Abstract
The Escherichia coli RhaR protein activates expression of the rhaSR operon in the presence of its effector, l-rhamnose. The resulting RhaS protein (plus l-rhamnose) activates expression of the l-rhamnose catabolic and transport operons, rhaBAD and rhaT, respectively. Here, we further investigated our previous finding that rhaS deletion resulted in a threefold increase in rhaSR promoter activity, suggesting RhaS negative autoregulation of rhaSR. We found that RhaS autoregulation required the cyclic AMP receptor protein (CRP) binding site at rhaSR and that RhaS was able to bind to the RhaR binding site at rhaSR. In contrast to the expected repression, we found that in the absence of both RhaR and the CRP binding site at the rhaSR promoter, RhaS activated expression to a level comparable with RhaR activation of the same promoter. However, when the promoter included the RhaR and CRP binding sites, the level of activation by RhaS and CRP was much lower than that by RhaR and CRP, suggesting that CRP could not fully coactivate with RhaS. Taken together, our results indicate that RhaS negative autoregulation involves RhaS competition with RhaR for binding to the RhaR binding site at rhaSR. Although RhaS and RhaR activate rhaSR transcription to similar levels, CRP cannot effectively coactivate with RhaS. Therefore, once RhaS reaches a relatively high protein concentration, presumably sufficient to saturate the RhaS-activated promoters, there will be a decrease in rhaSR transcription. We propose a model in which differential DNA bending by RhaS and RhaR may be the basis for the difference in CRP coactivation.
The Escherichia coli rhaSR operon encodes two l-rhamnose-responsive members of the AraC/XylS family of transcription activator proteins, RhaS and RhaR (28). In the presence of l-rhamnose, RhaS and RhaR are required to activate transcription of the operons encoding the l-rhamnose catabolic enzymes (rhaBAD) (see Fig. 1) and the l-rhamnose uptake protein (rhaT). The sole identified function of RhaR is to activate transcription of rhaSR and thereby increase RhaS protein concentration in the presence of l-rhamnose, while RhaS directly activates transcription of the rhaBAD and rhaT operons (11, 12, 27-30). Cyclic AMP receptor protein (CRP) is required for full expression of all three of the rha operons, but it functions as a coactivator that substantially activates transcription only when RhaS or RhaR also binds to the promoter regions (11, 15, 31). CRP activation at the rhaSR operon was shown to require the RNA polymerase (RNAP) α-subunit C-terminal domain (α-CTD) (33). It is likely that CRP coactivation also involves contacts with α-CTD at the rhaBAD and rhaT promoters. RhaS or RhaR may be required to bend the DNA to allow CRP to coactivate by contacting α-CTD, similar to other promoters (8).
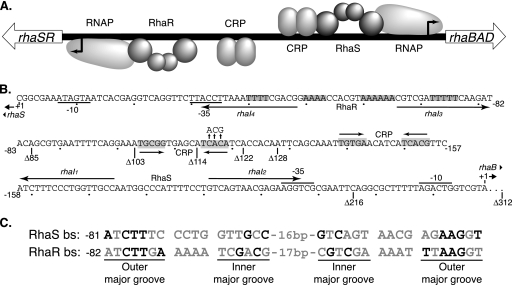
FIG. 1.
rhaSR-rhaBAD intergenic region. (A) Schematic representation of the rhaSR-rhaBAD intergenic region. The relative positions of the RNA polymerases (RNAPs) and the activator proteins RhaS, RhaR, and CRP are shown. The activators and sites shown above the line all influence rhaBAD expression, while the activators and sites shown below the line influence rhaSR expression. (B) The DNA sequence between the rhaBAD and rhaSR transcription start sites. The positions of the RhaS and RhaR binding sites are shown by arrows labeled with the half-site names (rhaI1 through rhaI4). The CRP binding site positions are shown as inverted arrows with some shaded residues, and the substitutions in the CRP binding site consensus positions are shown with vertical arrows pointing up above the CRP site. The −10 and −35 regions of the promoters are marked. Binding sites important at the rhaBAD promoter are shown above the line, while deletion endpoints (marked Δ), binding sites, and distances relative to the rhaSR promoter are shown below the line. The 70-bp sequence between the rhaBAD transcription start site and the rhaSR −312 position is not shown. (C) The sequences of the RhaS and RhaR binding sites (bs) are aligned. The black letters indicate those base pairs found important for sequence recognition by the activators, with all other letters in gray. The outer and inner major grooves of the inverted repeat half sites are labeled.
RhaS and RhaR are 30% identical to each other and likely arose by gene duplication. Both proteins function as homodimers and comprise two domains, an N-terminal dimerization and l-rhamnose binding domain and a C-terminal DNA binding domain (34; G. K. Hunjan, A. Kolin, and S. Egan, unpublished data). Flexible linkers connect the RhaS and RhaR domains; however, the sequences of the linkers do not appear to be critical for function (18). The RhaS and RhaR DNA binding sites consist of two imperfect 17-bp inverted repeat half sites that are separated by 16 or 17 bp (12, 29). The downstream half sites overlap the −35 hexamer by 4 bp, placing RhaS and RhaR in position to make protein-protein contacts with RNAP σ70 to activate transcription (6, 32). The RhaR binding site upstream of rhaSR contains four phased A tracts (14) and is bent to an estimated angle of 70° in the absence of RhaR and to an estimated angle of 160° upon RhaR binding (29). A single 4-bp A tract is present in the RhaS binding site, suggesting that this DNA sequence is likely less dramatically bent.
Our previous results suggested that in addition to activation of rhaBAD and rhaT, RhaS likely also negatively autoregulates its own expression. Deletion of rhaS resulted in a threefold increase in expression of a rhaS-lacZ translational fusion (extending from 312 bp upstream of the transcriptional start point through codon 20 of rhaS), while overexpression of rhaS resulted in a decrease in expression of the same fusion (11). It was proposed that this RhaS autoregulation could, in principle, occur by the following: formation of inactive RhaS-RhaR heterodimers; a DNA looping mechanism in which RhaS bound to its site at the rhaBAD promoter would inhibit activation by RhaR bound to its site at the rhaSR promoter; or RhaS competing for binding to the RhaR binding site at the rhaSR promoter (11). Here, we have further investigated the hypothesis that RhaS negatively autoregulates its own expression as well as the mechanism of this rhaSR autoregulation. Our results suggest a somewhat novel mechanism in which RhaS itself is capable of activating rhaSR expression from the RhaR binding site to nearly as high a level as RhaR. However, the CRP contribution to rhaSR activation is substantially lower when RhaS is the primary activator than when RhaR is the primary activator, resulting in a relative decrease in rhaSR expression. Differences in DNA bending by RhaS and RhaR may play a role in the differential ability of CRP to coactivate with RhaS versus RhaR. RhaS negative autoregulation likely functions to limit positive autoregulation of rhaSR by RhaR.
