovery of this protein that interacts directly with GPIIbIIIa provides new reagents for studies of GPIIbIIIa function and the potential for development of new antithrombotic inhibitors.
FIG. 9.
Diagrammatic representation of the partner molecules involved in the interactions between S. gordonii and human platelets leading to potential thrombus formation. Streptococcus surface glycoprotein Hsa interacts with sialic acid residues present on platelet glycoprotein GPIb mediating adherence and slowing of platelets (rolling) under shear. Antigen I/II family proteins SspA and SspB act in concert with Hsa to mediate platelet aggregation, although the platelet receptors for SspA and SspB are currently unknown. Streptococcus surface protein PadA interacts directly with platelet integrin GPIIbIIIa, also a receptor for fibrinogen and vWf, in an interaction that is blocked by inhibitors RGDS and Abciximab (ReoPro). There is also evidence that S. gordonii glycoprotein Hsa may interact with the α subunit of GPIIbIIIa (36).
Supplementary Material
Acknowledgments
We are grateful to Lindsay Dutton and Jane Brittan for excellent technical assistance and to Angela Nobbs and Sarah Maddocks for help with artwork and sequence analyses. We thank Nick Jakubovics, Jose Lopez, Niamh Moran, and Kelly Aylward for provision of strains and reagents.
This work was supported by a Wellcome Trust Research Training Fellowship (no. 084979) awarded to H.J.P. and by a Health Board of Ireland grant (no. RP/2006/211) awarded to S.W.K. and H.F.J.
Editor: A. Camilli
Footnotes
Published ahead of print on 2 November 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Barrau, K., A. Boulamery, G. Imbert, J. P. Casalta, G. Habib, T. Messana, J. L. Bonnet, E. Rubinstein, and D. Raoult. 2004. Causative organisms of infective endocarditis according to host status. Clin. Microbiol. Infect. 10:302-308. [DOI] [PubMed] [Google Scholar]
- 3.Bateman, A., M. T. G. Holden, and C. Yeats. 2005. The G5 domain: a potential N-acetylglucosamine recognition domain involved in biofilm formation. Bioinformatics 21:1301-1303. [DOI] [PubMed] [Google Scholar]
- 4.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 5.Bensing, B. A., J. A. Lopez, and P. A. Sullam. 2004. The Streptococcus gordonii surface proteins GspB and Hsa mediate binding to sialylated carbohydrate epitopes on the platelet membrane glycoprotein Ibα. Infect. Immun. 72:6528-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beynon, R. P., V. K. Bahl, and B. D. Prendergast. 2006. Infective endocarditis. BMJ 333:334-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demuth, D. R., Y. Duan, H. F. Jenkinson, R. Mcnab, S. Gil, and R. J. Lamont. 1997. Interruption of the Streptococcus gordonii M5 sspA/sspB intergenic region by an insertion sequence related to IS1167 of Streptococcus pneumoniae. Microbiology 143:2047-2055. [DOI] [PubMed] [Google Scholar]
- 8.Fitzgerald, J. R., T. J. Foster, and D. Cox. 2006. The interaction of bacterial pathogens with platelets. Nat. Rev. Microbiol. 4:445-457. [DOI] [PubMed] [Google Scholar]
- 9.Herzberg, M. C., G. D. Macfarlane, K. E. Gong, N. N. Armstrong, A. R. Witt, P. R. Erickson, and M. W. Meyer. 1992. The platelet interactivity phenotype of Streptococcus sanguis influences the course of experimental endocarditis. Infect. Immun. 60:4809-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakubovics, N. S., S. W. Kerrigan, A. H. Nobbs, N. Stromberg, C. J. van Dolleweerd, D. M. Cox, C. G. Kelly, and H. F. Jenkinson. 2005. Functions of cell surface-anchored antigen I/II family and Hsa polypeptides in interactions of Streptococcus gordonii with host receptors. Infect. Immun. 73:6629-6638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jakubovics, N. S., A. W. Smith, and H. F. Jenkinson. 2000. Expression of the virulence-related Sca (Mn2+) permease in Streptococcus gordonii is regulated by a diphtheria toxin metallorepressor-like protein ScaR. Mol. Microbiol. 38:140-153. [DOI] [PubMed] [Google Scholar]
- 12.Jakubovics, N. S., N. Stromberg, C. J. van Dolleweerd, C. G. Kelly, and H. F. Jenkinson. 2005. Differential binding specificities of oral streptococcal antigen I/II family adhesins for human or bacterial ligands. Mol. Microbiol. 55:1591-1605. [DOI] [PubMed] [Google Scholar]
- 13.Jenkinson, H. F. 1986. Cell-surface proteins of Streptococcus sanguis associated with cell hydrophobicity and coaggregation properties. J. Gen. Microbiol. 132:1575-1589. [DOI] [PubMed] [Google Scholar]