MATERIALS AND METHODS
Culture media and conditions.
Escherichia coli cultures for β-galactosidase assays were grown in MOPS [3-(N-morpholino)propanesulfonic acid]-buffered minimal growth medium supplemented with 0.4% glycerol, 0.2% Casamino Acids, and 0.002% thiamine (5) using the protocol of Neidhardt et al. (20). Tryptone broth (TB) (0.8% tryptone, 0.05% NaCl [pH 7.0]) supplemented with 0.2% maltose was used to grow cultures for bacteriophage λ infection. Tryptone-yeast extract (TY) liquid medium (0.8% tryptone, 0.05% yeast extract, and 0.05% NaCl [pH 7.0]) was used to grow cells for most other experiments and was supplemented with 5 mM CaCl2 to grow cultures for bacteriophage P1 infection. Antibiotics were used as indicated at 200 μg/ml for ampicillin and 25 μg/ml for chloramphenicol. All cultures were grown at 37°C with aeration.
General methods.
Standard methods were used for restriction endonuclease digestion and ligation. Transformation was carried out using chemically induced competent cells of E. coli, and plasmid DNA was purified by alkaline lysis (7) or using an EZNA plasmid minikit I (Omega Bio-Tek, Inc., Norcross, GA). Oligonucleotides were synthesized by MWG-Biotech (High Point, NC). The Northwestern University Genomics Core (Chicago, IL) performed DNA sequencing reactions. The DNA sequence of both strands was determined for the entire cloned region of all cloned DNA fragments. The Expand high-fidelity PCR system (Roche, Indianapolis, IN) was used to amplify nonmutagenized DNA fragments for cloning. The QIAquick PCR purification kit (Qiagen, Chatsworth, CA) was used to clean up PCR products. β-Galactosidase assays were performed by the method of Miller (19) as modified by Bhende and Egan (5). Specific activities were averaged from at least two independent assays, with two replicates per assay. RhaS fold repression values were calculated by dividing vector-only values by rhaS+ values in rhaR+ strains, while RhaS fold activation values were calculated by dividing rhaS+ values by vector-only values in ΔrhaR strains.
Strains, phage, and plasmids.
All strains in this study were derived from E. coli ECL116 (1). Plasmid-borne RhaS expression in all experiments was from promoters that were independent of RhaS, RhaR, and l-rhamnose. The promoter was either the lac promoter (pHG165 and pSU18) or a constitutive promoter with the rhaSR −10 element, a near-perfect −35 element (TTGACT), and no known upstream binding sites.
The lacZ fusions are named such that “Φ” stands for “fusion,” and the upstream endpoint of each fusion relative to the transcription start site (for example, −84, but without the minus sign) is given after the “Δ.” The downstream endpoint of all fusions, unless otherwise indicated, was within codon 20 of rhaS, at position +84 relative to the transcription start site. The exceptions had downstream endpoints within codon 20 of rhaR, at position +904 relative to the rhaSR transcription start site. Most lacZ fusions were translational fusions, unless otherwise noted. These were first constructed in plasmid pRS414, while transcriptional fusions were first constructed in pRS415 (25). The desired insert was generated by high-fidelity PCR of the promoter region of interest. Oligonucleotide 896 (11) was the downstream primer used to amplify rhaSR promoter regions with downstream endpoints at position +84. Oligonucleotide 2832 (5′-GCGGGATCCTTATTCGCAATATGGCGTAC-3′) was the downstream primer for the fusions with downstream endpoints at position +904. The lacZ fusions in pRS414 or pRS415 were recombined onto the genome of bacteriophage λ and integrated into the E. coli chromosome as single-copy lysogens (25). Several single-copy lysogen candidates were tested using β-galactosidase assays to distinguish likely single-copy lysogens from multiple lysogens based on activity levels. Single-copy lysogens were confirmed using a PCR test as well as the Ter test (13, 21). Phage P1 transduction was used to introduce the ΔrhaSR::kan or ΔrhaS rhaR+ (linked to zih-35::Tn10) alleles into strains as required.
Three new rhaSR promoter regions fused to lacZ, each with a different upstream endpoint, were constructed to identify the DNA elements required for RhaS autoregulation. These rhaS-lacZ fusions were constructed as described above using oligonucleotides 2727 (5′-GTCGAATTCTTTCCTGAAAATTCACGCTG-3′), 2726 (5′-GTCGAATTCTGCTCACCGCATTTCCTG-3′), and 2725 (5′-GGCGAATTCTGATGTGATGCTCACCGC-3′) in combination with oligonucleotide 896 to make Φ(rhaS-lacZ)Δ103, Φ(rhaS-lacZ)Δ114, and Φ(rhaS-lacZ)Δ122, respectively.
Two variant rhaSR promoters positioned the native rhaSR CRP binding site at positions −92.5 and −93.5, similar to the CRP site at the rhaBAD promoter (11). Oligonucleotides 2867 (5′-CACGAATTCTGTGATGCTCACCGCAGTATCTTGAAAAATCGACG-3′) and 2868 (5′-CACGAATTCTGTGATGCTCACCGCATGTATCTTGAAAAATCGACG-3′) were used with oligonucleotide 896, which placed the CRP binding site (underlined) 2 bp and 3 bp upstream of the RhaR binding site, respectively. The RhaS binding site at rhaBAD was also replaced with a RhaR binding site (with or without an upstream CRP site) by using a natural EcoRI site just downstream of the RhaS binding site.
Random mutagenesis of rhaS was performed as previously described (17). Briefly, the rhaS gene was PCR amplified with Taq polymerase under standard reaction conditions (36) and cloned into pHG165 (26).
Electrophoretic mobility shift assays.
Electrophoretic mobility shift assays were performed as previously described (15), with the following modifications. The DNA fragments were generated by PCR with one primer labeled with 32P at the 5′ end (using T4 polynucleotide kinase) and one unlabeled primer. The 49-bp DNA fragments each contained one 17-bp RhaS or RhaR binding half site (rhaI1, rhaI2, rhaI3, or rhaI4) flanked by the same 16-bp sequences for each half site. The flanking sequences were the same as the previously published half-site fusions with lacZ (34). The mobility shift assay buffer did not contain Nonidet P-40 or cyclic AMP and contained 0.5 mM Tris[2-carboxyethyl]-phosphine (TCEP) instead of 1 mM dithiothreitol. His6-RhaS(163-278) was purified as previously described (34). After electrophoresis, the gels were dried, exposed to a phosphor screen, and analyzed using the Cyclone storage phosphor system (Packard).
DNase I footprinting.
The DNA substrate for footprinting reactions was prepared by PCR amplification of the rhaSR-rhaBAD intergenic region using primers 2371 and 2409 (33) with one or the other of the primers 32P labeled in separate footprinting reactions. DNase I footprinting was performed as previously described (12).
RESULTS AND DISCUSSION
RhaS autoregulation of rhaSR requires the CRP binding site but not the RhaS binding site at rhaBAD.
Our first goal was to identify the DNA element(s) in the rhaSR-rhaBAD intergenic region required for RhaS autoregulation (Fig. 1). To do this, we assayed the ability of RhaS to repress rhaSR expression from a number of single-copy lacZ fusions with different upstream endpoints in a strain carrying an in-frame deletion of the majority of the rhaS gene, ΔrhaS rhaR+ (11). Here and throughout this work, we expressed RhaS from a moderate-copy-number plasmid to increase the repression by RhaS and thereby enable detection of otherwise relatively small effects. The strains in which these assays were performed are represented schematically in Fig. 2, lines 2 and 3. Given the precedence of AraC protein forming a repressing DNA loop (reviewed in reference 22), one question this experiment addressed was whether RhaS autoregulation might involve a DNA looping mechanism in which RhaS, bound to its site at the rhaBAD promoter, interacts with and inhibits activation by RhaR bound at the rhaSR promoter. The longest promoter fusion in these experiments was the same one that initially provided evidence of RhaS autoregulation (11). It extended upstream to position −312 (relative to the rhaSR transcription start site) and included the RhaS binding site at the divergent rhaBAD promoter (Fig. 1).
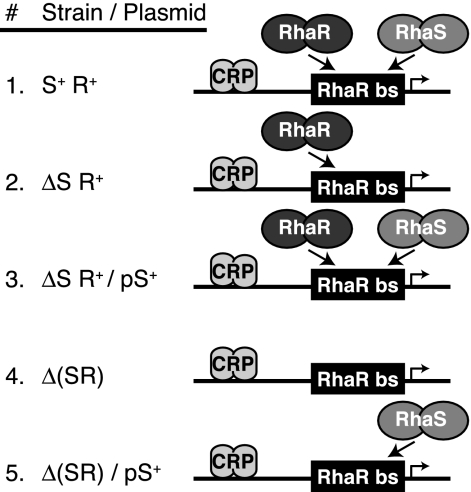
FIG. 2.
Schematic representations of strains used in this study. The relevant strain backgrounds used in many of these studies are represented schematically. In the strain designations shown in the figure, rhaS and rhaR are S and R, respectively; deletions of rhaS or both rhaS and rhaR are written as ΔS and Δ(SR), respectively; plasmid-borne rhaS is pS+. DNA is a horizontal line. The RhaR binding site is a black box labeled “RhaR bs.” The RhaS, RhaR, and CRP proteins are pairs of gray ovals, as labeled. The direction of transcription is indicated by the direction of the bent arrow. RhaS or RhaR binding to the RhaR binding site is represented by arrows, with competition represented by two proteins attempting to bind to the same site.
We found that expression of RhaS from a plasmid resulted in 30-fold or greater decreases in rhaSR promoter expression (Table 1) with fusions that had upstream endpoints in the range from positions −122 to −312 (Fig. 1). The RhaS binding site at rhaBAD is located between the −216 and −128 deletion endpoints; however, its deletion did not result in a significant change in RhaS autoregulation. This indicates that the RhaS binding site at rhaBAD is not required for RhaS negative autoregulation of rhaSR and rules out a mechanism in which RhaS represses rhaSR expression by forming a DNA loop from its site at rhaBAD.
TABLE 1.
RhaS repression of various rhaSR promoter fusions
| Φ(rhaS-lacZ) promoter truncation | Avg β-galactosidase activity (Miller units)a in a strain carrying: |
Fold repression by RhaS | |
|---|---|---|---|
| Vector onlyb | rhaS+ plasmidb | ||
| Δ85 | 8.9 | 7.0 | 1.3 |
| Δ103 | 10 | 2.7 | 3.7 |
| Δ114 | 12 | 2.9 | 4.1 |
| Δ122 | 155 | 5.3 | 29 |
| Δ128 | 204 | 6.9 | 30 |
| Δ216 | 234 | 7.1 | 33 |
| Δ312 | 203 | 4.9 | 41 |
| Δ312 CRP− | 22 | 3.5 | 6.3 |
β-Galactosidase activity was assayed from single-copy rhaS-lacZ fusions with the upstream promoter endpoints indicated. The Δ312 CRP− promoter has point mutations in three consensus positions of the rhaSR promoter CRP binding site. Cultures were grown in MOPS-buffered minimal growth medium containing ampicillin and l-rhamnose. The strain background was ΔrhaS rhaR+ zih-35::Tn10 recA::cat. The standard errors were less than 12% of the average activities.