- 14.Jenkinson, H. F., S. D. Terry, R. Mcnab, and G. W. Tannock. 1993. Inactivation of the gene encoding surface protein SspA in Streptococcus gordonii DL1 affects cell interactions with human salivary agglutinin and oral actinomyces. Infect. Immun. 61:3199-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerrigan, S. W., N. S. Jakubovics, C. Keane, P. Maguire, K. Wynne, H. F. Jenkinson, and D. Cox. 2007. Role of Streptococcus gordonii surface proteins SspA/SspB and Hsa in platelet function. Infect. Immun. 75:5740-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawson, J. W., and H. Gooder. 1970. Growth and development of competence in group H streptococci. J. Bacteriol. 102:820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leblanc, D. J., L. N. Lee, and J. M. Inamine. 1991. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase aad9 determinant from Enterococcus faecalis. Antimicrob. Agents Chemother. 35:1804-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loferer, H., M. Bott, and H. Hennecke. 1993. Bradyrhizobium japonicum TlpA, a novel membrane anchored thioredoxin-like protein involved in the biogenesis of cytochrome aa3 and development of symbiosis. EMBO J. 12:3373-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lombardo, V. T., E. Hodson, J. R. Roberts, T. J. Kunicki, T. S. Zimmerman, and Z. M. Ruggeri. 1985. Independent modulation of von Willebrand factor and fibrinogen binding to the platelet membrane glycoprotein IIb/IIIa complex as demonstrated by monoclonal antibody. J. Clin. Invest. 76:1950-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loughman, A., J. R. Fitzgerald, M. P. Brennan, J. Higgins, R. Downer, D. Cox, and T. J. Foster. 2005. Roles for fibrinogen, immunoglobulin and complement in platelet activation promoted by Staphylococcus aureus clumping factor A. Mol. Microbiol. 57:804-818. [DOI] [PubMed] [Google Scholar]
- 21.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWweese-Scott, L. Y. Geer, M. Gwadz, S. Q. He, D. I. Hurwitz, J. D. Jackson, Z. X. Ke, C. J. Lanczycki, C. A. Liebert, C. L. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. C. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishio, K., Y. Fujimura, S. Nishida, I. Takeda, A. Yoshioka, H. Fukui, Y. Tomiyama, and Y. Kurata. 1991. Antiplatelet glycoprotein-Ib monoclonal antibody (Op-F1) totally abolishes ristocetin-induced von Willebrand factor binding, but has minimal effect on the botrocetin-induced binding. Haemostasis 21:353-359. [DOI] [PubMed] [Google Scholar]
- 23.Nobbs, A. H., B. H. Shearer, M. Drobni, M. A. Jepson, and H. F. Jenkinson. 2007. Adherence and internalization of Streptococcus gordonii by epithelial cells involves β1 integrin recognition by SspA and SspB (antigen I/II family) polypeptides. Cell. Microbiol. 9:65-83. [DOI] [PubMed] [Google Scholar]
- 24.Ruggeri, Z. M. 2003. Von Willebrand factor, platelets and endothelial cell interactions. J. Thromb. Haemost. 1:1335-1342. [DOI] [PubMed] [Google Scholar]
- 25.Ruggeri, Z. M. 2009. Platelet adhesion under flow. Microcirculation 16:58-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruggeri, Z. M., and G. L. Mendolicchio. 2007. Adhesion mechanisms in platelet function. Circ. Res. 100:1673-1685. [DOI] [PubMed] [Google Scholar]
- 27.Sadler, J. E. 1998. Biochemistry and genetics of von Willebrand factor. Annu. Rev. Biochem. 67:395-424. [DOI] [PubMed] [Google Scholar]
- 28.Savage, B., S. J. Shattil, and Z. M. Ruggeri. 1992. Modulation of platelet function through adhesion receptors. A dual role for glycoprotein IIb-IIIa (integrin αIIbβ3) mediated by fibrinogen and glycoprotein Ib-von Willebrand factor. J. Biol. Chem. 5:11300-11306. [PubMed] [Google Scholar]
- 29.Schneewind, O., D. Mihaylovapetkov, and P. Model. 1993. Cell-wall sorting signals in surface proteins of gram-positive bacteria. EMBO J. 12:4803-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sulavik, M. C., and D. B. Clewell. 1996. Rgg is a positive transcriptional regulator of the Streptococcus gordonii gtfG gene. J. Bacteriol. 178:5826-5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi, Y., A. L. Sandberg, S. Ruhl, J. Muller, and J. O. Cisar. 1997. A specific cell surface antigen of Streptococcus gordonii is associated with bacterial hemagglutination and adhesion to α2-3-linked sialic acid-containing receptors. Infect. Immun. 65:5042-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi, Y., A. Yajima, J. O. Cisar, and K. Konishi. 2004. Functional analysis of the Streptococcus gordonii DL1 sialic acid-binding adhesin and its essential role in bacterial binding to platelets. Infect. Immun. 72:3876-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takamatsu, D., B. A. Bensing, H. Cheng, G. A. Jarvis, I. R. Siboo, J. A. Lopez, J. M. Griffiss, and P. M. Sullam. 2005. Binding of the Streptococcus gordonii surface glycoproteins GspB and Hsa to specific carbohydrate structures on platelet membrane glycoprotein Ibα. Mol. Microbiol. 58:380-392. [DOI] [PubMed] [Google Scholar]
- 34.Vuillard, L., C. Braunbreton, and T. Rabilloud. 1995. Non-detergent sulfobetaines—a new class of mild solubilization agents for protein purification. Biochem. J. 305:337-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner, C. L., M. A. Mascelli, D. S. Neblock, H. F. Weisman, B. S. Coller, and R. E. Jordan. 1996. Analysis of GPIIb/IIIa receptor number by quantification of 7E3 binding to human platelets. Blood 88:907-914. [PubMed] [Google Scholar]
- 36.Yajima, A., Y. Takahashi, and K. Konishi. 2005. Identification of platelet receptors for the Streptococcus gordonii DL1 sialic acid-binding adhesin. Microbiol. Immunol. 49:795-800. [DOI] [PubMed] [Google Scholar]
- 37.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.