RhaS expression reduced expression from rhaSR promoter fusions with endpoints between positions −114 and −85 by only 1.3- to 4.1-fold (Table 1). The upstream endpoint of the CRP binding site required for full rhaSR activation is at −119; therefore, the Δ114 fusion did not include the entire CRP binding site, whereas the Δ122 fusion did (Fig. 1). This suggested that the CRP binding site is required for maximal repression by RhaS. To further test this hypothesis, we assayed a fusion with an upstream endpoint at −312 and carrying point mutations at three consensus positions in the CRP binding site (Fig. 1) that we previously found greatly reduced CRP coactivation of rhaSR (34). We found that the CRP binding site point mutations reduced RhaS repression of the Δ312 fusion by more than sixfold to a level similar to that of the Δ114 fusion (Table 1, Δ312 CRP−). Taken together, these results support the hypothesis that the CRP binding site at rhaSR, but not the RhaS binding site at rhaBAD, is required for the RhaS-dependent decrease in rhaSR expression, or negative autoregulation.
RhaS binds to the RhaR binding site at rhaSR.
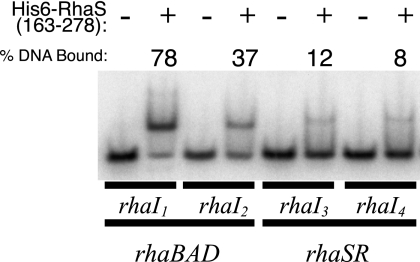
We next considered the hypothesis that RhaS might compete with RhaR for binding to the RhaR binding site at the rhaSR promoter. As previously noted (5, 12), the DNA sequences of the outer major grooves of the RhaS and RhaR binding sites at the rhaBAD and rhaSR promoters, respectively, are nearly identical (Fig. 1C), raising the possibility that RhaS might be capable of binding to the RhaR binding site. We tested this using in vitro electrophoretic mobility shift assays with 49-bp DNA fragments, each containing one of the 17-bp half sites for RhaR or RhaS binding and containing identical flanking DNA sequences. Given that full-length RhaS protein severely aggregates when overexpressed, we used the purified C-terminal DNA binding domain of RhaS [His6-RhaS(163-278), previously called His6-RhaS-CTD (34)] for these assays. His6-RhaS(163-278) was previously found capable of in vitro DNA binding and transcription activation (34). Using electrophoretic mobility shift assays, we found that His6-RhaS(163-278) was able to bind in vitro to DNA fragments carrying each of the RhaR half sites at rhaSR, although 3- to 10-fold less tightly than to the RhaS half sites at rhaBAD (Fig. 3). This apparent weaker binding is consistent with the expectation that an autoregulation mechanism would involve RhaS binding to the RhaR binding site only at RhaS protein concentrations above those necessary to saturate the RhaS binding sites at the rhaBAD and rhaT promoters. DNase I footprinting confirmed that His6-RhaS(163-278) bound to the expected RhaR half sites (data not shown).
FIG. 3.
Electrophoretic mobility shift assays of His6-RhaS(163-278) binding to RhaS and RhaR half sites. Electrophoretic mobility shift reactions were carried out in the absence (−) or presence (+) of 6 μM purified His6-RhaS(163-278) with 49-bp 32P-labeled DNA fragments including each of the RhaS half sites at rhaBAD and the RhaR half sites at rhaSR. The direction of electrophoresis was from the top down, as shown. The percent DNA bound was averaged from four independent assays.
RhaS activates rhaSR expression, but CRP coactivation is reduced.
The finding that RhaS can bind to the RhaR binding site at rhaSR suggested a model in which, upon reaching saturating levels in the cell, RhaS might compete for binding to the RhaR binding site and repress expression of rhaSR. However, the promoter-proximal RhaR half site at rhaSR is identically positioned relative to the promoter as the similar RhaS half site at rhaBAD, suggesting that RhaS might activate rather than repress transcription from this site. To test whether RhaS was able to activate rhaSR expression, we assayed expression from single-copy rhaS-lacZ fusions in a strain lacking RhaR (ΔrhaSR strain background), with RhaS expressed from the vector pSU18 (2) (plasmid pSE273 [34]). It was not possible to use a rhaS+ ΔrhaR strain, since rhaS expression requires RhaR. For comparison, we also assayed the same fusions in a strain expressing RhaR from the chromosome (ΔrhaS rhaR+ strain background), again with RhaS expressed from plasmid pSE273 (34). In the ΔrhaS rhaR+ strain, the vector-only samples represent the level of activation by RhaR, while the addition of the RhaS-encoding plasmid shows the RhaS-mediated reduction of the RhaR activation (Fig. 2, lines 2 and 3). In the ΔrhaSR strain, the vector-only samples represent the basal level of expression from the fusions, and the addition of the RhaS-encoding plasmid shows any ability of RhaS to activate expression of the fusions (Fig. 2, lines 4 and 5).
Similar to the previous experiment (Table 1), in the rhaR+ strain there was a 45-fold repression by RhaS from the fusion that included the CRP binding site [(rhaS-lacZ)Δ128] but only a 1.5-fold repression from the fusion that did not include the CRP binding site [(rhaS-lacZ)Δ85] (Table 2). We found that RhaS activated transcription of the (rhaS-lacZ)Δ85 fusion to a level similar to that of RhaR (4.5 and 6.4 Miller units, respectively) but activated the (rhaS-lacZ)Δ128 fusion to a much lower level than did RhaR (17 and 630 Miller units, respectively). The increased expression from the longer of these fusions is due to the contribution of CRP to the activation (15, 34), which in this case was nearly 100-fold in combination with RhaR but was less than fourfold in combination with RhaS. These results indicate that when RhaS binds to the RhaR binding site, RhaS itself is able to activate transcription nearly as well as RhaR. However, the contribution to activation by CRP at the promoter activated by RhaS was greatly reduced compared with the CRP-RhaR combination, thereby resulting in a relative repression of rhaSR expression upon activation by RhaS.
TABLE 2.
Repression and activation of rhaSR expression by RhaS
| Φ(rhaS-lacZ) promoter truncation | ΔrhaSrhaR+ straina |
ΔrhaSR straina |
||||
|---|---|---|---|---|---|---|
| Avg β-galactosidase activity (Miller units)b in strain carrying: |
Fold repression by RhaS | Avg β-galactosidase activity (Miller units) in strain carrying: |
Fold activation by RhaS | |||
| Vector onlyc | rhaS+ plasmidc | Vector only | rhaS+ plasmid | |||
| Φ(rhaS-lacZ)Δ85 | 6.4 | 4.4 | 1.5 | 0.17 | 4.5 | 26 |
| Φ(rhaS-lacZ)Δ128 | 630 | 14 | 45 | 0.84 | 17 | 20 |
The strain background was either ΔrhaS rhaR+zih-35::Tn10 or Δ(rhaSR)::kan.
β-Galactosidase activity was measured from single-copy lacZ fusion strains grown in MOPS-buffered minimal growth medium containing chloramphenicol and l-rhamnose. The standard errors were less than 30% of the average activities.
Upon addition of the RhaS-expressing plasmid, the expression levels from each fusion were nearly the same regardless of whether RhaR was present (4.4 versus 4.5 Miller units and 14 versus 17 Miller units) (Table 2). This suggests that under these conditions RhaR does not significantly contribute to the activation and that RhaS may fully outcompete RhaR for binding to the RhaR binding site. We do not expect RhaS will fully outcompete RhaR under physiological conditions (without RhaS overexpression). However, given the values in Table 2 and the three- to fourfold increase in rhaSR expression upon deletion of chromosomal rhaS (11), we calculate that RhaS represents approximately 67% of the protein bound at the rhaSR promoter in wild-type cells in the presence of l-rhamnose.
RhaS is more highly expressed than RhaR.
Given our finding that RhaS binds relatively weakly to the RhaR binding site (Fig. 3), we were surprised by the estimate that RhaS outcompetes RhaR for binding at rhaSR two-thirds of the time (at steady-state levels in the presence of l-rhamnose). One potential explanation for this finding is that, although rhaS and rhaR are transcribed together, the level of protein synthesized from the two genes might differ. Inspection of the Shine-Dalgarno (SD) sequences upstream of rhaS and rhaR suggests this might be the case, as the rhaS SD sequence is predicted to be strong (5′-AGGAGGC-3′), while the rhaR gene has a SD sequence predicted to be much weaker (5′-GCCAGGG-3′) relative to the consensus sequence (5′-AGGAGGT-3′) (24).
To test the hypothesis that rhaS and rhaR are translated at different levels, we constructed three new lacZ fusions driven by the rhaSR promoter in addition to the (rhaS-lacZ)Δ128 translational fusion used in previous experiments. The first new lacZ fusion had the same promoter region endpoints at positions −128 and +84 as the previous fusion, but it was a transcriptional fusion. We refer here to the fusions with endpoints at −128 and +84 as “short” fusions. The other two were a transcriptional fusion and a translational fusion, both with endpoints at −128 and +904 (“long” fusions). The +904 downstream endpoint is within codon 20 of rhaR; therefore, the long fusions measure the relative transcription and translation of rhaR. The expression levels from the short and long transcriptional fusions were essentially the same as each other, as expected (Table 3). However, the short translational fusion (which was a measure of rhaS translation) was 28-fold more highly expressed than the long translational fusion (which was a measure of rhaR translation). These results suggest that RhaS is expressed to a significantly higher level than RhaR and may explain how RhaS is able to dominate in the competition for binding to the RhaR binding site at steady-state levels in the presence of l-rhamnose.
TABLE 3.
Comparison of transcription and translation of the rhaS and rhaR genes
| Fusion type | Φ(rhaS-lacZ) promoter truncationa | Avg β-galactosidase activity (Miller units)b |
|
|---|---|---|---|
| − Rha | + Rha | ||
| Transcriptional | Φ(rhaS-lacZ)(−128 → +84) | 20 | 1,330 |
| Φ(rhaSR-lacZ)(−128 → +904) | 26 | 1,540 | |
| Translational | Φ(rhaS-lacZ)(−128 → +84) | 0.04 | 170 |
| Φ(rhaSR-lacZ)(−128 → +904) | 0.02 | 6.0 | |
The strain background was E. coli ECL116 for all promoter truncations shown (1).
β-Galactosidase activity was measured from single-copy lacZ fusion strains grown in MOPS-buffered minimal growth medium with or without l-rhamnose, as indicated. The standard errors were less than 8% of the average activities, except for values less than 1.0, where standard error was less than 25%.
RhaS and RhaR differ in their optimal CRP binding site positions.
It is likely relevant that although the RhaS and RhaR binding sites at rhaBAD and rhaSR are identically positioned (Fig. 1), the CRP (TGTGA motifs [4]) and RhaS sites at rhaBAD are separated by 3 bp, while the CRP and RhaR sites at rhaSR are separated by 21 bp (12, 16). In both cases, there is little or no activation by CRP in the absence of RhaS or RhaR (Table 2) (16); presumably, CRP requires DNA bending by the primary activator to enable contacts with α-CTD (8). We have shown that CRP coactivation requires α-CTD at the rhaSR promoter (34). Tobin and Schleif (29) found that the RhaR binding site DNA at rhaSR was bent to approximately 70 degrees when alone (presumably due to the four phased A tracts within the RhaR binding site [Fig. 1]) but that this bend was increased to approximately 160 degrees upon RhaR binding. There is at most one A tract in and around the RhaS binding site (Fig. 1), suggesting that the rhaBAD promoter DNA is likely to be significantly less bent. Taken together, these results suggest that, in addition to its own DNA bending (23), CRP may require DNA bending by a second activator protein in order to coactivate transcription at rhaSR and rhaBAD but that there might be differences in the extent of the DNA bending required at the two promoters.
We hypothesized that RhaS and RhaR differ in the position of the upstream CRP binding site that is optimal for coactivation. To test this hypothesis, we wished to move the CRP binding site at rhaSR to the position of the CRP binding site at rhaBAD. However, two possible lines of reasoning could identify the optimal position for the CRP site at rhaSR. The first was that the position of the CRP binding site relative to the transcription start site was the important parameter. We therefore constructed a rhaS-lacZ fusion in which the CRP site was positioned at −92.5 relative to the transcription start site, the same position as the site at rhaBAD (11). This placed the CRP site 2 bp upstream of the RhaR binding site [(rhaS-lacZ)CRP−2]. The second possibility was that the distance upstream of the RhaR binding site was the key parameter. Since the CRP site at rhaBAD is 3 bp upstream of the RhaS binding site, we placed the CRP site in the second rhaS-lacZ fusion 3 bp upstream of the RhaR binding site [at −93.5, (rhaS-lacZ)CRP−3].
We compared the expression from the rhaS-lacZ fusions with CRP sites 2 and 3 bp upstream of the RhaR binding site to expression from a fusion with no CRP binding site [(rhaS-lacZ)Δ85] and a fusion with a CRP site at the wild-type position [(rhaS-lacZ)Δ128] (Table 4). The expression levels in the ΔrhaSR strain indicated that the basal expression levels were similar in all cases. In the ΔrhaS rhaR+ strain, where RhaR but not RhaS can activate, we found that the wild-type CRP binding site [(rhaS-lacZ)Δ128] enabled a 30-fold increase in activation compared with no CRP binding site [(rhaS-lacZ)Δ85] (216 versus 6.9 Miller units, respectively). In contrast, the addition of the CRP sites 2 or 3 bp upstream of the RhaR binding site did not result in increased expression compared to (rhaS-lacZ)Δ85, indicating that there was no contribution of CRP to activation in combination with RhaR from these CRP site positions. This finding indicates that CRP can function as a coactivator from a site 21 bp, but not 2 or 3 bp, upstream of RhaR, and is consistent with the hypothesis that RhaS and RhaR differ in the position of the CRP binding site that is optimal for coactivation.
TABLE 4.
Influence of CRP binding site position on CRP coactivation of rhaSR
| Φ(rhaS-lacZ) promoter truncation | Avg β-galactosidase activity (Miller units) in straina: |
||
|---|---|---|---|
| ΔrhaSR | ΔrhaS rhaR+ | Δ(rhaSR) prhaS+ | |
| Φ(rhaS-lacZ)Δ85 | 0.20 | 6.9 | 7.4 |
| Φ(rhaS-lacZ)Δ128 | 0.38 | 216 | 31 |
| Φ(rhaS-lacZ)CRP−2 | 0.40 | 4.3 | 1.1 |
| Φ(rhaS-lacZ)CRP−3 | 0.15 | 7.1 | 26 |
β-Galactosidase activity was measured from single-copy lacZ fusion strains grown in MOPS-buffered minimal growth medium containing ampicillin and l-rhamnose. The CRP−2 and CRP−3 constructs replaced the native CRP site (20 bp upstream of the RhaR binding site) with the same sequence 2 or 3 bp upstream as described in the text. The standard errors were less than 23% of the average activities. The strain background was either ΔrhaS rhaR+zih-35::Tn10 or Δ(rhaSR)::kan. Each strain was transformed with empty vector, pHG165 (26), or plasmid pSE289, which is pHG165 rhaS+ [Δ(rhaSR)prhaS+] (17).
We also tested the ability of CRP to coactivate with RhaS at the CRP−2 and CRP−3 constructs by using a ΔrhaSR strain expressing RhaS from a plasmid (Table 4). When RhaS was the primary activator, a CRP site 2 bp upstream of the RhaR binding site resulted in a sevenfold reduction in expression [compared with no CRP binding site, (rhaS-lacZ)Δ85]. This suggests that CRP binding may interfere with RhaS binding at this promoter (a small reduction in expression was also seen with RhaR as the primary activator at this construct). However, the CRP site 3 bp upstream of RhaS (bound at the RhaR binding site) enabled a 3.5-fold increase in expression relative to no CRP binding site [(rhaS-lacZ)Δ85]. This was very similar to the CRP contribution to expression with the CRP site at the wild-type position [(rhaS-lacZ)Δ128] and RhaS as the primary activator. Therefore, although we expected the CRP site in the CRP−3 construct to be optimally positioned for CRP coactivation with RhaS, there was no increase in CRP coactivation compared with the wild-type CRP position at rhaSR. One explanation for this finding might be that the four phased A tracts within the RhaR binding site (Fig. 1) increase the extent of DNA bending by RhaS at rhaSR compared with RhaS binding at rhaBAD. This increase in DNA bending could change the position/geometry of the CRP protein and potentially decrease the ability of CRP to contact α-CTD.
These results support the hypothesis that the optimal CRP binding site position differs for RhaS versus RhaR. The position of the CRP site at rhaBAD (−92.5) is a fairly common class III activator position and is similar to the position of CRP at araBAD as well as the optimal upstream CRP site position in studies with tandem CRP sites at class I and class II positions (3, 35). The position of the CRP site at rhaSR (−111.5) is less typical, and in the tandem CRP site studies, CRP sites near this position made little, if any, contribution to activation (3). As such, we hypothesize that the four phased A tracts may contribute to the ability of CRP to activate well from such a distant site at rhaSR. Although the A tracts may explain why CRP did not coactivate well with RhaS at (rhaS-lacZ)CRP−3, they do not explain why CRP also did not coactivate well with RhaS at (rhaS-lacZ)Δ128, despite coactivating well with RhaR at this promoter. This finding indicates that there must be a difference between RhaS and RhaR that contributes to their respective optimal CRP site positions, perhaps a difference in the extent of DNA bending by the two proteins.
RhaS variants with increased activation at rhaSR.
Given the 30% amino acid sequence identity and 62% similarity between RhaS and RhaR, we attempted to identify the difference between RhaS and RhaR that dictates their difference in optimal CRP binding site position. We tested whether it would be possible to obtain increased CRP coactivation at rhaSR in combination with RhaS by screening for RhaS variants with increased activation at (rhaS-lacZ)Δ128, but without any increase in activation at (rhaS-lacZ)Δ85. In this way, we expected to eliminate from consideration substitutions that simply increased RhaS binding to the RhaR binding site, for example. We screened 40 independent pools of rhaS genes that had been PCR mutagenized and cloned into plasmid pHG165 as previously described (17). Although we previously isolated apparent gain-of-function mutants from these pools (17), here we were unable to isolate any RhaS variants that met both of these criteria among the approximately 38,000 clones screened. However, we characterized three (of many) RhaS variants with increased activation at both fusions. These RhaS variants, RhaS H205R (isolated twice) and RhaS H253Y, activated transcription at each of the rhaS-lacZ fusions to levels 20- to 60-fold higher than wild-type RhaS did (Table 5). RhaS H205R and RhaS H253Y are located at positions five and three of the recognition helices of the first and second helix-turn-helix (HTH) motifs of RhaS, respectively (5). In each case, the substitutions replaced the RhaS residue with the wild-type RhaR residue at the aligned position. This suggests that the substitutions increase RhaS binding to the RhaR binding site.
TABLE 5.
RhaS variants with increased activation at rhaSR
| Activator | Avg β-galactosidase activity (Miller units) in strain:a |
|
|---|---|---|
| Φ(rhaS-lacZ)Δ85 | Φ(rhaS-lacZ)Δ128 | |
| RhaR | 13 | 1,013 |
| RhaS | 7 | 22 |
| RhaS H205R | 160 | 440 |
| RhaS H253Y | 438 | 679 |
β-Galactosidase activity was measured from single-copy lacZ fusion strains grown in MOPS-buffered minimal growth medium containing ampicillin and l-rhamnose. The standard errors were less than 10% of the average activities. The strain background was Δ(rhaSR)::kan recA::cat. Wild-type RhaS, wild-type RhaR, or the RhaS variants were expressed from plasmid pHG165 (26) as previously described (17).
The inability to isolate RhaS variants that enabled increased CRP coactivation at rhaSR suggests that the difference that allows RhaR to enable CRP coactivation at rhaSR may involve more than one or two simple amino acid substitutions relative to RhaS. On the other hand, we were able to easily isolate RhaS variants with increased activity at both promoter fusions. This result and the finding that these variants had substitutions that made the RhaS amino acid sequence more like that of RhaR further support our conclusion that RhaS autoregulates rhaSR expression by binding to the RhaR binding site.
Summary of RhaS autoregulation model.
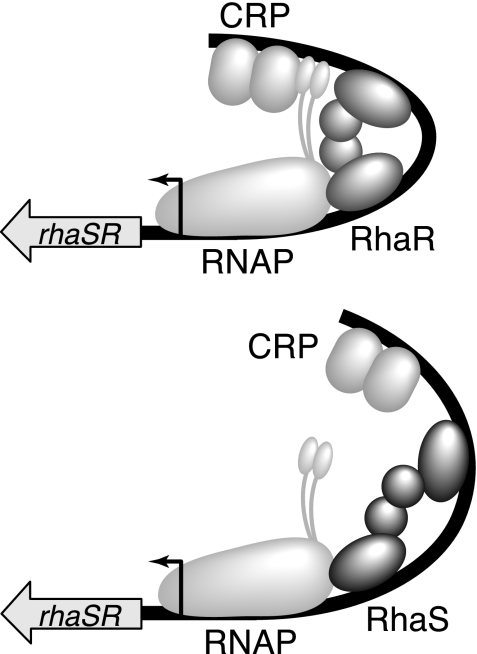
Our overall model for RhaS autoregulation is as follows (Fig. 4). Upon encountering l-rhamnose, RhaR binds upstream of rhaSR and activates transcription. The resulting RhaS protein then binds to its sites at the rhaBAD and rhaT promoters and activates transcription. As the RhaS protein concentration increases, RhaS also competes with RhaR for binding at the rhaSR promoter. CRP is unable to efficiently coactivate rhaSR transcription in combination with RhaS, likely as a result of inefficient contacts with α-CTD in this context. The result is a decrease in rhaSR expression by three- to fourfold. As RhaS protein levels fluctuate, this autoregulation would compensate by increasing or decreasing expression of rhaSR to return the RhaS protein concentration to its optimal level. We hypothesize that RhaS may not bend the DNA appropriately for CRP to coactivate rhaSR transcription as described above and illustrated in Fig. 4. We have not, however, ruled out the possibility that RhaS somehow interferes with CRP binding to its DNA site at rhaSR, but this seems unlikely, given the 21-bp spacing between the sites.
FIG. 4.
Model for RhaS negative autoregulation. Expression of the rhaSR operon in the presence of l-rhamnose. (Top) At relatively low RhaS protein concentrations, RhaR and CRP both contribute to rhaSR activation; therefore, expression is at its maximal level. CRP contacts α-CTD (shown as two small ovals connected by a flexible linker to RNAP). (Bottom) At relatively high RhaS protein concentrations, RhaS binds to the RhaR binding site, thereby replacing RhaR. RhaS contributes to rhaSR activation, but CRP does not effectively coactivate; therefore, rhaSR expression is reduced. CRP does not effectively contact α-CTD.
The RhaS autoregulation mechanism is unusual in that a transcriptional activator protein functions to decrease expression of an operon by activating transcription, but not allowing a second activator protein—in this case CRP—to coactivate transcription. The rhaSR mechanism is not unique among characterized regulatory schemes, however, with one similar system being regulation of the E. coli napF promoter (9, 10). At napF, the NarL protein competes with the 44% identical NarP protein for binding to a common DNA site. NarL binding to the site results in reduced napF expression relative to NarP binding, including a reduced contribution to activation by the Fnr protein (related to CRP) from its DNA site upstream of the NarP/NarL site (9, 10).
Acknowledgments
We thank Gurpreet Kaur Hunjan and Bria Wilkins for helpful comments on the manuscript.
This work was supported by NIH grant GM55099 from the National Institute of General Medical Sciences, NIH grant P20 RR17708 from the Institutional Development Award (IDeA) Program of the National Center for Research Resources, and the Department of Molecular Biosciences, University of Kansas, all to S.M.E.
Footnotes
Published ahead of print on 23 October 2009.
REFERENCES
- 1.Backman, K., Y.-M. Chen, and B. Magasanik. 1981. Physical and genetic characterization of the glnA-glnG region of the Escherichia coli chromosome. Proc. Natl. Acad. Sci. U. S. A. 78:3743-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartolome, B., Y. Jubee, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 3.Belyaeva, T. A., V. A. Rhodius, C. L. Webster, and S. J. W. Busby. 1998. Transcription activation at promoters carrying tandem DNA sites for the Escherichia coli cyclic AMP receptor protein: organisation of the RNA polymerase α subunits. J. Mol. Biol. 277:789-804. [DOI] [PubMed] [Google Scholar]
- 4.Berg, O. G., and P. H. von Hippel. 1988. Selection of DNA binding sites by regulatory proteins. Trends Biochem. Sci. 13:207-211. [DOI] [PubMed] [Google Scholar]
- 5.Bhende, P. M., and S. M. Egan. 1999. Amino acid-DNA contacts by RhaS: an AraC family transcription activator. J. Bacteriol. 181:5185-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhende, P. M., and S. M. Egan. 2000. Genetic evidence that transcription activation by RhaS involves specific amino acid contacts with sigma 70. J. Bacteriol. 182:4959-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Browning, D. F., and S. J. Busby. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2:57-65. [DOI] [PubMed] [Google Scholar]
- 9.Darwin, A. J., and V. Stewart. 1995. Nitrate and nitrite regulation of the Fnr-dependent aeg-46.5 promoter of Escherichia coli K-12 is mediated by competition between homologous response regulators (NarL and NarP) for a common DNA-binding site. J. Mol. Biol. 251:15-29. [DOI] [PubMed] [Google Scholar]
- 10.Darwin, A. J., E. C. Ziegelhoffer, P. J. Kiley, and V. Stewart. 1998. Fnr, NarP, and NarL regulation of Escherichia coli K-12 napF (periplasmic nitrate reductase) operon transcription in vitro. J. Bacteriol. 180:4192-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egan, S. M., and R. F. Schleif. 1993. A regulatory cascade in the induction of rhaBAD. J. Mol. Biol. 234:87-98. [DOI] [PubMed] [Google Scholar]
- 12.Egan, S. M., and R. F. Schleif. 1994. DNA-dependent renaturation of an insoluble DNA binding protein. Identification of the RhaS binding site at rhaBAD. J. Mol. Biol. 243:821-829. [DOI] [PubMed] [Google Scholar]
- 13.Gottesman, M. E., and M. B. Yarmolinsky. 1968. The integration and excision of the bacteriophage lambda genome. Cold Spring Harbor Symp. Quant. Biol. 33:735-747. [DOI] [PubMed] [Google Scholar]
- 14.Haran, T. E., and U. Mohanty. 2009. The unique structure of A-tracts and intrinsic DNA bending. Q. Rev. Biophys. 42:41-81. [DOI] [PubMed] [Google Scholar]
- 15.Holcroft, C. C., and S. M. Egan. 2000. Interdependence of activation at rhaSR by cyclic AMP receptor protein, the RNA polymerase alpha subunit C-terminal domain and RhaR. J. Bacteriol. 182:6774-6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holcroft, C. C., and S. M. Egan. 2000. Roles of cyclic AMP receptor protein and the carboxyl-terminal domain of the α subunit in transcription activation of the Escherichia coli rhaBAD operon. J. Bacteriol. 182:3529-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolin, A., V. Balasubramaniam, J. M. Skredenske, J. R. Wickstrum, and S. M. Egan. 2008. Differences in the mechanism of the allosteric L-rhamnose responses of the AraC/XylS family transcription activators RhaS and RhaR. Mol. Microbiol. 68:448-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolin, A., V. Jevtic, L. Swint-Kruse, and S. M. Egan. 2007. Linker regions of the RhaS and RhaR proteins. J. Bacteriol. 189:269-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powell, B. S., M. P. Rivas, D. L. Court, Y. Nakamura, and C. L. Turnbough, Jr. 1994. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 22:5765-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schleif, R. 2003. AraC protein: a love-hate relationship. Bioessays 25:274-282. [DOI] [PubMed] [Google Scholar]
- 23.Schultz, S. C., G. C. Shields, and T. A. Steitz. 1991. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science 253:1001-1007. [DOI] [PubMed] [Google Scholar]
- 24.Shine, J., and L. Dalgarno. 1974. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. U. S. A. 71:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 26.Stewart, G. S. A. B., S. Lubinsky-Mink, C. G. Jackson, A. Cassel, and J. Kuhn. 1986. pHG165: a pBR322 copy number derivative of pUC8 for cloning and expression. Plasmid 15:172-181. [DOI] [PubMed] [Google Scholar]
- 27.Tate, C. G., J. A. R. Muiry, and P. J. F. Henderson. 1992. Mapping, cloning, expression, and sequencing of the rhaT gene which encodes a novel L-rhamnose-H+ transport protein in Salmonella typhimurium and Escherichia coli. J. Biol. Chem. 287:6923-6932. [PubMed] [Google Scholar]
- 28.Tobin, J. F., and R. F. Schleif. 1987. Positive regulation of the Escherichia coli L-rhamnose operon is mediated by the products of tandemly repeated regulatory genes. J. Mol. Biol. 196:789-799. [DOI] [PubMed] [Google Scholar]
- 29.Tobin, J. F., and R. F. Schleif. 1990. Purification and properties of RhaR, the positive regulator of the L-rhamnose operons of Escherichia coli. J. Mol. Biol. 211:75-89. [DOI] [PubMed] [Google Scholar]
- 30.Tobin, J. F., and R. F. Schleif. 1990. Transcription from the rha operon psr promoter. J. Mol. Biol. 211:1-4. [DOI] [PubMed] [Google Scholar]
- 31.Via, P., J. Badia, L. Baldoma, N. Obradors, and J. Aguilar. 1996. Transcriptional regulation of the Escherichia coli rhaT gene. Microbiology 142:1833-1840. [DOI] [PubMed] [Google Scholar]
- 32.Wickstrum, J. R., and S. M. Egan. 2004. Amino acid contacts between sigma 70 domain 4 and the transcription activators RhaS and RhaR. J. Bacteriol. 186:6277-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wickstrum, J. R., T. J. Santangelo, and S. M. Egan. 2005. Cyclic AMP receptor protein and RhaR synergistically activate transcription from the l-rhamnose-responsive rhaSR promoter in Escherichia coli. J. Bacteriol. 187:6708-6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wickstrum, J. R., J. M. Skredenske, A. Kolin, D. J. Jin, J. Fang, and S. M. Egan. 2007. Transcription activation by the DNA-binding domain of the AraC family protein RhaS in the absence of its effector-binding domain. J. Bacteriol. 189:4984-4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, X., and R. Schleif. 1998. Catabolite gene activator protein mutations affecting activity of the araBAD promoter. J. Bacteriol. 180:195-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou, Y. H., X. P. Zhang, and R. H. Ebright. 1991. Random mutagenesis of gene-sized DNA molecules by use of PCR with Taq DNA polymerase. Nucleic Acids Res. 19:6052. [DOI] [PMC free article] [PubMed] [Google Scholar]